Abstract
A large body of evidence from the past decade supports the existence, in membrane from animal and yeast cells, of functional microdomains playing important roles in protein sorting, signal transduction, or infection by pathogens. In plants, as previously observed for animal microdomains, detergent-resistant fractions, enriched in sphingolipids and sterols, were isolated from plasma membrane. A characterization of their proteic content revealed their enrichment in proteins involved in signaling and response to biotic and abiotic stress and cell trafficking suggesting that these domains were likely to be involved in such physiological processes. In the present study, we used 14N/15N metabolic labeling to compare, using a global quantitative proteomics approach, the content of tobacco detergent-resistant membranes extracted from cells treated or not with cryptogein, an elicitor of defense reaction. To analyze the data, we developed a software allowing an automatic quantification of the proteins identified. The results obtained indicate that, although the association to detergent-resistant membranes of most proteins remained unchanged upon cryptogein treatment, five proteins had their relative abundance modified. Four proteins related to cell trafficking (four dynamins) were less abundant in the detergent-resistant membrane fraction after cryptogein treatment, whereas one signaling protein (a 14-3-3 protein) was enriched. This analysis indicates that plant microdomains could, like their animal counterpart, play a role in the early signaling process underlying the setup of defense reaction. Furthermore proteins identified as differentially associated to tobacco detergent-resistant membranes after cryptogein challenge are involved in signaling and vesicular trafficking as already observed in similar studies performed in animal cells upon biological stimuli. This suggests that the ways by which the dynamic association of proteins to microdomains could participate in the regulation of the signaling process may be conserved between plant and animals.
The plasma membrane of eukaryotes delineates the interface between the cell and the environment. Thus it is particularly involved in environmental signal recognition and their transduction into intracellular responses, playing a crucial role in many essential functions such as cell nutrition (involving transport of solutes in and out of the cell) or response to environmental modifications (including defense against pathogens).
Over the last 10 years, a new aspect of the plasma membrane organization has arisen from biophysical and biochemical studies performed with animal cells. Evidence has been given that the various types of lipids forming this membrane are not uniformly distributed inside the bilayer but rather spatially organized (1). This leads in particular to the formation of specialized phase domains, also called lipid rafts (2, 3). Recently a consensus emerged on the characteristics of these domains. Both proteins and lipids contribute to the formation and the stability of membrane domains that should be called “membrane rafts” and are envisaged as small (10–200-nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes (4). Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions (5). Because of their particular lipidic composition (enrichment in sterol, sphingolipids, and saturated fatty acids), these domains form a liquid ordered phase inside the membrane. This structural characteristic renders them resistant to solubilization by non-ionic detergents, and this property has been widely used to isolate lipid rafts as detergent-resistant membranes (DRMs)1 for further analysis (1). The most important hypothesis to explain the function of these domains is that they provide for lateral compartmentalization of membrane proteins and thereby create a dynamic scaffold to organize certain cellular processes (5). This ability to temporally and spatially organize protein complexes while excluding others conceivably allows for efficiency and specificity of cellular responses. In yeasts and animal cells, the association of particular proteins with these specialized microdomains has emerged as an important regulator of crucial physiological processes such as signal transduction, polarized secretion, cytoskeletal organization, generation of cell polarity, and entry of infectious organisms in living cells (6). Much of the early evidence for a functional role of lipid rafts came from studies of hematopoietic cells in which multichain immune receptors including the high affinity IgE receptor (FcεRI), the T cell receptor, and the B cell receptor (BCR) translocate to lipid rafts upon cross-linking (7). Moreover this signaling involves the relocalization of several proteins; for instance the ligation of the B cell antigen receptor with antigen induced a dissociation of the adaptor protein ezrin from lipid rafts (8). This release of ezrin acts as a critical trigger that regulates lipid raft dynamics during BCR signaling.
In plants, the investigations of the presence of such microdomains are very recent and limited to a reduced number of publications (for a review, see Ref. 9). A few years ago, Peskan et al. (10) reported for the first time the isolation of Triton X-100-insoluble fractions from tobacco plasma membrane. Mongrand et al. (11) provided a detailed analysis of the lipidic composition of such a detergent-resistant fraction indicating that it was highly enriched in a particular species of sphingolipid (glycosylceramide) and in several phytosterols (stigmasterol, sitosterol, 24-methylcholesterol, and cholesterol) compared with the whole plasma membrane from which it originates. Similar results were then obtained with DRMs prepared from Arabidopsis thaliana cell cultures (12) and from Medicago truncatula roots (13). So the presence in plant plasma membrane of domains sharing with animal rafts a particular lipidic composition, namely strong enrichment in sphingolipids together with free sterols and sterol conjugates, the latter being specific to the plant kingdom (11, 13), now seems established. In plant only a few evidences suggest in vivo the role of dynamic clustering of plasma membrane proteins, and they refer to plant-pathogen interaction. A cell biology study reported the pathogen-triggered focal accumulation of components of the plant defense pathway in the plasma membrane (PM), a process reminiscent of lipid rafts (14). Consistently a proteomics study of tobacco DRMs led to the identification of 145 proteins among which a high proportion were linked to signaling in response to biotic stress, cellular trafficking, and cell wall metabolism (15). This suggests that these domains are likely to constitute, as in animal cells, signaling platforms involved in such physiological functions.
Cryptogein belongs to a family of low molecular weight proteins secreted by many species of the oomycete Phytophthora named elicitins that induce a hypersensitivity-like response and an acquired resistance in tobacco (16). To understand molecular processes triggered by cryptogein, its effects on tobacco cell suspensions have been studied for several years. Early events following cryptogein treatment include fixation of a sterol molecule (17, 18); binding of the elicitor to a high affinity site located on the plasma membrane (19); alkalinization of the extracellular medium (20); efflux of potassium, chloride, and nitrate (20, 21); fast influx of calcium (22); mitogen-activated protein kinases activation (23, 24); nitric oxide production (25, 26); and development of an oxidative burst (27, 28). We previously identified NtrbohD, an NADPH oxidase located on the plasma membrane, as responsible for the reactive oxygen species (ROS) production occurring a few minutes after challenging tobacco Bright Yellow 2 (BY-2) cells with cryptogein (29). The fact that most of these very early events involve proteins located on the plasma membrane and that one of them, NtrbohD, has been demonstrated as exclusively associated to DRMs in a sterol-dependent manner (30) prompted us to analyze the modifications of DRM proteome after cryptogein treatment. In the present study, we aimed to confirm the hypothesis that, as observed in animal cells, the dynamic association to or exclusion of proteins from lipid rafts could participate in the signaling process occurring during biotic stress in plants.
To achieve this goal, we had to set up a quantitative assay allowing a precise comparison of the amounts of each protein in DRMs extracted from either control or cells treated with cryptogein. Among several technologies, we excluded DIGE (31), recently used to analyze whole cell proteome variations in plants (32–34), because membrane proteins are poorly soluble in the detergents used for two-dimensional electrophoresis; this limitation is all the more marked for proteins selected on the basis of their insolubility in non-ionic detergent, the criteria for DRMs isolation. Stable isotope labeling of proteins or peptides combined with MS analysis represents alternative strategies for accurate, relative quantification of proteins on a global scale (35, 36). In these approaches, proteins or peptides of two different samples are differentially labeled with stable isotopes, combined in an equal ratio, and then jointly processed for subsequent MS analysis. Relative quantification of proteins is based on the comparison of signal intensities or peak areas of isotope-coded peptide pairs extracted from the respective mass spectra. Stable isotopes can be introduced either chemically into proteins/peptides via derivatization of distinct functional groups of amino acids or metabolically during protein biosynthesis (for a review, see Ref. 37). Metabolic labeling strategies are based on the in vivo incorporation of stable isotopes during growth of organisms. Nutrients or amino acids in a defined medium are replaced by their isotopically labeled (15N, 13C, or 2H) counterparts eventually resulting in uniform labeling of proteins during the processes of cell growth and protein turnover (38). As a consequence, differentially labeled cells or organisms can be combined directly after harvesting. This minimizes experimental variations due to separate sample handling and thus allows a relative protein quantification of high accuracy.
14N/15N labeling has been recently proved to be suitable for comparative experiments performed with whole plants (39–42) and in plant suspension cells where the level of incorporation is equal to the isotopic purity of the salt precursor (43, 44). It has been used successfully to analyze some variations induced in A. thaliana plasma membrane proteome following heat shock (45) or cadmium exposure (46) and to compare phosphorylation levels of plasma membrane proteins after challenge of Arabidopsis cells with elicitors of defense reaction (47). In the present study, we used a mineral 14N/15N metabolic labeling of tobacco BY-2 cells before treatment with cryptogein and subsequent isolation of DRMs. The DRM proteins were further analyzed by one-dimensional SDS-PAGE and digested by trypsin, and peptides were subjected to microcapillary high performance LC-MS/MS (nano-LC-MS/MS). This metabolic method allowed a complete labeling of the proteome, and consequently a major drawback of this method is probably the difficulty to perform an exhaustive analysis of the very large amount of data generated. To solve this problem, a new quantification module of the MFPaQ software (48) was developed, allowing the automatic quantification of the identified peptides. The results derived from the program were validated through a comparison with manual quantification. Thus, we achieved the complete analysis of the DRM proteome variation and identified four proteins whose abundance in DRMs was decreased and one that was enriched in DRMs upon elicitation. The biological relevance of these results, which indicate that, in plant as in animals, the dynamic association of proteins to membrane domains is part of a signaling pathway, will be further discussed.
EXPERIMENTAL PROCEDURES
Materials
BY-2 cells (Nicotiana tabacum cv. Bright Yellow 2) were grown in Murashige and Skoog medium, pH 5.6, containing Murashige and Skoog salt (49), 1 mg/liter thiamine-HCl, 0.2 mg/liter 2,4-dichlorophenylacetic acid, 100 mg/liter myo-inositol, 30 g/liter sucrose, 200 mg/liter KH2PO4, and 2 g/liter MES. Cells were maintained by weekly dilution (2:80) into fresh medium.
Cell Labeling
For quantitative proteomics experiments, the labeling was achieved by substituting 15NH415NO3 (98% 15N) and K15NO3 (99% 15N) to the equivalent concentration of these salts (20 mm) in the culture medium for at least four passages over 4 weeks.
ROS Determination
Cells were harvested 6 days after subculture, filtered, and resuspended (1 g for 10 ml) in a 2 mm MES buffer, pH 5.90, containing 175 mm mannitol, 0.5 mm CaCl2, and 0.5 mm K2SO4. After a 3-h equilibration on a rotary shaker (150 rpm) at 25 °C, cells were treated with 50 nm cryptogein. The production of ROS was determined by chemiluminescence using luminol and a luminometer (BCL Book). Every 10 min, a 250-μl aliquot of the cell suspension was added to 50 μl of 0.3 mm luminol and 300 μl of the assay buffer (175 mm mannitol, 0.5 mm CaCl2, 0.5 mm K2SO4, and 50 mm MES, pH 6.5).
Preparation and Purity of Tobacco Plasma Membrane
All steps were performed at 4 °C. Cells were collected by filtration, frozen in liquid N2, and homogenized with a Waring Blendor in grinding medium (50 mm Tris-MES, pH 8.0, 500 mm sucrose, 20 mm EDTA, 10 mm DTT, and 1 mm PMSF). The homogenate was centrifuged at 16,000 × g for 20 min. After centrifugation, supernatants were collected, filtered through two successive screens (63 and 38 μm), and centrifuged at 96,000 × g for 35 min. This microsomal fraction was purified by partitioning in an aqueous two-phase system (polyethylene glycol 3350/dextran T-500; 6.6% each) to obtain the plasma membrane fraction (50). Marker activities used to evaluate the contamination of the plasma membrane fraction were as follows: azide-sensitive ATPase activity at pH 9 for mitochondria, nitrate-sensitive ATPase activity at pH 6 for tonoplast, antimycin-insensitive NADH cytochrome c reductase for endoplasmic reticulum, and analysis of lipid monogalactosyldiacylglycerol contents for chloroplasts (11).
Isolation of Detergent-resistant Membranes
Plasma membranes were resuspended in a buffer A containing 50 mm Tris-HCl, pH 7.4, 3 mm EDTA, and 1 mm 1,4-dithiothreitol and treated with 1% Triton X-100 (w/v) for 30 min on ice with very gentle shaking every 10 min. Solubilized membranes were placed at the bottom of a centrifuge tube and mixed with 60% sucrose (w/w) to reach a final concentration of 48% (w/w) and overlaid with a discontinuous sucrose gradient (40, 35, 30, and 20%, w/w). After a 20-h centrifugation at 100,000 × g, a ring of Triton X-100-insoluble membranes was recovered at the 30–35% interface, diluted in buffer A, and centrifuged for 4 h at 100,000 × g. The pellet was resuspended in buffer A, and protein concentrations were determined using the Bradford reagent with BSA as the standard.
Protein Separation by SDS-PAGE
DRM proteins were solubilized in a buffer consisting of 6 m urea, 2.2 m thiourea, 5 mm EDTA, 0.1% SDS, 2% N-octyl glucoside, and 50 mm Tris-HCl. Samples were first incubated at room temperature for 15 min and then in a sonic bath for another 15 min. After a centrifugation at 16,000 × g for 15 min, no pellet was observed. An aliquot of solubilized proteins (15 μg) was added to Laemmli buffer before being deposited on an 8% acrylamide gel and separated by SDS-PAGE. Proteins were visualized by Coomassie Blue staining. Each lane was systematically cut into 20 bands of similar volume for MS/MS protein identification.
Protein Digestion
Each band was incubated in 25 mm ammonium bicarbonate and 50% ACN until destaining. Gel pieces were dried in a vacuum SpeedVac (45 °C), further rehydrated with 30 μl of a trypsin solution (10 ng/μl in 50 mm NH4HCO3), and finally incubated overnight at 37 °C. The resulting peptides were extracted from the gel as described previously (51). The trypsin digests were dried in a vacuum SpeedVac and stored at −20 °C before LC-MS/MS analysis.
Nano-LC-MS/MS Analysis
The trypsin digests were separated and analyzed by nano-LC-MS/MS using an Ultimate 3000 system (Dionex, Amsterdam, the Netherlands) coupled to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The peptide mixture was loaded on a C18 precolumn (300-μm-inner diameter × 15 cm PepMap C18, Dionex) equilibrated in 95% solvent A (5% acetonitrile and 0.2% formic acid) and 5% solvent B (80% acetonitrile and 0.2% formic acid). Peptides were eluted using a 5–50% gradient of solvent B during 80 min at 300 nl/min flow rate. The LTQ-Orbitrap was operated in data-dependent acquisition mode with the Xcalibur software (version 2.0.6, Thermo Fisher Scientific). Survey scan MS spectra were acquired in the Orbitrap on the 300–2000 m/z range with the resolution set to a value of 60,000. The five most intense ions per survey scan were selected for CID fragmentation, and the resulting fragments were analyzed in the linear trap (LTQ). Dynamic exclusion was used within 60 s to prevent repetitive selection of the same peptide. To automatically extract peak lists from Xcalibur raw files, the ExtractMSN macro provided with Xcalibur was used through the Mascot Daemon interface (version 2.2.0.3, Matrix Science, London, UK). The following parameters were set for creation of the peak lists: parent ions in the mass range 400–4500, no grouping of MS/MS scans, and threshold at 1000. A peak list was created for each fraction analyzed (i.e. gel slice), and individual Mascot searches were performed for each fraction.
Database Search
MS/MS spectra were processed by Mascot against a subset of SOL Genomic Networks (SGN; download November 2007), which is an integrated genome database for Solanacae (52). The Unigene subset of this database is built by assembling together in contigs expressed sequence tag sequences that are ostensibly fragments of the same gene. We put together the tomato (Lycopersicum esculentum) and tobacco (N. tabacum) Unigene sequences in our database subset (60,227 entries). Tomato expressed sequence tags were selected for two reasons: because of the phylogenetic closeness between the genus Nicotiana and the genus Lycopersicum on the one hand and for the great number of entries in the database (34,829) on the other hand.
The following search parameters were applied: trypsin as cleaving enzyme, “ESI-Trap” as instrument, peptide mass tolerance of 10 ppm, MS/MS tolerance of 0.5 Da, and one missed cleavage allowed. Methionine oxidation and asparagine and glutamine deamination were chosen as variable modifications. 15N metabolic labeling was chosen as a quantitative method for Mascot database searching, allowing identification of labeled and unlabeled peptides within the same database search.
Bioinformatics Analysis
We used the MFPaQ program (48) version 4 to validate the data. This software is a Web application that allows fast and user-friendly verification of Mascot result files as well as data quantification using either isotopic labeling or label-free methods. It provides an interactive interface with Mascot results. It is based on three modules, the Mascot File Parser module, the quantification module, and a third module designed for differential analysis in which validated protein lists are compared. Version 4 of the program has a new data storage system (based on SQLite) and an improved quantification module that now handles the 15N labeling approach. The module is compatible with LTQ-Orbitrap .raw files and .mzXML files. Because the software is written in the Perl programming language, it may be installed on a wide range of operating systems (it has only been tested under Windows XP/2003 and Linux Ubuntu). An automated installer (for the Windows platform) and the source code of this release are available for download from the MFPaQ Website.
The protein validation was performed according to user-defined criteria based on the number, score, and rank of identified peptides. A protein has been validated if it has at least one top ranking peptide with a score greater than 48 (p value <0.001), two top ranking peptides with a score greater than 35 (p value <0.03), or three top ranking peptides with a score greater than 31 (p value <0.1). The above criteria were adjusted to obtain a false positive rate of about 1% at the protein level. To evaluate the false positive rate in these large scale experiments, all the initial database searches were performed using the “decoy” option of Mascot, i.e. the data were searched against a combined database containing the real specified protein sequences (target database) and the corresponding reversed protein sequences (decoy database). MFPaQ used the same criteria to validate decoy and target hits, computed the false discovery rate (FDR = number of validated decoy hits/(number of validated target hits + number of validated decoy hits) × 100) for each gel slice analyzed, and calculated the average of FDR for all slices belonging to the same gel lane (i.e. to the same sample). The FDRs found for the analysis of samples A and B were 1 and 1.2%, respectively.
The MFPaQ software is able to detect highly homologous Mascot protein hits, i.e. proteins identified with some top ranking MS/MS queries also assigned to another protein hit of higher score (i.e. red, non-bold peptides). These homologous protein hits have been validated only if they have been additionally assigned a specific top ranking (red and bold) peptide of score higher than 48.
The MFPaQ quantification module has been improved to handle the 15N labeling approach. The MS data processing and the results obtained using this quantification module are presented under “Results.”
RESULTS
Characterization of Biological Material
Metabolic 15N labeling was achieved by growing BY-2 cells on a basal salt medium containing 15N (or 14N) as a sole nitrogen source. After four subcultures of 7 days, the incorporation rates of isotopes were maximum (98% 15N) (supplemental Data S1).
Treatment of these cells with the fungal elicitor cryptogein was performed in independent experiments carried out to create an inverse labeling: on the one hand 15N-labeled cells were treated with cryptogein, whereas 14N-labeled cells were untreated ([15N]Cry/14N); on the other hand 14N-labeled cells were treated with cryptogein, whereas 15N-labeled cells remained untreated ([14N]Cry/15N).
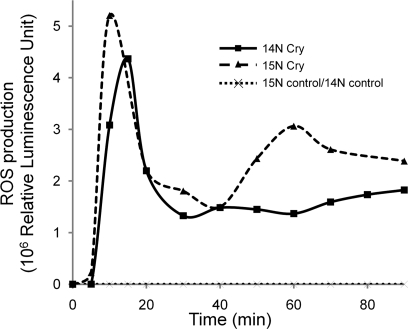
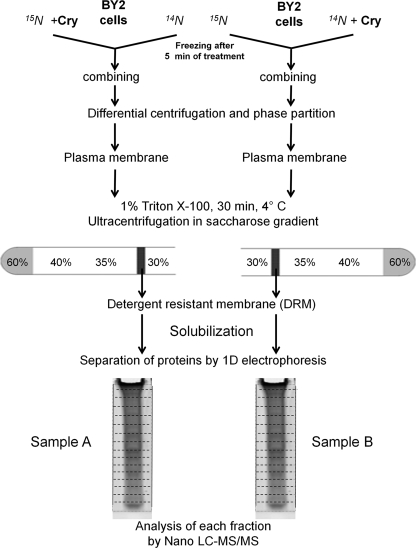
To control that the physiological responses were not affected by the isotope labeling and were comparable in the different experiments, a typical marker of elicitation, the production of ROS, was monitored during 90 min on aliquots of cells. As indicated in Fig. 1, both kinetics and intensity of ROS production were similar in the two experiments particularly in the early phase (0–30 min). Our purpose was to test the dynamics of protein association/dissociation to DRMs during cryptogein-induced signal transduction. As this kind of event typically occurs upstream of the signaling pathways, cell metabolism was stopped by freezing after 5 min of cryptogein treatment, which corresponds to the onset on ROS production (Fig. 1). Control and treated cells of each experiment were then mixed at equal weight, leading to two samples: [15N]Cry/14N-control (sample A) and [14N]Cry/15N-control (sample B). Each of these samples was submitted to subcellular fractionation, leading to DRM isolation according to the procedure indicated in Fig. 2. We selected the conventional phase partition procedure to obtain a highly enriched PM fraction. Typical preparations were enriched by a factor of 7–8 in the PM marker vanadate-sensitive ATPase activity compared with the starting material consisting of a crude microsomal fraction. Biochemical characterization of this PM fraction revealed that it was virtually free of mitochondrial contamination. Endoplasmic reticulum marker enzyme was depleted by a factor 8, and tonoplast was depleted by a factor 20 between microsomes and PM. These results indicate that the contamination by other endomembranes was low, and thus the PM fraction constitutes a suitable starting material for extraction of DRMs and for further proteomics analysis. It is well known that, in biological membranes, the formation of microdomains correlates with resistance to solubilization by non-ionic detergents, and this property has been widely used for biochemical characterization of these domains (1). Here plasma membranes were incubated at 4 °C with Triton X-100 (final concentration, 1%) at a detergent/protein ratio of 15, previously established as the most suitable for DRM extraction from this material (11). In these conditions, the amount of proteins recovered in the DRM fraction was around 5% (w/w) of the initial quantity present in the plasma membrane fraction.
Fig. 1.
Chemiluminescence detection of ROS accumulation triggered by cryptogein in BY-2 cells. At zero time, cryptogein (50 nm) was added to the cell suspension. Every 10 min (from 0 to 120 min), ROS accumulation was determined by chemiluminescence with luminol as described under “Experimental Procedures.” Control values are very close for both 14N and 15N BY-2 cells, so only one curve is presented in the graph. 14N (squares) and 15N (triangles) BY-2 cells show a similar accumulation of H2O2 from 5 min after elicitation with cryptogein. Values are expressed in arbitrary units of chemiluminescence.
Fig. 2.
Work flow of the experimental labeling experiments. Two experiments were carried out in parallel. In one case, 15N cells were subjected to cryptogein treatment, and 14N cells were used as control. In the second case, 14N cells were subjected to cryptogein treatment, and 15N cells were used as control. For each experiment, control and treated cells were frozen after 5 min of treatment, combined at equal weight after treatment, and subjected to the DRM isolation procedure described under “Experimental Procedures.” Proteins were separated by one-dimensional (1D) electrophoresis, and each lane was cut into 20 bands of equal volume for MS/MS protein identification.
Identification and Quantification of BY-2 DRM Proteins by Mass Spectrometry
Tobacco plasma membrane DRMs were found to be soluble in a buffer consisting of both non-ionic (N-octyl glucoside) and ionic (SDS) detergents and high concentration of chaotropic agents. As sample complexity rendered a prefractionation necessary, we then chose to separate the protein mixture by one-dimensional electrophoresis (SDS-PAGE) (Fig. 2). This separation also made samples compatible with subsequent LC-MS/MS analysis. The electrophoresis lane was cut into 20 pieces, and digests obtained from each piece after trypsin addition were separately analyzed by nano-HPLC coupled to an LTQ-Orbitrap mass spectrometer. The raw data were searched by the Mascot software using the SGN database subset of tobacco and tomato contigs (called Unigenes) chosen to improve the number of matching peptides. The redundancy between MS/MS spectra corresponding to the same peptide is managed by the search engine (Mascot). The mass spectrometer thus allowed the identification of 11,540 total peptides among which 2015 were unique (1719 unique peptides for sample A and 1788 for sample B). This led to the identification of 748 Unigenes for sample A and 728 for sample B.
In this study, we used the MFPaQ software to characterize DRM proteins and the variation of protein expression profile in response to cryptogein stimulation. The quantification module of the MFPaQ software (48) has been improved to manage the 15N labeling strategy. We briefly describe here the algorithm implemented in the module. First, the software extracts Mascot search results obtained using the 15N metabolic labeling quantitative method. Then labeled and unlabeled peptides are grouped into peptide pairs. If only one of the forms is present, the software automatically computes the number of nitrogens for the corresponding peptide sequence and then predicts the monoisotopic mass of the missing form. Intensities of peptide pairs are then extracted from the MS survey scans of raw data files in batch mode, and heavy/light ratios are computed for each peptide pair. The ratios of all validated peptides are averaged for each protein within a gel slice, and a coefficient of variation is calculated for the ratio of the proteins that have been quantified with several peptides. Peptide ratio outliers for a given protein are automatically excluded from the protein average ratio. The outlier detection method is based on the box plot analysis and used if a minimum of four peptide pairs has been quantified. When a protein is identified and quantified several times in consecutive gel slices, a final protein ratio is computed by averaging the several ratios found for this protein in the different fractions, and a global coefficient of variation is calculated. Finally the program is able to export all of the quantification results including peptide and protein information in Excel spreadsheets.
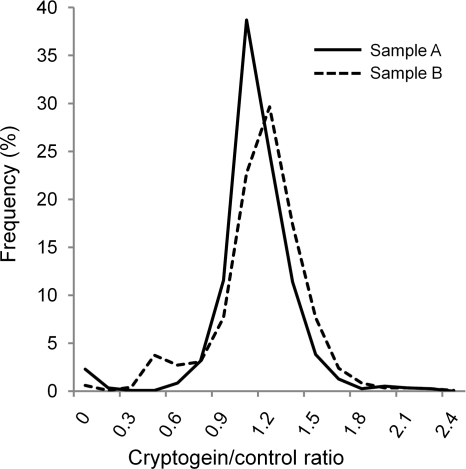
Automatic analysis using MFPaQ was performed on the peptides identified in the two samples. The distribution of their intensity ratio in treated/control sample (i.e. 15N/14N for sample A and 14N/15N for sample B) is presented in Fig. 3. The median value of the ratio is 1.07 for sample A and 1.10 for sample B.
Fig. 3.
Distribution of peptide abundance ratios calculated automatically. All peptides common to the two samples (1183) were automatically quantified by the module developed in MFPaQ. The ratios represented correspond to (the intensity of a given peptide in DRMs extracted from elicited cells)/(its intensity in DRMs extracted from control cells) (i.e. 15N/14N for sample A and 14N/15N for sample B).
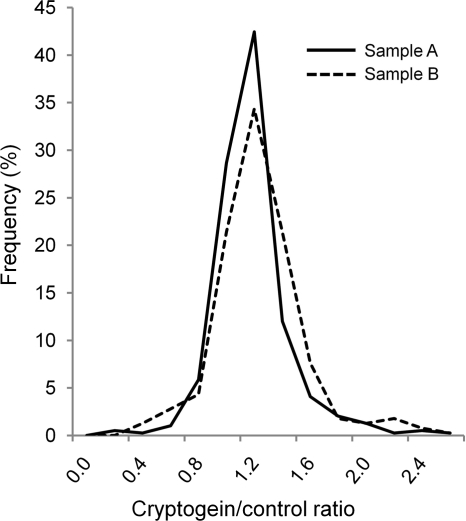
To validate the results obtained with the software, a manual quantification was performed on a subset of the identification results (585 peptides). The ion currents resulting from 14N and 15N monoisotopic peaks of these peptides were measured using the Xcalibur software (version 2.0.6). The peptides were selected if the score Mascot for one of the two isotopes was valid (score >48). The evaluation of ion current of the other isotope was performed for the exact mass (nearly 5 ppm) and an identical retention time. The quantification was made by calculating, for each peptide, the ratio of peak areas corresponding to the two samples (treated versus control). This manual analysis performed on 585 peptides led to the distribution exposed in Fig. 4. The median value of this distribution is 1.13 giving a difference of 0.06 compared with the value obtained with the software results. It has to be noted that the median value, using automatic quantification results, is 1.15 if it is based on the same peptide subset as the one used for the manual procedure. Thus, both techniques indicate a global medium ratio of intensity in treated/control cells DRMs slightly above 1 and a good coincidence between global data obtained by manual and automatic quantification.
Fig. 4.
Distribution of peptide abundance ratios calculated manually. For 583 peptides chosen randomly, a manual quantification was performed by calculating the area of the corresponding peaks. The ratios represented correspond to (the intensity of a given peptide in DRMs extracted from elicited cells)/(its intensity in DRMs extracted from control cells) (i.e. 15N/14N for sample A and 14N/15N for sample B).
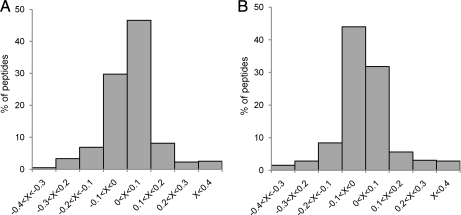
The correlation between the ratios obtained with the two techniques for a single peptide was then examined, calculating the relative difference between the values obtained for 383 peptides common to samples A and B. The result, presented in Fig. 5, is identical for the two samples and indicated that for 76% of these peptides the relative difference between the two methods is below 10% (and for 90% of them is it below 20%).
Fig. 5.
Distribution of relative distance between manual and automatic peptide quantifications. Shown is the comparison of peptide intensity ratios as determined by MFPaQ and by manual inspection is expressed by the relative difference: (Automatic quantification − Manual quantification)/Manual quantification. A, sample A; B, sample B.
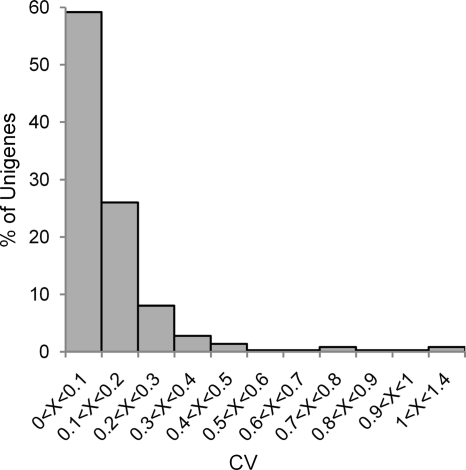
We further analyzed the coefficient of variation among the treated/control ratios, calculated by MFPaQ, of the different peptides identifying a single Unigene. As indicated on Fig. 6, this coefficient of variation is inferior to 0.1 for 60% of the Unigenes and below 0.2 for 85% of them. The mean value of these coefficients for all the Unigenes is 0.13.
Fig. 6.
Distribution of coefficients of variation (CV) among cryptogein/control ratios of peptides identifying a single protein. For all proteins identified with at least two peptides this coefficient of variation was obtained by dividing the standard deviation of the ratios of intensity (cryptogein/control) of the different identifying peptides by the mean value of these ratios.
In the database used, SGN numbers correspond to contigs that may belong to the same protein. To eliminate this redundancy, we used the “bulk search” tool available on the SGN Website that allows mapping between SGN identifiers and National Center for Biotechnology Information (NCBI) identifying GI numbers. Unigenes corresponding to the same GI number were grouped under the same protein identification. In this way, the final protein lists contain some proteins from other species than those used to build the SGN database subset (N. tabacum and L. esculentum). By applying the rules of elimination of the redundancy described above, we obtained a list of 350 proteins identified and quantified that corresponds to the smallest set of proteins explaining the identified peptides presence (supplemental Data S2 and S3). Only 61% of proteins have been identified and quantified in both experiments A and B. The main reason is the well known bias of LC-MS/MS experiments (“shotgun” analysis) of a peptide sample issued from a complex mixture of proteins with a dynamic range of concentrations comprising several orders of magnitude. If the number of ionized peptides is greater than the mass spectrometer can analyze (because of its sequencing speed) then the peptides in small quantities are not systematically identified. Quite similar results indicating 60% of identified plant DRM proteins found in two independent analyses have been published recently (44).
The fact that our biological material corresponds to a plant species, the genome of which is not fully sequenced, led us to use the SGN database to increase the amount of identifications. However, this added a supplemental step (described above) of Unigene grouping to reach the final protein identification. We thus further analyzed the suitability of the MFPaQ software by processing our MS/MS spectra directly against the UniProt database (downloaded on October 2008). As expected, the number of identified and quantified proteins was lower (291), but the mean value of the coefficient of variations of the treated/control ratios for the different peptides identifying a single protein was quite similar to the one described above for the Unigenes (data not shown). Thus, all the validation steps indicate the suitability of the MFPaQ software to analyze the data from a quantitative proteomics experiment using 14N/15N labeling.
Cryptogein-induced Modification of Protein Association to Tobacco DRMs
These results described above prompted us to use the automatic quantification procedure to analyze the relative abundance of the proteins identified in DRMs extracted from control or elicited cells in the two reciprocal experiments.
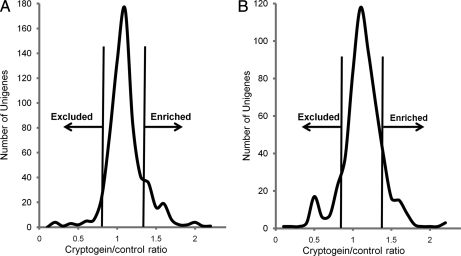
Quantification of Unigenes using MFPaQ led to a similar distribution of individual ratios treated/control in the two samples; the median value is 1.07 for sample A and 1.04 for sample B (Fig. 7). A similar analysis performed on proteins identified using the UniProt database, thus giving direct protein quantification, yielded quite comparable results (1.075 for sample A and 1.03 for sample B).
Fig. 7.
Distribution of cryptogein/control protein ratios in samples A and B. In the two samples, ratios were calculated for each protein as the mean value of the ratios of its identifying peptides. Vertical lines indicate the thresholds corresponding to the 10% of proteins exhibiting the highest (enriched) or the lowest (excluded) ratios in each experiment. The values of these thresholds are identical in the two experiments: 0.8 for exclusion and 1.4 for enrichment.
In the two samples, 80% of the Unigenes quantified using MFPaQ exhibited a ratio of treated/control close to the median value of the experiment (between 0.8 and 1.4), indicating that the association of most of the proteins to DRMs is not significantly modified by cryptogein. Furthermore a control experiment mixing equally control 15N- and control 14N-labeled cells led to a quite similar dispersion of the 15N/14N ratios; 80% of them were between 0.9 and 1.3.
We thus analyzed the 20% of Unigenes that were out of this set in the two experiments: they were equally distributed in the two samples above (10%) and below (10%) the median value, leading to threshold values that were identical in the two experiments (0.8 for exclusion and 1.4 for enrichment). The Unigenes exhibiting a similar modification of their association to DRMs after cryptogein treatment in the two experiments were then selected and submitted to the procedure of grouping using the “bulk search tool” to eliminate redundancy between Unigenes corresponding to a single protein as described above.
This led to the identification of five proteins: the abundance of four of them decreased in DRMs extracted from cryptogein treated cells, whereas one of them was more abundant in DRMs after elicitation (Table I). Interestingly the four proteins excluded from DRMs after cryptogein treatment are linked to vesicular trafficking (Dynamin-1A, Dynamin-1E, Dynamin-2A, and Dynamin-2B), whereas the protein that was more abundant in DRMs after elicitation is a signaling proteins (a 14-3-3 protein).
Table I. Proteins differentially quantified in DRMs extracted from either control or elicited cells.
Among the 10% of proteins exhibiting the highest or the lowest cryptogein/control intensity ratios, those indicated in this table behaved similarly in the two experiments. Sp., plant species abbreviated as follows: N.t., N. tabacum; A.t., A. thaliana. Acc. no., accession number in GenBankTM database. CV, coefficient of variation among the treated/control ratios of peptides identifying a single protein.
| Acc. no. | Sp. | Protein name | Number of Unigenes | Sample A |
Sample B |
||||
|---|---|---|---|---|---|---|---|---|---|
| Global ratio | Number of peptides | CV | Global ratio | Number of peptides | CV | ||||
| Vesicular trafficking | |||||||||
| GI:5931765 | N.t. | Dynamin-1A | 2 | 0.41 | 13 | 3.13 | 0.68 | 20 | 10.7 |
| GI:18411520 | A.t. | Dynamin-1E | 3 | 0.45 | 15 | 10.7 | 0.71 | 12 | 5.6 |
| GI:15218486 | A.t. | Dynamin-2A | 1 | 0.394 | 5 | 16 | 0.79 | 4 | 16 |
| GI:15218837 | A.t. | Dynamin-2B | 1 | 0.524 | 4 | 4.5 | 0.73 | 2 | 4.7 |
| Signaling | |||||||||
| GI:3023189 | N.t. | 14-3-3 C | 1 | 1.43 | 5 | 5.1 | 2.15 | 4 | 3.4 |
DISCUSSION
Automatic Procedure for Quantitative Analysis of Protein Abundance following 15N/14N Labeling
The number of proteins identified in the present study (350) is higher than the number found in our previous work (15). This can be explained by the fact that we used a more powerful mass spectrometer (LTQ-Orbitrap) here than in the previously published study (nanospray LCQ Deca XP Plus ion trap mass spectrometer) and that we performed the database search against a Solanacae-specific database allowing a higher level of identification. However, both the identities of proteins and their functional groups are consistent between the two studies; the proteins involved in signaling and response to stress were in both cases the most represented (supplemental data S3). In counterpart, the nano-LC-MS/MS analysis performed on 14N/15N-labeled samples generated a large amount of data. This in turn necessitated appropriate bioinformatics tools for data analysis. Here we used the MFPaQ software (48) that was initially developed to perform quantification of ICAT and SILAC experiments. The quantification module has been extended to manage 15N labeling experiments, and the results obtained with the software were validated by manual assessment on a subset of peptides (Fig. 5). The results obtained indicate a close correlation between manual and automatic peptide quantification and a low deviation of ratios for peptides identifying a single protein.
A few other tools exist for performing quantitative analysis of 15N labeling experiments. Andreev et al. (53) developed the Quantitation of N-15/14 (QN) algorithm based on the Trans-Proteomic Pipeline (54) for 14N/15N quantification of identification results obtained with the Sequest search engine and therefore not compatible with Mascot. Moreover some modules of this program are written in MATLAB and thus require the installation of this package. Another module has been developed for the Trans-Proteomic Pipeline by Palmblad et al. (55). It uses Mascot identification results as input and any type of raw files converted to the mzXML format. The Mascot search must be run twice, once against 14N masses and once against 15N masses, resulting in two Mascot result files for each raw file that must be subsequently renamed using an external script. Finally the MSQuant program (56), often used for SILAC experiments, can also perform 14N/15N quantification starting from Mascot results and LTQ-Orbitrap raw files and was recently described for such applications (44). However, this standalone software also needs extra steps (Mascot results export and conversion using manual configuration and execution of scripts). The MFPaQ software offers a very user-friendly alternative for integrated protein validation and 14N/15N quantification through a Web browser interface. It can read directly non-processed RAW files from an LTQ-Orbitrap or standard mzXML files and is also well integrated with the Mascot search engine (direct access to the identification results on the Mascot server). MFPaQ provides an interactive interface allowing the user to organize and compile results from shotgun experiments and proven to be efficient for data validation and quantification after 15N labeling, protein fractionation, analysis of consecutive fractions by several nano-LC-MS/MS runs, and multisearch with the Mascot engine. It was used successfully in this study to point out variations of protein abundances in the DRM compartment of plant cells following stimulation with cryptogein.
Quantitative Analysis of the Tobacco Cell DRM Proteome: Effect of Cryptogein Stimulation
In animal cells, a few studies report a quantitative analysis of compositional changes in DRM protein content upon biological stimulus. MacLellan et al. (57), using ICAT, reported an increase of 23 proteins in DRMs isolated from human smooth muscle cells after stimulation with platelet-derived growth factor. Using the same technique of chemical labeling, Gupta et al. (8) found three proteins enriched and one depleted in DRMs isolated from B cells following ligation of the BCR with antigens. Using the two-dimensional DIGE system Kobayashi et al. (58) identified 20 proteins, the abundance of which increased in DRMs following T cell receptor stimulation in T lymphocytes. Very recently, a quantitative analysis of macrophage DRM proteome using stable isotope labeling of amino acids indicated an enrichment of about 15 proteins in response to lipopolysaccharide (59). Thus, although a deep modification in the composition of lipid raft proteome has been proposed following T cell antigen receptor triggering (60), the above cited studies are in favor of a limited modification of protein association to DRMs upon biological stimulus. This is consistent with our results indicating one protein enriched and four depleted in tobacco DRMs following cryptogein treatment. Moreover enrichment or depletion factors (ranging in our study from 0.4 to 2.1) are in agreement with studies performed in animal cells indicating factors ranging from 0.8 to 1.85 in macrophage DRMs in response to lipopolysaccharides (59), from 0.37 to 3.5 in DRMs from B cells (8), or from 1.3 to 1.8 in DRMs from human smooth muscles challenged with growth factor (57). Finally it has to be noted that times of treatment with the biological stimulus in these studies (5 min (8, 59) or 15 min (57)) are quite comparable to those used in this study (5 min of cryptogein treatment).
Although metabolic labeling and quantitative proteomics were used to analyze the sterol dependence of protein association to A. thaliana DRMs (44), only one very recent study has so far been performed to determine the variation of protein content of plant DRMs upon a biological stimulus (61). However, this study has been performed comparing DRMs from control plants and plants acclimated to low temperatures for several days and thus does not refer to an early signaling process. The data presented here indicate that a process of discrete modification of DRM protein content seems to occur very early in a similar manner in plant and animal cells in a context of biological stimulation. It has to be noted that, in many cases, functional studies have confirmed the biological relevance of the data obtained by quantitative proteomics studies. For instance, the protein ezrin had been shown using quantitative proteomics to be excluded from DRMs of B cells upon ligation of antigen receptor: functional studies using cells transformed with dominant positive mutants of this protein demonstrated that it was an inhibitor of lipid rafts coalescence, indicating that the release of ezrin from lipid rafts regulates their dynamics during signaling (8). In a similar manner, histochemical analysis confirmed that some proteins enriched in T cell DRMs after activation were indeed redistributed from cytosol to microdomains of the membrane during this process. All these data seem to confirm the hypothesis that plant microdomains could play, like their animal counterpart, a key role in the signaling process leading to the establishment of cell responses.
Biological Significance of Variations of Protein Content Observed in DRMs of Cryptogein-elicited Tobacco Cells
The proteins, the abundance of which is modified in DRMs after elicitation of tobacco cells with cryptogein, belong to two functional families: vesicular trafficking and signaling. Similar results have already been obtained in animal cells: upon BCR engagement, apart from proteins corresponding to the BCR complex, all proteins that exhibited modifications of their association to DRMs were linked to the cytoskeleton or vesicle trafficking (8). Consistently proteins undergoing modification of their association to human smooth muscle cell DRMs after treatment with growth factor are essentially cytoskeletal proteins and endocytosis-related proteins (57). These two examples come from quantitative proteomics studies of the DRM fraction; however, a huge amount of the data probing the association to DRMs of particular proteins using immunological tools clearly indicates that signal transduction and membrane trafficking are two physiological process essentially linked to DRMs (62–65). In tobacco the functional grouping of DRM and plasma membrane proteins indicated that proteins involved in signaling and cell trafficking undergo an increase of their relative importance in DRMs compared with plasma membrane (15). This suggested that, like their animal counterpart, plant DRMs could be involved in the regulation of such a physiological function, which is confirmed by the results presented in this study.
The fungal elicitor cryptogein has been proven in tobacco cells to trigger numerous physiological events, obviously corresponding to a signaling cascade. Thus, the identification in this study of a DRM-associated protein related to signaling is quite relevant. This protein is a 14-3-3 protein enriched in DRMs after cryptogein treatment. Studies over the past 20 years have proven 14-3-3 to be ubiquitous; it is found in most eukaryotic organisms and tissues (66). In animals, these proteins play a central role in the regulation of many cellular processes such as control of the cell cycle, differentiation, apoptosis, targeting of proteins to different cellular locations, and coordination of multiple signal transduction pathways (67–70). These proteins could achieve such functions by directly regulating the activity of proteins involved in a signal transduction cascade, promoting the formation of multiprotein complexes, or modulating the expression of particular genes by regulating the activity or localization of transcription factors. In a previous study, we used a two-hybrid screen to find proteins able to specifically interact with NtrbohD, the tobacco oxidase that has been proven to be responsible for ROS production in tobacco cells elicited with cryptogein (29). These experiments led to the isolation of a cDNA encoding a protein belonging to the family of 14-3-3 proteins (Nth14-3-3, AJ309008). When BY-2 cell lines were transformed with antisense constructs of Nth14-3-3, the expression of the antisense transgene was correlated with a strong inhibition of ROS production following elicitation with cryptogein (72). This demonstrated the involvement of a 14-3-3 protein in the regulation of ROS production. The 14-3-3 protein identified in the present study is extremely close to Nth14-3-3 (89% identity and 94% homology), making quite plausible a functional redundancy between these two isoforms (72). Indeed the question of the specificity of function of this isoform remains because the tobacco genome comprises 17 isoforms of 14-3-3 proteins, and studies on the binding of different isoforms of 14-3-3 to different isoforms of H+-ATPase in A. thaliana indicated no absolute specificity (73). However, it has to be noted that among the different isoforms of 14-3-3 identified in this study several exhibited a treated/control ratio above 1.4 in one experiment and slightly below in the other. Thus although we only considered one isoform as strictly above the threshold in the two experiments, the hypothesis of an association of several 14-3-3 proteins to DRMs upon cryptogein treatment cannot be excluded. NtrbohD has been proven to be exclusively associated to the tobacco DRM fraction in a sterol-dependent manner (30), and the enrichment in this fraction of a 14-3-3 protein able to act as a positive regulator of this oxidase at a timing corresponding to the onset of ROS production would be particularly biologically relevant.
Recently our group demonstrated that among the very early events triggered by cryptogein a clathrin-mediated endocytosis process occurred 5 min after stimulation (74). It is thus noteworthy that the four proteins identified as involved in cell trafficking belong to the dynamin-related protein (DRP) family. Dynamins are high molecular weight GTPases that play a central role in dynamics of membrane biogenesis and maintenance in eukaryotic cells that require a constant turnover of membrane constituents mediated by the processes of endocytosis and exocytosis (for a review, see Ref. 75). The basic feature of this group of proteins is that they form helical structures able to wrap around the membranes and either tubulate them or pinch them off of larger membrane sheets. In A. thaliana, DRP1A expressed as a functional DRP1A-GFP fusion protein under the control of its native promoter formed discrete foci at the plasma membrane of root epidermal cells (76) consistent with a localization in membrane domains of this protein. The same study indicated that DRP1A-GFP dynamics are perturbed upon treatment with fenpropimorph, a pharmacological inhibitor of the sterol biosynthetic pathway in plants, or with compounds, such as tyrphostin A23, known to block clathrin-mediated endocytosis. Moreover a null mutant for DRP1A exhibited reduced endocytosis, indicating the involvement of this protein in such a process. DRP1A and DRP1E from A. thaliana share 80% homology, and no clear data are available concerning the physiological role of DRP1E. The DRP2 subfamily is characterized by the presence of a pleckstrin homology domain believed to bind to phosphoinositides with a broad range of specificity and affinity (77). In recent years, both phosphoinositides and phosphoinositide-binding proteins have been reported to display a restricted, rather than a uniform, distribution across intracellular membranes (78). A significant enrichment of BY-2 cell DRMs in phosphoinositides species has been observed.2 Moreover from a functional point of view, recent results obtained in plants indicate a crucial role of phosphatidylinositol in the setup of endocytosis triggered either by salt stress (79) or Rhizobium infection (80). Finally DRP2A is involved in clathrin-coated vesicle trafficking (81), and a colocalization of this protein with DRP2B has been reported (71). All these data are consistent both with the localization in tobacco DRMs of these four dynamin-related proteins and with their putative involvement in an endocytotic process already demonstrated in our model at a timing (5 min) in total agreement with the proteomics analysis conducted here. Finally the decrease observed for these proteins in their association to DRMs is, in this context, quite coherent with their internalization concomitant to the vesicle formation. We are currently analyzing further the role of these candidate proteins in the signaling process triggered by the elicitor using reverse genetics.
Conclusion
The development and validation of an automatic procedure for analyzing the data generated after in vivo labeling of tobacco cells allowed a quantitative analysis of the variation of a whole subcellular proteome after stimulation by an elicitor of defense reaction. The discrete variation of association to DRMs of some proteins upon cryptogein treatment indicate that plant microdomains could, like their animal counterpart, play a role in the signaling process underlying the setup of defense reaction in plants. Furthermore proteins identified as differentially associated to tobacco DRMs after cryptogein challenge are involved in signaling and vesicular trafficking as already observed in similar studies performed in animal cells upon biological stimuli. This suggests that the ways by which the dynamic association of proteins to microdomains could participate in the regulation of signaling process may be conserved between plant and animals. Finally this study led to the identification of five putative new actors of the cryptogein signaling pathway. Future investigation will have to determine the position of this dynamics of association to microdomains in the signaling cascade triggered by the elicitor and the precise role of these proteins in this process in link with physiological events already identified.
Supplementary Material
Acknowledgments
We are grateful to M. Ponchet and B. Industrie for the gift of cryptogein and to N. Leborgne-Castel and Sebastien Mongrand for critical reading of the manuscript.
Footnotes
* This work was supported in part by the Plant Health division of INRA (Ph.D. grant to T. S.), by the Conseil Régional de Bourgogne (Ph.D. grant to T. S. and J. M. and postdoctoral grant to S. V.), and by Agence Nationale de la Recherche (ANR) Grant ANR-05-JCJC-0209.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 S. Mongrand, personal communication.
1 The abbreviations used are:
- DRM
- detergent-resistant membrane
- BY-2
- Bright Yellow 2
- DRP
- dynamin-related protein
- PM
- plasma membrane
- BCR
- B cell receptor
- ROS
- reactive oxygen species
- nano-LC-MS/MS
- microcapillary high performance LC-MS/MS
- MFPaQ
- Mascot file parsing and quantification
- LTQ
- linear trap quadrupole
- SGN
- SOL Genomic Networks
- FDR
- false discovery rate
- Cry
- cryptogein
- SILAC
- stable isotope labeling with amino acids in cell culture
- GFP
- green fluorescent protein.
REFERENCES
- 1.Brown D. A., London E. (1998) Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol 164, 103–114 [DOI] [PubMed] [Google Scholar]
- 2.Rietveld A., Simons K. (1998) The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta 1376, 467–479 [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 4.Pike L. J. (2006) Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res 47, 1597–1598 [DOI] [PubMed] [Google Scholar]
- 5.Simons K., Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 6.Rajendran L., Simons K. (2005) Lipid rafts and membrane dynamics. J. Cell Sci 118, 1099–1102 [DOI] [PubMed] [Google Scholar]
- 7.Sedwick C. E., Altman A. (2002) Ordered just so: lipid rafts and lymphocyte function. Sci. STKE 2002, RE2. [DOI] [PubMed] [Google Scholar]
- 8.Gupta N., Wollscheid B., Watts J. D., Scheer B., Aebersold R., DeFranco A. L. (2006) Quantitative proteomics analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid rafts dynamics. Nat. Immunol 7, 625–633 [DOI] [PubMed] [Google Scholar]
- 9.Zappel N. F., Panstruga R. (2008) Heterogeneity and lateral compartmentalization of plant plasma membranes. Curr. Opin. Plant Biol 11, 632–640 [DOI] [PubMed] [Google Scholar]
- 10.Peskan T., Westermann M., Oelmüller R. (2000) Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem 267, 6989–6995 [DOI] [PubMed] [Google Scholar]
- 11.Mongrand S., Morel J., Laroche J., Claverol S., Carde J. P., Hartmann M. A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J. J. (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem 279, 36277–36286 [DOI] [PubMed] [Google Scholar]
- 12.Borner G. H., Sherrier D. J., Weimar T., Michaelson L. V., Hawkins N. D., Macaskill A., Napier J. A., Beale M. H., Lilley K. S., Dupree P. (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137, 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefebvre B., Furt F., Hartmann M. A., Michaelson L. V., Carde J. P., Sargueil-Boiron F., Rossignol M., Napier J. A., Cullimore J., Bessoule J. J., Mongrand S. (2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144, 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat R. A., Miklis M., Schmelzer E., Schulze-Lefert P., Panstruga R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. U.S.A 102, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morel J., Claverol S., Mongrand S., Furt F., Fromentin J., Bessoule J. J., Blein J. P., Simon-Plas F. (2006) Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5, 1396–1411 [DOI] [PubMed] [Google Scholar]
- 16.Ricci P. (1997) in Plant-Microbe Interactions ( Stacey G., Keen N. T. eds) pp. 53–75, Chapman and Hall, New York [Google Scholar]
- 17.Osman H., Vauthrin S., Mikes V., Milat M. L., Panabières F., Marais A., Brunie S., Maume B., Ponchet M., Blein J. P. (2001) Mediation of elicitin activity on tobacco is assumed by elicitin-sterol complexes. Mol. Biol. Cell 12, 2825–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vauthrin S., Mikes V., Milat M. L., Ponchet M., Maume B., Osman H., Blein J. P. (1999) Elicitins trap and transfer sterols from micelles, liposomes and plant plasma membranes. Biochim. Biophys. Acta 1419, 335–342 [DOI] [PubMed] [Google Scholar]
- 19.Wendehenne D., Binet M. N., Blein J. P., Ricci P., Pugin A. (1995) Evidence for specific, high-affinity binding sites for a proteinaceous elicitor in tobacco plasma membrane. FEBS Lett 374, 203–207 [DOI] [PubMed] [Google Scholar]
- 20.Blein J. P., Milat M. L., Ricci P. (1991) Response of cultured tobacco cells to cryptogein, a proteinaceous elicitor from Phytophthora cryptogea. Possible plasmalemma involvement. Plant Physiol 95, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendehenne D., Lamotte O., Frachisse J. M., Barbier-Brygoo H., Pugin A. (2002) Nitrate efflux is an essential component of the cryptogein signaling pathway leading to defense responses and hypersensitive cell death in tobacco. Plant Cell 14, 1937–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecourieux D., Mazars C., Pauly N., Ranjeva R., Pugin A. (2002) Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 14, 2627–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebrun-Garcia A., Ouaked F., Chiltz A., Pugin A. (1998) Activation of MAPK homologues by elicitors in tobacco cells. Plant J 15, 773–781 [DOI] [PubMed] [Google Scholar]
- 24.Zhang S., Du H., Klessig D. F. (1998) Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould K. S., Lamotte O., Klinguer A., Pugin A., Wendehenne D. (2003) Nitric oxide production in tobacco leaf cells: a generalized stress response? Plant Cell Environ 26, 1851–1862 [Google Scholar]
- 26.Lamotte O., Gould K., Lecourieux D., Sequeira-Legrand A., Lebrun-Garcia A., Durner J., Pugin A., Wendehenne D. (2004) Analysis of nitric oxide signaling functions in tobacco cells challenged by the elicitor cryptogein. Plant Physiol 135, 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusterucci C., Stallaert V., Milat M. L., Pugin A., Ricci P., Blein J. P. (1996) Relationship between active oxygen species, lipid peroxidation, necrosis, and phytoalexin production induced by elicitins in Nicotiana. Plant Physiol 111, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon-Plas F., Rustérucci C., Milat M. L., Humbert C., Montillet J. L., Blein J. P. (1997) Active oxygen species production in tobacco cells elicited by cryptogein. Plant Cell Environ 20, 1573–1579 [Google Scholar]
- 29.Simon-Plas F., Elmayan T., Blein J. P. (2002) The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J 31, 137–147 [DOI] [PubMed] [Google Scholar]
- 30.Roche Y., Gerbeau-Pissot P., Buhot B., Thomas D., Bonneau L., Gresti J., Mongrand S., Perrier-Cornet J. M., Simon-Plas F. (2008) Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J 22, 3980–3991 [DOI] [PubMed] [Google Scholar]
- 31.Tonge R., Shaw J., Middleton B., Rowlinson R., Rayner S., Young J., Pognan F., Hawkins E., Currie I., Davison M. (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1, 377–396 [DOI] [PubMed] [Google Scholar]
- 32.Amme S., Matros A., Schlesier B., Mock H. P. (2006) Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot 57, 1537–1546 [DOI] [PubMed] [Google Scholar]
- 33.Bohler S., Bagard M., Oufir M., Planchon S., Hoffmann L., Jolivet Y., Hausman J. F., Dizengremel P., Renaut J. (2007) A DIGE analysis of developing poplar leaves subjected to ozone reveals major changes in carbon metabolism. Proteomics 7, 1584–1599 [DOI] [PubMed] [Google Scholar]
- 34.Casati P., Zhang X., Burlingame A. L., Walbot V. (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol. Cell. Proteomics 4, 1673–1685 [DOI] [PubMed] [Google Scholar]
- 35.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 36.Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 37.Thelen J. J., Peck S. C. (2007) Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beynon R. J., Pratt J. M. (2005) Metabolic labeling of proteins for proteomics. Mol. Cell. Proteomics 4, 857–872 [DOI] [PubMed] [Google Scholar]
- 39.Bindschedler L. V., Palmblad M., Cramer R. (2008) Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics: an oxidative case study. Phytochemistry 69, 1962–1972 [DOI] [PubMed] [Google Scholar]
- 40.Hebeler R., Oeljeklaus S., Reidegeld K. A., Eisenacher M., Stephan C., Sitek B., Stühler K., Meyer H. E., Sturre M. J., Dijkwel P. P., Warscheid B. (2008) Study of early leaf senescence in Arabidopsis thaliana by quantitative proteomics using reciprocal 14N/15N labeling and difference gel electrophoresis. Mol. Cell. Proteomics 7, 108–120 [DOI] [PubMed] [Google Scholar]
- 41.Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 860–881 [DOI] [PubMed] [Google Scholar]
- 42.Nelson C. J., Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Implication of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 [DOI] [PubMed] [Google Scholar]
- 43.Engelsberger W. R., Erban A., Kopka J., Schulze W. X. (2006) Metabolic labeling of plant cell cultures with K(15)NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kierszniowska S., Seiwert B., Schulze W. X. (2009) Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8, 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmblad M., Mills D. J., Bindschedler L. V. (2008) Heat-shock response in Arabidopsis thaliana explored by multiplexed quantitative proteomics using differential metabolic labelling. J. Proteome Res 7, 780–785 [DOI] [PubMed] [Google Scholar]
- 46.Lanquar V., Kuhn L., Lelièvre F., Khafif M., Espagne C., Bruley C., Barbier-Brygoo H., Garin J., Thomine S. (2007) 15N-metabolic labeling for comparative plasma membrane proteomics in Arabidopsis cells. Proteomics 7, 750–754 [DOI] [PubMed] [Google Scholar]
- 47.Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 48.Bouyssié D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., Girard J. P., Monsarrat B. (2007) Mascot file parsing and quantification (MFPaQ), a new software to parse, validate and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics 6, 1621–1637 [DOI] [PubMed] [Google Scholar]
- 49.Murashige T., Skoog F. (1962) A revised method for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497 [Google Scholar]
- 50.Larsson C., Sommarin M., Widell S. (1994) in Aqueous Two-Phase Systems ( Walter H., Johansson G. eds) Vol. 228, pp. 451–459, Academic Press Inc., San Diego, CA [Google Scholar]
- 51.Borderies G., Jamet E., Lafitte C., Rossignol M., Jauneau A., Boudart G., Monsarrat B., Esquerré-Tugayé M. T., Boudet A., Pont-Lezica R. (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis 24, 3421–3432 [DOI] [PubMed] [Google Scholar]
- 52.Mueller L. A., Solow T. H., Taylor N., Skwarecki B., Buels R., Binns J., Lin C., Wright M. H., Ahrens R., Wang Y., Herbst E. V., Keyder E. R., Menda N., Zamir D., Tanksley S. D. (2005) The SOL genomics network. A comparative resource for Solanacae biology and beyond. Plant Physiol 138, 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreev V. P., Li L., Rejtar T., Li Q., Ferry J. G., Karger B. L. (2006) New algorithm for 15N/14N quantitation with LC-ESI-MS using an LTQ-FT mass spectrometer. J. Proteome Res 5, 2039–2045 [DOI] [PubMed] [Google Scholar]
- 54.Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palmblad M., Bindschedler L. V., Cramer R. (2007) Quantitative proteomics using uniform 15N-labeling, MASCOT, and the trans-proteomic pipeline. Proteomics 7, 3462–3469 [DOI] [PubMed] [Google Scholar]
- 56.Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 57.MacLellan D. L., Steen H., Adam R. M., Garlick M., Zurakowski D., Gygi S. P., Freeman M. R., Solomon K. R. (2005) A quantitative proteomic analysis of growth factor-induced compositional changes in lipid rafts of human smooth muscle cells. Proteomics 5, 4733–4742 [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi M., Katagiri T., Kosako H., Iida N., Hattori S. (2007) Global analysis of dynamic changes in lipid raft proteins during T-cell activation. Electrophoresis 28, 2035–2043 [DOI] [PubMed] [Google Scholar]
- 59.Dhungana S., Merrick B. A., Tomer K. B., Fessler M. B. (2009) Quantitative proteomics analysis of macrophage rafts reveals compartmentalized activation of the proteasome and proteasome-mediated ERK activation in response to lipopolysaccharide. Mol. Cell. Proteomics 8, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bini L., Pacini S., Liberatori S., Valensin S., Pellegrini M., Raggiaschi R., Pallini V., Baldari C. T. (2003) Extensive temporally regulated reorganization of the lipid raft proteome following T-cell antigen receptor triggering. Biochem. J 369, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minami A., Fujiwara M., Furuto A., Fukao Y., Yamashita T., Kamo M., Kawamura Y., Uemura M. (2009) Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimatation. Plant Cell Physiol 50, 341–359 [DOI] [PubMed] [Google Scholar]
- 62.Alonso M. A., Millán J. (2001) The role of lipid rafts in signalling and membrane trafficking in T-lymphocytes. J. Cell Sci 114, 3957–3965 [DOI] [PubMed] [Google Scholar]
- 63.Dykstra M., Cherukuri A., Sohn H. W., Tzeng S. J., Pierce S. K. (2003) Location is everything: lipid rafts and immune cell signaling. Annu. Rev. Immunol 21, 457–481 [DOI] [PubMed] [Google Scholar]
- 64.Holowka D., Gosse J. A., Hammond A. T., Han X., Sengupta P., Smith N. L., Wagenknecht-Wiesner A., Wu M., Young R. M., Baird B. (2005) Lipid segregation and IgE receptor signaling: a decade of progress. Biochim. Biophys. Acta 1746, 252–259 [DOI] [PubMed] [Google Scholar]
- 65.Pike L. J. (2003) Lipid rafts: bringing order to chaos. J. Lipid Res 44, 655–667 [DOI] [PubMed] [Google Scholar]
- 66.DeLille J. M., Sehnke P. C., Ferl R. J. (2001) The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol 126, 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finnie C., Borch J., Collinge D. B. (1999) 14-3-3s: eukaryotic regulatory proteins with many functions. Plant. Mol. Biol 40, 545–554 [DOI] [PubMed] [Google Scholar]
- 68.Fu H., Subramanian R. R., Masters S. C. (2000) 14-3-3s: eukaryotic regulatory proteins with many functions. Annu. Rev. Pharmacol. Toxicol 40, 617–647 [DOI] [PubMed] [Google Scholar]
- 69.Palmgren M. G., Fuglsgang A. T., Jahn T. (1998) Deciphering the role of 14-3-3s. Exp. Biol. Online 3 [Google Scholar]
- 70.Roberts M. R. (2000) Regulatory 14-3-3 protein-protein interactions in plant cells. Curr. Opin. Plant Biol 3, 400–405 [DOI] [PubMed] [Google Scholar]
- 71.Fujimoto M., Arimura S., Nakazono M., Tsutsumi N. (2008) Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep 27, 1581–1586 [DOI] [PubMed] [Google Scholar]
- 72.Elmayan T., Fromentin J., Riondet C., Alcaraz G., Blein J. P., Simon-Plas F. (2007) Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant Cell Environ 30, 722–732 [DOI] [PubMed] [Google Scholar]
- 73.Alsterfjord M., Sehnke P. C., Arkell A., Larsson H., Svennelid F., Rosenquist M., Ferl R. J., Sommarin M., Larsson C. (2004) Plasma membrane H+-ATPase and 14-3-3 isoforms of Arabidopsis leaves: evidence for isoform specificity in the 14-3-3/H+-ATPase interaction. Plant Cell Physiol 45, 1202–1210 [DOI] [PubMed] [Google Scholar]
- 74.Leborgne-Castel N., Lherminier J., Der C., Fromentin J., Houot V., Simon-Plas F. (2008) The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in Bright Yellow-2 tobacco cells. Plant Physiol 146, 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verma D. P., Hong Z. (2005) The ins and outs in membrane dynamics: tubulation and vesiculation. Trends Plant Sci 10, 159–165 [DOI] [PubMed] [Google Scholar]
- 76.Konopka C. A., Bednarek S. Y. (2008) Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol 147, 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salim K., Bottomley M. J., Querfurth E., Zvelebil M. J., Gout I., Scaife R., Margolis R. L., Gigg R., Smith C. I., Driscoll P. C., Waterfield M. D., Panayotou G. (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J 15, 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- 78.Carlton J. G., Cullen P. J. (2005) Coincidence detection in phosphoinositide signaling. Trends Cell Biol 15, 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.König S., Ischebeck T., Lerche J., Stenzel I., Heilmann I. (2008) Salt-stress-induced association of phosphatidylinositol 4,5-bisphosphate with clathrin-coated vesicles in plants. Biochem. J 415, 387–399 [DOI] [PubMed] [Google Scholar]
- 80.Peleg-Grossman S., Volpin H., Levine A. (2007) Root hair curling and Rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. J. Exp. Bot 58, 1637–1649 [DOI] [PubMed] [Google Scholar]
- 81.Hong Z., Bednarek S. Y., Blumwald E., Hwang I., Jurgens G., Menzel D., Osteryoung K. W., Raikhel N. V., Shinozaki K., Tsutsumi N., Verma D. P. (2003) A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol 53, 261–265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.