Abstract
A decade after its inception, MALDI imaging mass spectrometry has become a unique technique in the proteomics arsenal for biomarker hunting in a variety of diseases. At this stage of development, it is important to ask whether we can consider this technique to be sufficiently developed for routine use in a clinical setting or an indispensable technology used in translational research. In this report, we consider the contributions of MALDI imaging mass spectrometry and profiling technologies to clinical studies. In addition, we outline new directions that are required to align these technologies with the objectives of clinical proteomics, including: 1) diagnosis based on profile signatures that complement histopathology, 2) early detection of disease, 3) selection of therapeutic combinations based on the individual patient's entire disease-specific protein network, 4) real time assessment of therapeutic efficacy and toxicity, 5) rational redirection of therapy based on changes in the diseased protein network that are associated with drug resistance, and 6) combinatorial therapy in which the signaling pathway itself is viewed as the target rather than any single “node” in the pathway.
MS has become a versatile tool that we are familiar with in large part due to important electronic and informatics advancements. The ability to obtain the molecular weight is one of the first steps in the identification of a molecule. With the addition of primary structural information mass spectrometry has become a useful technique to identify molecules within complex mixtures.
Biological specimens, such as tissues, urine, or plasma, are complex and highly heterogeneous, which makes them inherently difficult to analyze. Further research and developments are necessary to achieve reliable biological models for understanding and studying pathologies. Therefore, it is of primary importance to identify the constituents of these systems and subsequently understand how they function within the framework of the tissue. With regard to clinical proteomics, there is the added dimension of disease, and therefore, the main goal is to characterize the cellular circuitry with a focus on the impact of the disease and/or therapy on these cellular networks.
Mass spectrometry has become a centerpiece technology predominantly in the field of proteomics. Nonetheless a more comprehensive understanding of the constituents of biological systems will be aided by determining the constituent distribution. This anatomical dimension has been added through mass spectrometry imaging (MSI)1 especially using MALDI-MSI.
MALDI is an ion source that is well compatible with the introduction of raw materials and surfaces. Shortly after its introduction, MALDI was used for direct tissue profiling. The first applications were neurobiological studies on dissected organs from the mollusk Lymnaea stagnalis (1–8), crustaceans (9), and other mollusks (10, 11). More recently, MALDI was used to generate profiles from tissue sections and ion images using a scanning method to analyze the surface (12) (Fig. 1). This led to the first MALDI MS tissue section imaging micrographs in 1997 (13–15). These studies were followed by 10 years of intense efforts to improve the sensitivity, reproducibility, data processing, tissue preservation, and preparation treatments to fully characterize the proteome leading to a clear improvement of molecular images (16–39) (Fig. 2).
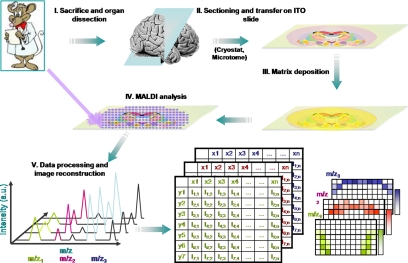
Fig. 1.
Schematic representation of the MALDI-MSI work flow. After tissue sectioning and transfer onto a conductive and transparent sample plate, the MALDI matrix is deposited, and data are acquired by recording mass spectra according to a raster of points covering the surface to be analyzed. Mass spectra recorded with their coordinates on the tissue are processed, and molecular images of the localization of molecules can be reconstructed. a.u., arbitrary units; ITO, idium tin oxide.
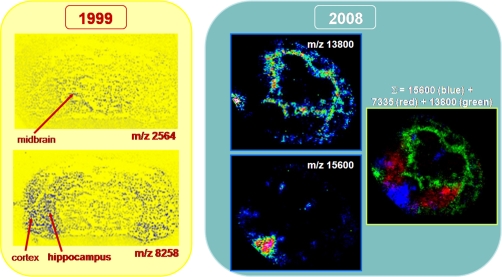
Fig. 2.
Ten years' evolution from one of the first MALDI images presented in 1999 at the 47th ASMS Conference on Mass Spectrometry and Allied Topics (left) (reprinted with permission of Caprioli and co-workers (84)) and molecular images obtained by our group for mouse stem cells injected in brain tissue sections (right) (M. Wisztorski, C. Meriaux, M. Salzet, and I. Fournier, unpublished results).
These developments led to clinical studies using MALDI-MSI technology. Clinical proteomics has many objectives including 1) diagnosis based on signatures as a complement to histopathology, 2) early disease detection, 3) individualized selection of therapeutic combinations that best target the patient's entire disease-specific protein network, 4) real time assessment of therapeutic efficacy and toxicity, 5) rational redirection of therapy based on changes in the diseased protein network that are associated with drug resistance, and 6) combinatorial therapy in which the signaling pathway itself is viewed as the target rather than any single “node” in the pathway.
Based on these key objectives, can we consider MALDI-MSI a mature technology for use in clinical studies? What is the potential impact of this technology in anatomy/pathology and disease? By reviewing each objective, do we have sufficient evidence that MALDI-MSI satisfies the criteria imposed by clinical proteomics? We will now specifically address each of these key points.
DIAGNOSIS BASED ON SIGNATURES AS A COMPLEMENT TO HISTOPATHOLOGY
In some cases, diagnosis or tissue classification cannot be easily achieved through standard histological staining. Further refinements based on molecular signatures and statistical data, which are currently missing, are crucial for improved diagnostics. The development of rapid and reliable screening of human tissues for diagnostics (e.g. biopsies or smears) has been improved with modern proteomics. By using MALDI-MSI, a molecular diagnosis could be done on tissue directly in the environment of the tumors. MALDI-MSI could help to detect the tumor boundary or infiltration of adjacent normal tissue that presents a normal histology. It could also help to detect the early stage of pathology that presents no histological modifications and to prevent tumor recurrence at the site of surgical resection. One of the major advances of MSI is the correlation of the MALDI images with histological information. MALDI-MSI software (for a review, see Ref. 40) superimposes the MALDI images over a macroscopic or microscopic optical image of the sample taken before MALDI measurement. Although the primary macroscopic optical image is sufficient to recognize the outline of the tissue and define the measurement area, it is not usually possible to observe histological features in the image (in contrast to microscopic images). For a histological interpretation, it is necessary to use stained tissue sections. Two approaches have been used to correlate histology with MALDI-MSI results: performing MALDI-MSI and histological staining on consecutive sections (41, 42) or staining the sample after MALDI measurement (43). The latter technique has been successfully used by pathologists (Fig. 3) (44), which suggests that combining MALDI-MSI and classic histological staining provides pathologists with more information to make better diagnoses. The next step is not only to perform a diagnosis based on m/z signatures but also on molecular data generated from identification of specific biomarkers that have been characterized as pathological signatures.
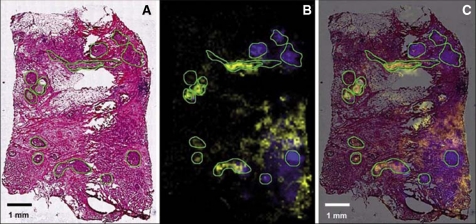
Fig. 3.
Mass species representing molecular features of preinvasive and invasive lesions of the breast. A, optical microscopic image of hematoxylin- and eosin-stained tissue section showing several carcinomatous in situ regions (outlined in green). The staining was done after MALDI measurement of the tissue section. This allows an unambiguous correlation with MALDI imaging results. B, visualization of ion density images of two selected masses (m/z 9750 shown in yellow and m/z 4519 shown in blue). C, overlay of hematoxylin and eosin staining and MALDI molecular image. The distribution of these two masses suggests a divergent clonal evolution of the preinvasive lesions. These two masses are also present in the invasive cancer cells surrounding some carcinomas in situ (right site). Scanning resolution, 80 μm. Scale bar, 1 mm. (Reprinted with permission of Walch et al. (44).)
However, another challenge for pathologists is tissue classification, which is required to catalogue tumors or benign tissues. The major technological improvement that MALDI-MSI provides is the direct identification of novel markers within an in situ context from fixed sections/biopsy embedded in paraffin (e.g. archived material) (42). Several studies on cancer and neurodegenerative diseases have demonstrated that MALDI-MSI is a key technology for identifying biomarkers, assessing their localization, and cross-validation (29, 45–51). The use of archived, formalin-fixed, paraffin-embedded tissue from hospital pathology departments represents a “gold mine” of existing data (42, 52–54). The application of MALDI MS imaging to archived materials could lead to the creation of an international disease marker database that would facilitate the development of early diagnostics for various pathologies as well as for follow-up examination of disease progression.
Therefore, the addition of statistical analysis will be very important for the comparison of the different tissue components (e.g. tumor versus benign or healthy). Each tissue type depends upon the nature of its composition of cells. Thus, biocomputational methods are absolutely necessary to identify individualized molecular patterns to aid in diagnosis and prognosis.
The advantage of MALDI-MSI is the ability to obtain a large collection of mass spectra spread out over a tissue section while retaining the absolute spatial location of these measurements for subsequent analysis and imaging. One of the statistical techniques to reduce the complexity of the information in multidimensional data sets in MALDI-MSI is principal component analysis (PCA) (55). PCA is a multivariate preanalysis tool that allows for the correlation and identification of the major spatial and mass-related trends in the data that guide further downstream analysis (56). PCA reduces the dimensionality of the data set but does not classify the spectra. This is a transformation of the original coordinate system defined by peak intensities to a coordinate system that better explains the variance within the data set. This has been recently used in a prostate cancer study (43).
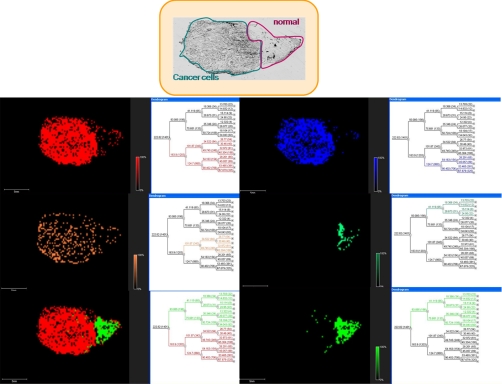
The next required step is the hierarchical clustering of the tissue based on PCA statistical analyses that reflect the most important variance of ions within the tissue (57). Dendrograms can be constructed, and each branch represents ions present in the same group of cells (e.g. epithelial cancer cells versus benign cells). Thus, this representation provides access to huge numbers of individual spectra and reduces the complexity of the data set. It can also be correlated with histology as previously used for mouse kidney (Fig. 4) (44), gastric (58), and ovarian cancer (Fig. 5).
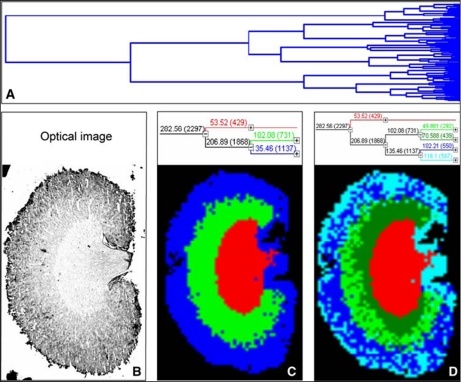
Fig. 4.
Hierarchical clustering of a mouse kidney data set achieved by MALDI-MSI. A, full dendrogram of all spectra in a mouse kidney data set. B, optical image of the mouse kidney analyzed by MALDI-MSI. C and D, reconstruction of selected dendrogram branches and corresponding images. The three main branches reflect the renal cortex (blue), medulla (green), and pelvis (red). C, the medulla branch separates into two distinct areas, whereas the cortex branch further differentiates into fat and connective tissue of the renal capsule and hilus and the actual cortex (D). (Reprinted with permission of Walch et al. (44).)
Fig. 5.
Hierarchical clustering using the ClinProt tool (Bruker Daltonics) after PCA of a stage 4 mucous ovarian carcinoma section covered with ionic matrix using the Shimadzu CHIP 1000 microspotter. a, optical image of the ovarian carcinoma section. b–f, reconstructed selected dendrograms and corresponding images. The two main branches reflect the carcinoma (red; a) and the healthy (green; f) parts in the section. b and c are two carcinoma subclasses, and d is a subclass of the healthy part. e represents a merge of the two branches.
EARLY DISEASE DETECTION
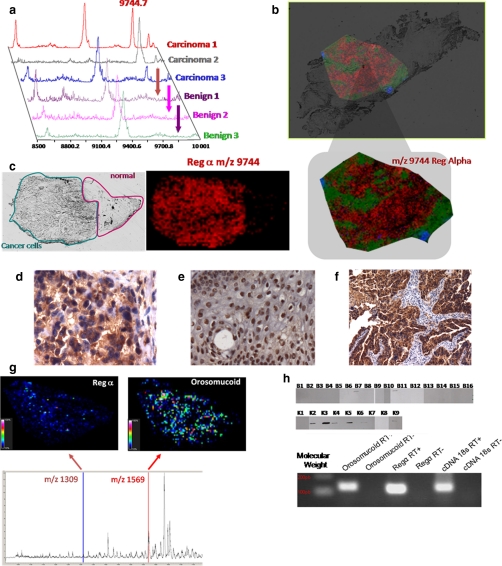
Based on MALDI MS profiling (Fig. 6a) and imaging strategies (Fig. 6, b and c), several biomarkers have been identified in various cancer studies. In stage III and IV ovarian cancer, a highly prevalent (80%) biomarker has been identified using MALDI MS and nano-LC-nano-ESI MS using MS and MS/MS after separation by reverse phase HPLC and trypsin enzymatic digestion. This marker with an m/z of 9744 corresponds to an 84-amino acid fragment from the 11 S proteasome activator complex (PA28 α or REG-α) (33). This biomarker was validated using MALDI MSI (Fig. 6, b and c), classic immunohistochemistry with an antibody raised against the C-terminal part of the protein containing the fragment of interest (Fig. 6, d, e, and f), specific MALDI-MSI using the Tag-mass concept (Fig. 6g) (59), quantitative PCR, and Western blot (Fig. 6h). Recently, we confirmed the REG-α expression in an ovarian epithelial cell line by quantitative PCR.2 Immunohistochemistry confirmed the epithelial expression of this fragment with a nuclear localization in benign epithelial cells and a cytoplasmic localization in carcinoma cells (Fig. 6, d and e). This localization pattern indicates that this antibody can be used to discriminate borderline tumor cases, which are the most difficult to diagnose. Thus, a specific antibody that discriminates between cells transitioning from benign to malignant will be an asset for early diagnosis. Taken together, these studies indicate that direct tissue analysis and specific MALDI-MSI strategies facilitate biomarker identification and validation.
Fig. 6.
Validation of C-terminal fragment of the immunoproteasome REG-α and orosomucoid as ovarian biomarkers. a, MALDI-MS profiles of three ovarian carcinomas versus benign tumor samples. b, MALDI-MSI molecular image of REG-alpha fragment (m/z 9744) at a resolution of 50 μm from an ovarian carcinoma tissue section. c, optical image of the tissue section with the region of interest defined (cancerous versus healthy part) and MALDI-MSI of the REG-α fragment showing its presence exclusively in the cancer part. d–f, immunocytochemical data obtained after the antigen retrieval technique and H&E coloration with the anti-C-terminal REG-α antibody: d, cytoplasm localization of the anti-C-terminal REG-α labeling in ovary carcinoma; e, nucleus localization of the anti-C-terminal REG-α labeling in ovary benign tumor; f, epithelial cells labeled with the anti-C-terminal REG-α in ovarian carcinoma. g, specific MALDI imaging analysis using the Tag-mass concept with anti-C-terminal REG-α and an anti-human IgG tag (reporter m/z 1309) and anti-orosomucoid and anti-human monoclonal antibody (reporter m/z 1569). h, top, Western blot analyses with the anti-C-terminal REG-α (immunoproteasome 11 S) of the 16 benign tumors and nine carcinomas (33); bottom, quantitative PCR validation of REG-α and orosomucoid from the SKVO3 ovarian cancer epithelial cell line.
In addition, data can be obtained from fundamental studies by analyzing the ontogeny of protein expression during morphogenesis and tumorigenesis, and proteins that could potentially serve as biomarkers for diagnosing diseases can be identified as demonstrated by an MSI study on murine prostate cancer development (60). Murine prostate during development (1–5 weeks of age), at sexual maturation (6 weeks of age), and in adulthood (at 10, 15, or 40 weeks of age) was compared with prostate tumors from 15-week-old mice genetically engineered to express the large T antigen gene under the control of the prostate-specific probasin promoter (LPB-Tag mice). This approach identified proteins that were differentially expressed at specific time points during prostate development. The expression of probasin and spermine-binding protein, which are associated with prostate maturation, decreased during prostate tumor formation (60). This study was the first use of MALDI-MSI to follow ontogeny to tumorigenesis (60).
There is no doubt of the usefulness of MALDI-MSI in biomarker development for early diagnosis. However, MALDI-MSI is still not being routinely used in a clinical setting and has not yet been adjusted to conform to clinical proteomics procedures. Only a limited number of international groups have used this technology effectively in clinical settings; however, the number of clinical studies applying MSI has dramatically increased in the past 2 years (43, 44, 54, 61–66).
INDIVIDUALIZED SELECTION OF THERAPEUTIC COMBINATIONS THAT BEST TARGET THE PATIENT'S ENTIRE DISEASE-SPECIFIC PROTEIN NETWORK
MALDI-MSI is highly advantageous for in situ drug tracking. In fact, it enables the detection of both endogenous and exogenous compounds present in tissues with molecular specificity and preserves their spatial orientation. This unique combination coupled with excellent sensitivity and rapid analysis presents potential advantages for a wide range of applications in diverse biological fields. As described previously, recent advances have demonstrated that the technique can be applied to cancer research, neuroscience, and pharmaceutical development (67). MALDI-MSI can be used in clinical studies to provide a molecular ex vivo view of resected organs. This allows for the label-free tracking of both endogenous and exogenous compounds with spatial resolution and molecular specificity (67–77). Several examples support the idea that MALDI-MSI technology will become a key tool in drug development (67–73), including novel drug design through the ability to analyze metabolic pathways directly in tissues (e.g. through in situ multiplex metabolite analysis), as well as in the elucidation of secondary effects and unexpected feedback loops (78). Currently MSI of biomolecules and chemical compounds in cell-based assays and highly complex tissue sections is used in parallel with classic mass spectrometry ionization techniques to identify chemical compounds interfering with enzymatic function, receptor-ligand binding, or molecules modulating a protein-protein interaction.
There is evidence supporting MSI as a key technique that can be used in combination with other therapeutic technologies. Recently the efficacy of combining radiation (XRT) with a dual epidermal growth factor receptor (EGFR)/vascular endothelial growth factor receptor inhibitor, AEE788, in prostate cancer models with different levels of EGFR expression was analyzed using Doppler sonography, tumor blood vessel destruction (visualized by immunohistochemistry), and MSI (76). Tumor xenografts established from DU145 or PC-3 prostate cancer cell lines inoculated into the hind limbs of athymic nude mice were assigned to four treatment groups: 1) control, 2) AEE788, 3) XRT, and 4) AEE788 and XRT. AEE788 had a radiosensitization effect in human umbilical vein endothelial cells and increased their susceptibility to apoptosis. Therefore, concurrent AEE788/XRT treatment compared with either treatment alone led to a significant delay in tumor growth in animals bearing DU145 tumors. Conversely there was no effect on the growth of PC-3 tumors with combination therapy. In DU145 tumors, there was a significant decrease in tumor blood flow with combination therapy as assessed by using Doppler sonography and tumor blood vessel destruction. MSI demonstrated that AEE788 is bioavailable and heterogeneously distributed in DU145 tumors receiving therapy, supporting the efficacy of the combination of AEE788 and XRT in vitro and in vivo in DU145-based models. In contrast, in PC-3-based models, the tumors were adequately treated with XRT alone without any added benefit from combination therapy. These findings correlated with differences in EGFR expression. Overall this study demonstrated the effects of therapeutics on both tumor cell proliferation and vascular destruction using complementary technologies, including MALDI-MSI in a clinical proteomics protocol.
REAL TIME ASSESSMENT OF THERAPEUTIC EFFICACY AND TOXICITY
MSI technology will also help significantly advance the analysis of novel therapeutics and may provide deeper insight into therapeutic and toxicological processes, revealing the mechanism of efficacy or side effects at the molecular level (79). A study by Atkinson et al. (80) using AQ4N (banoxatrone) (1,4-bis-5,8-dihydroxyanthracene-9,10-dione) as a prodrug demonstrated that MSI can be used for both drug and clinical development. In hypoxic cells, AQ4N is reduced to AQ4 (cytotoxic form), which is a topoisomerase II inhibitor. By inhibiting topoisomerase II within these hypoxic areas, AQ4N sensitizes tumors to existing chemo- and radiotherapy treatments. The distribution of AQ4N and AQ4 in treated H460 human tumor xenografts has been examined by MALDI-MSI, and images of the distribution of AQ4N and AQ4 show little overlap (80). The distribution of ATP in the tumor xenografts was studied as an endogenous marker of hypoxia because concentrations of ATP are known to decrease with hypoxia. The ATP distribution was similar to that of AQ4N, suggesting that in regions with abundant ATP expression (i.e. normoxic tissue) there was no evidence of conversion of AQ4N to AQ4. This indicates that the cytotoxic metabolite AQ4 is confined to hypoxic regions of the tumor (80).
RATIONAL REDIRECTION OF THERAPY BASED ON CHANGES IN THE DISEASED PROTEIN NETWORK THAT ARE ASSOCIATED WITH DRUG RESISTANCE
There are no studies that have used MALDI-MSI to redirect therapy. However, this objective will be the next challenge for this research field.
COMBINATORIAL THERAPY IN WHICH THE SIGNALING PATHWAY ITSELF IS VIEWED AS THE TARGET RATHER THAN ANY SINGLE NODE IN THE PATHWAY
Similar to the fifth objective, there are no studies on combinatorial therapy that focus on a signaling pathway as a whole because of the infancy of this technology.
CONCLUSION
MALDI MSI emerged only 10 years ago, so it is still a young technology that is continuously evolving. Further developments are still needed to establish this technology in a clinical setting. For example, standardization of protocols must be undertaken between tissue collection, storage, preparation, and data acquisition. Moreover improvements in resolution will be sought because the minimum area that can be examined is currently in the range of a few cells. The actual resolution that can be routinely achieved while keeping good sensitivity (i.e. sufficient ion yields) is 50 × 50 μm2. To achieve increased resolution it is necessary to reduce the surface area irradiated by the laser beam, which most obviously can be achieved by decreasing the laser beam diameter. However, under 30–40-μm laser beam diameter a decrease in ion production resulting in decreased sensitivity is observed. Thus, active research to decrease the size of the irradiated area without loss of sensitivity or attempts to better understand and optimize the ionization processes involved in MALDI are underway. From our own work, we have developed masks that achieve a resolution of 30 × 30 μm without reducing the production of ions and result in an increased sensitivity of 2–3-fold (81). A new generation of masks at 10 × 10 μm is now being developed. Nonetheless the present levels of sensitivity allow the detection of a small group of cells but are not sufficient to detect discrete modification at a single cell level. However, detection limits are the same as those obtained for the analysis of a complex mixture using classical MALDI-TOF procedures. Contrary to the predictions that the most abundant proteins are the only ones to be detected with MALDI, it has been observed that ionization efficiency is an important parameter and that a low abundance molecule that is well ionized can also be detected. The detection of low abundance proteins can also be improved with the development of statistical software allowing the treatment of large cohorts of patient with large data sets for biomarkers, tissue classification, and stage of disease development. Other limitations include the detectable mass range, which is typically between m/z 400 and 30,000. The lower limit is due to the use of matrix that masks the analysis below this m/z. The upper limitation is not understood, and various protocols are being tested to overcome this limitation, such as new tissue treatments, new matrices, or development of new ion sources to generate multicharged ions. One difficulty that has been overcome is the direct identification of biomarkers on tissues. Bottom-up strategies using on-tissue trypsin digestion have been developed for frozen (31) and formalin-fixed, paraffin-embedded (42, 54) tissues. Ideally the ability to perform on-tissue top-down protein characterization is one of our future objectives. MALDI imaging will ultimately provide high resolution molecular imaging but will also result in direct biomarker identification with statistical validation such that it will become an essential proteomics tool in clinical histopathology.
Additional needed developments will be three-dimensional reconstruction to obtain tumor maps (82). MALDI-MSI will improve tissue classification necessary to perform retrospective studies, will assist clinical studies from the bench to bedside, and will provide a remarkable follow-up procedure. Improved tissue classification using MALDI-MSI on the same tissues used by pathologists for diagnosis will speed up the process of molecular diagnosis. Molecular tissue classification after MALDI-MSI based on known biomarkers or using unsupervised multivariable analyses can positively affect patient treatment. For example, borderline ovarian cancers are difficult to detect clinically until they are advanced in size or stage. The most common presenting symptoms are abdominal pain, increasing girth or abdominal distension, and abdominal mass. Approximately 23% of patients are asymptomatic. With such tumors, correct diagnosis is difficult to reach, and the molecular profiles provided by MALDI-MSI may facilitate classification and aid the development of a treatment strategy. Moreover depending on the nature of the malignancy (e.g. serous or mucous with or without cell infiltration), the therapeutic strategy is different. Should MALDI-MSI tumor classification libraries be created, these could permit clinicians to individually tailor patient treatments in a practical manner. Based on the tissue biomarkers identified by MSI, it could be important to follow up the evolution of the malignancy during treatment, for example after cisplatin treatment to define whether resistance to the treatment may appear. Furthermore in cases where traditional biomarkers cannot be clearly detected in biopsies, MALDI-MSI could become critical to the outcome. This points to the importance of establishing tumor MSI libraries to facilitate multicenter studies and the creation of MSI classification maps.
At this point, it is obvious that further clinical studies using MALDI-MSI technology are required. Nonetheless MALDI-MSI has opened the door to molecular tissue classification, which could be of great use to pathologists with regard to diagnosis but also in drug development and diagnosis coupled with magnetic resonance imaging (MRI) technology. A major advance for MALDI-MSI will be its coupling with positron emission tomography, x-ray, computed tomography instrumentation, and MRI for both preclinical and clinical research. The complementarities between non-invasive techniques and molecular data obtained from MALDI MS imaging will result in a more precise diagnosis. In clinical studies, the need for information on the spatial localization of pathologically gene-encoded products has become more pressing. The three-dimensional volume reconstructions generated by MALDI-MSI data (83) now offer the possibility to compare the molecular data with data obtained using positron emission tomography or MRI. These associations will enhance the use of MALDI-MSI. Ultimately comparing the MRI image of a tumor and the image generated by MALDI-MSI at a molecular level will provide a comprehensive data set for diagnosis and treatment selection.
Footnotes
* This work was supported by grants from the CNRS Département de la politique industrielle (to M. S. and I. F.), Ministère de L'Education Nationale, de L'Enseignement Supérieur et de la Recherche, Institut National du Cancer (to I. F.), Agence National de la recherche (to I. F.), the Institut National du Cancer (to I. F.), and the Canadian Institutes of Health Research (to R. D. and M. S.).
2 M. El Ayel, D. Bonnel, I. Fournier, and M. Salzet, unpublished data.
1 The abbreviations used are:
- MSI
- mass spectrometry imaging
- PCA
- principal component analysis
- XRT
- x-ray therapy
- EGFR
- epidermal growth factor receptor
- MRI
- magnetic resonance imaging.
REFERENCES
- 1.van Veelen P. A., Jiménez C. R., Li K. W., Wildering W. C., Geraerts W. P., Tjaden U. R., van der Greef J. (1993) Direct peptide profiling of single neurons by matrix-assisted laser desorption ionization mass spectrometry. Org. Mass Spectrom 28, 1542–1546 [DOI] [PubMed] [Google Scholar]
- 2.Jiménez C. R., van Veelen P. A., Li K. W., Wildering W. C., Geraerts W. P., Tjaden U. R., van der Greef J. (1994) Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption ionization mass spectrometry of single neurons. J. Neurochem 62, 404–407 [DOI] [PubMed] [Google Scholar]
- 3.Li K. W., Hoek R. M., Smith F., Jiménez C. R., van der Schors R. C., van Veelen P. A., Chen S., van der Greef J., Parish D. C., Benjamin P. R. (1994) Direct peptide profiling by mass spectrometry of single identified neurons reveals complex neuropeptide-processing pattern. J. Biol. Chem 269, 30288–30292 [PubMed] [Google Scholar]
- 4.Li K. W., Jiménez C. R., Van Veelen P. A., Geraerts W. P. (1994) Processing and targeting of a molluscan egg-laying peptide prohormone as revealed by mass spectrometric peptide fingerprinting and peptide sequencing. Endocrinology 134, 1812–1819 [DOI] [PubMed] [Google Scholar]
- 5.Li K. W., van Golen F. A., van Minnen J., van Veelen P. A., van der Greef J., Geraerts W. P. (1994) Structural identification, neuronal synthesis, and role in male copulation of myomodulin-A of Lymnaea: a study involving direct peptide profiling of nervous tissue by mass spectrometry. Brain Res. Mol. Brain Res 25, 355–358 [DOI] [PubMed] [Google Scholar]
- 6.Dreisewerd K., Kingston R., Geraerts W. P., Li K. W. (1997) Direct mass spectrometric peptide profiling and sequencing of nervous tissues to identify peptides involved in male copulatory behavior in Lymnaea stagnalis. Int. J. Mass Spectrom 169, 291–299 [Google Scholar]
- 7.Jiménez C. R., Burlingame A. L. (1998) Ultramicroanalysis of peptide profiles in biological samples using MALDI mass spectrometry. Exp. Nephrol 6, 421–428 [DOI] [PubMed] [Google Scholar]
- 8.Jiménez C. R., Li K. W., Dreisewerd K., Spijker S., Kingston R., Bateman R. H., Burlingame A. L., Smit A. B., van Minnen J., Geraerts W. P. (1998) Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry 37, 2070–2076 [DOI] [PubMed] [Google Scholar]
- 9.Redeker V., Toullec J. Y., Vinh J., Rossier J., Soyez D. (1998) Combination of peptide profiling by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and immunodetection on single glands or cells. Anal. Chem 70, 1805–1811 [DOI] [PubMed] [Google Scholar]
- 10.Rubakhin S. S., Li L., Moroz T. P., Sweedler J. V. (1999) Characterization of the Aplysia californica cerebral ganglion F cluster. J. Neurophysiol 81, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 11.Sweedler J. V., Li L., Floyd P., Gilly W. (2000) Mass spectrometric survey of peptides in cephalopods with an emphasis on the FMRFamide-related peptides. J. Exp. Biol 203, 3565–3573 [DOI] [PubMed] [Google Scholar]
- 12.Caprioli R. M., Farmer T. B., Gile J. (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem 69, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 13.Stoeckli M., Chaurand P., Hallahan D. E., Caprioli R. M. (2001) Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med 7, 493–496 [DOI] [PubMed] [Google Scholar]
- 14.Chaurand P., Schwartz S. A., Caprioli R. M. (2002) Imaging mass spectrometry: a new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr. Opin. Chem. Biol 6, 676–681 [DOI] [PubMed] [Google Scholar]
- 15.Fournier I., Day R., Salzet M. (2003) Direct analysis of neuropeptides by in situ MALDI-TOF mass spectrometry in the rat brain. Neuro Endocrinol. Lett 24, 9–14 [PubMed] [Google Scholar]
- 16.Caldwell R. L., Caprioli R. M. (2005) Tissue profiling by mass spectrometry: a review of methodology and applications. Mol. Cell. Proteomics 4, 394–401 [DOI] [PubMed] [Google Scholar]
- 17.Jurchen J. C., Rubakhin S. S., Sweedler J. V. (2005) MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom 16, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 18.Maddalo G., Petrucci F., Iezzi M., Pannellini T., Del Boccio P., Ciavardelli D., Biroccio A., Forlì F., Di Ilio C., Ballone E., Urbani A., Federici G. (2005) Analytical assessment of MALDI-TOF imaging mass spectrometry on thin histological samples. An insight in proteome investigation. Clin. Chim. Acta 357, 210–218 [DOI] [PubMed] [Google Scholar]
- 19.Aerni H. R., Cornett D. S., Caprioli R. M. (2006) Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem 78, 827–834 [DOI] [PubMed] [Google Scholar]
- 20.Altelaar A. F., Klinkert I., Jalink K., de Lange R. P., Adan R. A., Heeren R. M., Piersma S. R. (2006) Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal. Chem 78, 734–742 [DOI] [PubMed] [Google Scholar]
- 21.Crossman L., McHugh N. A., Hsieh Y., Korfmacher W. A., Chen J. (2006) Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom 20, 284–290 [DOI] [PubMed] [Google Scholar]
- 22.Lemaire R., Tabet J. C., Ducoroy P., Hendra J. B., Salzet M., Fournier I. (2006) Solid ionic matrixes for direct tissue analysis and MALDI imaging. Anal. Chem 78, 809–819 [DOI] [PubMed] [Google Scholar]
- 23.Lemaire R., Wisztorski M., Desmons A., Tabet J. C., Day R., Salzet M., Fournier I. (2006) MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal. Chem 78, 7145–7153 [DOI] [PubMed] [Google Scholar]
- 24.Stauber J., Lemaire R., Wisztorski M., Ait-Menguellet S., Lucot J. P., Vinatier D., Desmons A., Deschamps M., Proess G., Rudolf I., Salzet M., Fournier I. (2006) New developments in MALDI imaging mass spectrometry for pathological proteomic studies: introduction to a novel concept, the specific MALDI imaging. Mol. Cell. Proteomics 5, S247–249 [Google Scholar]
- 25.Wiseman J. M., Ifa D. R., Song Q., Cooks R. G. (2006) Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem. Int. Ed. Engl 45, 7188–7192 [DOI] [PubMed] [Google Scholar]
- 26.Wisztorski M., Brunet L., Dreiserwer K., Hillenkamp F., Berkenkamp S., Salzet M., Fournier I. (2006) Effect of metals coating for UV MALDI-a-TOF mass spectrometry imaging (MALDI IMS) and direct tissue analysis in UV/IR MALDI-o-TOF mass spectrometry, in Proceedings of the 54th ASMS Conference on Mass Spectrometry and Allied Topics, Seattle, May 30–June 2, 2006, Abstract No. ThP 328, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 27.Chaurand P., Schriver K. E., Caprioli R. M. (2007) Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J. Mass Spectrom 42, 476–489 [DOI] [PubMed] [Google Scholar]
- 28.Cornett D. S., Reyzer M. L., Chaurand P., Caprioli R. M. (2007) MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods 4, 828–833 [DOI] [PubMed] [Google Scholar]
- 29.Dreisewerd K., Lemaire R., Pohlentz G., Salzet M., Wisztorski M., Berkenkamp S., Fournier I. (2007) Molecular profiling of native and matrix-coated tissue slices from rat brain by infrared and ultraviolet laser desorption/ionization orthogonal time-of-flight mass spectrometry. Anal. Chem 79, 2463–2471 [DOI] [PubMed] [Google Scholar]
- 30.Garrett T. J., Prieto-Conaway M. C., Kovtoun V., Bui H., Izgarian N., Stafford G., Yost R. A. (2007) Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. Int. J. Mass Spectrom 260, 166–176 [Google Scholar]
- 31.Groseclose M. R., Andersson M., Hardesty W. M., Caprioli R. M. (2007) Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom 42, 254–262 [DOI] [PubMed] [Google Scholar]
- 32.Hankin J. A., Barkley R. M., Murphy R. C. (2007) Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom 18, 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemaire R., Menguellet S. A., Stauber J., Marchaudon V., Lucot J. P., Collinet P., Farine M. O., Vinatier D., Day R., Ducoroy P., Salzet M., Fournier I. (2007) Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, Reg alpha fragment, is a new potential ovary cancer biomarker. J. Proteome Res 6, 4127–4134 [DOI] [PubMed] [Google Scholar]
- 34.McLean J. A., Ridenour W. B., Caprioli R. M. (2007) Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom 42, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 35.Taban I. M., Altelaar A. F., van der Burgt Y. E., McDonnell L. A., Heeren R. M., Fuchser J., Baykut G. (2007) Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom 18, 145–151 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y., Allegood J., Liu Y., Wang E., Cachón-Gonzalez B., Cox T. M., Merrill A. H., Jr., Sullards M. C. (2008) Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem 80, 2780–2788 [DOI] [PubMed] [Google Scholar]
- 37.Seeley E. H., Oppenheimer S. R., Mi D., Chaurand P., Caprioli R. M. (2008) Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J. Am. Soc. Mass Spectrom 19, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taira S., Sugiura Y., Moritake S., Shimma S., Ichiyanagi Y., Setou M. (2008) Nanoparticle-assisted laser desorption/ionization based mass imaging with cellular resolution. Anal. Chem 80, 4761–4766 [DOI] [PubMed] [Google Scholar]
- 39.Trim P. J., Atkinson S. J., Princivalle A. P., Marshall P. S., West A., Clench M. R. (2008) Matrix-assisted laser desorption/ionisation mass spectrometry imaging of lipids in rat brain tissue with integrated unsupervised and supervised multivariant statistical analysis. Rapid Commun. Mass Spectrom 22, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 40.Jardin-Mathé O., Bonnel D., Franck J., Wisztorski M., Macagno E., Fournier I., Salzet M. (2008) MITICS (MALDI Imaging Team Imaging Computing System): a new open source mass spectrometry imaging software. J. Proteomics 71, 332–345 [DOI] [PubMed] [Google Scholar]
- 41.Chaurand P., Schwartz S. A., Billheimer D., Xu B. J., Crecelius A., Caprioli R. M. (2004) Integrating histology and imaging mass spectrometry. Anal. Chem 76, 1145–1155 [DOI] [PubMed] [Google Scholar]
- 42.Lemaire R., Desmons A., Tabet J. C., Day R., Salzet M., Fournier I. (2007) Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res 6, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 43.Schwamborn K., Krieg R. C., Reska M., Jakse G., Knuechel R., Wellmann A. (2007) Identifying prostate carcinoma by MALDI-Imaging. Int. J. Mol. Med 20, 155–159 [PubMed] [Google Scholar]
- 44.Walch A., Rauser S., Deininger S. O., Höfler H. (2008) MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem. Cell Biol 130, 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown L. M., Helmke S. M., Hunsucker S. W., Netea-Maier R. T., Chiang S. A., Heinz D. E., Shroyer K. R., Duncan M. W., Haugen B. R. (2006) Quantitative and qualitative differences in protein expression between papillary thyroid carcinoma and normal thyroid tissue. Mol. Carcinog 45, 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaurand P., Sanders M. E., Jensen R. A., Caprioli R. M. (2004) Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am. J. Pathol 165, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson M. D., Floyd J. L., Caprioli R. M. (2006) Proteomics in diagnostic neuropathology. J. Neuropathol. Exp. Neurol 65, 837–845 [DOI] [PubMed] [Google Scholar]
- 48.Marko-Varga G., Lindberg H., Löfdahl C. G., Jönsson P., Hansson L., Dahlbäck M., Lindquist E., Johansson L., Foster M., Fehniger T. E. (2005) Discovery of biomarker candidates within disease by protein profiling: principles and concepts. J. Proteome Res 4, 1200–1212 [DOI] [PubMed] [Google Scholar]
- 49.Meistermann H., Norris J. L., Aerni H. R., Cornett D. S., Friedlein A., Erskine A. R., Augustin A., De Vera Mudry M. C., Ruepp S., Suter L., Langen H., Caprioli R. M., Ducret A. (2006) Biomarker discovery by imaging mass spectrometry: transthyretin is a biomarker for gentamicin-induced nephrotoxicity in rat. Mol. Cell. Proteomics 5, 1876–1886 [DOI] [PubMed] [Google Scholar]
- 50.Sköld K., Svensson M., Nilsson A., Zhang X., Nydahl K., Caprioli R. M., Svenningsson P., Andrén P. E. (2006) Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J. Proteome Res 5, 262–269 [DOI] [PubMed] [Google Scholar]
- 51.Stoeckli M., Knochenmuss R., McCombie G., Mueller D., Rohner T., Staab D., Wiederhold K. H. (2006) MALDI MS imaging of amyloid. Methods Enzymol 412, 94–106 [DOI] [PubMed] [Google Scholar]
- 52.Groseclose M. R., Massion P. P., Chaurand P., Caprioli R. M. (2008) High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 8, 3715–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronci M., Bonanno E., Colantoni A., Pieroni L., Di Ilio C., Spagnoli L. G., Federici G., Urbani A. (2008) Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: application to MALDI-TOF imaging MS investigations. Proteomics 8, 3702–3714 [DOI] [PubMed] [Google Scholar]
- 54.Stauber J., Lemaire R., Franck J., Bonnel D., Croix D., Day R., Wisztorski M., Fournier I., Salzet M. (2008) MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res 7, 969–978 [DOI] [PubMed] [Google Scholar]
- 55.Van de Plas R., Ojeda F., Dewil M., Van Den Bosch L., De Moor B., Waelkens E. (2007) Prospective exploration of biochemical tissue composition via imaging mass spectrometry guided by principal component analysis. Pac. Symp. Biocomput 458–469 [PubMed] [Google Scholar]
- 56.Djidja M. C., Carolan V., Loadman P. M., Clench M. R. (2008) Method development for protein profiling in biological tissues by matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rapid Commun. Mass Spectrom 22, 1615–1618 [DOI] [PubMed] [Google Scholar]
- 57.McCombie G., Staab D., Stoeckli M., Knochenmuss R. (2005) Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem 77, 6118–6124 [DOI] [PubMed] [Google Scholar]
- 58.Deininger S. O., Ebert M. P., Fütterer A., Gerhard M., Röcken C. (2008) MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J. Proteome Res 7, 5230–5236 [DOI] [PubMed] [Google Scholar]
- 59.Lemaire R., Stauber J., Wisztorski M., Van Camp C., Desmons A., Deschamps M., Proess G., Rudlof I., Woods A. S., Day R., Salzet M., Fournier I. (2007) Tag-mass: specific molecular imaging of transcriptome and proteome by mass spectrometry based on photocleavable tag. J. Proteome Res 6, 2057–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaurand P., Rahman M. A., Hunt T., Mobley J. A., Gu G., Latham J. C., Caprioli R. M., Kasper S. (2008) Monitoring mouse prostate development by profiling and imaging mass spectrometry. Mol. Cell. Proteomics 7, 411–423 [DOI] [PubMed] [Google Scholar]
- 61.Din S., Lennon A. M., Arnott I. D., Hupp T., Satsangi J. (2007) Technology insight: the application of proteomics in gastrointestinal disease. Nat. Clin. Pract. Gastroenterol. Hepatol 4, 372–385 [DOI] [PubMed] [Google Scholar]
- 62.Fournier I., Wisztorski M., Salzet M. (2008) Tissue imaging using MALDI-MS: a new frontier of histopathology proteomics. Expert Rev. Proteomics 5, 413–424 [DOI] [PubMed] [Google Scholar]
- 63.Francese S., Dani F. R., Traldi P., Mastrobuoni G., Pieraccini G., Moneti G. (2009) MALDI mass spectrometry imaging, from its origins up to today: the state of the art. Comb. Chem. High Throughput Screen 12, 156–174 [DOI] [PubMed] [Google Scholar]
- 64.Norris J. L., Cornett D. S., Mobley J. A., Andersson M., Seeley E. H., Chaurand P., Caprioli R. M. (2007) Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int. J. Mass Spectrom 260, 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tilz G. P., Wiltgen M., Demel U., Faschinger C., Schmidinger H., Hermetter A. (2007) Insights into molecular medicine: development of new diagnostic and prognostic parameters. Wien. Med. Wochenschr 157, 122–129 [DOI] [PubMed] [Google Scholar]
- 66.Wisztorski M., Lemaire R., Stauber J., Menguelet S. A., Croix D., Mathé O. J., Day R., Salzet M., Fournier I. (2007) New developments in MALDI imaging for pathology proteomic studies. Curr. Pharm. Des 13, 3317–3324 [DOI] [PubMed] [Google Scholar]
- 67.Reyzer M. L., Caprioli R. M. (2007) MALDI-MS-based imaging of small molecules and proteins in tissues. Curr. Opin. Chem. Biol 11, 29–35 [DOI] [PubMed] [Google Scholar]
- 68.Hsieh Y., Casale R., Fukuda E., Chen J., Knemeyer I., Wingate J., Morrison R., Korfmacher W. (2006) Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun. Mass Spectrom 20, 965–972 [DOI] [PubMed] [Google Scholar]
- 69.Hsieh Y., Chen J., Korfmacher W. A. (2007) Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J. Pharmacol. Toxicol. Methods 55, 193–200 [DOI] [PubMed] [Google Scholar]
- 70.O'Brien E., Dedova I., Duffy L., Cordwell S., Karl T., Matsumoto I. (2006) Effects of chronic risperidone treatment on the striatal protein profiles in rats. Brain Res 1113, 24–32 [DOI] [PubMed] [Google Scholar]
- 71.Reyzer M. L., Hsieh Y., Ng K., Korfmacher W. A., Caprioli R. M. (2003) Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom 38, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 72.Rubakhin S. S., Jurchen J. C., Monroe E. B., Sweedler J. V. (2005) Imaging mass spectrometry: fundamentals and applications to drug discovery. Drug Discov. Today 10, 823–837 [DOI] [PubMed] [Google Scholar]
- 73.Wang H. Y., Jackson S. N., McEuen J., Woods A. S. (2005) Localization and analyses of small drug molecules in rat brain tissue sections. Anal. Chem 77, 6682–6686 [DOI] [PubMed] [Google Scholar]
- 74.Dekker L. J., van Kampen J. J., Reedijk M. L., Burgers P. C., Gruters R. A., Osterhaus A. D., Luider T. M. (2009) A mass spectrometry based imaging method developed for the intracellular detection of HIV protease inhibitors. Rapid Commun. Mass Spectrom 23, 1183–1188 [DOI] [PubMed] [Google Scholar]
- 75.Hopfgartner G., Varesio E., Stoeckli M. (2009) Matrix-assisted laser desorption/ionization mass spectrometric imaging of complete rat sections using a triple quadrupole linear ion trap. Rapid Commun. Mass Spectrom 23, 733–736 [DOI] [PubMed] [Google Scholar]
- 76.Huamani J., Willey C., Thotala D., Niermann K. J., Reyzer M., Leavitt L., Jones C., Fleishcher A., Caprioli R., Hallahan D. E., Kim D. W. (2008) Differential efficacy of combined therapy with radiation and AEE788 in high and low EGFR-expressing androgen-independent prostate tumor models. Int. J. Radiat. Oncol. Biol. Phys 71, 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trim P. J., Henson C. M., Avery J. L., McEwen A., Snel M. F., Claude E., Marshall P. S., West A., Princivalle A. P., Clench M. R. (2008) Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem 80, 8628–8634 [DOI] [PubMed] [Google Scholar]
- 78.Reyzer M. L., Caldwell R. L., Dugger T. C., Forbes J. T., Ritter C. A., Guix M., Arteaga C. L., Caprioli R. M. (2004) Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res 64, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 79.Khatib-Shahidi S., Andersson M., Herman J. L., Gillespie T. A., Caprioli R. M. (2006) Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem 78, 6448–6456 [DOI] [PubMed] [Google Scholar]
- 80.Atkinson S. J., Loadman P. M., Sutton C., Patterson L. H., Clench M. R. (2007) Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom 21, 1271–1276 [DOI] [PubMed] [Google Scholar]
- 81.Wisztorski M., Verplanck N., Thomy V., Stauber J., Camart J. C., Salzet M., Fournier I. (2007) Use of Masks in MALDI-IMS: an easy tool for increasing spatial resolution of images by decreasing irradiated area, in Proceedings of the 55th ASMS Conference on Mass Spectrometry and Allied Topics, Indianapolis, June 4–7, 2007, Abstract No. WPD-066, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 82.Sinha T. K., Khatib-Shahidi S., Yankeelov T. E., Mapara K., Ehtesham M., Cornett D. S., Dawant B. M., Caprioli R. M., Gore J. C. (2008) Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat. Methods 5, 57–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Andersson M., Groseclose M. R., Deutch A. Y., Caprioli R. M. (2008) Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat. Methods 5, 101–108 [DOI] [PubMed] [Google Scholar]
- 84.Stoeckli M., Chaurand P., Caprioli R. M. (1999) Applications of MALDI MS imaging to biological samples, in Proceedings of the 47th ASMS Conference on Mass Spectrometry and Allied Topics, Dallas, June 13–17, 1999, Abstract No. TPG-264, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]