Abstract
The nuclear pore complex (NPC) is a macromolecular assembly embedded within the nuclear envelope that mediates bidirectional exchange of material between the nucleus and cytoplasm. Our recent work on the yeast NPC has revealed a simple modularity in its architecture and suggested a common evolutionary origin of the NPC and vesicle coating complexes in a progenitor protocoatomer. However, detailed compositional and structural information is currently only available for vertebrate and yeast NPCs, which are evolutionarily closely related. Hence our understanding of NPC composition in a full evolutionary context is sparse. Moreover despite the ubiquitous nature of the NPC, sequence searches in distant taxa have identified surprisingly few NPC components, suggesting that much of the NPC may not be conserved. Thus, to gain a broad perspective on the origins and evolution of the NPC, we performed proteomics analyses of NPC-containing fractions from a divergent eukaryote (Trypanosoma brucei) and obtained a comprehensive inventory of its nucleoporins. Strikingly trypanosome nucleoporins clearly share with metazoa and yeast their fold type, domain organization, composition, and modularity. Overall these data provide conclusive evidence that the majority of NPC architecture is indeed conserved throughout the Eukaryota and was already established in the last common eukaryotic ancestor. These findings strongly support the hypothesis that NPCs share a common ancestry with vesicle coating complexes and that both were established very early in eukaryotic evolution.
Nearly all eukaryotic cells possess an extensive endomembrane system that is principally responsible for protein targeting and modification (1). The nucleus, the defining eukaryotic feature, is separated from the cytoplasm by a double bilayered nuclear envelope (NE)1 that is contiguous with the rest of this endomembrane system via connections to the endoplasmic reticulum. Nuclear pore complexes (NPCs) fenestrate the NE, serving as the exclusive sites mediating exchange between the nucleoplasmic and cytoplasmic compartments. Macromolecules are chaperoned through the NPC by numerous transport factors. It has been proposed that the endomembrane system and nucleus have an autogenous origin (i.e. evolving from invaginations of an ancestral plasma membrane) and were established early in eukaryotic evolution (2).
The composition of the NPC has been cataloged at ∼30 distinct nucleoporins (Nups) (3) for the yeast Saccharomyces cerevisiae (4) and vertebrates (5), two members of the Opisthokonta (animals, fungi, and closely related protists). Ultrastructural studies have identified objects morphologically similar (at a first approximation) to opisthokont NPCs in the other major eukaryote supergroups (6–8). However, very few data are available concerning the detailed NPC molecular composition and architecture for nearly all eukaryotic lineages, leaving a relatively narrow view of the “typical” NPC and its origins. A few examples of potential Nup orthologs beyond the opisthokonts have been reported, leading to the suggestion that substantial portions of the NPC may have an ancient, pre-last common eukaryotic ancestor (LCEA) origin (9). However, a more extensive study has concluded that LCEA possessed a primitive ancestral NPC that passed few components to its modern descendants (10).
In yeast and vertebrates, the NPC consists of an eight-spoked core surrounding a central tube that serves as the conduit for macromolecular exchange. Each spoke can be divided into two similar nucleoplasmic and cytoplasmic halves. The eight spokes connect to form several coaxial rings: the membrane rings, the two outer rings at the nucleoplasmic and cytoplasmic periphery, and the two adjacent inner rings (11). Groups of Nups that we term “linker Nups” are attached between both sets of outer and inner rings. Another group of related proteins, collectively termed phenylalanine-glycine (FG) Nups, are largely exposed on the inner surface of the spokes and anchored either to the inner rings or to the linker Nups (11).
Opisthokont Nups can be grouped into three structural classes (11, 12). The first class comprises membrane-bound proteins that anchor the NPC into the NE. The second class is the core scaffold Nups; these proteins constitute the bulk of the NPC mass, form the central tube, and provide the scaffold for the deployment of the third class of Nups across both faces of the NPC. The core scaffold Nups are remarkably restricted at the structural level and contain only three distinct arrangements of 2-fold types: proteins dominated by an α-solenoid fold (also termed a helix-turn-helix repeat domain), proteins consisting of a β-propeller fold, and finally proteins composed of an amino-terminal β-propeller fold followed by a carboxyl-terminal α-solenoid fold (which we here term a β-α structure) (12). FG Nups comprise the third class. These Nups carry multiply repeated degenerate “Phe-Gly” motifs (FG repeats) separated by hydrophilic or charged residues that form large unstructured domains. Each FG Nup also contains a small structured domain (often a coiled coil motif) that serves as the anchor site for interaction with the remainder of the NPC.
Many transport factors belong to a structurally related protein family collectively termed karyopherins (Kaps) (13, 14). Transport across the NPC depends on the interactions between Kaps, cargo molecules, and the disordered repeat domains of FG Nups; the latter are thought to form the selective barrier for nucleocytoplasmic transport, guiding the Kap·cargo complexes (and other transport factors) through the central tube while excluding other macromolecules (for reviews, see Refs. 3 and 15–22).
Significantly we have previously noted that the fold composition and arrangement of many of the core scaffold Nups are shared with proteins that form coating structures that participate in the generation and transport of vesicles between different endomembrane compartments; significantly many vesicle coating complex proteins and NPC scaffold Nups share an α-solenoid fold, β-propeller fold, or β-α structure (12, 23–28). These similarities gave rise to the “protocoatomer hypothesis,” which suggests a common ancestry for the NPC and these vesicle coat complexes. However, it is unclear how many, if any, of these particular core scaffold Nups are widely conserved, and hence it is unclear how general this potential relationship is throughout the Eukaryota. Thus, two scenarios are possible. The first is that the coatomer-like proteins are only found in a subset of the eukaryotes (including the opisthokonts), indicating that they are a relatively recent acquisition of only some eukaryotes and are not a general feature of all NPCs. The second is that the coatomer-like proteins are conserved in all eukaryotes, providing strong support to the protocoatomer hypothesis. To directly address this issue we characterized the NPC of Trypanosoma brucei, a highly divergent but experimentally tractable organism, using proteomics. The resulting data indicate an ancient origin for the majority of the NPC components and shed light on the origin of LCEA itself.
EXPERIMENTAL PROCEDURES
Proteomics Analysis of the T. brucei Nuclear Pore Complex-enriched Preparation (TbNEP)
The overall strategy for the identification of the T. brucei Nups (TbNups) is depicted in Fig. 1. The TbNEP was isolated as described previously (29). To reduce complexity and dynamic range within the sample and maximize the number of identifications, we used five distinct fractionation strategies against the TbNEP (Fig. 1 and supplemental data). These included (i) SDS-PAGE with MALDI-MS (30, 31); (ii) hydroxyapatite chromatography fractionation prior to SDS-PAGE and MALDI-MS; (iii) binding TbNEP to a C4 cartridge, digestion with trypsin, and analysis by LC-MS; (iv) differential enrichment of TbNEP proteins by chemical extraction prior to trypsin digestion and LC-MS (32); and (v) hydroxyapatite chromatography coupled to trypsin digestion and LC-MS. Peak lists were generated from the raw data using “Extract_msn” in Thermo Electron Xcalibur version 2.0 using default settings without enhancement or filters. The peak lists were submitted to X!Tandem (33) (version 2006.06.01.1) and searched against an in-house curated T. brucei protein database (generated July 5, 2005 using data from the genome sequencing project; the database was searched in its entirety). The X!Tandem search parameters were set as follows: missed cleavages permitted, 1; precursor ion tolerance, 4.0 Da; fragment ion tolerance, 0.4 Da; fixed modifications, carbamidomethylation of cysteine; variable modifications, oxidation of methionine. To reduce the possibility of false positives, only those individual MS/MS spectra with an expectation score better than 10−2 were considered.
Fig. 1.
Summary flow chart of biochemical, mass spectrometric, and bioinformatics methods used to identify putative T. brucei nucleoporins and transport factors. Strategies 1–5 are indicated by the red, blue, green, purple, and black colored arrows, respectively. The boxes are colored as follows: gold, protein recovery steps; light blue, protein separation steps; and brown, mass spectrometry techniques. Following mass spectrometry, the bioinformatics strategy outlined here identified 30 putative TbNPC-associated proteins from the initial pool of 757 identified proteins in the TbNEP. SDS-PAGE of fractions from a representative hydroxyapatite separation of the nuclear envelope fraction is shown at the top left. FW, flow-through and wash. Concentrations of phosphate in the elution buffer are indicated above the gel lanes, and apparent molecular masses (in kDa) are shown to the left of the gel. SDS-PAGE of T. brucei NE proteins that were subjected to chemical extraction is shown at the top right. The three extractions (base, salt and detergent (Det.), and heparin) are separated by vertical dashed lines. The pellet (P) and supernatant (S) are indicated. The number of Nups versus the total number of proteins identified with each successive strategy is depicted in the scatter plot (bottom right). Although the total number of proteins identified increases dramatically with further experimentation, the number of NPC-associated proteins levels off after four strategies. 1-D, one-dimensional.
Bioinformatics Analysis of the TbNEP Data Set
ORFs within the TbNEP data set were queried against GeneDB to obtain annotations, functional assignments, structural information, and sequence relationships to additional predicted gene products. ORFs were also analyzed and characterized by pairwise sequence alignments (basic local alignment search tool (BLAST) (34), PSI-BLAST using three iterations (35), and FASTA (36)) against the National Center for Biotechnology Information (NCBI) non-redundant database and in-house nuclear envelope protein databases (primarily Homo sapiens, Rattus norvegicus, and S. cerevisiae sequences). Unless otherwise noted, all algorithms were used with default search parameters. To search for the presence of conserved structural domains, a hidden Markov model (HMMer (37)) alignment to the Pfam HMM profile database of domain families was conducted (38). Following the in silico analysis, functionally unassigned ORFs present within the TbNEP data set were analyzed for several secondary structure elements, including β-sheets and α-helices (PSIPRED (39)), transmembrane helices (Phobius (40)), natively unfolded regions (Disopred (41)), and coiled coil regions (COILS (42)). Natively unfolded FG repeat domains were identified using a pattern recognition algorithm developed in-house (PROWL). Multiple sequence alignments were conducted with ClustalX (43). In some instances, multiple alignments were also subjected to phylogenetic analysis using MrBayes (44).
In Situ Tagging and Visualization
Open reading frames of interest were in situ tagged using the pMOTag4G and pMOTag4H vectors (45); see supplemental data for details and primer sequences. The linear PCR products were purified and sterilized by ethanol precipitation. T. brucei Lister 427 procyclic stage cells were transfected by electroporation with 10–25 μg of PCR product and cultured in SDM-79 (46, 47) supplemented with 10% fetal bovine serum and 0.25% hemin. Following transfection, 25 μg/ml hygromycin was added, and clones were screened by limiting dilution. After 3 weeks at least three colonies were assayed for correct insertion and expression using PCR and/or Western blotting (supplemental Fig. S1). For fluorescence microscopy tagged cell lines (suspended at 1 × 107 cells ml−1) were fixed with 2% formaldehyde for 5 min at room temperature and allowed to settle onto a coverslip treated with (3-aminopropyl)-triethoxysilane. Nonattached cells were washed away with PBS, and the coverslip was then mounted in 50% glycerol and 0.4 μg/ml 4′,6-diamino-2-phenylindole dihydrochloride in PBS. Immunofluorescence microscopy was conducted similarly as above except that after washing with PBS the attached cells were permeabilized with 0.1% Nonidet P-40 in PBS. Subsequently the coverslips were blocked for 20 min in PBG (PBS with 0.2% cold fish gelatin (Sigma) and 0.5% BSA) prior to incubation for 90 min with antibody (rabbit anti-Nup107 diluted to 1:100 (48)). After extensive washing with PBG, cells were incubated for 1 h with TRITC-conjugated secondary antibody (mouse anti-rabbit, 1:500). Images were acquired either with the DeltaVision Image Restoration microscope (Applied Precision/Olympus) using an Olympus 100×/1.40 numerical aperture objective or a Leica TCS-NT with a 63×/1.40 numerical aperture objective. GFP was either imaged directly using FITC emission and excitation filters with a 2-s exposure or labeled as above with anti-GFP at 1:3000 (30) and then secondarily labeled with goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Molecular Probes) at 1:1000. At least 15 Z-stacks (0.15-μm thickness) were acquired. Raw images were manipulated using a deconvolution algorithm (softWoRxTM v3.5.1, Applied Precision, enhanced additive setting). γ levels and false colors were adjusted to enhance contrast only, and final images were assembled in Adobe Photoshop.
RESULTS
Identification of Putative T. brucei Nups
Subfractionation of T. brucei yields two fractions highly enriched in NPCs, namely an NE fraction and an NPC/lamina-enriched fraction (29). Here we performed a comprehensive proteomics analysis of these TbNEPs using multiple complementary approaches that identified a total of 757 proteins (Fig. 1, Table I, supplemental Table S1, and supplemental information). As anticipated, the high sequence divergence between eukaryote Nups precluded facile identification of orthologs based only on primary sequence comparisons (9, 10). Hence we used a combination of experimental and in silico approaches to parse the TbNEP data set. First 448 proteins could be excluded on the basis that sequence homology searches clearly predicted a function that is unassociated with the TbNPC, such as ribosomal, endoplasmic reticulum, and cytosolic proteins. The remaining 309 proteins were parsed for features associated with known Nups. These criteria were based on predicted fold types, the presence of sequence motifs, predicted molecular weight, and predicted secondary structures. We used a secondary structure prediction algorithm (PSIPRED) to identify proteins with regions of predicted secondary structure consistent with the eight major fold types present within the vertebrate and yeast Nups (12). We also searched for motifs that are found within the NPC and NE, including transmembrane helices, natively unfolded regions (including those containing the FG repeats unique to nucleoporins), and coiled coil regions (12). This filtered search was based on the hypothesis that the trypanosomatid NPC shares many architectural features with that of the opisthokonts and would only miss those components that are species-specific or too divergent to recognize. However, should this hypothesis prove incorrect, we would fail to identify the majority of the NPC components.
Table I. Putative TbNPC-associated proteins.
| Accession number | Annotation | Mass kDa | log(e) | No. of unique identified peptides | Sequence coverage% | Category | Domains or fold typea | GFP localized? |
|---|---|---|---|---|---|---|---|---|
| Tb09.160.0340 | TbMlp-2 | 92.3 | −2.2 | 3 | 6.2 | Mlp | CC: 88–200, 206–283, 294–368, 416–596 | SPBb during anaphase |
| Tb11.03.0810 | TbMlp-1 | 109.6 | −23.8 | 13 | 19.5 | Mlp | CC: 292–336, 383–426, 436–496, 638–671, 689–748, 852–881, 884–974 | Yes |
| Tb10.61.2630 | TbSec13 | 41.6 | −14.5 | 4 | 12.0 | Nup | β-Propeller | Yes |
| Tb11.02.2120 | TbNup48 | 48.4 | −15.9 | 5 | 14.1 | Nup | β-Propeller | Yes |
| Tb09.211.4780 | TbNup82 | 82.3 | −35.0 | 16 | 30.4 | Nup | α-Solenoid | Yes |
| Tb11.02.0460 | TbNup89 | 89 | −52.8 | 19 | 32.6 | Nup | α-Solenoid | Yes |
| Tb10.6k15.3670 | TbNup96 | 96.4 | −74.6 | 23 | 39.9 | Nup | α-Solenoid | Yes |
| Tb11.01.7630 | TbNup109 | 108.6 | −21.9 | 9 | 10.8 | Nup | β-Propeller α-solenoid | Yes |
| Tb927.7.2300 | TbNup132 | 132.2 | −30.8 | 14 | 14.6 | Nup | β-Propeller α-solenoid | Yes |
| Tb10.6k15.2350 | TbNup144 | 144.2 | −70.9 | 27 | 30.5 | Nup | β-Propeller α-solenoid | Yes |
| Tb10.6k15.1530 | TbNup181 | 181.4 | −15.7 | 7 | 6.7 | Nup | α-Solenoid | Yes |
| Tb927.4.2880 | TbNup225 | 225.4 | −35.9 | 20 | 19.5 | Nup | α-Solenoid | Yes |
| Tb11.01.7200 | TbNup53a | 52.7 | −27.6 | 8 | 31.5 | Nup FG | CC: 407–443; FG (GFG): 16–263 | Yes |
| Tb927.3.3540 | TbNup53b | 52.8 | −36.0 | 9 | 34.2 | Nup FG | CC: 159–194, 248–262, 364–378; FG (GFG): 10–72 | Yes |
| Tb11.02.0270 | TbNup59 | 58.7 | −24.3 | 6 | 14.4 | Nup FG | CC: 452–509, 617–638; FG (FGFG): 194–299 | Not tagged |
| Tb927.4.5200 | TbNup62 | 62.4 | −26.0 | 9 | 29.9 | Nup FG | FG (GGFGA): 8–349; CC: 453–486, 493–521 | Not tagged |
| Tb927.4.4310 | TbNup64 | 64.1 | −52.6 | 13 | 27.7 | Nup FG | CC: 149–228; FG (FSFG): 331–583 | Yes |
| Tb927.8.8050 | TbNup75 | 74.7 | −3.2 | 2 | 4.0 | Nup FG | CC: 150–237; FG (FSFG): 317–684 | Yes |
| Tb927.3.3180 | TbNup98 | 98 | −129.9 | 20 | 27.6 | Nup FG | FG (FSFG): 321–986 | Yes |
| Tb11.01.2885 | TbNup140 | 140.2 | −20.2 | 9 | 17.6 | Nup FG | FG ((A/V)FGQ): 209–1432 | Yes |
| Tb11.01.2880 | TbNup149 | 149.1 | −2.9 | 2 | 2.9 | Nup FG | FG (VFGT): 267–388, 1007–1288 | Yes |
| Tb11.03.0140 | TbNup158 | 158.2 | −99.7 | 33 | 35.7 | Nup FG | FG (GGFGQ): 5–550; β-sandwich: 713–851; α-solenoid | Yes |
| Tb927.7.5760 | TbNTF2 | 15.8 | −2.7 | 3 | 45.9 | Transport factor | Not tagged | |
| Tb11.02.0870 | Ran-binding protein 1 | 17.6 | −13.0 | 3 | 24.8 | Transport factor | Not tagged | |
| Tb927.3.1120 | TbRTB2 | 24.3 | −109.9 | 23 | 83.4 | Transport factor | Not tagged | |
| Tb09.160.2360 | TbGLE2 | 38.3 | −8.4 | 4 | 14.6 | Transport factor | β-Propeller | Not tagged |
| Tb927.6.2640 | TbKap60 | 58 | −18.6 | 6 | 18.3 | Transport factor | Not tagged | |
| Tb10.70.4720 | TbKap95 | 95 | −8.5 | 4 | 9.5 | Transport factor | Not tagged | |
| Tb10.6k15.3020 | TbKap104 | 103.8 | −2.5 | 2 | 5.4 | Transport factor | Transportin2-like | Not tagged |
| Tb11.01.7010 | TbKap123 | 117.8 | −16.6 | 4 | 7.9 | Transport factor | Not tagged |
a The residue boundaries of the domains are listed along with the domain identifier: CC, coiled coil; FG, FG repeat. The most abundant FG repeat motif is listed within parentheses.
b SPB, spindle pole body.
Using these approaches, we identified a total of 22 candidate TbNups (Table I and supplemental data). Each candidate TbNup was identified in at least two proteomics analyses, suggesting that this cohort represents enriched and relatively abundant proteins within the NPC-containing fractions consistent with their assignment as candidate NPC-associated proteins. Five considerations suggest that we identified most TbNups. (i) Five ORFs in the T. brucei genome, Tb10.61.2630, Tb10.6k15.2350, Tb10.6k15.3670, Tb11.03.0140, and Tb927.4.5200, are annotated as putative TbNups based on sequence similarity; the products of all five were identified by our proteomics analysis. (ii) Every recognizable FG repeat-containing polypeptide encoded by the trypanosome genome was detected in the proteome. (iii) Eight transport factor homologs were identified, indicating that even transiently NPC-associated proteins were present in our preparations. (iv) We used proteomics strategies with progressively increasing dynamic ranges, allowing the identification of progressively less abundant proteins, the last of which more than doubled the total number of proteins in the data set but identified no additional candidate TbNups (Fig. 1). (v) Given the conserved morphology, size, and symmetry of the trypanosome NPC (29), one would expect a number of trypanosome NPC components (22 identified nucleoporins) similar to that in yeast (30 nucleoporins, or 26 excluding yeast-specific gene duplications) and vertebrates (28 nucleoporins) (3). These criteria indicate that identification of NPC components within the TbNEP preparation was thorough, capturing the majority of the trypanosome nucleoporins.
Localization of T. brucei Candidate Nucleoporins
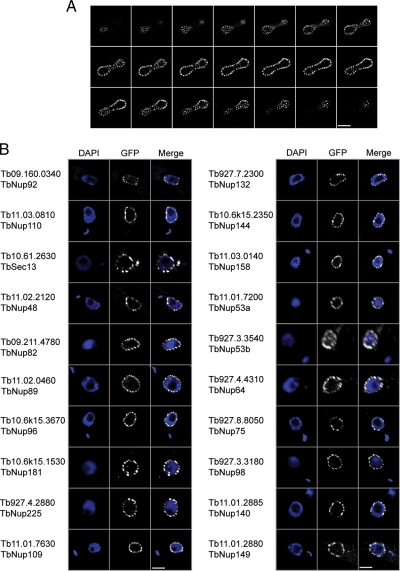
The candidate TbNups were localized by genomic tagging and fluorescence microscopy (Table I and Figs. 2 and 3). Almost all the GFP-tagged candidate TbNups displayed a similar punctate decoration restricted to the rim of the nucleus (Fig. 2). The puncta displayed a relatively homogeneous intensity and distribution; the average density of fluorescent puncta was 5.1 puncta/μm2 (n = 10, σ = 0.8) with an average of 93 puncta (σ = 16) per nucleus (see Fig. 2A for an example). Such patterns are considered highly characteristic for Nups in all other eukaryotic taxa examined (49–53), and indeed all four of the annotated Nup homologs that we tested, Tb10.61.2630, Tb10.6k15.2350, Tb10.6k15.3670, and Tb11.03.0140, displayed this pattern. We confirmed using double labeling with a cross-reacting anti-Nup antibody that this pattern represents NPC localization (Fig. 3A) (48). In total, 20 of the 22 putative TbNups displayed such punctate rim staining, identifying them as bona fide TbNups (Fig. 2B). Multiple attempts to tag the two remaining candidate TbNups, Tb11.02.0270 and Tb927.4.5200, failed to generate positive clones. Seven additional proteins in the data set are not classified as TbNups because they localized as diffuse or speckled staining in the cytosol or nucleus (supplemental Fig. S2). Such localizations may be false negatives due to disrupted protein targeting upon carboxyl-terminal epitope tagging or alternatively may represent truly non-NPC-associated proteins.
Fig. 2.
Validation of candidate T. brucei Nups. A, one copy of open reading frame Tb11.03.0140 (TbNup158) was genomically tagged at the COOH terminus with GFP. A montage of 21 confocal planes from the analysis of a TbNup158-tagged trypanosome in late anaphase is shown; each z-slice is 150 nm thick. There are ∼150 puncta associated with the nuclear envelope in this example. B, fluorescent microscopy gallery of COOH-terminal genomically labeled TbNups and corresponding 4′,6-diamino-2-phenylindole dihydrochloride (DAPI) fluorescence to visualize the DNA. Apart from TbSec13, which was labeled using the 3×HA epitope and visualized with a mouse monoclonal anti-hemagglutinin antibody at 1:1000, all other open reading frames were tagged with GFP. Scale bars, 2 μm.
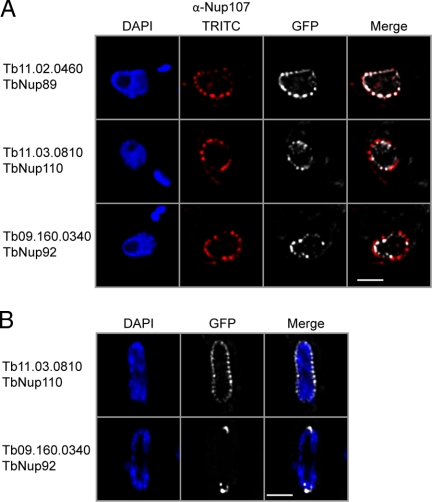
Fig. 3.
TbNup92 exhibits cell cycle-dependent localization. A, a rabbit polyclonal antibody against HsNup107 (48) was used to stain a trypanosome cell bearing tagged TbNup89. Co-localization of these signals further supports assignment of the puncta as the trypanosome NPC (top). Two coiled coil TbNups, TbNup110 and TbNup92, only partially co-localize with this antibody and are found immediately to the nuclear side of the NPCs and adjacent to them, suggesting association with the nuclear basket of the NPC and consistent with potential similarity to Tpr (bottom). B, TbNup110-GFP and TbNup92-GFP, visualized in mitotic cells, demonstrate that although TbNup110 remains associated with the NPC throughout mitosis TbNup92 relocates to opposite poles in a region similar to the spindle attachment site. Scale bar, 2 μm. DAPI, 4′,6-diamino-2-phenylindole dihydrochloride.
Structural Classification of TbNups
β-Propeller and α-Solenoid Fold Type-containing TbNups
A well conserved family of opisthokont Nups consists mainly of a β-propeller fold type (54). We found two clear examples in trypanosomes, Sec13p and also an ALADIN ortholog (TbNup48). ALADIN is also present in metazoa and plants but not S. cerevisiae (Fig. 4 and supplemental Fig. S3A) (55). Significantly, a homolog of Seh1p (a β-propeller Nup in opisthokonts) is conspicuously absent from the proteome.
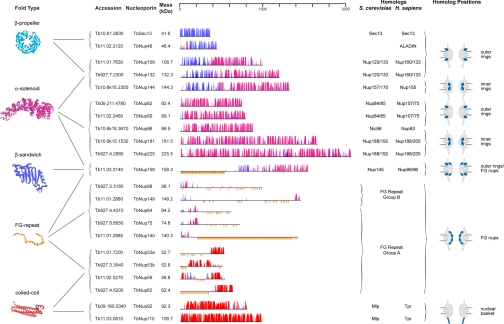
Fig. 4.
Predicted secondary structure features, fold, and location for validated TbNups. The ruler at the top indicates residue number. Within a map, the horizontal black line represents the polypeptide length of the Nup with the NH2 terminus to the left. The y axis indicates the confidence score of the predicted secondary structure element. Predicted α-helices are shaded in magenta, predicted β-sheets are in blue, and predicted coiled coil regions are in red. The vertical orange lines below the primary structure indicate FG dipeptides. Representative models of the Nup domains, colored according to their fold type, are shown to the left. The TbNups are binned according to their predicted fold type, and thus probable function, within the TbNPC; possible yeast and human homologs are indicated in the right-most column. Predicted positions of each Nup or Nup structural class within the NPC are shown at right based on the architecture as determined for S. cerevisiae.
There are five T. brucei α-solenoid Nups (Fig. 4); the number and mass of these proteins appear to have remained essentially unchanged between the Opisthokonta and trypanosomes. There are three smaller plus two larger α-solenoid Nups in S. cerevisiae (ScNup84, ScNup85, and ScNic96; ScNup188 and ScNup192), humans (HsNup107, HsNup75, and HsNup93; HsNup188 and HsNup205), and now trypanosomes (TbNup82, TbNup89, and TbNup96; TbNup181 and TbNup225). In most cases there is low sequence similarity between trypanosome, yeast, plant, or human α-solenoid Nups (supplemental Fig. S3B). For example, the nucleoporin-interacting component domain of ScNic96/HsNup93 is greatly diverged in trypanosomes, and the Pfam expect values for alignment between the consensus nucleoporin-interacting component domain and trypanosome TbNup96 is 10−5 compared with 10−177 (HsNup93) and 10−166 (ScNic96).
Proteins containing either β-propeller or α-solenoid fold types are ubiquitous (56). However, proteins with an amino-terminal β-propeller fold and carboxyl-terminal α-solenoid fold (β-α structure) architecture are restricted to the endomembrane system and are important components of the coats in coated vesicles and the scaffold of the NPC (25). Trypanosomes have homologs (TbNup109 and TbNup132) for the two smaller β-α structure Nups of S. cerevisiae (ScNup120 and ScNup133) and humans (HsNup133 and HsNup160). There is also a larger β-α structure trypanosome Nup (TbNup144) that is orthologous to HsNup155 and the two S. cerevisiae HsNup155 paralogs (ScNup157 and ScNup170) that arose from a yeast lineage-specific genome-wide duplication (57). With respect to primary structure, HsNup155, ScNup157, and ScNup170 are the only β-α structure Nups that are significantly conserved between opisthokonts and trypanosomes (supplemental Fig. S3C).
A Conserved β-Sandwich Domain
TbNup158 has a distinct and conserved domain structure. A highly conserved β-sandwich domain is situated between an FG repeat domain and an α-solenoid fold type (Fig. 4), which unambiguously identifies this gene product as an ortholog of HsNup98-96 and ScNup145. In the opisthokonts, however, the β-sandwich domain displays an autoproteolytic activity that initiates self-cleavage at a conserved H(F/Y)(S/T) tripeptide (58, 59). Although the β-sandwich domain is very highly conserved in T. brucei and the related excavate Giardia lamblia, both protist homologs lack the catalytic residues required for cleavage (supplemental Fig. S5). Consistent with this finding, we found that the trypanosome homolog TbNup158 does not cleave and instead functions as the full-length protein based on both Western blotting (supplemental Fig. S1) and mass spectrometry.
FG Repeat-containing TbNups
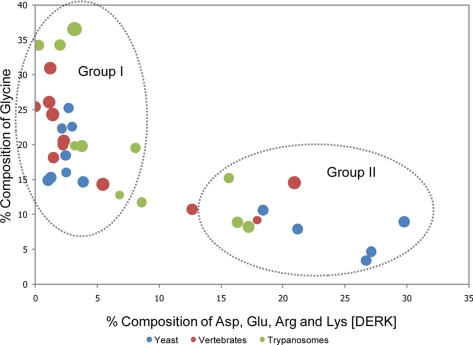
Like their opisthokont counterparts, the FG regions of trypanosome FG Nups are predicted to be natively unfolded. An extraordinarily high rate of amino acid substitution within FG Nups (60, 61) results in huge sequence divergence (supplemental Table S2A), confounding in silico identification of homology. A high level of genomic plasticity may be a common feature among FG Nups. An example of such plasticity may be TbNup140 and TbNup149, which are encoded by adjacent genes with an abnormally small intergenic region; whereas Northern and Western blotting suggests two separately transcribed messages (supplemental Figs. S1 and S9), in the related kinetoplastid Leishmania major, the ortholog LmjF28.3030 is apparently expressed as a single polypeptide. The vertebrate, S. cerevisiae, and trypanosome FG repeat domains generally have a similar frequency of Phe residues approximately ∼3-fold higher than the mean occurrence in their respective proteomes. Additionally these domains are generally depleted in large side chain amino acids and enriched in small side chain residues. This compositional bias is likely a general feature for natively unfolded regions (60, 62). The abundance of Gly varies considerably between FG repeat domains and displays a clear inverse correlation to the acidic and basic residues Asp, Glu, Arg, and Lys (Fig. 5 and supplemental Fig. S4). Thus, Nup FG repeat domains generally fall into two groups: group I contains Gly-enriched, DERK-deficient sequences, and group II contains significantly less Gly than group A and substantially more DERK residues (Fig. 5). Among the FG Nups, the homologs of TbNup158 can be uniquely identified because of the characteristic nature of their characteristic domains (see above). It is noteworthy that the FG regions of all the homologs of TbNup158 fall into group I, suggesting that the function of a given FG domain is conserved even if its sequence is not. In yeast and vertebrates, FG Nups that are symmetrically localized tend to fall into group I, whereas Nups with an asymmetric localization fall into group II albeit with some exceptions. Although the locations of these trypanosome Nups are currently not known, it will be of significant interest to ascertain whether this compositional feature is a potential predictor for FG Nup location. There is also some conservation in the structured domains of the FG Nups; TbNup53a, TbNup53b, TbNup59, and TbNup62 all possess a putative coiled coil domain, which as it does in their yeast and vertebrate counterparts likely serves to anchor these Nups to the NPC (Fig. 4) (12).
Fig. 5.
Correlation between the frequency of glycine and charged residues in trypanosome, yeast, and human FG repeat Nups. The percent composition of Gly is plotted against Asp, Glu, Arg, and Lys (DERK) residue frequency. Each data point represents an FG Nup from either S. cerevisiae (blue) or H. sapiens (red) or a candidate FG Nup from T. brucei (green). The diameter of each data point is directly proportional to the phenylalanine concentration within the respective Nup. FG Nups tend to cluster into two groups: high Gly, low DERK (Group I) and low Gly, high DERK (Group II). The average natural occurrence (in vertebrates) for Phe is ∼4% and for Gly is ∼7%, and the sum natural occurrence for the charged residues is ∼23%.
Nuclear Basket
Two members of the validated TbNup cohort, TbNup110 and TbNup92, exhibited highly characteristic localizations distinct from the other TbNups. Both partially co-localize with the NPCs (Fig. 3A) but are also found between NPCs at the inner face of the NE. Both proteins also have large predicted coiled coil domains (Table I). Their location and domain architecture are highly reminiscent of metazoan Tpr and its homologs S. cerevisiae Mlp1p/Mlp2p and Schizosaccharomyces pombe Nup211p and Alm1p (although at the sequence level they have undergone extensive species-specific divergence or may not share common ancestry) (Fig. 4 and supplemental Fig. S3D). These proteins appear to be components of the nuclear basket (63–68). Significantly although TbNup110 maintains a NPC location throughout the cell cycle, TbNup92 relocalizes during late mitosis to NE regions opposite the division plane where the mitotic spindle is likely anchored (Fig. 3B) (69). Localization to the spindle pole body is observed for one each of the S. pombe and S. cerevisiae Tpr homologs, Alm1p and Mlp2p, respectively, remarkably similar in behavior to TbNup92. This suggests, together with the structural data, that TbNup92 is an Mlp2 analog (64, 65) and that TbNup92 and TbNup110 are components of the basket structure at the trypanosome NPC nuclear face (29).
Integral Membrane Proteins
The membrane trypanosome Nups remain unidentified. Of the unannotated proteins within the TbNEP, 30% are predicted to contain at least one transmembrane helix (supplemental Table S1), but none contain a domain structure characteristic of opisthokont membrane Nups (i.e. cadherin-like domains for Pom152 or gp210 or NE constituents). One possibility is that we failed to recognize the integral membrane Nups; given the extremely low similarity between yeast and vertebrate membrane Nups this would not be surprising.
Transport Factors
In addition to 22 TbNups, we identified nine transport factors in the proteome (Table I). These proteins generally prove easier to identify by sequence homology searches than the TbNups because of a relatively high sequence similarity retained across the Eukaryota (supplemental Fig. S6). This sequence conservation is possibly due to the large number of interactions that these molecules must support, although additional factors may also be important.
Divergent Features of the TbNPC
The TbNEP did not contain any obvious homologs for several Nups found in S. cerevisiae or vertebrates. These include HsNup358, ScNup2, HsNup214/ScNup159, Seh1, and HsNup88/ScNup82. It is unlikely that these proteins have been overlooked as all have readily observable fold type, domain, and motif signatures; e.g. HsNup88/ScNup82 contains a β-propeller fold. It is therefore likely that these Nups have been either lost or diverged such that even in silico domain prediction fails. The presence of homologs of these Nups, as well as any trypanosomatid-specific Nups, will be elucidated with further investigations (potentially by co-immunoprecipitation or similar strategies).
Modular Duplications in the NPC
Each of the S. cerevisiae NPC spokes can be divided into two columns in which almost every Nup in one column has a counterpart of similar size, fold, and position in the adjacent column, and it is almost certain this holds true for the vertebrate NPC as well (11). We show here that this relationship also extends to trypanosomes (supplemental Fig. S8), indicating that an underlying 16-fold symmetry is likely universal. We previously proposed that a simpler module underwent ancient duplication and divergence events to generate the current NPC (11). The folds and orthologous relationships detected for trypanosomes (supplemental Fig. S8) fully support this modular duplication, which must have occurred prior to LCEA.
DISCUSSION
During the transition from prokaryote to eukaryote, cells gained a cytoskeleton, an elaborate endomembrane system, and a nucleus. The order in which these events occurred has been challenging to infer; there is no primitive state among extant eukaryotes (69, 70), and any reconstruction of evolutionary history has relied on the assumption that all modern eukaryotes derived from an LCEA. Because the NPC, a nuclear component in all eukaryotes, functions to maintain the distinct compositions of the nucleoplasm and cytoplasm, it is likely that the NPC co-evolved with the nuclear envelope. The NPC also retains distant relationships to intracellular transport systems (11, 12, 25).
Degree of Conservation of the NPC among Eukaryotes
We believe that we have identified the majority of the trypanosome nucleoporins (see “Results”), certainly enough to permit meaningful comparisons with the nucleoporin composition of opisthokont NPCs. Thus, by comparing validated sets of trypanosome and opisthokont Nups we are able to access the degree of conservation of NPC architecture across the Eukaryota, providing insight into both the LCEA and relationships between the NPC and endomembrane trafficking factors. Significantly, trypanosome NPC components share a remarkable level of architectural and compositional complexity with opisthokont Nups. Moreover, except for the transmembrane domain Nups that remain cryptic, homologs of all major classes of NPC proteins could be identified despite great levels of sequence divergence. Rather than primary sequences, eukaryotes appear to preserve the detailed fold arrangements within their NPC components.
This high level of conservation indicates an ancient origin for much of the NPC structure. The opisthokont NPC core scaffold comprises almost entirely β-propeller and α-solenoid fold types (11, 71). Eleven TbNups contain these folds, representing a remarkable degree of concordance between number, molecular weight, and architecture when compared against opisthokont core scaffold counterparts (Fig. 4 and supplemental Fig. S8). Given the evolutionary distance between these lineages, this concordance strongly suggests a near universal conservation of the basic NPC architecture. Further, although the sequences of trypanosome FG Nups are highly divergent compared with opisthokonts, they all share (i) extensive regions bearing Phe repeats, (ii) flanking of Phe by a small amino acid (usually Gly), and (iii) composition of the spacer residues particularly in respect to charge. These highly conserved features (together with the observed conservation of transport factors) also point to a conserved mechanism for mediating nucleocytoplasmic transport (72).
A further conserved NPC component appears to be the nuclear basket (29, 49, 73). Two putative T. brucei basket components, TbNup92 and TbNup110, consist of coiled coil domains and localize to the NPC but present negligible sequence similarity to ScMlp1p, ScMlp2p, or HsTpr. Furthermore TbNup92 and TbNup110 are clearly nonparalogous unlike ScMlp1p and ScMlp2p. However, similar to ScMlp1, TbNup110 localizes to the NPC throughout the cell cycle, whereas TbNup92 localizes to a position proximal to the spindle pole during mitosis analogous to ScMlp2 (67). S. pombe possesses a configuration similar to trypanosomes: two Mlp analogs of which only one exhibits differential localization during mitosis (64, 65). Only one such protein, Tpr, is present in metazoa. Our data do not allow unequivocal assignment of TbNup92 and TbNup110 as nuclear basket proteins, but a trypanosome nuclear basket has been visualized (29), and the overall architecture and behavior during mitosis of these proteins is highly suggestive of analogous function and hence location. If TbNup92 and TbNup110 are indeed components of the trypanosome nuclear basket this would indicate that basket proteins share essentially no sequence similarity and are potentially the products of lineage-specific gene duplications. These duplications may represent an instance of convergent evolution. Retention of the basket structure itself, however, would point to its importance in the overall mechanism of nuclear transport, likely at the level of RNA export (3).
Despite conservation of the NPC, homologs of membrane-bound Nups were not identified. It seems unlikely that such proteins were depleted from the TbNEP as we readily identified a great many transmembrane domain-containing proteins within this material. This may imply that although both the core and FG Nups are conserved membrane-associated Nups are unrecognizable by our algorithms. Alternatively the fact that pore membrane proteins are apparently dispensable for NPC function and assembly in Aspergillus (74) might indicate that membrane proteins are not a necessary component of the trypanosome NPC. Similarly prominent peripheral opisthokont Nups are also absent from our proteome; again these may be unidentified, truly absent, or replaced by trypanosome-specific analogs. Finally vertebrates carry three additional β-propeller Nups when compared with S. cerevisiae. Two possibilities could account for this: their ancestor had a simpler NPC that was elaborated in vertebrates or yeast lost these proteins (75). The presence of one of these additional β-propeller Nups (ALADIN) in trypanosomes clearly favors the secondary loss model.
The Protocoatomer Hypothesis for the Origin of NPC and Coated Vesicles
The similarity between the core scaffold Nups and components of vesicle coatomer complexes in both yeast and metazoa led to the suggestion that a pre-LCEA primitive membrane deforming complex evolved into both the NPC and the diverse set of membrane coat systems in extant eukaryotic taxa (11, 12, 25). Significantly if general membrane deforming complexes were the first components to arise, the model would then suggest that the basic α-solenoid/β-propeller architecture predates emergence of the NPC/NE (25). A key test of this protocoatomer hypothesis is therefore that these structural features must be retained by the contemporary NPC of all eukaryotes; however, prior in silico analysis has failed to provide unequivocal evidence (10).
The presence of an extensive trypanosome repertoire of β-propeller, α-solenoid, and β-α structure proteins, all abundant in vesicle coating complexes and restricted to the eukaryotic endomembrane system, plus clear conservation of a large proportion of the opisthokont NPC core by the trypanosome NPC strongly supports the protocoatomer hypothesis for the origin of eukaryotic endomembrane systems (12, 25). Evidence in favor includes the similar inventory, predicted molecular weight, and domain structure of the core Nups; the similar number and conserved amino acid composition of the FG Nups; the markedly similar morphology of NPCs across the Eukaryota; conservation of soluble transport factors, which suggests a conserved nuclear transport mechanism; and detectable sequence similarity between a minority of trypanosome and opisthokont Nups, including the highly conserved β-sandwich autoproteolytic domain of TbNup158 (supplemental data). Others have suggested that LCEA possessed an ancestral NPC with little resemblance to the modern one, passing few components to its descendants (10). However, the evidence here leads us to reject this model and instead robustly supports a model positing a common origin from a complex NPC followed by extensive divergent evolution (Fig. 6). It therefore follows that the LCEA likely possessed an NPC that was structurally analogous to the contemporary NPCs found in extant taxa, revealing its ancient relationship with vesicle coating complexes.
Fig. 6.
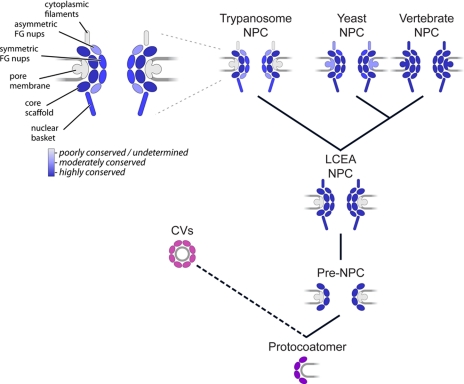
A model for the evolutionary origin of the NPC. A primitive coating complex (bottom, purple) evolved into numerous vesicle coating complexes (pink) and a simpler pre-NPC, which through duplication and divergence of its constituents produced a complex and elaborate NPC in the LCEA. The composition and architecture of the contemporary NPC throughout the Eukaryota is largely conserved with species-specific adaptations arising primarily by divergent evolution. The inferred degrees of conservation of the indicated different architectural elements of the trypanosome, yeast, and vertebrate NPC (with vertebrate set as the standard) is shown in shades of blue based on the analysis presented here. CVs, coated vesicles.
Supplementary Material
Acknowledgments
We acknowledge members of the B. T. Chait, M. P. Rout, and G. A. M. Cross laboratories for assistance and discussions. We thank Alison North and the Rockefeller University Bio-Imaging Resource Center for invaluable help with imaging. Numerous colleagues, including J. B. Dacks, J. S. Glavy, D. Fenyö, M. Niepel, J. C. Padovan, and B. Ueberheide have offered assistance and discussion to which the authors are indebted.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants RR00862 (to B. T. C.), GM062427 (to M. P. R.), and RR022220 (to M. P. R. and B. T. C.). This work was also supported by the Tri-Institutional Training Program in Chemical Biology (to J. A. D.) and Wellcome Trust Grant 082813/Z/07/Z (to M. C. F. and M. P. R.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- NE
- nuclear envelope
- FG
- phenylalanine-glycine
- GFP
- green fluorescent protein
- Kap
- karyopherin
- LCEA
- last common eukaryotic ancestor
- NPC
- nuclear pore complex
- Nup
- nucleoporin
- TbNEP
- T. brucei nuclear pore complex-enriched preparation
- TbNup
- T. brucei Nup
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Dacks J. B., Field M. C. (2007) Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci 120, 2977–2985 [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. (1975) The origin of nuclei and of eukaryotic cells. Nature 256, 463–468 [DOI] [PubMed] [Google Scholar]
- 3.Suntharalingam M., Wente S. R. (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775–789 [DOI] [PubMed] [Google Scholar]
- 4.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol 148, 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. (2002) Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol 158, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akey C. W., Radermacher M. (1993) Architecture of the Xenopus nuclear-pore complex revealed by 3-dimensional cryoelectron microscopy. J. Cell Biol 122, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim R. Y., Aebi U., Stoffler D. (2006) From the trap to the basket: getting to the bottom of the nuclear pore complex. Chromosoma 115, 15–26 [DOI] [PubMed] [Google Scholar]
- 8.Lim R. Y., Fahrenkrog B. (2006) The nuclear pore complex up close. Curr. Opin. Cell Biol 18, 342–347 [DOI] [PubMed] [Google Scholar]
- 9.Bapteste E., Charlebois R. L., MacLeod D., Brochier C. (2005) The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol 6, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mans B. J., Anantharaman V., Aravind L., Koonin E. V. (2004) Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3, 1612–1637 [DOI] [PubMed] [Google Scholar]
- 11.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 12.Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A 103, 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moroianu J., Blobel G., Radu A. (1995) Previously Identified Protein of Uncertain Function is Karyopherin-Alpha and Together with Karyopherin-Beta Docks Import Substrate at Nuclear-Pore Complexes. Proc. Natl. Acad. Sci. U.S.A 92, 2008–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radu A., Blobel G., Moore M. S. (1995) Identification of a Protein Complex that is Required for Nuclear-Protein Import and Mediates Docking of Import Substrate to Distinct Nucleoporins. Proc. Natl. Acad. Sci. U.S.A 92, 1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becskei A., Mattaj I. W. (2005) Quantitative models of nuclear transport. Curr. Opin. Cell Biol 17, 27–34 [DOI] [PubMed] [Google Scholar]
- 16.Macara I. G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev 65, 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pemberton L. F., Paschal B. M. (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6, 187–198 [DOI] [PubMed] [Google Scholar]
- 18.Peters R. (2005) Translocation through the nuclear pore complex: Selectivity and speed by reduction-of-dimensionality. Traffic 6, 421–427 [DOI] [PubMed] [Google Scholar]
- 19.Rout M. P., Aitchison J. D. (2001) The nuclear pore complex as a transport machine. J. Biol. Chem 276, 16593–16596 [DOI] [PubMed] [Google Scholar]
- 20.Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T. (2003) Virtual gating and nuclear transport: The hole picture. Trends Cell Biol 13, 622–628 [DOI] [PubMed] [Google Scholar]
- 21.Quimby B. B., Dasso M. (2003) The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol 15, 338–344 [DOI] [PubMed] [Google Scholar]
- 22.Weis K. (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441–451 [DOI] [PubMed] [Google Scholar]
- 23.Boehmer T., Jeudy S., Berke I. C., Schwartz T. U. (2008) Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell 30, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debler E. W., Ma Y., Seo H. S., Hsia K. C., Noriega T. R., Blobel G., Hoelz A. (2008) A fence-like coat for the nuclear pore membrane. Mol. Cell 32, 815–826 [DOI] [PubMed] [Google Scholar]
- 25.Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., Sali A., Rout M. P. (2004) Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dokudovskaya S., Williams R., Devos D., Sali A., Chait B. T., Rout M. P. (2006) Protease accessibility laddering: a proteomic tool for probing protein structure. Structure 14, 653–660 [DOI] [PubMed] [Google Scholar]
- 27.Hsia K. C., Stavropoulos P., Blobel G., Hoelz A. (2007) Architecture of a coat for the nuclear pore membrane. Cell 131, 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrader N., Koerner C., Koessmeier K., Bangert J. A., Wittinghofer A., Stoll R., Vetter I. R. (2008) The crystal structure of the Ran-Nup153ZnF2 complex: a general Ran docking site at the nuclear pore complex. Structure 16, 1116–1125 [DOI] [PubMed] [Google Scholar]
- 29.Rout M. P., Field M. C. (2001) Isolation and characterization of subnuclear compartments from Trypanosoma brucei. Identification of a major repetitive nuclear lamina component. J. Biol. Chem 276, 38261–38271 [DOI] [PubMed] [Google Scholar]
- 30.Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 31.Tackett A. J., Dilworth D. J., Davey M. J., O'Donnell M., Aitchison J. D., Rout M. P., Chait B. T. (2005) Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol 169, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 33.Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 34.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 35.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson W. R., Lipman D. J. (1988) Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A 85, 2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eddy S. R. (1998) Profile hidden Markov models. Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 38.Sonnhammer E. L., Eddy S. R., Birney E., Bateman A., Durbin R. (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26, 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuffin L. J., Bryson K., Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 40.Käll L., Krogh A., Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 41.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 42.Lupas A., Van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 43.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huelsenbeck J. P., Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 45.Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 46.Brun R., Schönenberger M. (1979) Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop 36, 289–292 [PubMed] [Google Scholar]
- 47.Field M. C., Horn D., Carrington M. (2008) Analysis of small GTPase function in trypanosomes. Methods Enzymol 438, 57–76 [DOI] [PubMed] [Google Scholar]
- 48.Glavy J. S., Krutchinsky A. N., Cristea I. M., Berke I. C., Boehmer T., Blobel G., Chait B. T. (2007) Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 subcomplex. Proc. Natl. Acad. Sci. U.S.A 104, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beck M., Förster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. (2004) Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 50.Davis L. I., Blobel G. (1986) Identification and characterization of a nuclear-pore complex protein. Cell 45, 699–709 [DOI] [PubMed] [Google Scholar]
- 51.Davis L. I., Fink G. R. (1990) The Nup1 gene encodes an essential component of the yeast nuclear-pore complex. Cell 61, 965–978 [DOI] [PubMed] [Google Scholar]
- 52.De Souza C. P., Horn K. P., Masker K., Osmani S. A. (2003) The SONBNUP98 nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165, 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whalen W. A., Yoon J. H., Shen R., Dhar R. (1999) Regulation of mRNA export by nutritional status in fission yeast. Genetics 152, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E. C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84, 265–275 [DOI] [PubMed] [Google Scholar]
- 55.Cronshaw J. M., Matunis M. J. (2003) The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc. Natl. Acad. Sci. U.S.A 100, 5823–5827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrade M. A., Perez-Iratxeta C., Ponting C. P. (2001) Protein repeats: structures, functions, and evolution. J. Struct. Biol 134, 117–131 [DOI] [PubMed] [Google Scholar]
- 57.Wolfe K. H., Shields D. C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 [DOI] [PubMed] [Google Scholar]
- 58.Fontoura B. M., Blobel G., Matunis M. J. (1999) A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol 144, 1097–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenblum J. S., Blobel G. (1999) Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. U.S.A 96, 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. (2003) Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. U.S.A 100, 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Denning D. P., Rexach M. F. (2007) Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell. Proteomics 6, 272–282 [DOI] [PubMed] [Google Scholar]
- 62.Weathers E. A., Paulaitis M. E., Woolf T. B., Hoh J. H. (2004) Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett 576, 348–352 [DOI] [PubMed] [Google Scholar]
- 63.Byrd D. A., Sweet D. J., Panté N., Konstantinov K. N., Guan T., Saphire A. C., Mitchell P. J., Cooper C. S., Aebi U., Gerace L. (1994) Tpr, a large coiled-coil protein whose amino-terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear-pore complex. J. Cell Biol 127, 1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X. Q., Du X., Liu J., Balasubramanian M. K., Balasundaram D. (2004) Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast 21, 495–509 [DOI] [PubMed] [Google Scholar]
- 65.Jiménez M., Petit T., Gancedo C., Goday C. (2000) The alm1(+) gene from Schizosaccharomyces pombe encodes a coiled-coil protein that associates with the medial region during mitosis. Mol. Gen. Genet 262, 921–930 [DOI] [PubMed] [Google Scholar]
- 66.Krull S., Thyberg J., Björkroth B., Rackwitz H. R., Cordes V. C. (2004) Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 15, 4261–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B. T., Rout M. P. (2005) The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol 170, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strambio-de-Castillia C., Blobel G., Rout M. P. (1999) Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol 144, 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., McCourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. F. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol 52, 399–451 [DOI] [PubMed] [Google Scholar]
- 70.Dacks J. B., Walker G., Field M. C. (2008) Implications of the new eukaryotic systematics for parasitologists. Parasitol. Int 57, 97–104 [DOI] [PubMed] [Google Scholar]
- 71.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 72.Rexach M., Blobel G. (1995) Protein import into nuclei—association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692 [DOI] [PubMed] [Google Scholar]
- 73.Kiseleva E., Goldberg M. W., Daneholt B., Allen T. D. (1996) RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol 260, 304–311 [DOI] [PubMed] [Google Scholar]
- 74.Liu H. L., De Souza C. P., Osmani A. H., Osmani S. A. (2009) The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84–120 complex. Mol. Biol. Cell 20, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Q., Rout M. P., Akey C. W. (1998) Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1, 223–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.