Abstract
Lethal 3 malignant brain tumor 1 (L3MBTL1), a homolog of the Drosophila polycomb tumor suppressor l(3)mbt, contains three tandem MBT repeats (3xMBT) that are critical for transcriptional repression. We recently reported that the 3xMBT repeats interact with mono- and dimethylated lysines in the amino termini of histones H4 and H1b to promote methylation-dependent chromatin compaction. Using a series of histone peptides, we now show that the recognition of mono- and dimethylated lysines in histones H3, H4 and H1.4 (but not their trimethylated or unmodified counterparts) by 3xMBT occurs in the context of a basic environment, requiring a conserved aspartic acid (D355) in the second MBT repeat. Despite the broad range of in vitro binding, the chromatin association of L3MBTL1 mirrors the progressive accumulation of H4K20 monomethylation during the cell cycle. Furthermore, transcriptional repression by L3MBTL1 is enhanced by the H4K20 monomethyltransferase PR-SET7 (to which it binds) but not SUV420H1 (an H4K20 trimethylase) or G9a (an H3K9 dimethylase) and knockdown of PR-SET7 decreases H4K20me1 levels and the chromatin association of L3MBTL1. Our studies identify the importance of H4K20 monomethylation and of PR-SET7 for L3MBTL1 function.
Keywords: cell cycle, chromatin, L3MBTL1, lysine monomethylation, PR-SET7
Introduction
Covalent histone modifications create docking sites for effector proteins that influence cellular behavior. Although some histone modifications (for example, histone H3 and H4 N-terminal acetylation) collectively influence gene expression, others (for example, methylation) have site-specific signaling potential (Lachner and Jenuwein, 2002; Huebert and Bernstein, 2005). Histone lysine methylation is coupled with both transcriptional competence (H3K4me3, H3K36me3 and H3K79me2) and silencing (H3K9me3, H3K27me3). Furthermore, the methylation status (mono-, di- or tri- (me1/me2/ me3)) of specific lysines, extends the options for signaling to chromatin (Peters et al., 2003; Rice et al., 2003) as exemplified for H4K20 methylation. H4K20me1 is associated with facultative heterochromatin (Fang et al., 2002; Martens et al., 2005; Sims et al., 2006), and the inactive X chromosome (Kohlmaier et al., 2004). In contrast, H4K20me3 marks constitutive heterochromatin (Schotta et al., 2004; Mikkelsen et al., 2007).
Specialized protein domains recognize modified histones, with preference for specific amino acids and modification states (Seet et al., 2006; Turner, 2007). The H3K4me2/3 mark is recognized by motifs such as the chromo, tudor, PHD and WD40 domains (Ruthenburg et al., 2007). The chromodomains of HP1 (Jacobs and Khorasanizadeh, 2002) and CHD1 (Flanagan et al., 2005; Pray-Grant et al., 2005), and the tudor domains of JMJD2A (Huang et al., 2006), bind preferentially to trimethylated lysines, but also interact with lower methylation states (mono- and di-). In contrast, the tudor domain of 53BP1, can discriminate between the di- and trimethyl state of H4K20, preferring the dimethyl form (Botuyan et al., 2006; Kim et al., 2006). Tudor and chromodomains are members of a conserved ‘Royal’ family of motifs and are related to PWWP, Agenet and MBT domains (Maurer-Stroh et al., 2003).
We have been studying L3MBTL1, a human homolog of the Drosophila lethal (3) malignant brain tumor (D-l(3)mbt) polycomb protein, that functions as a tumor suppressor in the larval brain (Gateff et al., 1993; Wismar et al., 1995). Human L3MBTL1 is located on chromosome 20q within a region commonly deleted in hematological malignancies (MacGrogan et al., 2004). L3MBTL1 contains three tandem MBT repeats (3xMBT), a C2HC zinc finger and an SPM oligomerization domain (Figure 1a). Each of the three MBT repeats contains a conserved, three β-stranded, core element (Wang et al., 2003), a structural arrangement seen in other ‘Royal’ domains.
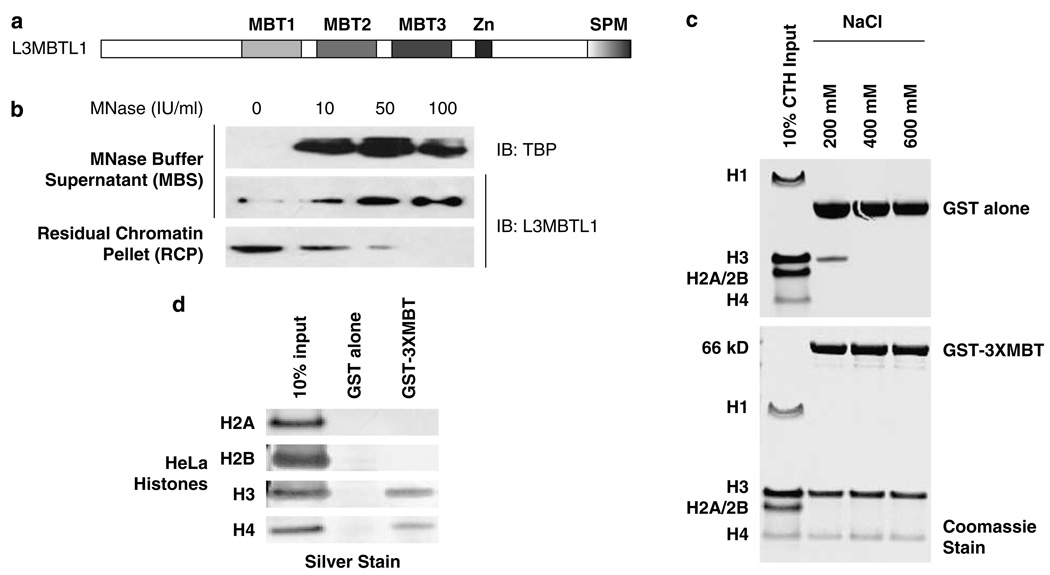
Figure 1.
Chromatin association of L3MBTL1 (and its 3xMBT repeats) in vivo and in vitro. (a) Schematic representation of the domain architecture of the human L3MBTL1 protein. (b) Western blots show the progressive release of endogenous L3MBTL1 from a K562 chromatin pellet (RCP) (third panel) into the micrococcal nuclease (MNase) buffer supernatant (MBS) (second panel) with increasing MNase input. The concomitant release of a chromatin-associated factor, TATA box-binding protein (TBP), into the MBS served as control (top panel). (c) In vitro binding of recombinant GST (top) and GST-3xMBT (bottom) to post-translationally modified calf thymus histones (CTH) analysed in pull down experiments at indicated NaCl concentrations in binding and wash buffers. (d) Binding of individually purified HeLa core histones H3 and H4 (but not H2A or H2B) to GST-3xMBT (at 600mm NaCl).
We have shown that L3MBTL1 functions as a transcriptional repressor (Boccuni et al., 2003) and that L3MBTL1 is complexed with Rb and can bind to E2F target genes such as c-myc and cyclin E (Trojer et al., 2007). Furthermore, changes in L3MBTL1 expression have been shown to alter normal mitotic progression (Koga et al., 1999; Yohn et al., 2003). Several recent studies have identified the methyl-lysine binding properties of the MBT domains in L3MBTL1 and D-Sfmbt; however, the specificity of such binding has not been well addressed (Kim et al., 2006; Klymenko et al., 2006). We have shown that histone lysine methylation promotes 3xMBT-mediated chromatin compaction in vitro (Trojer et al., 2007), and now provide evidence that the cell cycle-regulated chromatin association of L3MBTL1 occurs as the H4K20me1 mark accumulates, and that the repressive properties of L3MBTL1 are enhanced by the H4K20 monomethylase PR-SET7.
Results
The 3xMBT region of L3MBTL1 is sufficient for its chromatin association in vivo and in vitro
To test whether L3MBTL1 is chromatin-associated in vivo, we isolated nuclei from K562 erythroleukemia cells that express appreciable levels of L3MBTL1 (MacGrogan et al., 2004), and generated soluble (nucleoplasm) and insoluble (chromatin) fractions, as described (Mendez and Stillman, 2000). L3MBTL1 protein is found in the chromatin fraction (residual chromatin pellet; lane 1, Figure 1b). Digestion of the pellet with increasing concentrations of micrococcal nuclease (MNase) releases L3MBTL1 (and the TATA box-binding protein (TBP)) into the MNase buffer supernatant, identifying L3MBTL1 as a chromatin-associated factor. Using similar techniques, we determined that the 3xMBT repeats by themselves are capable of chromatin association (Supplementary Figures S1A, B, and C). Transiently expressed full-length L3MBTL1 or the 3xMBT repeat region, both localized to chromatin (in 293T cells) whereas the N terminus of L3MBTL1 did not (despite good nuclear accumulation; Supplementary Figure S1B). A deletion mutant of L3MBTL1 lacking the 3xMBT region (L3MBTL1-ΔMBT) did localize to the chromatin fraction. However, its chromatin association may be indirect, as the protein contains the SPM dimerization domain. Nevertheless, the 3xMBT region of L3MBTL1, in isolation, associates with chromatin in vivo.
To define the basis for the chromatin association of the 3xMBT repeats, we performed in vitro pull-down assays using recombinant 3xMBT repeats fused to a GST tag (GST-3xMBT) and calf thymus histones (Figure 1c). GST-3xMBT consistently interacted with histones H3 and H4, under stringent conditions (600mm NaCl in the binding and wash buffers) (lower panel, Figure 1c), whereas histones H1, H2A or H2B did not bind GST-3xMBT, even at low stringency. To exclude the possibility that the simultaneous binding to H3 and H4 is a consequence of their hetero-oligomerization, acid extracted histones H2A, H2B, H3 and H4 were individually purified from unsynchronized HeLa cells. Native histones H3 and H4, but not H2A or H2B, bound GST-3xMBT (at 600mm NaCl; Figure 1d). In contrast, no interaction was detected between 3xMBT and recombinant (unmodified) histones H3 and H4 (not shown).
L3MBTL1 (andthe 3xMBT region) preferentially bind the lower methylatedstates of lysines
To identify the specific post-translational modifications responsible for the interaction of 3xMBT with histones H3 and H4, we used fluorescence polarization (FP) measurements to quantify the binding of 3xMBT to a panel of differentially methylated histone peptides (Table 1), as described (Jacobs et al., 2004). 3xMBT bound several methylated lysines in the histones H3, H4 and H1.4 tails and displayed a modest preference for the H4K20 site (KD: 6µm for the H4K20me1 peptide) over the H3K4, H3K9, H3K27, H3K36 and H1.4K26 sites (KD: 12–31 µm for the monomethylated peptides; Table 1). However, strong discrimination toward the mono- and dimethylated states of specific lysines over their trimethylated and unmodified states was seen (Figures 2a and b), with at least an order of magnitude lower affinity. To exclude the possibility that the promiscuity of interactions with different mono- and dimethylated lysines in histones H3 and H4 peptides was due to the use of an isolated region (3xMBT), we examined the binding preferences for the full-length L3MBTL1 protein and obtained virtually identical results (Table 1). The affinity of 3xMBT (and L3MBTL1) for lower methylated states of histone lysines occurs in the µm range, similar to the affinity of chromodomains for trimethylated lysines (KD: 4–20 µm).
Table 1.
Binding affinities of 3xMBT and L3MBTL1 for indicated histone peptides
| Histone | Peptide sequence | Iso-electric point (pI) | Modification | 3xMBT KD (µm) |
L3MBTL1 KD (µm) |
|---|---|---|---|---|---|
| H3-NTD | ARTKQTARKSTGGKA | 12 | H3K4 unmodified | >750 | >1000 |
| H3K4me1 | 19 | 14 | |||
| H3K4me2 | 150 | >150 | |||
| H3K4me3 | >700 | >1000 | |||
| ARTKQTARKSTGGKA | 12 | H3K9 unmodified | >1000 | >700 | |
| H3K9me1 | 9 | 26 | |||
| H3K9me2 | 15 | 24 | |||
| H3K9me3 | >600 | >200 | |||
| TAGASRKGKQRKTAT | 12 | H3K9me1 (scrambled) | 35 | 35 | |
| QLATKAARKSAPATGGV | 11.2 | H3K27 unmodified | >500 | >700 | |
| H3K27me1 | 31 | 140 | |||
| H3K27me2 | 50 | 150 | |||
| H3K27me3 | >300 | >1000 | |||
| SAPATGGVKKPHRYRP | 11.1 | H3K36 unmodified | >1000 | >1000 | |
| H3K36me1 | 15 | 15 | |||
| H3K36me2 | 15 | 32 | |||
| H3K36me3 | >280 | >1000 | |||
| H4-NTD | KGGAKRHRKVLRDNIQ | 11.7 | H4K20 unmodified | >750 | >750 |
| H4K20me1 | 6 | 9 | |||
| H4K20me2 | 8 | 14 | |||
| H4K20me3 | >200 | >750 | |||
| LNRQDIAGKGKHVKRR | 11.7 | H4K20me1(scrambled) | 22 | 41 | |
| H4-CTD | EETRGVLKVFLENVI | 4.8 | H4K59me1 | >1000 | NT |
| YTEHAKRKTVTAMDV | 8.5 | H4K79me1 | >750 | NT | |
| H3-CTD | REIAQDFKTDLRFQS | 6.1 | H3K79me1 | >1000 | NT |
| H1.4-NTD | PVKKKARKSAGAAKR | 12 | H1.4K26 unmodified | >500 | NT |
| H1.4K26me1 | 13 | NT | |||
| H1.4K26me2 | 16 | NT | |||
| H1.4K26me3 | >300 | NT |
Abbreviations: CTD, C terminal domain; NTD, N terminal domain; NT, not tested.
Single site-binding constants (KD in µm) of 3xMBT or full-length L3MBTL1 for indicated histone N- and C-terminal peptides containing the specified modifications were obtained using fluorescence polarization measurements. Modified lysines are indicated in bold and underlined. Binding curves for each peptide were analysed by nonlinear least squares fitting using the equation A=[Af–(Ab–Af)]×[MBT]/(KD+[MBT]), where A=measured anisotropy at a given protein concentration and Af and Ab correspond to the anisotropy of free and bound peptides, respectively.
Values represent averages of at least three experiments.
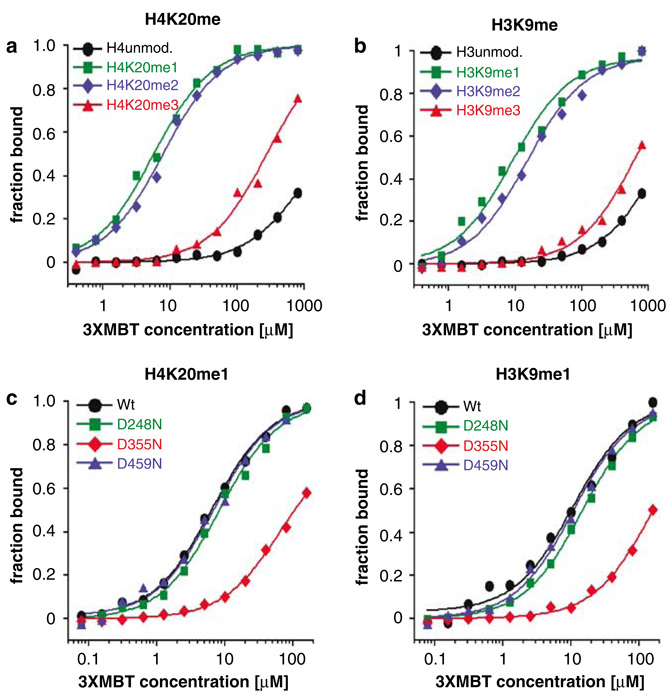
Figure 2.
3xMBT preferentially binds lower methylation states of lysine residues in histones H3 and H4 tails. Interaction of the 3xMBT region of L3MBTL1 with unmodified or differentially methylated (a) H4K20 and (b) H3K9 peptides was quantified using fluorescence polarization (FP). Binding curves represent averages from at least three independent experiments. Binding of 3xMBT proteins, with mutations in the ligand-binding pockets (D248N, D355N or D459N), to (c) H4K20me1 and (d) H3K9me1 peptides assayed using FP (KD for binding to H3K9me1 peptide; Wt 11 µm, for D248N 15 µm, for D355N 163 µm, for D459N 11 µm).
To examine how the peptide sequence affected the binding of 3xMBT to mono- and dimethylated lysine residues, we synthesized peptides containing a monomethylated lysine with the same composition as the natural H3 or H4 peptides, but with randomized amino acids surrounding the modified lysine. Only a slight decrement in affinity was observed, compared to the wild-type peptides (H3 and H4 scrambled in Table 1). However, when we tested additional methylated lysine residues (H4K59, H4K79 and H3K79) (Zhang et al., 2003), which are not embedded in basic sequence environments (pI <10), we found no binding of 3xMBT (Table 1, H3- and H4-C-terminal domain peptides). Thus, the binding of 3xMBT to mono- and dimethylated lysines is only seen using ‘basic’ peptides (pI>11, Table 1).
Using isothermal calorimetry (Supplementary Figures S2A and B); we confirmed the preference for H4K20me1 (KD: 17µm) over the H3K9me1 (KD: 32µm) site, and identified single sites of interaction (n~1) for both peptides. Thus, the 3xMBT region of L3MBTL1 can bind mono- and dimethylated lysines within the H3 and H4 peptides in a 1:1 stoichiometry.
To assess whether the 3xMBT domain binds H4K20me1 in a transcriptionally repressive or activating context, we immunoblotted the HeLa histone H4 species pulled down by GST-3xMBT with a panel of modification-specific antibodies. H4 species recognized by GST-3xMBT were devoid of the acetylation marks H4K5 and H4K16 (but enriched for H4K20me1). HeLa histone H3 recognized by GST-3xMBT lacked H3K9 acetylation (Supplementary Figure S4), supporting the idea that L3MBTL1 may be more commonly found in repressive environments.
A conserved aspartic acid in the second MBT repeat of 3xMBT mediates recognition of both H3K9me1 and H4K20me1 peptides
The MBT repeats each contain a ligand-binding pocket lined by aromatic residues (Wang et al., 2003) and a conserved aspartic acid (D248, D355 and D459). As a D215N mutation in the Drosophila sex comb on midleg protein results in developmental defects (Bornemann et al., 1998), we generated GST-3xMBT constructs with D (aspartic acid) to N (asparagine) substitutions in each of the MBT repeats. A single mutation (D355N) in the second MBT repeat abolished binding to both H4K20me1 and H3K9me1 peptides, whereas the corresponding mutations in the first and third repeats did not perturb the 3xMBT–methyl–lysine interaction (Figures 2c and d). We conclude that 3xMBT contains a single binding site for the recognition of mono- (and di) methylated H3, H4 (and H1b (Trojer et al., 2007)) histone peptides.
The chromatin association of L3MBTL1 mirrors the progressive accumulation of H4K20me1 during the cell cycle
The H4K20me1 modification is cell cycle regulated although the timing of its appearance and peak levels have varied among different cell types and experimental strategies (Fang et al., 2002; Rice et al., 2002; Pesavento et al., 2008). Therefore, we examined the levels of L3MBTL1 during distinct phases of the cell cycle, and using synchronized K562 cells, we found high levels of L3MBTL1 protein in G1/S and S phase cells, but barely detectable levels in G2/M cells (not shown). Using immunofluorescence and confocal microscopy to define the cellular localization of L3MBTL1 in K562 cells, we found that L3MBTL1 localized to the nucleus in a diffuse non-speckled pattern, and that L3MBTL1 staining decreased as cells undergo mitosis (that is, in those with an emerging tubulin spindle; Figure 3a).
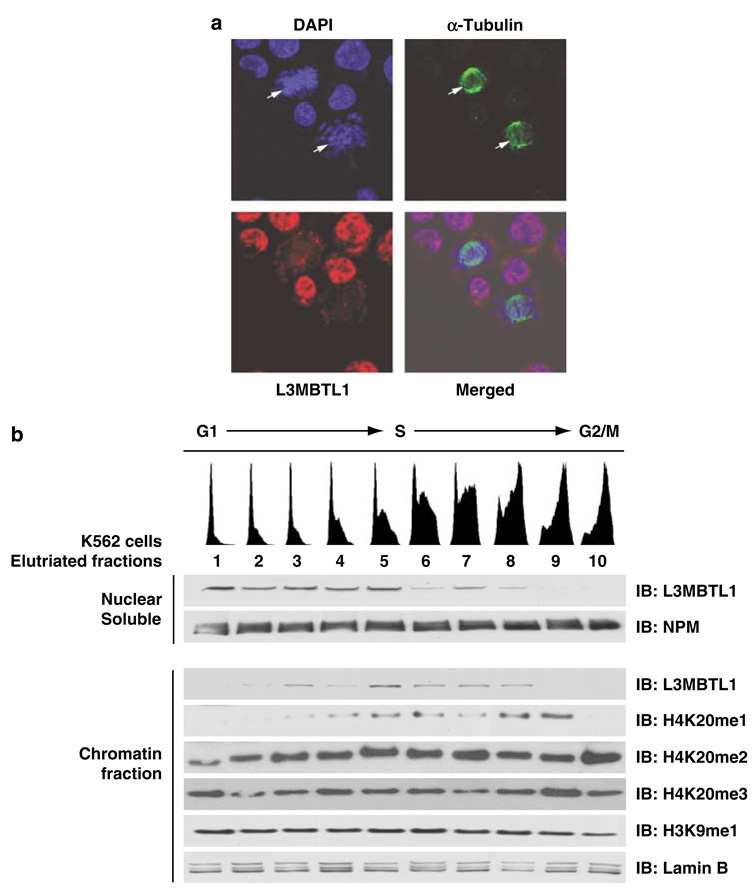
Figure 3.
The chromatin association of L3MBTL1 reflects the progressive accumulation of H4K20me1 during the cell cycle. (a) Confocal micrographs of K562 cells stained with anti-tubulin-α (green) and anti-L3MBTL1 (red) antibodies. DNA was counterstained with 4’-6-diamidino-2-phenylindole (blue). L3MBTL1 shows a diffuse nuclear distribution but is absent from mitotic cells (indicated by arrows). (b) Asynchronous cultures of K562 cells were size fractionated using centrifugal elutriation. Cell cycle status of each fraction was assessed by propidium iodide staining (indicated on top). Nuclear soluble and chromatin pellets isolated from each of the 10 fractions were analysed by immunoblotting using the indicated antibodies. Levels of L3MBTL1 and nucleophosmin (NPM; loading control) in the nuclear soluble fractions (panels 1 and 2) were assessed, as were the relative levels of L3MBTL1, H4K20 methylation (mono-, di- and tri-), H3K9 monomethylation and lamin B (loading control) in the chromatin pellet fractions (panels 3–8).
We also size-separated asynchronous K562 cells into 10 fractions by density centrifugal elutriation, as previously described (Koff et al., 1992), and determined the cell cycle profile (G1, S and G2/M fractions) for each of the 10 pools by propidium iodide staining and FACS analysis (top portion of Figure 3b). We prepared nuclear soluble fractions and chromatin pellets from each fraction (as above) and observed a decrease in L3MBTL1 protein levels in the G2/M fractions (Figure 3b, fractions 9 and 10). During late S-phase, L3MBTL1 was predominantly in the chromatin pellet (versus the nucleoplasm; Figure 3b, fractions 5–8), whereas the bulk of the protein was in the nuclear soluble fraction during G1 and early S phases (Figure 3b, fractions 1–5). Immunoblotting of the chromatin fractions, using a panel of antibodies specific for the H4K20me1, me2 and me3 marks, showed progressive accumulation of the H4K20me1 mark (during the S phase; fractions 4–8) leading to its peak in early G2 (fraction 9). In contrast, levels of H4K20 di- and trimethylation and of H3K9 monomethylation did not vary with cell cycle phase (Figure 3b, panels 5, 6 and 7). Thus, the chromatin association of L3MBTL1 during the cell cycle reflects the progressive accumulation of H4K20 monomethylation.
L3MBTL1 interacts with the H4K20 monomethyltransferase PR-SET7 in vivo and in vitro
The ability of proteins that recognize histone modifications to interact with the enzymes that catalyse those post-translational modifications is well documented (Stewart et al., 2005). Therefore, we investigated whether L3MBTL1 interacts with PR-SET7, the only known H4K20 monomethyltransferase (Fang et al., 2002; Nishioka et al., 2002). We transiently over-expressed HA-tagged L3MBTL1 and myc-tagged PR-SET7 in 293T cells. Reciprocal immunoprecipitation studies using anti-HA or anti-myc antibodies confirmed the specific interaction of L3MBTL1 and PR-SET7 (Figure 4a). We next generated a K562 cell line that stably expresses HA-tagged L3MBTL1 (K562-L3MBTL1), and following an anti-HA immunoprecipitation step detected the interaction of HA-L3MBTL1 with endogenous PR-SET7, but not with methyltransferases PRMT1, G9a (Figure 4b), ESET, EZH1 or EZH2 (not shown).
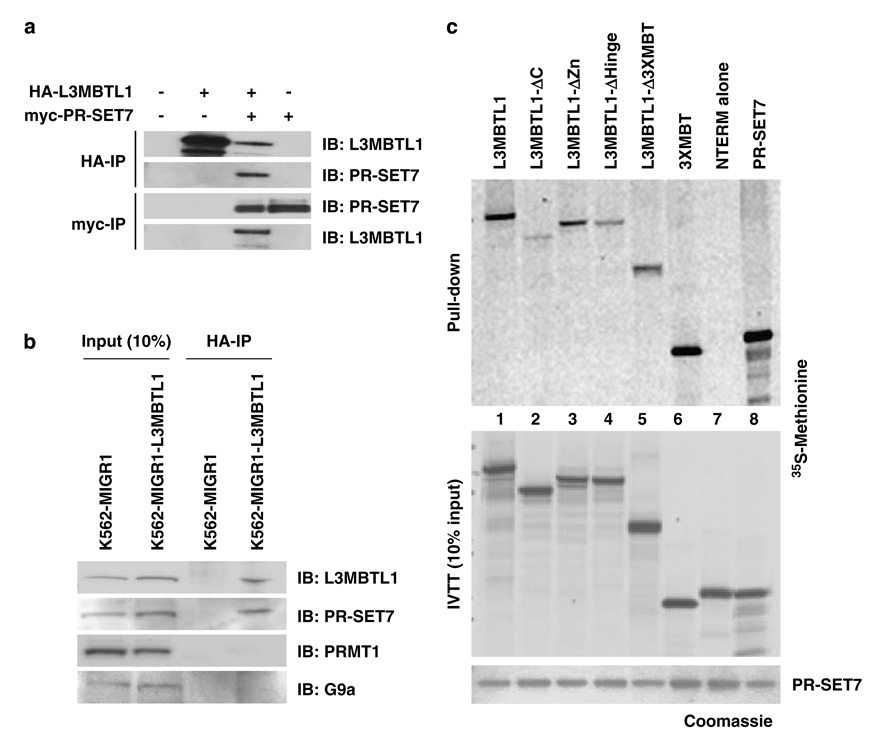
Figure 4.
L3MBTL1 interacts with the H4K20 monomethyltransferase PR-SET7 in vivo and in vitro. (a) Reciprocal immunoprecipitations (anti-HA and anti-myc) from nuclear extracts of 293T cells transiently transfected with plasmids encoding myc-tagged PR-SET7 and HA-tagged L3MBTL1 were analysed by western blotting using anti-PR-SET7 and anti-L3MBTL1 antibodies. (b) K562 cells engineered to stably express HA-L3MBTL1 (K562-L3MBTL1) or empty vector (control) were used in immunoprecipitation (IP) experiments using anti-HA affinity beads. Immobilized protein complexes (HA-IP) as well as 10% of input nuclear extracts were analysed by western blotting, using the indicated antibodies. (c) Bacterially expressed His-tagged PR-SET7 (5 µg; lower panel) was bound to Ni affinity beads and incubated with 35S-methionine-labeled in vitro-translated (IVTT) full-length L3MBTL1, the indicated L3MBTL1 deletional mutants or PR-SET7 (which forms homodimers). Ten percent of the IVTT input is shown (middle panel). Coomassie stained gels were dried and subjected to autoradiography (upper panels).
Using bacterially expressed PR-SET7 and in vitro translated L3MBTL1 (wild-type and deletion mutant proteins; Supplementary Figure S1A), we examined their direct interaction in vitro (Figure 4c). Full-length L3MBTL1, a mutant lacking the zinc finger and the 3xMBT region (in isolation) strongly interacted with recombinant PR-SET7 (lanes 1, 3 and 6), whereas the N-terminal region alone did not (lane 7). The known homodimerization of PR-SET7 was used as positive control (lane 8). Given the decreased affinity of PR-SET7 for L3MBTL1 mutants that lack the C-terminus or hinge region (Figure 4c, top panel), we conclude that L3MBTL1 contacts PR-SET7 through both the 3xMBT repeats and the hinge region.
The 3xMBT repeats mediate H4K20me1 targeted repression by L3MBTL1
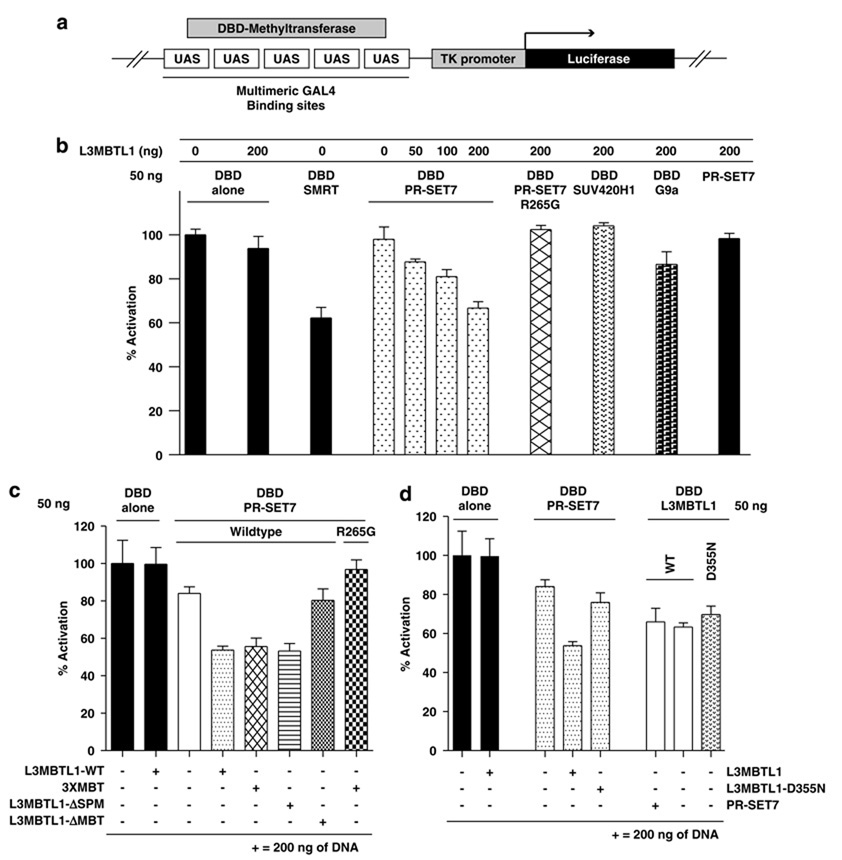
To examine the importance of H4K20me1 (and PR-SET7 H4K20 monomethylase activity) for the repressor function of L3MBTL1, we utilized the HEK293-TK22 cell line (Ishizuka and Lazar, 2003) that contains a stably integrated luciferase reporter gene downstream of multimeric GAL4 sites and a TK promoter (Figure 5a). In this system, GAL4 DNA-binding domain PR-SET7 fusion protein (DBD-PR-SET7) alone did not significantly repress luciferase expression. However, coexpression of L3MBTL1 with DBD-PR-SET7 resulted in dose-dependent repression comparable to that of the corepressor SMRT (DBD-SMRT) (Zamir et al., 1997) at multiple time points, with repression at 36 h post-transfection shown in Figure 5b. Significantly, expression of a catalytically dead form of PR-SET7 (R265G mutation; DBD-PR-SET7-R265G) (Nishioka et al., 2002) with L3MBTL1 did not repress luciferase expression. Furthermore, neither DBD-SUV420H1 (an H4K20 trimethyltransferase (Schotta et al., 2004)) nor DBD-G9a (an H3K9 mono-/dimethyltransferase (Rice et al., 2003)) caused significant repression when coexpressed with L3MBTL1 (Figure 5b).
Figure 5.
PR-SET7 promotes, and the 3xMBT repeats mediate, H4K20me1-targeted repression by L3MBTL1. (a) Schematic representation of the stably integrated luciferase reporter gene, downstream of multimeric GAL4-binding sites and a minimal thymidine kinase (TK) promoter, in the HEK293-TK22 cell line. (b–d) Plasmid constructs (amounts in ng) with or without fusion to the DNA-binding domain of GAL4 (DBD) were transfected into the reporter cell line. Luciferase activity in relation to the DBD alone control is shown (% activation). Averages from experiments performed in quadruplicate were calculated and plotted. The transcriptional repressor SMRT (DBD-SMRT) served as positive control (in panel b).
Although the L3MBTL1-ΔMBT mutant localizes to chromatin, it did not synergize with DBD-PR-SET7 (Figure 5c), nor did the L3MBTL1-D355N mutant (Figure 5d). However, PR-SET7 did cooperatively repress luciferase activity with both the 3xMBT repeats and the L3MBTL1 mutant that lacks the SPM domain (L3MBTL1-ΔSPM) (Figure 5c). As L3MBTL1-D355N interacts with wild-type PR-SET7, and the PR-SET7-R265G mutant interacts with L3MBTL1 (not shown), we conclude that recognition (or generation) of H4K20me1 clearly promotes transcriptional repression by L3MBTL1.
Binding of 3xMBT repeats to chromatin reflects the levels of H4K20 but not H3K9 methylation
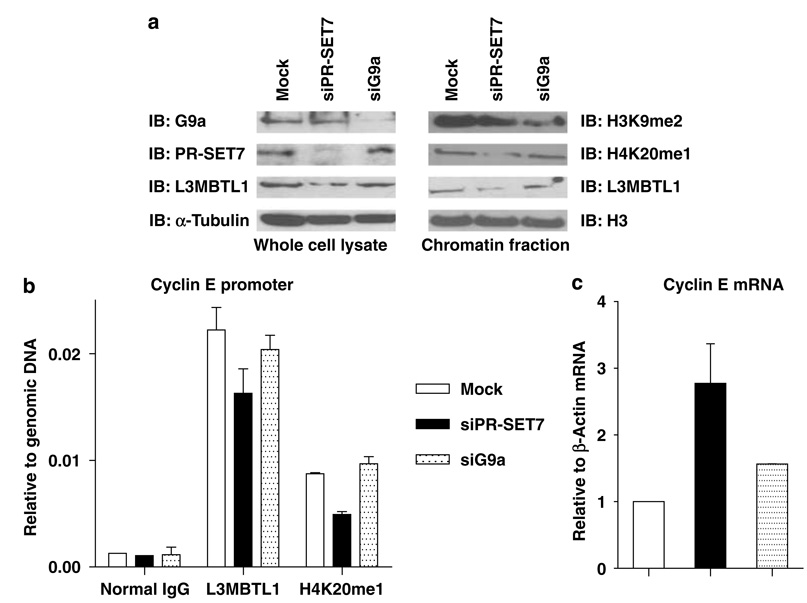
To validate the importance of the H4K20me1 mark, we transiently transfected K562 cells with siRNAs directed against the PR-SET7 or G9a. A significant reduction of target mRNA (>80%) and protein levels was achieved in each case (Supplementary Figures S3A and B). Notably, the cell cycle profile of siPR-SET7-treated K562 cells was not different from control cells at 24 or 48 h post-transfection (not shown). Although H4K20me1 levels and the chromatin association of L3MBTL1 decreased following the fall in PR-SET7 expression, the decrease in H3K9me2 in the G9a knockdown cells did not perturb the chromatin localization of L3MBTL1 (Figure 6a, right panels).
Figure 6.
The chromatin association of L3MBTL1 is regulated by siRNA-mediated knockdown of PR-SET7. (a) Protein levels of G9a, PR-SET7, L3MBTL1 and α-tubulin (left side) in whole cell lysates of transiently transfected K562 cells (mock, siPR-SET7, or siG9a) were assessed by immunoblotting. Chromatin pellets were assessed for the levels of H3K9me2, H4K20me1, L3MBTL1 and total H3 by immunoblotting (right side). (b) Quantitative assessment of the impact of H4K20me1 status on the in vivo localization of L3MBTL1 to the cyclin E promoter by chromatin immunoprecipitation followed by quantitative-PCR using K562 cells transfected with siRNA directed against PR-SET7 or G9a (compared to mock-transfected cells). (c) Real-time PCR was used to quantify cyclin E mRNA levels in mock-, siPR-SET7- or siG9a-transfected K562 cells.
L3MBTL1 localizes to the cyclin E promoter in association with the H4K20me1 mark
We have recently shown that L3MBTL1 is bound to the cyclin E promoter (Trojer et al., 2007). As D-l(3)mbt interacts with a repressive E2F (Lewis et al., 2004), and as we find binding of L3MBTL1 to several E2Fs (Gurvich NG et al., unpublished data), we performed chromatin immunoprecipitation assays to determine whether L3MBTL1 and H4K20me1 colocalize to the E2F binding sites in the cyclin E promoter. We found significant enrichment of L3MBTL1 and H4K20me1 (compared to normal immunoglobulin G control) in K562 cells, and knockdown of PR-SET7, but not of G9a, led to a concomitant reduction in the levels of H4K20me1 and L3MBTL1 on the promoter (Figure 6b). Quantitative reverse transcriptase–PCR analysis confirmed an increase in cyclin E mRNA levels in the siPR-SET7, but not mock or siG9a, transfected cells (Figure 6c). As we did not see any cell cycle defects in siPR-SET7-treated K562 cells, this change in cyclin E levels is unlikely due to difficulty in S-phase progression as reported for U2OS cells with knockdown of PR-SET7 (as reported by Jorgensen et al. (2007) and Tardat et al. (2007)). Using several siRNAs against L3MBTL1, we have been able to achieve less than a 50% knockdown in K562 cells. We did achieve a 50% reduction of L3MBTL1 levels in two cell lines (T98G and U2OS), using stable shRNA-based knockdown. This level of L3MBTL1 knockdown resulted in only a 1.5-fold increase in cyclin E expression. Greater knockdown of L3MBTL1 may not be tolerable to these cells. Furthermore, other proteins that recognize the H4K20me1 mark (including other MBT-containing proteins) may persist and prevent a higher level of cyclin E expression. Nonetheless, we conclude that L3MBTL1 localizes to and represses a target gene promoter, in association with the H4K20me1 mark.
Discussion
Our detailed in vitro and in vivo analysis of human L3MBTL1, the survey of chromatin-binding domains by Kim et al. (2006), and the recruitment of D-Sfmbt to chromatin (Klymenko et al., 2006), identify the MBT domain as a novel methyl-lysine-binding motif. The MBT domain prefers lower methylation states and fully discriminates against the trimethylated and unmodified lysines, in stark contrast to most other methyl recognition motifs. Although specialized domains display binding specificity for distinct methyl-lysine marks, in few cases have they been extensively tested against different sites of histone lysine methylation. Despite the apparent preference for the H4K20me1 mark, the 3xMBT repeats exhibit a clear preference for methyl-lysines embedded in the basic sequences of histone tails (pI>11), over those in the globular domain (pI <10).
A single amino-acid substitution in the second MBT domain (D355N) abolished interactions with both H3K9me1 and H4K20me1. Furthermore, the H3K9me1 and H4K20me1 peptides compete with each other for binding to 3xMBT (unpublished observations), demonstrating that these modified lysines bind exclusively to the second MBT domain. The 3xMBT domains of L3MBTL1 may function similar to the tandem tudor domains of 53BP1/Crb2, where methyllysine recognition is attributed to a critical aspartic acid in one domain, with residues in the other domains providing stabilization (Botuyan et al., 2006). Indeed, the report of methyl-lysine recognition by a recombinant fragment of L3MBTL1 containing only the MBT2 and MBT3 domains (Kim et al., 2006) does suggest that some specificity is generated by the tandem arrangement.
While previous studies have relied on qualitative assays to address methylation site specificity of the MBT domains of L3MBTL1, we have used a quantitative approach, paying attention to peptide length and composition. The reported binding of H4K20me2 and H3K4me1, but not H4K20me1 or other lower methylation states of histone H3 (K9, K27 or K36) by Kim et al. (2006) is intriguing, as the peptide lengths are comparable to those used in our study. The presence of all three repeats in our studies, versus their use of the MBT2 and MBT3 repeats alone in a different peptide screening approach, may account for the observed differences. The lack of binding of 3xMBT to methylated lysines in histone H3 (K4, K9 or K27), in a pull-down assay in our previous report (Trojer et al., 2007), could reflect the use of histone peptides that were shorter in length (<10 amino acids) and less basic than the 15 mers used in this study. However, it is worth noting that a shorter peptide length did not seem to affect the binding of 3xMBT to H4K20me1, and may reflect the preservation of the basic amino-acid composition. In our previous study, we observed binding to H1b-K26 me1 (also abolished by a D355 mutation in the second MBT repeat) using a peptide that was 16 amino acids in length (with a centrally placed modified lysine) and had a pI>12. We have examined the binding of 3xMBT to an H1.4-K26me1 peptide and find affinities comparable to the H3K9me1 peptide, but weaker than H4K20me1 (Table 1). Despite this, we found no binding to histone H1 in our pull-down assay (Figure 1c). However, we cannot rule out the possibility that this reflects the low levels of H1.4K26me1 in the calf thymus histone preparation.
The binding constants of L3MBTL1, and indeed other effectors, are rather weak, suggesting that stabilization of binding may occur via interaction with other proteins. Binding of HP1 to SUV39H1 appears to be important for its chromatin localization on H3K9me3 (Bannister et al., 2001; Lachner et al., 2001; Schotta et al., 2002). The interaction of L3MBTL1 with PR-SET7 and the functional link to its methyltransferase activity on H4K20 may represent a similar, stabilizing interaction.
While 3xMBT can interact with both histones H3 and H4 in vitro, our functional studies identify PR-SET7 H4K20 monomethyltransferase activity as being a strong promoter of its transcriptional repressor activity. The cell cycle-regulated chromatin association of L3MBTL1 mimics the emergence of the H4K20me1 mark (but not its peak), and the significant decrease in L3MBTL1 levels that precede the onset of mitosis is likely critical for proper cell cycle progression. Notably, the absence of PR-SET7 (and the H4K20me1 mark) also results in defects in cell cycle progression (Karachentsev et al., 2005; Jorgensen et al., 2007; Tardat et al., 2007). The absence of L3MBTL1 in the premitotic fraction 9 (Figure 3b) when H4K20me1 is abundant suggests that additional post-translational modifications of histones, or of L3MBTL1, may target the protein for premitotic destruction. In this regard, Pesavento et al. (2008) and Julien and Herr (2004) have demonstrated that H4K20me1 is targeted for conversion to H4K20me2 by an unknown methylase at this stage of the cell cycle (Talasz et al., 2005; Vakoc et al., 2006). L3MBTL1 may function to protect H4K20me1 from progression to H4K20me2 until some mitotis-associated enzymatic activity becomes functional. Knockdown of PR-SET7 in K562 cells (>80%) led to a decrease in global H4K20me1 levels resulting in a comparable decrease in the chromatin-associated L3MBTL1. The suboptimal drop in H4K20me1 levels in siPR-SET7-treated K562 cells (similar to that reported by Pesavento et al., 2008) may indicate the existence of uncharacterized H4K20 monomethylases or stable persistence of the mark. Our data suggest that recruitment of L3MBTL1 to the promoter regions of genes that it represses, is regulated by the H4K20me1 mark.
Initial reports highlighted the importance of H4K20me1 in ‘facultative’ heterochromatin and X-inactivation (Fang et al., 2002; Nishioka et al., 2002; Kohlmaier et al., 2004; Karachentsev et al., 2005; Martens et al., 2005; Sims et al., 2006). Recently, this exclusivity has been questioned by studies that suggest a role in transcriptional competence and elongation (Talasz et al., 2005; Vakoc et al., 2006; Barski et al., 2007; Pesavento et al., 2008). The recent finding that H4K20me1 is further partitioned into species that carry mono-, di- or tetraacetylated marks on H4K16 (Talasz et al., 2005; Vakoc et al., 2006; Pesavento et al., 2008) suggests several site-specific roles for this mark in gene regulation. Of relevance for L3MBTL1 is our finding that the HeLa H4 histones recognized by GST-3xMBT, which are enriched for H4K20me1, are largely devoid of acetylation (Supplementary Figure S4). This implies that L3MBTL1 mediates repression in the context of an H4K20me1 mark in facultative heterochromatin (that lacks acetylation). The nuclear distribution of L3MBTL1 and H4K20me1, and the identification of HP1γ, a protein linked to facultative heterochromatin, in the L3MBTL1 complex further support our inference (Sims et al., 2006; Dialynas et al., 2007; Trojer et al., 2007).
L3MBTL1 can bind the PR-SET7 H4K20 monomethylase and it appears that this enzymatic activity promotes its repressor function. Our data implies that the H4K20me1 mark is required for repression by L3MBTL1, but to formally prove this would require mutating this lysine. As this is lethal in eukaryotes, such proof is not feasible. Post-translational modifications of L3MBTL1, such as phosphorylation and/or ubiquitination, may lead to the dissociation of L3MBTL1 from chromatin in premitotic cells when H4K20me1 peaks. Domains in L3MBTL1 other than the 3xMBT region (for example, the SPM, zinc finger and the hinge region) may further influence the binding of L3MBTL1 to nucleosomes, its chromatin compaction, the facilitation or removal of additional histone modifications and the recruitment of other effector proteins. Further studies should clarify the biological significance of this family of proteins.
Materials and methods
Plasmids, cell lines and protein expression
K562 cell lines that stably express L3MBTL1 were generated using MIGR1-HA-L3MBTL1-IRES-GFP retroviral vector. L3MBTL1 and 3xMBT were cloned into the pBEn-SBP vector (Stratagene, La Jolla, CA, USA). Protein expression was performed as previously described (Nishioka et al., 2002; Wang et al., 2003).
Chromatin association assays
Chromatin association assays were performed as previously described (Mendez and Stillman, 2000). In vitro GST pull-down assays were performed using buffer containing 10mm HEPES (pH 8.0), 3mm EDTA, 0.05% Tween and 600mm NaCl.
Histone peptides
Histone peptides (~15 amino acids long) were synthesized incorporating unmodified, mono-, di- or trimethylated lysines. Peptides were labeled using 5-carboxyfluorescein, succinimidyl ester (5-FAM, SE) (Molecular probes, Carlsbad, CA, USA), purified by reverse HPLC on a C18 column (Vydac, Hesperia, CA, USA) and verified by MALDI mass spectrometry.
Fluorescence polarization assays
Fluorescence polarization binding assays were performed in 20mm Tris (pH 8.0), 20mm NaCl using 100 nm fluorescein-labeled peptide as previously described (Jacobs et al., 2004) and data collected on a Hidex Chameleon reader.
Coimmunoprecipitations and antibodies
Immunoprecipitation and washing buffers contained 0.05% NP-40, 0.05% Tween, 20mm Tris (pH 8), 1mm EDTA, 1mm DTT, 250mm NaCl and protease inhibitors (Calbiochem, San Diego, CA, USA). The L3MBTL1 antibody has been described previously (Boccuni et al., 2003). Other antibodies were from Upstate (Lake Placid, NY, USA) (H3K9me1, H4K20me1, 2 or 3, G9a, PR-SET7 and PRMT1), Sigma (St Louis, MO, USA) (HA, lamin-B, tubulin-α and NPM) and Santa Cruz (Santa Cruz, CA, USA) (TBP and myc).
Luciferase reporter assays
HEK293-TK22 cells (Ishizuka and Lazar, 2003) were transfected in six-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with indicated amounts of plasmids, 5 ng of TK-Renilla, and pUC19 plasmids to reach a final concentration of 4 µg. Relative luminescence was measured 36 h post-transfection using the Dual Luciferase Reporter assay kit (Promega, Madison, WI, USA).
siRNA-mediated knockdown of PR-SET7 and G9a in K562 cells
K562 cells (4 × 106 cells) were nucleofected with double-stranded duplex siRNAs (IDT, Coralville, IA, USA) directed against PR-SET7 or G9a (sequences in Supplementary data) using an Amaxa instrument and prescribed reagents (Amaxa Inc., Gaithersburg, MD, USA) and harvested at 36 h.
Chromatin immunoprecipitation analyses
Transfected K562 cells (mock, siPR-SET7 and siG9a) were used for chromatin immunoprecipitation experiments using established protocols. Enriched DNA was quantified using an ABI 7700 instrument and SYBR green and normalized to input DNA (Applied Biosystems, Foster City, CA, USA). Data were analysed by the standard curve method. Primer Recognition of histone methyl lysines by L3MBTL1 pairs for quantitative-PCR analysis of chromatin immunoprecipitation amplified sequences flanked the E2F sites in the cyclin E promoter.
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Acknowledgements
We dedicate this paper to the late Piernicola Boccuni, whose beautiful life as a physician–scientist and as a husband, father and friend, ended far too quickly. This work was made possible by grants from the National Institute of Health (SDN), the Leukemia and Lymphoma Society (SDN, XZ), the Herbert and Lee Friedman Fellowship Fund (XZ), the MSKCC Clinical Scholars Program (PB), the Damon Runyon Research Foundation (WF), and the Mel Stottlemyre Research Foundation (NK). We thank Drs C David Allis (The Rockefeller University, NY) for guidance and advice, Wooikoon Wang and Dinshaw Patel (MSKCC, NY) for structural insights; Tarun K Dam and C Fred Brewer (Albert Einstein Medical College, NY) for help with ITC experiments and Andrew Koff (MSKCC, NY) for scientific advice and assistance with centrifugal elutriation.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6) J Biol Chem. 2003;278:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- Bornemann D, Miller E, Simon J. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics. 1998;150:675–686. doi: 10.1093/genetics/150.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas GK, Terjung S, Brown JP, Aucott RL, Baron-Luhr B, Singh PB, et al. Plasticity of HP1 proteins in mammalian cells. J Cell Sci. 2007;120:3415–3424. doi: 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Gateff E, Loffler T, Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: developmental studies and molecular localization of the gene. Mech Dev. 1993;41:15–31. doi: 10.1016/0925-4773(93)90052-y. [DOI] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Huebert DJ, Bernstein BE. Genomic views of chromatin. Curr Opin Genet Dev. 2005;15:476–481. doi: 10.1016/j.gde.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Fischle W, Khorasanizadeh S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 2004;376:131–148. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–725. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrogan D, Kalakonda N, Alvarez S, Scandura JM, Boccuni P, Johansson B, et al. Structural integrity and expression of the L3MBTL gene in normal and malignant hematopoietic cells. Genes Chromosomes Cancer. 2004;41:203–213. doi: 10.1002/gcc.20087. [DOI] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol. 2008;28:468–486. doi: 10.1128/MCB.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, et al. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H, Lindner HH, Sarg B, Helliger W. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem. 2005;280:38814–38822. doi: 10.1074/jbc.M505563200. [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, III, Vaquero A, Kalakonda N, Boccuni P, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wismar J, Loffler T, Habtemichael N, Vef O, Geissen M, Zirwes R, et al. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- Yohn CB, Pusateri L, Barbosa V, Lehmann R. l(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics. 2003;165:1889–1900. doi: 10.1093/genetics/165.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I, Dawson J, Lavinsky RM, Glass CK, Rosenfeld MG, Lazar MA. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).