Abstract
Neuroproteomic studies of drug abuse offer the potential for a systems-level understanding of addiction. Understanding cocaine-responsive alterations in brain protein expression that persist even with extended abstinence may provide insight into relapse liability. In the current study, protein changes in the medial prefrontal cortex of cocaine self-administering rats following 1 and 100 days of enforced abstinence were quantified by 2D-DIGE. We have previously reported increased drug-seeking and drug-taking, as well as mRNA and epigenetic changes in this model even after 100 days of enforced abstinence. A number of statistically-significant changes in proteins relating to synapse function and neuronal remodeling were evident, including neurofilament medium and heat shock protein 73 (Hsp73) which increased at 1 day of abstinence, but returned to normal levels following 100 days of abstinence. −1 and synaptosome-associated protein 25 kDa (SNAP-25) were unchanged at 1 day of abstinence, but were significantly decreased after 100 days. These data demonstrate that while some protein changes return to normal levels following enforced cocaine abstinence, a number remain or become altered after long periods, up to 100 days, of cocaine abstinence. Those protein expression changes that do not reset to pre-cocaine exposure levels may contribute to the persistent relapse potential that occurs in response to cocaine abstinence.
Keywords: Cocaine, medial prefrontal cortex, proteomics, withdrawal, self-administration
INTRODUCTION
The societal burden caused by cocaine abuse has driven research to understand behavioral and molecular changes that underlie the drug craving and high relapse liability that have been observed in humans following cocaine abuse and periods of abstinence [1,2]. While cocaine acts acutely as a central nervous system stimulant by blocking dopamine-reuptake at the presynaptic nerve terminals of dopaminergic neurons, repeated cocaine use produces a psychological dependence that results in drug craving even after detoxification. Subsequently, re-exposure to the drug, environmental cues associated with the drug, and/or stress can increase drug craving and relapse potential [3,4]. This relapse potential is independent of the direct actions of cocaine, as it persists long after the detoxification, and therefore understanding the neurobiological basis of relapse liability is a key goal of addiction research. Additionally, preventing relapse is a prime point for pharmacotherpeutic intervention in addiction treatment [5].
To model persistent neurobiological changes after cocaine self-administration, animal models have been developed that demonstrate incubation, an increased drug-seeking or drug-taking behavior with abstinence (for review see [6,7]). We have previously reported a rodent model of increased cocaine reinforcing efficacy with enforced abstinence [8,9]. In this model, animals exposed to discrete trial (DT) self-administration followed by varying periods of enforced abstinence, revealed increased extinction responding following 30 and 100 days of enforced abstinence [9]. That is, following 30 or 100 days of enforced abstinence, rats work harder (as measured by pressing a previously-active lever) than rats with no abstinence, or shorter periods of abstinence, which is interpreted as an increase in drug-seeking behavior.
Several research teams, including ours, have explored alterations in gene and protein expression in these models of relapse liability [10,11,9]. Although there have been important advances, neuroproteomic approaches have not been applied to these models of relapse liability. Previously, we and others have described the proteomic technologies applicable to addiction studies [12,13].
In the current study, large-scale discovery proteomic techniques were used to gain a better understanding of the proteomic changes occurring with cocaine administration that persist with abstinence, and those occurring specifically during abstinence, in the medial prefrontal cortex (mPFC). The mPFC is a component of the mesolimbic reward pathway and has been clinically demonstrated to be activated in drug craving [14,15]. Persistent molecular alterations in the mPFC, even with extended abstinence may be a neurobiological contributer to this behavior. The elucidation of the molecular events related to cocaine self-administration and abstinence will contribute to the understanding of drug relapse, and may be useful in the development of clinically relevant measures to prevent or reduce the likelihood of relapse behaviors in the human population.
MATERIALS AND METHODS
Animals
Male Sprague-Dawley rats (Harlan Inc., IN), weighing approximately 300–400g at the start of the experiments, were used. Rats were maintained on a reverse light/dark cycle (lights on 3:00 pm to 3:00 am) with food (Ralston, Purina, St. Louis, MO) and water available ad libitum. All research was approved by the Wake Forest University School of Medicine and Penn State College of Medicine Animal Care and Use Committees and conducted according to the Guide for the Care and Use of Laboratory Animals, as promulgated by the National Institutes of Health.
Surgery
Rats were anesthetized with a ketamine (100 mg/kg, i.p.) and xylazine (8 mg/kg, i.p.) combination following a 3- to 7-day acclimation period. Anesthetized rats were implanted with a chronic indwelling Silastic cannula in the right jugular vein as described previously [16]. The cannula exited the skin on the dorsal surface of the scapulae region, and was attached to a counterbalanced, fluid swivel mounted above the operant chamber allowing free movement within the chamber. Tygon tubing connected the swivel to an infusion pump (Razel Scientific Instruments, Inc.) with a 5-rpm motor. The operant chambers were 25×25×25 cm in size, and served as both housing and testing chambers.
Cocaine self-administration
Cocaine hydrochloride was obtained from the National Institute on Drug Abuse (Rockville, MD), dissolved in sterile 0.9% saline and passed through a 0.2 μm filter. The dosages are expressed as the salt.
Rats were trained to self-administer cocaine following recovery from surgery through exposure to a fixed ratio 1 (FR1) schedule of reinforcement as described previously [8,9]. Briefly, a retractable lever was extended into the cage to signal session onset for each daily session. Response on the lever resulted in the delivery of 1.5 mg/kg cocaine over a 5 second period. At the start of each injection, the lever retracted and a stimulus light was activated for 20 seconds signaling a time-out period. Each training session lasted until 40 infusions had been self-administered, at which time the lever was retracted until the next daily session began.
Conditions were changed to a discrete-trials schedule after the rats had established a stable daily pattern of cocaine intake for at least 5 days (40 infusions within 6 hours, and no increasing or decreasing trends in time to finish the session). Subsequently, rats were given access to cocaine during 10-min discrete trials that were initiated at 15-min intervals with the introduction of a retractable lever into the chamber. A response resulted in an infusion of cocaine, which was signaled by the illumination of a stimulus light for 20 sec. The lever was retracted and the trial terminated if an injection (1.5 mg/kg/inj) was collected or if 10 minutes had elapsed. Rats received four discrete trials per hour (i.e., DT4), 24 hours per day for 10 days. Following 10 days of self-administration, animals were placed in standard polycarbonate cages for 1 or 100 days. Following this deprivation period, rats were sacrificed and the brains were rapidly removed for protein isolation and analysis.
Dissection
Brains were rapidly removed, cooled in ice-chilled saline and placed in an ice-chilled ASI brain slicer (ASI Instruments, Warren MI). The section from Bregma +4.2 to 2.2mm [17] was cut along the forceps minor and the cortex medial of this cut was collected. This is considered the medial prefrontal cortex, and includes the cingulated area, prelimbic cortex, and medial orbital cortex.
Protein preparation
Brain tissue was suspended in 10 volumes of protein solublization buffer (0.2% Triton-X-100, 25mM HEPES pH 8.0, 250mM sucrose, 100uM EDTA, 1mM DTT, and 1mg/mL protease inhibitor cocktail (Sigma)). Tissue was sonicated 3 times for 10 seconds (with one minute on ice between each sonication) and centrifuged at 10,000g for 10 minutes. Supernatant was collected and assayed using a bicinchoninic acid protein assay (Pierce Endogen).
2D-DIGE
2D-DIGE (2-dimensional differential in-gel electrophoresis) was performed as previously described [18]. All 2D-DIGE methods and results are provided in MIAPE-GE compliant form [19] (Supplementary Table 1). Protein from 6 animals in each of three groups (n=6; Naïve, 1 day of abstinence, and 100 days of abstinence) was purified from lipids and nucleic acids by precipitation (2D-Cleanup, GE Healthcare) and quantified using the 2D-Quant protein assay (GE Healthcare). Samples were brought to pH 8.5 and 50μg of each sample were randomly assigned to be labeled using Cy3 or Cy5 dye (GE Healthcare). Each animal was individually analyzed in this manner (n=6 for each of the three groups). A pool of equal amounts of each sample was labeled with Cy2 dye and 50μg of this pool was combined with a Cy3 and a Cy5 labeled sample and loaded onto each gel. A separate 200μg of unlabeled protein pool was used for a preparative gel for mass spectrometry identification of protein species. A higher protein load on this gel was used to facilitate identification by mass spectrometry. Samples were focused on 24cm pH 4–7 IEF gels (GE Healthcare) and separated by molecular weight on 10% polyacrylamide gels at 2W/gel overnight in 1X Tris/Glycine/SDS running buffer (BioRad). Upon completion of electrophoresis, the preparative gel was fixed in 30% methanol, 7.5% acetic acid, and stained with SyproRuby (Bio-Rad). Gels were imaged on a Typhoon 9410 scanner at a resolution of 100 microns (GE Healthcare). Analysis of gel images was performed using DeCyder 6.5 software (GE Healthcare) to detect protein spots and calculate relative expression values.
MALDI-ToF/ToF Mass Spectrometry
MALDI-ToF/ToF mass spectrometry was performed as described previously [18,20]. Protein spots with a significance of p<0.05 and fold change ± 1.1 were picked for identification by mass spectrometry. Protein spots were removed from the preparative gel using a robotic Ettan Spot Picker (GE Healthcare) and gel plugs were washed twice with 200mM ammonium bicarbonate/40% acetonitrile for 30 minutes and then dehydrated in 75% acetonitrile for 20 minutes. Gel plugs were then dried in a vacuum centrifuge at 30°C and incubated overnight at 37°C in 20μL of 0.02μg/μL trypsin in 40mM ammonium bicarbonate/10% acetonitrile. Digested proteins were extracted from gel plugs with the addition of 50% acetonitrile/0.1% TFA for 30 minutes. Extracted peptides were dried to completion in a vacuum centrifuge at 30°C and resuspended in 0.5% TFA. Samples were then desalted using C18 ZipTips (Millipore) according to the manufacturer’s protocol and as described previously [18] and eluted in 5 μL of 50% acetonitrile/0.1% TFA. 1.5μL of the resulting peptide fragments from each gel plug were spotted onto a 384-position MALDI sample plate with 0.7μL of 2mg/mL ACH-Cinnamic acid/10mM ammonium monobasic phosphate. MS and MS/MS spectrometry were performed in peptide mass fingerprinting approach using a 4800 Proteomics Analyzer (Applied Biosystems). The instrument was calibrated with Applied Biosystems Calibration Mixture 1. Measurements were taken in the positive ion mode between 800 and 4000 m/z with a signal to noise filter of 10, mass exclusion tolerance of 0.2Da, and a peak density filter of 50 peaks per 200Da. The 10 most intense ions with a minimum signal to noise of 75 that were not included on an exclusion list containing trypsin autolysis, matrix, and tryptic peptides of human keratin were subjected to MS/MS. MS/MS was performed without collision-induced decay in a mass range from 60Da to 20Da below the precursor mass with a fragment tolerance of 0.2Da for +1 charged ions. Data were submitted to a MASCOT (v2.0.00) search engine for protein identification with GPS Explorer 3.5 software (Applied Biosystems) using the Rattus NCBI database downloaded on Feb 16, 2008 (107,758 entries searched). Proteins were considered identified with a MASCOT combined MS and MS/MS protein score confidence interval of greater than 97%.
MIAPE Standards Data
In agreement with the developing proteomic standards promulgated by the MIAPE working groups [21], we have included all pertinent MIAPE standard information in Supplementary Table 1 for both the 2D-DIGE experiments (MIAPE-GE version 0.4) [21] and protein identification (MIAPE-MSI version 1.1) [19]. This data is presented in outline format for ease of use.
Western Blot Analysis
Immunoblot analysis was performed for proteins of interest to selectively confirm changes from 2D-DIGE analysis. 10 μg of Naïve, 1 day and/or 100 days abstinent protein samples (n=6) were resolved by denaturing SDS-PAGE gel on 4–12% Nu-PAGE pre-cast gels (Invitrogen). Protein was then transferred onto Immobilin-P transfer membranes (Millipore) and blocked in 5% milk; PBS+0.1% Tween 20. Blots were then probed with dynamin-1, SNAP-25, actin (Chemicon International) and neurofilament medium primary antibodies (Santa Cruz Biotechnologies). Membrane-bound antibodies were detected with horseradish peroxidase conjugated secondary antibodies (GE Healthcare) and visualized using enhanced chemiluminescence (Perkin-Elmer). Dynamin-1, SNAP-25, and neurofilament medium bands were quantified using Image Quant TL (GE Healthcare) and values were normalized to the quantity of actin present in each lane.
Statistics
Significance from 2D-DIGE analysis was determined with DeCyder 6.5 software (GE Healthcare) based on one-way analysis of variance (ANOVA) with a post hoc Student’s t-test. Statistical significance for the confirmatory western blot analyses was determined using a one-tailed t-test. Principle Components Analysis (PCA) was completed using the GeneSpring 7.3 software (Agilent Technologies) using expression data for the entire data set, as well as the 32 significantly changed proteins (p<0.05, ANOVA).
RESULTS
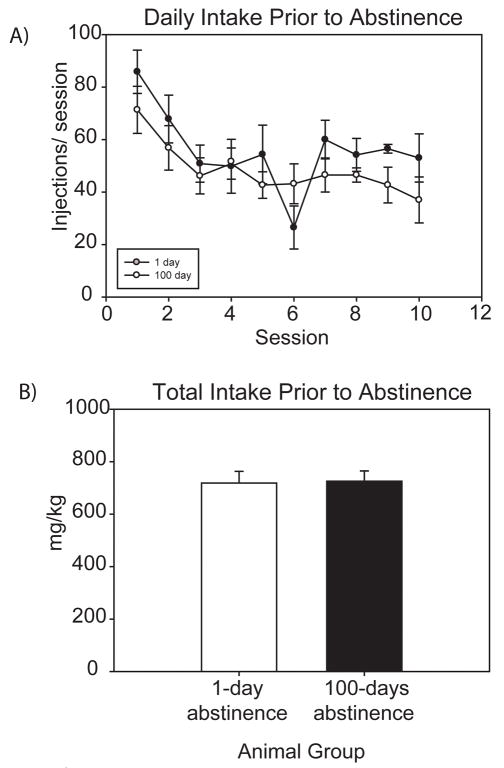
Behavioral data from the rats used in this study has been previously published [9]; however, specific data for those individual rats used in this study have been included. Rats placed on a DT4 schedule of cocaine self-administration establish stable patterns of intake over a 10-day period (Figure 1). There was no difference in the number of self-administration injections between the animals destined for the 1 and 100 day abstinent groups (Figure 1a). Total intake at the end of the 10 day self-administration protocol was not significantly different between the 1 and 100 day abstinent groups, with total intake values of 718 mg/kg and 725 mg/kg, respectively (Figure 1b). As previously described, 100 day abstinent animals will show increased extinction responding when compared to those after 1 day of abstinence [9]. The animals in this study were not subjected to an extinction session, however, to avoid any protein changes caused by responding during an extinction session. Therefore, the 1 and 100 day groups examined in this study have had the same drug self-administration history, but the 100 day animals will exhibit greater drug seeking if re-exposed to the opportunity for self-administration.
Figure 1. Cocaine intake of rats used for 2D-DIGE analysis.
A) Both groups of self-administering rats (1 and 100 days abstinence) demonstrated similar intakes across self-administration sessions (prior to abstinence). B) Total cocaine intake over the 10 day self-administration period was not significantly different between the groups. Average intake for the 1-day abstinent group was 718.3 mg/kg, while the 100-day abstinent group averaged 725.5 mg/kg.
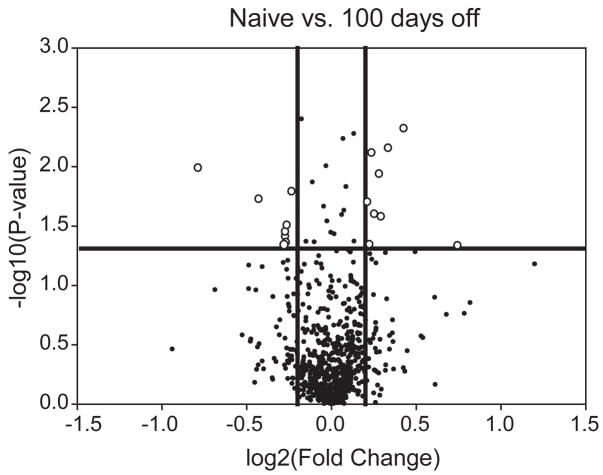
2D-DIGE analysis was performed on protein from naïve, 1 day and 100 days abstinent rats (n=6 in each group). These comparisons would provide the opportunity to identify changes occurring with cocaine self-administration that do not persist (changed naïve vs. 1 day), those changes that occur with cocaine self-administration and persist even with abstinence (changed both naïve vs. 1 day and naïve vs. 100 days), and changes which occur only with abstinence (changed naïve vs. 100 day). The latter two comparisons were of particular interest as these changes would track with the increased drug-seeking behavior. In the 2D-DIGE experiment, 735 protein spots were visualized (Figure 2). Spots with statistically significant changes via ANOVA (p<0.05) were examined. To better determine changes in protein expression, or modification, these changes were also subjected to a ± 1.1 fold change cut-off, yielding 50 changes between the three independent groups, corresponding to 32 spots. This approach of a statistical and fold change cut-off is adapted from the microarray field to reduce the number of false-positives, without overly conservative multiple testing corrections [22]. The fold change level was determined from the drug addiction literature, where small magnitude changes have been demonstrated to be behaviorally relevant (e.g., the landmark work by Self and colleagues demonstrating that a 22–40% change in GluR1 levels may promote extinction of cocaine-seeking [23]). Specifically, 20 changes were observed between Naïve and 1 day, 16 between naïve and 100 days and, 14 between 1 and 100 days. The remainder of the protein spots (703, 96%) did not exhibit a statistically-significant change. For visualization of long-term changes (changes persisting following 100 days of abstinence), a volcano plot was generated with a fold change and p-value cut-off, which includes 20 changed protein spots (Figure 3).
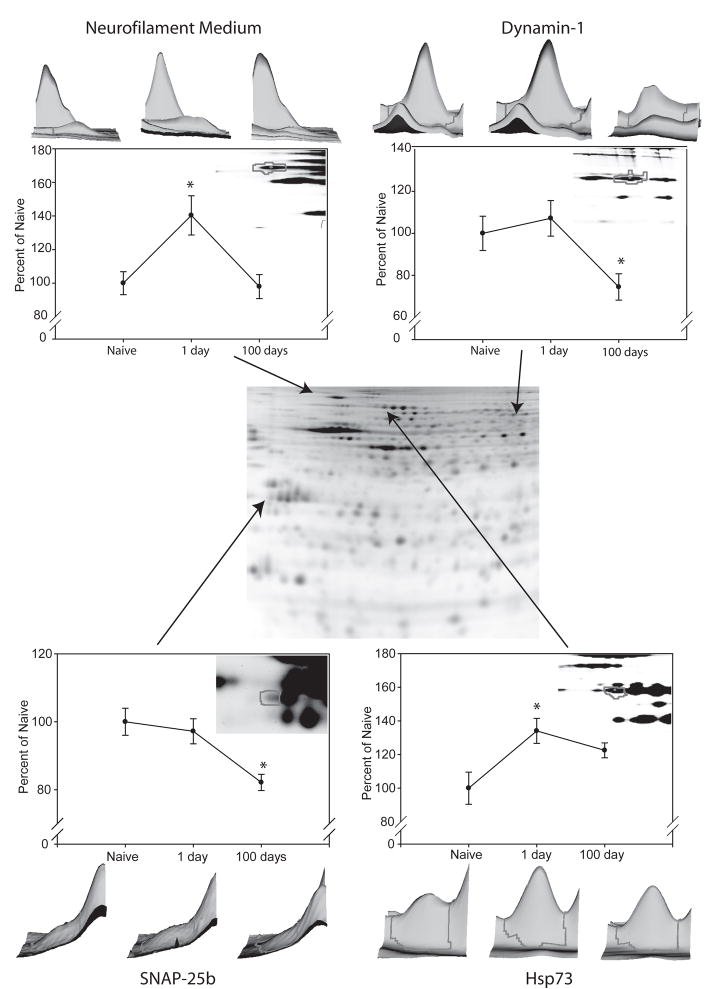
Figure 2. 2D-DIGE analysis with identified proteins of interest.
Samples from naïve, 1 day abstinent, and 100 day abstinent rats (n=6) were analyzed using 2D-DIGE. The center image is an example of a resulting gel, where separated proteins range in isoelectric point from pH 4–7 (left to right) and in molecular weight from ~140 to 20kDa (top to bottom). 4 proteins of particular interest are presented. 3-dimensional representations and graphs of each spot are created using DeCyder 6.5 software based on signal intensity and spot volume and are shown for neurofilament medium (upper left), SNAP-25b (lower left), dynamin-1 (upper right) and Hsp73 (lower right). *p<0.05, ANOVA followed by t-test.
Figure 3. Volcano plot analysis of persistent proteomic changes.
Significant changes were observed between naïve and 100 day abstinent rats (t-test p<0.05; above the horizontal line). Adding a fold-change cut-off of ± 1.20 (outside of the vertical lines) reveals 20 changes of greater than ±1.2 fold at a p-value of <0.05 (open circles).
Of the visualized protein spots, all 32 changes (ANOVA p<0.05) were examined by MALDI-ToF/ToF mass spectrometry for protein identification. Additionally, over 200 unchanged protein spots were examined by MALDI-ToF/ToF mass spectrometry for protein identification to provide identities for subsequent ontological analysis. Of those submitted for identification, 182 protein spots were identified with high confidence, including 20 of the 32 protein spots that were changed (Tables 1 and Supplementary Table 2). The protein identifications are separated into three groups, based on their profile over a time course: (1) changes that occur during drug administration that do not persist (naïve vs. 1 day); (2) changes that occur with drug administration and persist (naïve vs. 1 day and naïve vs. 100 days); and (3) proteins unchanged by drug administration but changed during abstinence (naïve vs. 100 days). Included in these changes are several proteins of interest, including neurofilament medium (1.20-fold increase naïve vs. 1 day, p=0.012); synaptosome-associated protein, 25kDa (SNAP-25; 1.20-fold decrease naïve vs. 100 day, p=0.031), dynamin-1 (1.34-fold decrease, naïve vs. 100 day, p=0.019), and heat shock protein 73 (HSP73) (average 1.32-fold increase, naïve vs. 1 day) (the average represents a number of separate protein spots that were identified as HSP73) (Figure 3).
Table 1.
Statistically Significant Changes in Protein Expression
| Proteins Changed with Drug Administration that DO NOT PERSIST following 100 days of abstinence | Change at 1 day | |
|---|---|---|
| Fold Change | P-value (t-test) | |
| Alpha-enolase | −1.15 | 0.011 |
| Alpha-internexin | +1.16 | 0.005 |
| Gamma-actin | +1.20 | 0.01 |
| Heat shock protein 73 | +1.32 | 0.030 |
| Neurofilament medium polypeptide | +1.20 | 0.012 |
| Proteins Changed with Drug Administration that PERSIST following 100 days of abstinence | Change | |||
|---|---|---|---|---|
| at 1 day | at 100 days | |||
| Fold Change | P-value (t-test) | Fold Change | P-value (t-test) | |
| Alpha-internexin | +1.16 | 0.005 | +1.10 | 0.020 |
| Creatine kinase | +1.23 | 0.010 | +1.10 | 0.049 |
| Dihydropyrimidase related protein-2 (DRP-2) | +1.14 | 0.015 | +1.35 | 0.003 |
| H+ transporting ATPase | −1.11 | 0.014 | −1.13 | 0.005 |
| Phosphatidylethanolamine-binding protein (PEBP) | +1.10 | 0.030 | +1.10 | 0.014 |
| Similar to capping protein beta subunit | −1.11 | 0.034 | −1.18 | 0.013 |
| Proteins UNCHANGED with Drug Administration and CHANGED BY ASTINENCE | Change at 100 days | |
|---|---|---|
| Fold Change | P-value (t-test) | |
| ATP synthase beta subunit | +1.18 | 0.004 |
| Dynamin-1 | −1.34 | 0.035 |
| Glyoxylase domain-containing protein 4 | −1.21 | 0.0076 |
| Malate dehydrogenase 1 | −1.21 | 0.039 |
| SNAP-25B | −1.20 | 0.031 |
| Tumor protein, translationally-controlled 1 | −1.11 | 0.012 |
All proteins shown have ANOVA p<0.05
Using the PANTHER 6.0 database (Applied Biosystems) for ontological analysis, the population of identified proteins was examined to determine if any particular protein classes or biological processes were over- or under-represented in the population [24,25,26]. Of the total population of proteins identified in this study, proteins related to cell structure are over-represented in the group of changes (p<0.05). Dynamin-1, alpha-internexin, and neurofilament medium are all proteins involved in cell structure and are part of a group that is over-represented in the group of changed proteins.
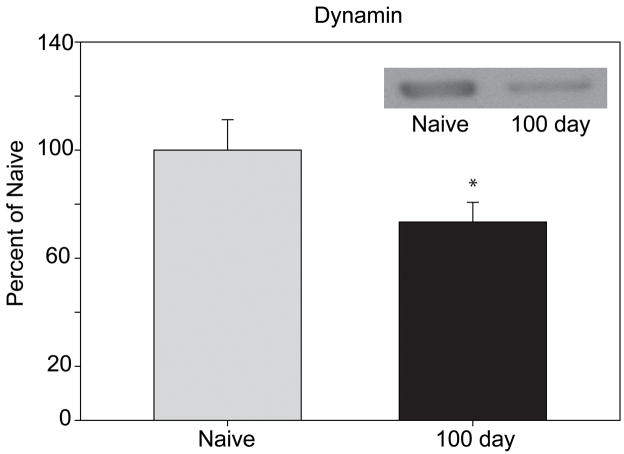
Selected post-hoc confirmation was performed to validate the findings of the 2D-DIGE results and determine if the changes were in total protein abundance or in a specific modification. As cell structure-related proteins were found to be over-represented by ontological analysis, dynamin-1 and neurofilament medium levels were examined by immunoblotting. A statistically significant decrease in dynamin-1 levels during abstinence (1.27-fold decrease, p=0.037) was confirmed by immunoblotting (Figure 4). This finding confirms not only the dynamin-1 quantitation but provides evidence for the validity of the other measurements in this study. Neurofilament medium showed a small magnitude increase between naïve and 1-day abstinent animals, similar to that observed via 2D-DIGE, however, this change failed to reach statistical significance (data not shown).
Figure 4. Western blot analysis of dynamin-1 levels.
Levels of dynamin-1 were examined via western blot. Dynamin-1 levels between naïve and 100 day abstinent animals (n=6/group) showed a 1.27 fold decrease (p=0.037, t-test). The image in the upper right corner shows representative signal from the resulting blot.
Total protein levels of SNAP-25 were also examined by immunoblotting and were not found to be significantly changed (data not shown). By 2D-DIGE analysis, multiple forms of SNAP-25 were found, however, only one was changed in a statistically significant manner. The 2D-DIGE and immunoblotting results suggest that the change in SNAP-25 is a post-translational modification which will not be observed on a 1-dimensional gel, which combines all isoforms into a single band. This also highlights the challenge to proteomic studies of confirming post-translational modifications, as this is both technically difficult and not amenable to high-throughput analyses. Confirmation of all the changes observed by 2D-DIGE was not undertaken, especially for those changes which did not persist with abstinence, as these are unlikely to be related to the increased drug-seeking behavior of interest.
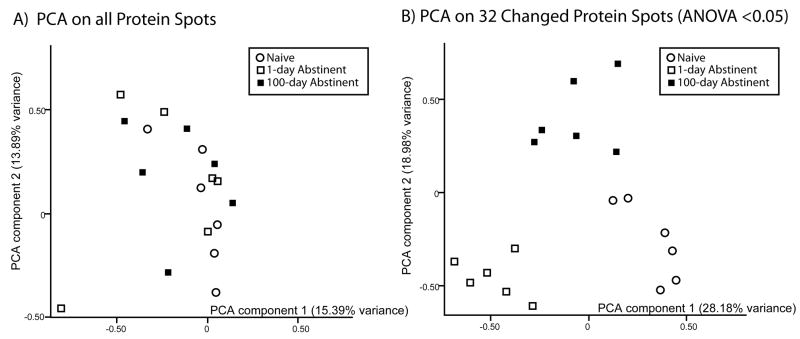
Finally, to visualize the relationship of the animal groups, a principal component analysis (PCA), was used to cluster the individual animals based on expression patterns. PCA, using all protein spots in the study (735), did not segregate the individual animals into their separate groups (Figure 5A, each spot on the graph represents the expression pattern of an individual animal). This confirms a portion of the basic hypothesis of the study that only specific proteins are changed in expression and/or modification and that there is not a major, global shift in the mPFC proteome with cocaine self-administration and abstinence. When the PCA analysis was performed again using the expression pattern of the 32 changed protein spots (ANOVA p<0.05) (Figure 5B), the individual animals separated into their respective groups. The 1 day and 100 day animals are resolved from the naïve animals along the X-axis (principal component 1), although the 100 day group is closer to the naïve. However, the 100 day group is also separated from the other groups on the Y-axis (principal component 2). The major finding of this visualization is that even after 100 days of abstinence from cocaine self-administration, the animals in the 100 days group do not return to a pattern identical to naïve animals, but rather remain in a distinct space.
Figure 5. Principal component analysis of changed proteins.
PCA analysis allows visualization of the protein expression patterns of individual rats. A) PCA analysis using data from all of the protein spots. Based on the expression profiles of all protein spots, individual rats do not segregate into groups. B) PCA analysis using data from changed proteins. Analysis of the expression profiles of changed proteins only (p<0.05) correctly separates individuals based on grouping: naïve (open circles), 1 day (open squares) and 100 days abstinent (closed squares).
DISCUSSION
Once established, maintaining drug abstinence is a primary goal of substance abuse treatment [1]. With this goal in mind, we have used a behavioral model of drug relapse liability to examine proteomic changes in the mPFC, a key activation site in drug craving and relapse [14,15]. Previously, we have demonstrated mRNA expression and epigenetic changes in the mPFC with this model that persist even with prolonged abstinence from cocaine self-administration [9]. Alterations in brain mRNA [9] and protein levels [27,11,28,29] have been reported in many model systems and may be regulated, in part, through epigenetic changes [9,30]. However, long-lasting protein changes have not been examined in this self-administration model of relapse liability.
In studies of cocaine self-administration and abstinence, persistent neuronal protein changes are of such specific interest because they may underlie the potential to relapse even after prolonged abstinence. Grimm and colleagues have shown increases in brain-derived neurotrophic factor (BDNF) protein in the VTA, NAc, and amygdala following 30 and 90 days of withdrawal from cocaine self-administration [29]. Conversely, increases in the levels of glutamate receptor subunits (GluR1 and NR1) have been observed in these brain areas and in the striatum after similar periods of abstinence [31,28,27]. Of specific interest, changes in glutamate receptor subunit expression have been linked behaviorally to cocaine craving and relapse. Changes in GluR1 and GluR2 in the nucleus accumbens have been independently identified to be associated with increased extinction responding and drug craving [23,10].
Although the aforementioned proteins were not observed in this proteomic screen, a number of additional proteins have been identified that may contribute to the development and/or expression of the increased drug-seeking and drug-taking behavior that occurs during abstinence. While many protein changes that occur during cocaine self-administration return to normal levels sometime between 1 and 100 days of enforced abstinence, many proteins levels remain changed even after 100 days. Others are altered during the abstinence period, after remaining unchanged during cocaine administration. These may be of importance to the incubation of cocaine seeking/taking. The proteins, identified in this study to be changed, create a physiological profile that, when visualized by PCA, allows for an accurate grouping of animals based on the biochemical consistency of the molecular fingerprint of cocaine withdrawal. The result of this is a separation of naïve, non-incubated (1 day), and incubated (100 days) groups strictly by proteomic profile. In a general context, while not producing a predictive model, this visualization tool demonstrates that the mPFC neuroproteome does not return to a naïve state even after extended abstinence from cocaine self-administration. This clustering is an effect that is seen when observing the profiles of changed proteins, but disappears when examining the expression profiles of all protein spots, indicating the importance of this specific subset of proteins.
Of the identified proteins, a number of metabolic proteins were persistently changed, indicating a possible role for altered metabolism associated with withdrawal from cocaine administration, an observation made previously in non-human primates [32]. In addition, PANTHER analysis of all the identified proteins revealed an over-representation of cell structure proteins among the changes. This list, including dynamin-1, neurofilament medium, and alpha-internexin, highlights the important role that these proteins may play in neuronal restructuring that occurs following cocaine abuse and abstinence (for a review, see [33]). A number of changed proteins have also been previously identified to play a role in substance abuse, including neurofilament medium, SNAP-25, and dynamin-1. HSP73 is also highlighted as a protein of interest based on its cellular role.
Neurofilament medium is a member of the neural intermediate filament family that also includes neurofilaments heavy and light. Present in the axons of neurons, neurofilaments play a role in axon growth. Although alterations in the levels of neurofilament medium do not persist into periods of abstinence, an increase in levels directly following self-administration could indicate a remodeling of the neuronal architecture. Additionally, a small-magnitude increase in alpha-internexin, a protein known to associate with neurofilaments, reinforces the possibility of increases in axonal growth that could be capable of producing long-lasting changes in neuronal structure. Previous studies have shown decreases in the levels of these neurofilaments in the VTA following repeated cocaine, morphine, or alcohol administration [34,35] as well as following chronic nicotine administration [36].
Changes in synaptic proteins are well represented in these findings, and demonstrate the importance of the role of synaptic communication in withdrawal-associated behaviors. Dynamin-1, a key player in endocytosis of synaptic vesicles, has also been implicated in substance abuse, and has been studied during periods of drug withdrawal. Garcia-Fuster and colleagues have reported increases in dynamin-1 levels in the cerebral cortex with chronic heroin and morphine administration, as well as heroin withdrawal [37]. Results from other groups have confirmed this finding following chronic morphine [38] and have shown that the localization of dynamin-1 at the post-synaptic density is also altered by morphine [39]. Changes in dynamin-1 levels associated with cocaine administration and withdrawal, however, have not been reported previously.
Another synapse-associated protein, SNAP-25, associated with the soluble NSF attachment receptor (SNARE) complex involved in vesicle exocytosis, is also altered by chronic morphine. Alterations in phosphorylation states of SNAP-25 have been associated with decreases in SNARE complex formation following chronic morphine [40]; however, this is the first demonstration of changes following cocaine administration. The changes observed in this study provide evidence of alterations in a post-translationally modified form of SNAP-25, which could possibly represent a phosphorylated state. Both dynamin-1 and SNAP-25 are unaltered after 1 day of enforced abstinence; however, both are decreased following 100 days of abstinence, possibly indicating a decrease in the ability for and/or efficiency of synaptic communication that develops during abstinence periods.
HSP73 belongs to the family of heat shock proteins that is involved in response to stressors within cells. Unlike many other heat shock proteins, HSP73 is constitutively expressed. Although changes in levels of HSP73 have been linked to cell death in Parkinson’s disease [41], HSP73 levels have not been linked to substance abuse. However, levels of HSP73 have been shown to decrease in the embryonic mouse brain upon exposure to ethanol [42]. Our observed increases in levels of HSP73 indicate a cocaine-induced stress upon cells in the mPFC following cocaine self-administration and acute abstinence.
Much as in the microarray field, there is an ongoing debate over the best statistical methods to minimize both Type I (false positive) and Type II (false negative) errors in discovery proteomic experiments. Power calculations for 2D-DIGE have been proposed that suggest the need for high samples numbers (N) to detect smaller magnitude changes [43]. The proposed power calculations reduce the likelihood of Type I errors, but retain the possibility of increased Type II error. Additionally, power calculations will be dependent on the species, organ system, and treatment studied. Previously, 2D-DIGE has identified consistent changes of less than 1.4-fold in the brain [44,45]. Our statistical analysis was based on our previous experience with functional genomic [9] and proteomic [18] studies of drug abuse as well as previous reports of behaviorally-relevant magnitudes of protein expression changes in the brain with cocaine-abuse [23] and using the standards in the microarray field as guidance [46]. Even with this rationale for our statistical analyses, we performed a confirmation experiment on dynamin-1 and validated that the 34% decrease observed by 2D-DIGE was recapitulated as a 27% decrease by immunoblotting. As the proteomic field continues to search for high-throughput confirmatory methods, similar to qPCR confirmation of microarray data, verification of proteomic discovery data currently presents a challenge due to the variety of post-translational modifications and isoforms identified, which are not quantified by standard western blotting. New methodologies, such as multiple reaction monitoring, offer potential solutions to this difficulty, but overcoming this challenge is a crucial next step in proteomics to compliment the advances in discovery approaches.
The limitations involved in this type of study revolve around the collective expression of several different cell types (neurons, astrocytes, microglia, oligodendrocytes), each with different gene and protein expression patterns [47]. Addiction studies are generally focused on the neuronal population, which comprises only 20–30% of the cellular population of the brain [48]. This mixture of cellular populations has a direct effect on quantitation resulting in a muting of expression differences that occur in a subset of cells. Improvements in laser-capture microdissection may allow for future proteomic studies to examine specific cell types [49].
Another challenge to neuroproteomic studies is that proteomic methods are limited in the number of proteins they can examine. Unlike gene expression studies which can examine tens of thousands of genes, proteomic studies, regardless of the specific techniques, can only examine a few thousand proteins at a time [12]. Without the amplification techniques of molecular biology, proteomic studies often examine only the most abundant proteins.
Unchanged proteins were identified in this study for the purpose of investigating the classes of proteins represented by whole tissue proteomics (Supplementary Table 2). Indeed, high abundance proteins, such as actin, tubulin, enolases, heat shock proteins, and mitochondrial respiration proteins compromised a large fraction of the over 150 proteins identified. Fractionation techniques, such as synaptosomal or post-synaptic density preparations, among others, may provide an approach to examine less abundant proteins related to cellular regions of interest [39].
The strength of this study lies in the extension of proteomic changes that occur with drug administration into the realm of withdrawal periods using a large-scale discovery proteomic platform. Many of the studies referenced above refer to protein levels while drug (cocaine, heroin, and morphine) is still present within the body (or immediately after cessation of drug administration), while few extend analysis into periods of abstinence. This study demonstrates that many protein changes do not persist into periods of abstinence; however, there are many changes that do persist, leaving cocaine-abstinent animals with an altered proteomic profile. Those changes that persist may contribute to specific functions related to relapse liability. To further understand the implications of the changes identified, future studies will focus on localization and confirmation of these changes to specific cellular populations. Furthermore, the need has been highlighted for technological advancements in neuroproteomic studies of drug abuse and abstinence-associated physiology. Additional methodologies will be needed to increase the coverage and sensitivity of neuroproteomic studies to identify proteins involved in psychiatric disorders, including drug abuse.
Supplementary Material
Supplementary Table 1. Supplementary Table 1: MIAPE Standards Data
Supplementary Table 2. MALDI-ToF/ToF data for identified proteins (Separate files)
Acknowledgments
This work was supported by grants R01DA013770-08 (KEV), F31-DA02281902 (MEL), R01 DA14030 (DCSR)and K01DA13957 (DM). The authors wish to thank the Penn State College of Medicine Proteomics Core for mass spectrometric analysis and technical assistance.
Abbreviations
- BDNF
Brain derived neurotrophic factor
- DT
Discrete trial
- FR
Fixed ratio
- HSP73
Heat shock protein 73
- LCM
Laser capture microdissection
- MDMA
Methylenedioxymethamphetamine
- mPFC
Medial prefrontal cortex
- NAc
Nucleus accumbens
- qRT-PCR
Quantitative RT-PCR
- SNAP-25
Synaptosome-associated protein 25
- SNARE
Soluble NSF attachment receptor
- VTA
Ventral tegmental area
References
- 1.O’Brien CP. A range of research-based pharmacotherapies for addiction. Science. 1997;278:66–70. doi: 10.1126/science.278.5335.66. [DOI] [PubMed] [Google Scholar]
- 2.Gawin FH, Kleber HD. Abstinence symptomatology and psychiatric diagnosis in cocaine abusers. Clinical observations. Arch Gen Psychiatry. 1986;43:107–113. doi: 10.1001/archpsyc.1986.01800020013003. [DOI] [PubMed] [Google Scholar]
- 3.de Wit H. Priming effects with drugs and other reinforcers. Exp Clin Psychopharmacol. 1996;4:5–10. [Google Scholar]
- 4.Sinha R. How does stress increase risk of drug abuse and relapse. Psychopharmacology (Berl) 2001;158:343–359. doi: 10.1007/s002130100917. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien CP. Anticraving medications for relapse prevention: a possible new class of psychoactive medications. Am J Psychiatry. 2005;162:1423–1431. doi: 10.1176/appi.ajp.162.8.1423. [DOI] [PubMed] [Google Scholar]
- 6.Lu L, Grimm JW, Hope BT, Shaham Y. Incubation of cocaine craving after withdrawal: a review of preclinical data. Neuropharmacology. 2004;47(Suppl 1):214–226. doi: 10.1016/j.neuropharm.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Morgan D, Roberts DC. Sensitization to the reinforcing effects of cocaine following binge-abstinent self-administration. Neurosci Biobehav Rev. 2004;27:803–812. doi: 10.1016/j.neubiorev.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Morgan D, Brebner K, Lynch WJ, Roberts DC. Increases in the reinforcing efficacy of cocaine after particular histories of reinforcement. Behav Pharmacol. 2002;13:389–396. doi: 10.1097/00008877-200209000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Freeman WM, Patel KM, Brucklacher RM, Lull ME, et al. Persistent Alterations in Mesolimbic Gene Expression with Abstinence from Cocaine Self-Administration. Neuropsychopharmacology. 2008;33:1807–1817. doi: 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad KL, Tseng KY, Uejima JL, Reimers JM, et al. Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature. 2008;454:118–121. doi: 10.1038/nature06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L, Dempsey J, Shaham Y, Hope BT. Differential long-term neuroadaptations of glutamate receptors in the basolateral and central amygdala after withdrawal from cocaine self-administration in rats. J Neurochem. 2005;94:161–168. doi: 10.1111/j.1471-4159.2005.03178.x. [DOI] [PubMed] [Google Scholar]
- 12.Freeman WM, Vrana KE. In: Quantitative functional genomics and proteomics of drug abuse. Madras BK, Rutter JL, Colvis CM, Shurtleff JD, Pollock JD, Von Zastrow M, editors. Cold Spring Harbor Laboratory Press; 2006. pp. 433–456. [Google Scholar]
- 13.Williams K, Wu T, Colangelo C, Nairn AC. Recent advances in neuroproteomics and potential application to studies of drug addiction. Neuropharmacology. 2004;47(Suppl 1):148–166. doi: 10.1016/j.neuropharm.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Grant S, London ED, Newlin DB, Villemagne VL, et al. Activation of memory circuits during cue-elicited cocaine craving. Proc Natl Acad Sci U S A. 1996;93:12040–12045. doi: 10.1073/pnas.93.21.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childress AR, Mozley PD, McElgin W, Fitzgerald J, et al. Limbic activation during cue-induced cocaine craving. Am J Psychiatry. 1999;156:11–18. doi: 10.1176/ajp.156.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts DC, Goeders NE. Drug self-administration: experimental methods and determinants. In: Boulton AA, Baker GB, Greenshaw AJ, editors. Humana. 1989. pp. 349–398. [Google Scholar]
- 17.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 5. 2005. [DOI] [PubMed] [Google Scholar]
- 18.Freeman WM, Brebner K, Amara SG, Reed MS, et al. Distinct proteomic profiles of amphetamine self-administration transitional states. Pharmacogenomics J. 2005;5:203–214. doi: 10.1038/sj.tpj.6500309. [DOI] [PubMed] [Google Scholar]
- 19.Robin X, Hoogland C, Appel RD, Lisacek F. MIAPEGelDB, a web-based submission tool and public repository for MIAPE gel electrophoresis documents. J Proteomics. 2008;71:249–251. doi: 10.1016/j.jprot.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Freeman WM, Lull ME, Guilford MT, Vrana KE. Depletion of abundant proteins from non-human primate serum for biomarker studies. Proteomics. 2006;6:3109–3113. doi: 10.1002/pmic.200500717. [DOI] [PubMed] [Google Scholar]
- 21.Taylor CF, Paton NW, Lilley KS, Binz PA, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Reid LH, Jones WD, Shippy R, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutton MA, Schmidt EF, Choi KH, Schad CA, et al. Extinction-induced upregulation in AMPA receptors reduces cocaine-seeking behaviour. Nature. 2003;421:70–75. doi: 10.1038/nature01249. [DOI] [PubMed] [Google Scholar]
- 24.Thomas PD, Kejariwal A, Campbell MJ, Mi H, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W, Wesley M, Freeman WM, Liang B, et al. Alterations in ionotropic glutamate receptor subunits during binge cocaine self-administration and withdrawal in rats. J Neurochem. 2004;89:1021–1033. doi: 10.1111/j.1471-4159.2004.02392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, Grimm JW, Shaham Y, Hope BT. Molecular neuroadaptations in the accumbens and ventral tegmental area during the first 90 days of forced abstinence from cocaine self-administration in rats. J Neurochem. 2003;85:1604–1613. doi: 10.1046/j.1471-4159.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 29.Grimm JW, Hope BT, Wise RA, Shaham Y. Neuroadaptation. Incubation of cocaine craving after withdrawal. Nature. 2001;412:141–142. doi: 10.1038/35084134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Choi KH, Renthal W, Tsankova NM, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Hope BT, Crombag HS, Jedynak JP, Wise RA. Neuroadaptations of total levels of adenylate cyclase, protein kinase A, tyrosine hydroxylase, cdk5 and neurofilaments in the nucleus accumbens and ventral tegmental area do not correlate with expression of sensitized or tolerant locomotor responses to cocaine. J Neurochem. 2005;92:536–545. doi: 10.1111/j.1471-4159.2004.02891.x. [DOI] [PubMed] [Google Scholar]
- 32.Porrino LJ, Smith HR, Nader MA, Beveridge TJ. The effects of cocaine: a shifting target over the course of addiction. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:1593–1600. doi: 10.1016/j.pnpbp.2007.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47(Suppl 1):33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 34.Beitner-Johnson D, Nestler EJ. Morphine and cocaine exert common chronic actions on tyrosine hydroxylase in dopaminergic brain reward regions. J Neurochem. 1991;57:344–347. doi: 10.1111/j.1471-4159.1991.tb02133.x. [DOI] [PubMed] [Google Scholar]
- 35.Nestler EJ. Molecular neurobiology of drug addiction. Neuropsychopharmacology. 1994;11:77–87. doi: 10.1038/npp.1994.37. [DOI] [PubMed] [Google Scholar]
- 36.Bunnemann B, Terron A, Zantedeschi V, Merlo PE, et al. Chronic nicotine treatment decreases neurofilament immunoreactivity in the rat ventral tegmental area. Eur J Pharmacol. 2000;393:249–253. doi: 10.1016/s0014-2999(00)00104-7. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Fuster MJ, Ferrer-Alcon M, Miralles A, Garcia-Sevilla JA. Modulation of Fas receptor proteins and dynamin during opiate addiction and induction of opiate withdrawal in rat brain. Naunyn Schmiedebergs Arch Pharmacol. 2003;368:421–431. doi: 10.1007/s00210-003-0801-9. [DOI] [PubMed] [Google Scholar]
- 38.Prokai L, Zharikova AD, Stevens SM., Jr Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: a quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J Mass Spectrom. 2005;40:169–175. doi: 10.1002/jms.736. [DOI] [PubMed] [Google Scholar]
- 39.Moron JA, bul-Husn NS, Rozenfeld R, Dolios G, et al. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol Cell Proteomics. 2007;6:29–42. doi: 10.1074/mcp.M600184-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Xu NJ, Yu YX, Zhu JM, Liu H, et al. Inhibition of SNAP-25 phosphorylation at Ser187 is involved in chronic morphine-induced down-regulation of SNARE complex formation. J Biol Chem. 2004;279:40601–40608. doi: 10.1074/jbc.M406896200. [DOI] [PubMed] [Google Scholar]
- 41.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, et al. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 42.Jing H, Li Y. Effects of ethanol on mouse embryonic brain development and heat shock protein 73 expression. Toxicol In Vitro. 2004;18:601–607. doi: 10.1016/j.tiv.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Karp NA, Lilley KS. Maximising sensitivity for detecting changes in protein expression: experimental design using minimal CyDyes. Proteomics. 2005;5:3105–3115. doi: 10.1002/pmic.200500083. [DOI] [PubMed] [Google Scholar]
- 44.Pinaud R, Osorio C, Alzate O, Jarvis ED. Profiling of experience-regulated proteins in the songbird auditory forebrain using quantitative proteomics. Eur J Neurosci. 2008;27:1409–1422. doi: 10.1111/j.1460-9568.2008.06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawlyk AC, Ferber M, Shah A, Pack AI, et al. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–2313. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- 46.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 47.Cahoy JD, Emery B, Kaushal A, Foo LC, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh SK, Clarke ID, Terasaki M, Bonn VE, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 49.Gutstein HB, Morris JS, Annangudi SP, Sweedler JV. Microproteomics: Analysis of protein diversity in small samples. Mass Spectrom Rev. 2008;27:316–330. doi: 10.1002/mas.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Supplementary Table 1: MIAPE Standards Data
Supplementary Table 2. MALDI-ToF/ToF data for identified proteins (Separate files)