Abstract
Delivering genes to mediate functions of cells is a crucial technology for both basic science and clinical applications. Though numerous non-viral gene delivery systems have been developed, the diversity of mammalian cells poses a great challenge to the material design. Here, we demonstrate that surface-induced mineralization represents a promising approach to systematically customize DNA delivery with respect to the characteristics of cells. We initially examined gene transfer in nine cell types derived from different tissues and organisms by surface-induced DNA/calcium carbonate nanocomposites derived from a library of mineral solutions. Subsequently, we correlated gene transfer efficiency with cellular uptake, pH responsiveness of nanocomposites, and phagosomal pH of individual cell types. Based on the correlation, we were able to optimize the DNA delivery to the cell types of interest. Surface-induced mineralization possesses great potential for customizing gene transfer in realizing gene- and cell-based therapy and probing functions of genes.
Keywords: Nanocomposite, tissue engineering, biomineralization, gene transfer
1. Introduction
Delivering DNA macromolecules to mammalian cells represents a promising therapeutic strategy and an effective tool for basic sciences. DNA molecules cannot efficiently diffuse across the intercellular and intracellular barriers alone and require the assistance of delivery vectors. Though viral-based vectors are more effective, concern over safety issues and difficulty in engineering viruses for specific cell types have limited their applications[1–3]. Non-viral delivery vectors are an attractive alternative strategy due to their safety, low cost and flexibility[4, 5]. However, a great challenge to the material design of gene delivery systems is the heterogeneity of cell types, which exhibit distinct characteristics of transporting materials across membranes, intracellular routing, and regulation of gene expression. Though combinatorial approaches have been developed to screen a vast library of polymeric materials for gene and siRNA delivery[6, 7], the usable library has so far been limited to a few number of cell types. Development of a versatile material design platform that allows customization of gene transfer with respect to an individual cell type could advance the applications of synthetic DNA delivery systems.
Recently, a surface-induced mineralization process has been developed to fabricate DNA/calcium carbonate nanocomposites that can effectively deliver DNA to bone and neural cells[8, 9]. The composition and morphology can be easily controlled and tuned through adjusting the composition of the mineral solutions and surface properties of substrates. Interestingly, it appeared that the gene transfer efficiency in different cell types can be tuned by adjusting the composition of mineral solutions[9]. In addition, cells can be directly grown on DNA/calcium carbonate nanocomposite-coated, cell-culture-friendly surfaces, which permit high throughput screening of mineral formulations or substrates for a given cell type.
In this study, we explored the possibility of establishing a platform based on the surface-induced mineralization process to customize the nanocomposites with respect to characteristics of individual cell types. By using a small library of mineral solutions, we initially examined gene transfer efficiency in nine different cell types derived from different tissues and organisms. Subsequently, we correlated the efficiency of gene transfer with the properties of nanocomposites and the physiological characteristics of individual cell types. Based on the correlation, we optimized the DNA delivery to two cells types.
2. Materials and methods
2.1. Formation of surface-induced DNA–doped nanocomposites
All reagents for mineralization were purchased from Sigma. Mineral solutions were prepared as previously described[10]. All components except CaCl2 were added together to Milli-Q water and CaCl2 was added last to prevent spontaneous precipitation. The resulting solution was buffered to pH 7.4 with Tris-HCl and filtered by a 0.2 µm pore-size filter and stored at 4°C. One milliliter of a given mineral solution with different formulations (Table 1) was mixed with 10 µl of a DNA solution (100 µg/ml) and added to 24-well tissue-culture plates (BD Biosciences; San Jose, CA). The reporter plasmid, gWIZ Beta-gal (Aldevron; Fargo, ND), encodes for the reporter enzyme β-galactosidase (β-gal). The mineralization was conducted at 37 °C in a humidified incubator for an indicated period of time.
Table 1.
Formulations of mineral solutions
| Concentration (mM) | A | B | G | H | G-Sr | G-F |
|---|---|---|---|---|---|---|
| CaCl2.2H2O | 2.5 | 3.7 | 2.5 | 2.5 | 2.5 | 2.5 |
| KH2PO4 | 1 | 1 | 1 | 1 | 1 | 1 |
| NaCl | 141 | 141 | 141 | 141 | 141 | 141 |
| KCl | 4 | 4 | 4 | 4 | 4 | 4 |
| MgSO4.6H2O | 0.5 | 0.5 | 0 | 0.5 | 0 | 0 |
| MgCl2.6H2O | 1 | 1 | 0 | 5 | 0 | 0 |
| NaHCO3 | 4.2 | 4.2 | 4.2 | 4.2 | 4.2 | 4.2 |
| SrCl2 | 0 | 0 | 0 | 0 | 3 | 0 |
| NaF | 0 | 0 | 0 | 0 | 0 | 3 |
For the formation of DNA-1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)-doped nanocomposites, 10 µl of DOTAP in Dulbecco’s phosphate buffered saline (DPBS) (0.6 or 1.2 mg/ml) was mixed with 10 µl of DNA solution (100 µg/ml) in DPBS. The resulting DNA-DOTAP complex was incubated at room temperature for 15 min. One milliliter of a given mineral solution with different formulations (Table 1) was mixed with 20 µl of the DNA-DOTAP complex and added to each well of 24-well plates. The mineralization was conducted at 37 °C in a humidified incubator for an indicated period of time.
2.2. Morphology and composition of nanocomposites
Scanning electron microscopy (SEM) was used to characterize the size and morphology of nanocomposites mineralized for 24 h. The samples were sputter-coated with 12 nm of platinum using a SPI Sputter™ Coater (Structure Probe, Inc.; West Chester, PA) and were analyzed with a JEOL 7000 SEM with a beam voltage of 10 keV (Electron Microscopy Center, University of Washington). Energy dispersive x-ray spectroscopy (EDX) was used to characterize the chemical compositions of nanocomposites. The samples were sputter-coated with carbon and analyzed with a beam voltage of 15 keV.
2.3. pH responsiveness of DNA–doped nanocomposites
Mineralization was carried out as described above for the indicated durations. After mineralization, the solution was removed and 1 ml fresh mineral solution A (simulated body fluid) was added to each well. Dissolution profiles of different minerals were determined by gradually decreasing the pH of each well using 0.1 M hydrochloric acid. The volume of acid added was less than 20 µl for each pH point. Samples were shaken on a rotary mixer to insure quick and even distribution and allowed to equilibrate for 2 minutes before sampling. Calcium released from nanocomposites was determined using a modified colorimetric method based on complex formation with ortho-cresolphthalein[11]. The pH at which 50% of the total calcium was released, designated pH50, was used as an indicator of pH responsiveness. The dissolution was conducted at room temperature.
2.4. Determination of the efficiency of DNA incorporation
During the course of mineralization, 50 µl of the supernatant in each well was sampled and assayed for the concentration of DNA. The quantity of DNA in the supernatant was determined using the Quant-iT PicoGreen dsDNA reagent (Invitrogen; Carlsbad, CA) following the manufacturer’s protocol. The amount of DNA precipitated was calculated by subtracting the amount of DNA remaining in the supernatants from the total DNA added to each well.
2.5. Cell culture
MG-63 cells were cultured in Dulbecco’s Modified Eagle Media (DMEM), supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (P-S), 1% L-glutamine and 1% sodium pyruvate; Saos-2 cells were cultured in DMEM, supplemented with 10% FBS, 1% P-S and 2% L-glutamine; EMT6 cells were cultured in DMEM, supplemented with 10% FBS, 1% P–S, 1% L-glutamine; Caco-2 cells were cultured in Modified Eagle Medium (MEM), supplemented with 20% FBS and 1% P–S; Ishikawa and Hela cells were cultured in MEM, supplemented with 10% FBS, 1% P–S and 1% L-glutamine; TC1 cells were cultured in RPMI Media 1640, supplemented with 10% FBS, 1% HEPES, 1% sodium pyruvate, 1% L-glutamine, 0.15% sodium bicarbonate and 0.45% glucose; B35 cells were cultured in DMEM, supplemented with 10% FBS and 1% L-glutamine; Hep G2 cells were cultured in MEM, supplemented with 10% FBS and 1% sodium pyruvate. All the cells were maintained in an incubator at 37 °C and 5% CO2.
2.6. Imaging of cells with bright-field light microscopy
Cells grown on surfaces coated with DNA-doped nanocomposites were monitored by a Nikon TE2000 inverted microscope with a 20× objective. Images were acquired with a CoolSnap ES2 charge-coupled device camera (Photometrics; Tucson, AZ).
2.7. Gene transfer of cells cultured on DNA-doped nanocomposites
1×105 cells in 500 µl tissue culture medium were directly plated on surfaces coated with DNA-doped nanocomposites mineralized for 8 h for MG-63, Saos-2, Caco-2, Ishikawa, TC1, Hela, B35 and Hep G2; 5×105 cells were plated for EMT6 per well. Cells were incubated at 37°C and 5% CO2 for 36 h before analysis of cell viability and transfection efficiency. A commercial transfection reagent, Lipofectamine 2000™ (Invitrogen; Carlsbad, CA), was used as a positive control following the manufacturer’s protocol.
2.8. Determination of gene transfer efficiency
Cells were grown on surfaces coated with DNA-doped nanocomposites for 36 h. The media was then removed, and samples were then lysed with 150 µl of a solution containing 10 µM 2-ME, 9 mM MgCl2 and 0.1% triton X-100 in DPBS for 15 min. Then, three freeze-thaw cycles between −80 °C and 37 °C were performed to ensure complete release of proteins from cells. 50 µl of the lysed cell solution was mixed with 50 µl solution containing 0.15 mM chlorophenol red-β-D-galactoside (CPRG), 10 µM 2-ME, 9 mM MgCl2 and 0.1% triton X-100 in DPBS and incubated at 37 °C and 5% CO2 for 30 min. The absorption was measured at 570 nm using a SpectraMax M5 microplate reader (Molecular Devices; Sunnyvale, CA). The quantity of β-gal produced by the cells was determined by using a standard curve constructed with known concentrations of β-gal. Gene transfer efficiency was expressed as ng of β-gal per mg of total protein. Total protein was measured using the Coomassie protein assay (Biorad; Hercules, CA). Briefly, 5 µl of the lysed cell solution was diluted with 5 µl DPBS and then mixed with 200 µl of Coomassie solution. The absorption at 595 nm was determined with a microplate reader. The quantity of protein was determined by using a standard curve with known concentration of bovine serum albumin protein.
2.9. Determination of the metabolic activity of cells cultured on DNA-doped nanocomposites
The metabolic activity of cells on DNA-doped nanocomposites was determined using the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric method, which is based on the reduction of the MTT salt into formazan crystals by viable cells. Herring sperm DNA (1 µg/ml) was used for mineralization. Briefly, cells were grown on surfaces coated with DNA-doped nanocomposites as above. 36 h later, 50 µl of a 5 mg/ml solution of MTT (Sigma; St. Louis, MO) in water was added and incubated at 37°C and 5% CO2 for an additional 2.5 h. Following centrifugation, the supernatant was removed and 1 ml dimethyl sulfoxide was added to cells and incubated at 37°C for 30 min to dissolve the formazan crystals. The absorption of the wells at 570 nm was determined using a microplate reader.
2.10. Quantification of DNA uptake by flow cytometry
0.5 µg fluorescein isothiocyanate-labeled DNA (2.7 kilobase pairs) (Mirus Bio; Madison, WI) was co-precipitated with 1 ml mineral solution for 8 h. The solution was removed and cells were plated at the same densities as above. 24 h after cells were cultured on surfaces coated with DNA-doped nanocomposites, the plate was washed with 0.25 ml Hank’s Balanced Salt Solutions (HBSS). Cells were detached using 250 µl of trypsin-EDTA (0.1% Trypsin, 0.4% EDTA·4Na) at 37°C for 15 min. Cells were then washed with a solution containing 1% FBS in DPBS twice and analyzed by flow cytometry immediately using a BD FACScan2 (Cell Analysis Facility, Department of Immunology, University of Washington). Trypan blue was used to quench the fluorescence associated with the cell surface. The data was analyzed with FlowJo (Treestar; Ashland, OR), and the arithmetic mean of fluorescence intensity (MFI) of each sample was determined. The uptake of DNA by cells was expressed as the MFI of cells cultured on surfaces coated with DNA-FITC-doped nanocomposites subtracted with the MFI of cells cultured on uncoated surface.
2.11. Measurement of phagosomal pH
100 nm beads with amine functional groups (Polysciences; Warrington, PA) were coupled with the pH-sensitive dye FITC and the pH-insenstive dye AlexaFluor 647 succinimidyl ester (AlexaFluor647) (Invitrogen; Carlsbad, CA). Beads were resuspended in 500 µl 0.1 M carbonate buffer with a pH of 9 at 1% w/v. 5 µl of FITC, and AlexaFlour647 from a 10 mg/ml stock in PBS were added to the beads and mixed. The beads were then incubated at 4°C for 16 h, washed 3 times with PBS, and resuspended 500 µl PBS. Cells were incubated with beads for 15 min, washed 3 times with PBS, and further incubated at 37 °C in media for 15 min and immediately analyzed by flow cytometry. The ratio of the mean fluorescence intensity of FITC and AlexaFluor647 for each sample was determined (FL1/FL3 ratio). Values were calibrated against a standard curve obtained by resuspending cells that were exposed to coupled beads for 2 h. Cells used for obtaining calibration curves were fixed with 4% PFA, permeabilized with 0.1% triton X-100, incubated in citrate buffers with defined pH and immediately analyzed by flow cytometry.
2.12. Statistical analysis
Triplicate samples were included for all experiments and expressed as the average. Experiments were repeated at least two times. The standard deviation is within 5% if not mentioned.
3. Results and discussion
3.1. Morphology and composition of surface-induced calcium carbonate nanocomposites
A library of six mineral solutions was chosen in this study based on variations from simulated body fluid (Table 1). Our initial choice of mineral compositions was based on our previous study[9]. Additionally, two mineral solutions, G-F and G-Sr, were included. In G-Sr, strontium (Sr) was added to mineral solution G. Previous reports have shown that DNA/strontium phosphate precipitates resulted in less lethal and differentiation-inducing effects on many epithelial cell types compared to calcium phosphate (CaP) preciptates[12]. In mineral solution G-F, fluoride (F) was added to mineral solution G. It has been demonstrated that replacing OH− ions with F− ions in hydroxyapatite decreases the solubility of the mineral phase, making it more resistant to acid-induced de-mineralization[13].
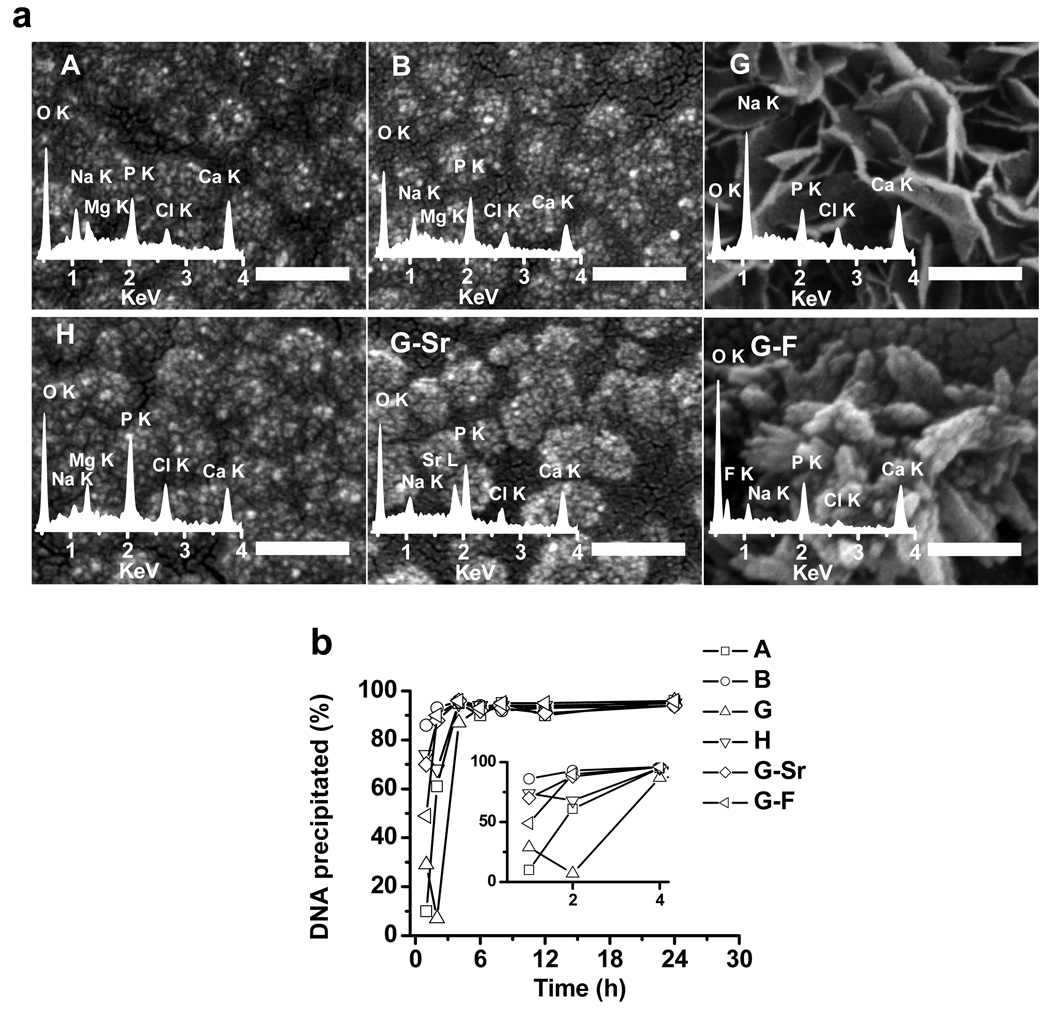
The mineralization was initiated on tissue culture-treated polystyrene surfaces. Nanocomposites were named after the mineral formulation they are derived from. Scanning electron microscopy (SEM) was used to examine the morphology of nanocomposites. As shown in Fig. 1a, nanocomposites A, B, H, and G-Sr exhibited similar morphology, in which clusters of minerals formed “grape-shaped” micro-domains with a size of 100 ~ 200 nm in diameter. In contrast, mineral solution G resulted in a thin, “plate-like” morphology and mineral solution G-F resulted in clusters of “needle-like” morphology. Energy-dispersive x-ray spectroscopy (EDX) was used to examine the composition of the nanocomposites. All of the mineral solutions resulted in structures containing calcium and phosphate (Fig. 1a, insets). For nanocomposites A, B, and H, magnesium was detected whereas it was absent in nanocomposite G, G-Sr and G-F. For nanocomposites G-Sr and G-F, strontium and fluoride, respectively, were detected indicating their incorporation into the mineral structures. Semi-quantitative analysis with EDX revealed varying calcium-to-phosphate (Ca/P) ratios for each mineral formulation (Supplemental Table 1). Among all the mineral solutions, formulation G, which lacks magenesium, resulted in the highest Ca/P ratio (1.83 ± 0.35) in nanocomposites. The incorporation of both magnesium (formulations A and B) and strontium (formulation G-Sr) resulted in a decrease of the Ca/P ratio, indicating that calcium in nanocomposites can be replaced by magnesium and strontium, respectively. Incorporation of fluoride (G-F) led to a slight decrease of the Ca/P ratio to 1.68 ± 0.34. Taken together, the composition of CaP nanocomposites, which may potentially affect their environment responsiveness[14] and thus gene transfer efficiency, can be easily tailored by adjusting the formulation of mineral solutions. This would offer great flexibility for rapid and high-throughput screening of a wide range of mineral formulations for a cell type of interest.
Figure 1. Characterization of DNA-doped nanocomposites.
a, SEM images of surface-induced nanocomposites from different mineral formulations. The insets are EDAX spectra from the corresponding nanocomposites. Scale bars are 200 nm. b, The kinetics of DNA precipitation. The inset is the kinetics of precipitation during the first 4 h of mineralization.
3.2. Efficiency of DNA incorporation into calcium carbonate nanocomposites
The efficiency of DNA incorporation into surface-induced nanocomposites by different mineral solutions was examined. Depending on the mineral composition, DNA molecules were incorporated at different rates within the first 4 h (Fig. 1b). For mineral solutions A and G, only 10% and 29% of total DNA molecules was precipitated in the first hour, respectively. With the addition of fluoride (G-F) or strontium (G-Sr), DNA molecules were precipitated much faster; 49% and 70% of total DNA were incorporated into nanocomposites, respectively. For mineral solution B, which contains the highest concentration of calcium, almost 90% of DNA molecules were precipitated in first 4 h. Despite the differences in the rate of DNA precipitation within the first 4 h, nearly 100% of the DNA was precipitated in nanocomposites at 8 h independent of mineral formulations. Thus, DNA was rapidly and efficiently incorporated into nanocomposites derived from the initial library of mineral solutions.
3.3. Biocompatibility of surface-induced DNA-doped nanocomposites
We have previously demonstrated that tissue culture surfaces coated with nanocomposites could support cell growth[9]. However, only three cell types and three different nanocomposites were examined. To establish a platform which could be used to screen and optimize the mineral compositions for any cells of interest, we examined the biocompatibility of nanocomposites derived from the current library in a variety of cell types, including fibroblast, epithelial, and neuronal cells (Table 2). These cells are derived from various tissues, including bone, breast, colon, endometrium, lung, cervix, liver, and brain of different organisms. For all the cell types we investigated, cells on the nanocomposite-coated surfaces displayed similar morphology as those on non-coated surfaces (Supplemental Figure 1). The majority of cell types covered similar surface areas on nanocomposite-coated surfaces as non-coated ones. Thus, nanocomposites did not adversely affect cell attachment and growth.
Table 2.
Cell lines used in this study
| Cell type | Organ | Organism | |
|---|---|---|---|
| MG-63 | fibroblast | bone | human |
| Saos-2 | epithelial | bone | human |
| EMT6 | epithelial | breast | mouse |
| Caco-2 | epithelial | colon | human |
| Ishikawa | epithelial | endometrium | human |
| TC1 | epithelial | lung | mouse |
| Hela | epithelial | cervix | human |
| Hep G2 | epithelial | liver | human |
| B35 | neuronal | nervous system | rat |
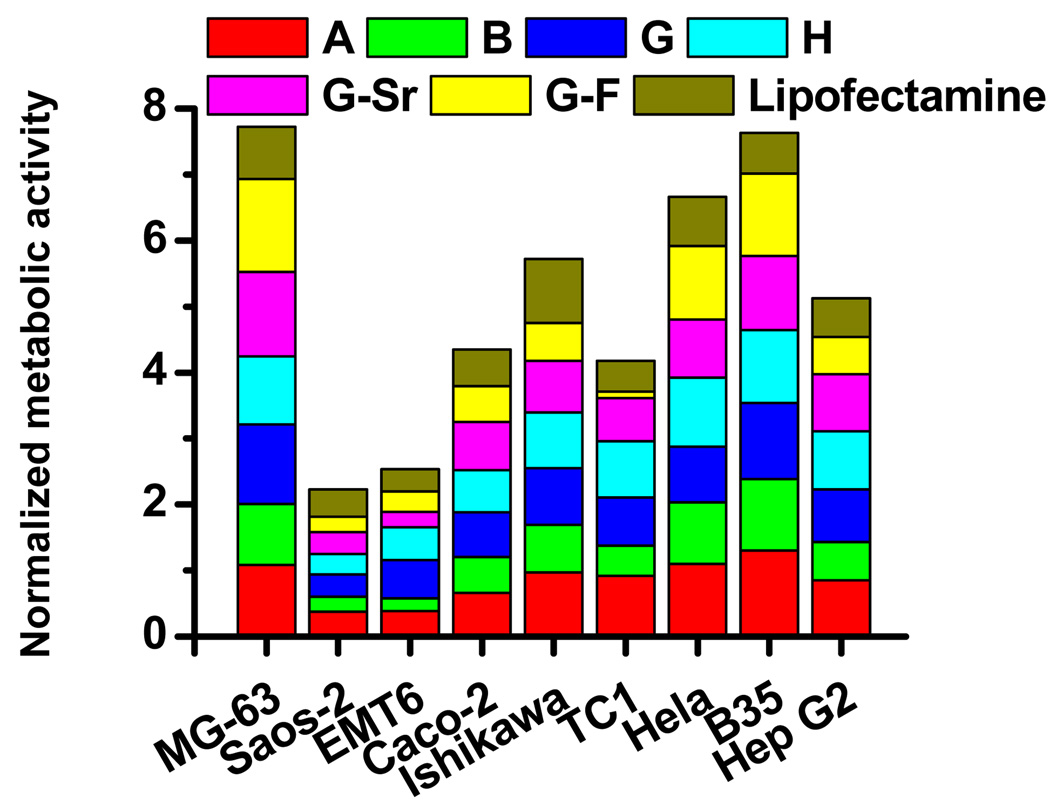
The cytotoxicity of nanocomposites was subsequently assessed by comparing the cellular metabolic activity of cells on nanocomposite-coated surfaces to that of cells on non-coated ones. DNA-doped nanocomposites did not induce significant levels of cell death and affect cell proliferation for the majority of cell types (Fig. 2). For MG-63, Caco-2, Ishikawa, TC1, Hela and Hep G2, cells proliferated normally on surfaces coated with nanocomposites derived from almost all the mineral formulations except G-F, on which there were 50% less viable cells than on non-coated surfaces for TC-1. For EMT6 cells, most mineral formulations significantly reduced the viability, except for G and H. For Saos-2, the relative metabolic activities of cells on all the six DNA-doped nanocomposites were only about 30% of that on non-coated surfaces. Previous studies have shown that the proliferation and differentiation of Saos-2, a human osteoblast-like cell line, was affected by mineral surfaces of different properties[15]. Therefore, nanocomposite-coated surfaces may induce Saos-2 differentiation and reduce proliferation, which is consistent with previous observations.
Figure 2. The relative metabolic activity of cells cultured on cell-culture surfaces coated with DNA-doped nanocomposites.
The metabolic activity of cells cultured on surfaces coated with nanocomposites formed from different mineral formulations was determined with the MTT assay after 36h and normalized to the value of cells on non-coated surfaces.
3.4. Gene transfer by DNA-doped nanocomposites in nine cell types
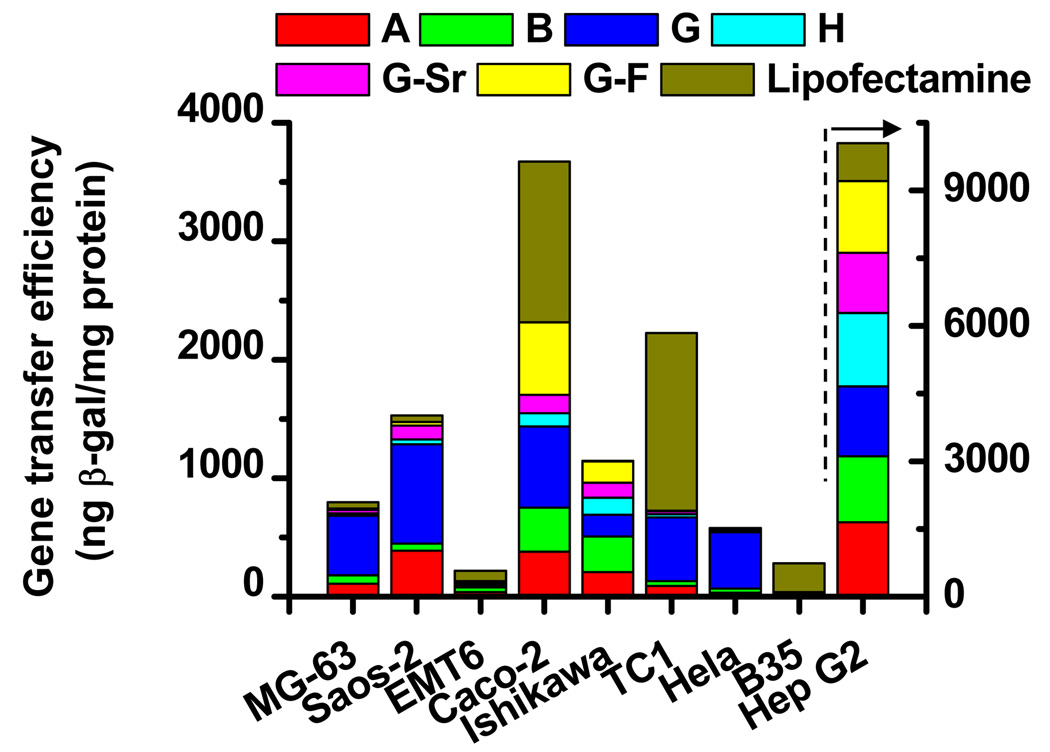
The ability of DNA-doped nanocomposites formed from the six mineral solutions to facilitate gene transfer to mammalian cells was tested on nine cell types (Fig. 3). DNA-doped nanocomposites derived from different mineral solutions yielded varying gene transfer efficiencies for all cell types. Interestingly, nanocomposite G, which lacks magnesium, resulted in the highest gene transfer efficiency in most of the cell types, with values ranging from 500 to 1500 ng β-gal/mg protein. In Ishikawa cells, nanocomposite B yielded the highest gene transfer efficiency, approximately 300 ng β-gal/mg protein. In contrast, nanocomposite H, which contains the highest level of magnesium, resulted in the lowest level of gene transfer efficiency in most cell types; nanocomposite G-Sr also yielded very low levels of gene transfer efficiency. In all the examined cell lines but EMT6 and B35, there existed an optimized composition of nanocomposites which could efficiently deliver genes to cells. The gene transfer efficiency in most cell types by the optimized mineral formulation is comparable or even higher than that of a commercial reagent, Lipofectamine 2000™.
Figure 3. The gene transfer efficiency in nine cell lines by DNA-doped nanocomposites.
Nanocomposites were mineralized incorporating the Lac Z reporter gene encoding β-galactosidase (β-gal) for 8h. The gene transfer efficiency of cells was expressed as the amount of the reporter enzyme β-gal normalized by total protein.
3.5. Correlation of gene transfer efficiency with the effectiveness of DNA uptake
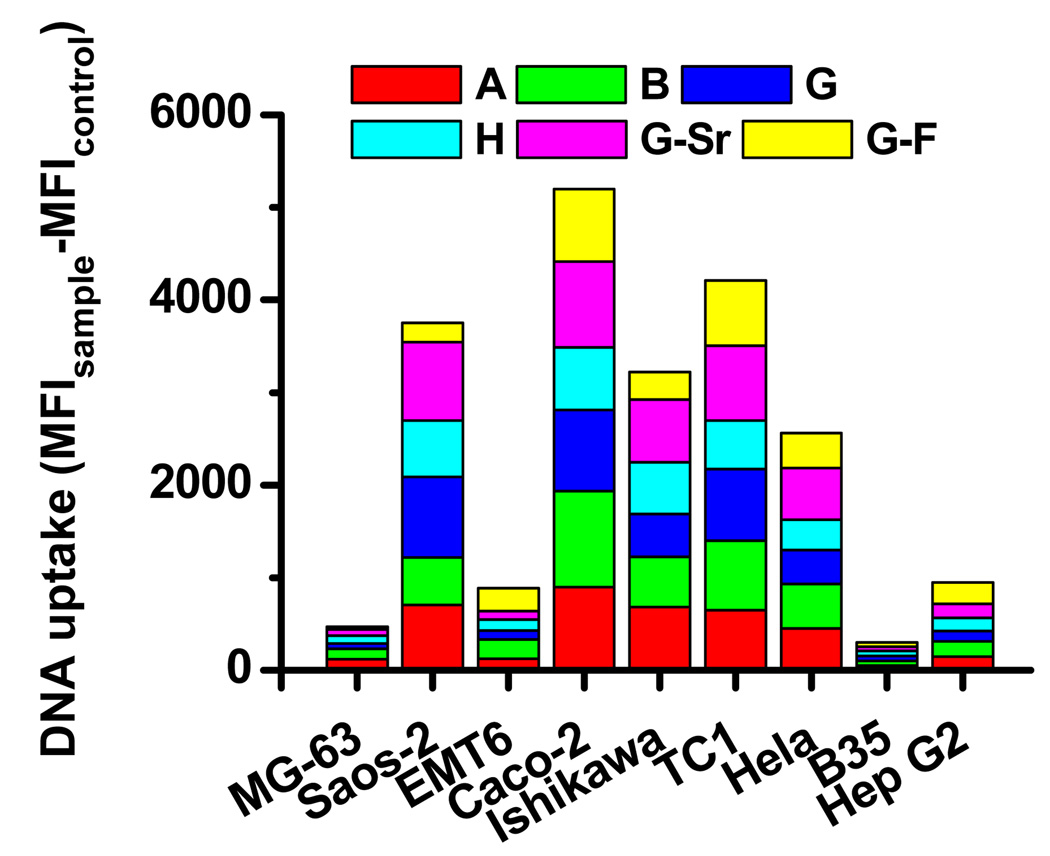
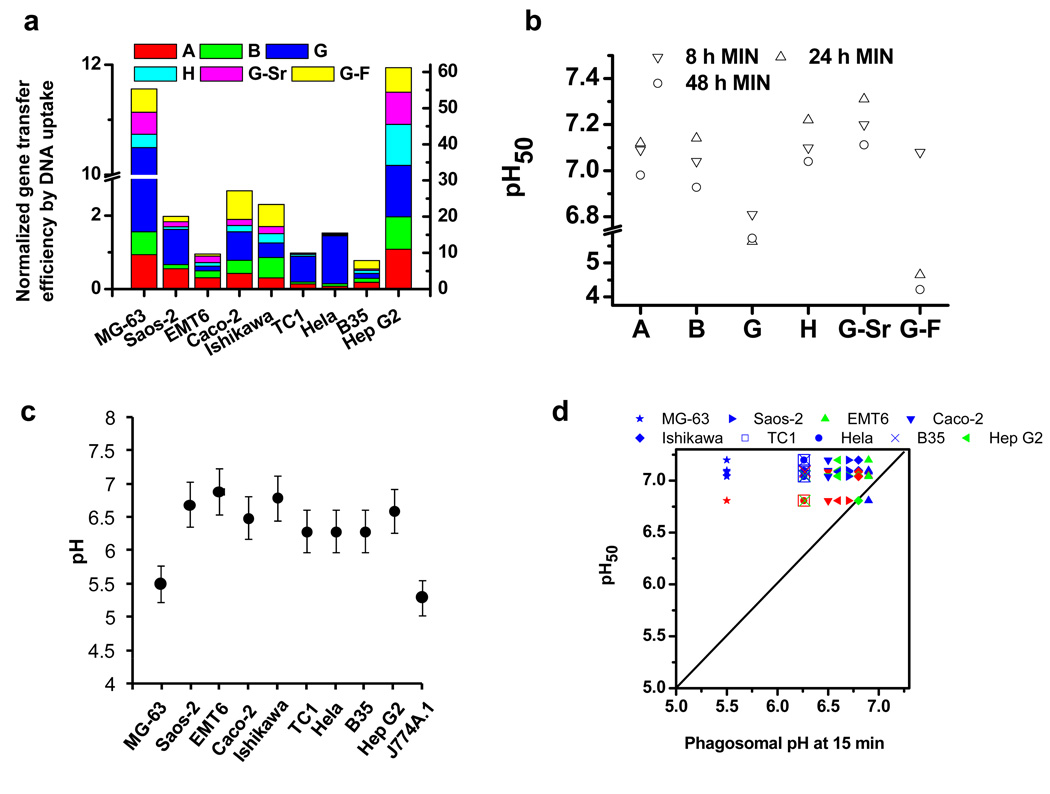
The nanocomposites formed from the initial library of mineral solutions and mineralization conditions achieved successful gene transfer in most of the cell types investigated except EMT6 and B35. Nanocomposites from different mineral solutions exhibited significant differences in the efficiency of gene transfer to a designated cell type. The cellular uptake and intracellular transport of DNA are two key steps for effective gene transfer. Initially, we examined how the gene transfer efficiency mediated by surface-induced nanocomposites correlated with the level of cellular uptake. The uptake of DNA by cells cultured on the surfaces coated with nanocomposites was determined using flow cytometry (Fig. 4). DNA was effectively delivered into Saos-2, Caco-2, Ishikawa, TC1, and Hela by nanocomposites derived from almost all the mineral formulations except those from formulation G-F. Correspondingly, high levels of gene transfer efficiency were observed in those cells, particularly when nanocomposites from formulation G were used. DNA was poorly transported into MG-63, EMT6, B35 and Hep G2 by nanocomposites formed from the current library of mineral formulations. For EMT6 and B35, the low gene transfer efficiency correlated with the low level of DNA uptake. However, we cannot exclude the possibility that the intracellular transport of DNA in both cell types may also be limited. Surprisingly, for MG-63 and Hep G2, though a much lower level of DNA was delivered into the cells compared to other cell types, the gene transfer efficiency was high, particularly in nanocomposite G.
Figure 4. The cellular uptake of DNA mediated by surface-induced nanocomposites.
Nanocomposites were mineralized with fluorescein-labeled DNA for 8h. The degree of DNA uptake, quantified as the mean fluorescence intensity (MFI) by flow cytometry, was expressed as the MFI of cells cultured on surfaces coated with DNA-doped nanocomposites subtracted that of cells on surfaces in the absence of DNA.
3.6. Correlation of gene transfer efficiency with the deviation of pH responsiveness of nanocomposites from early phagosomal pH of cells
The efficiency of gene transfer in general correlates with the intracellular level of DNA across cell types. However, for each individual cell type, there was not a direct correlation between the level of intracellular DNA and the gene transfer efficiency of nanocomposites derived from different mineral formulations. After normalizing the gene transfer efficiency based on the amount of intracellular DNA to exclude the effect of uptake of DNA (Fig. 5a), in a designated cell type, nanocomposites derived from different formulations exhibited varying gene transfer efficiency per endocytosed DNA; for most of cell types, nanocomposite G demonstrated the highest gene transfer efficiency. The results so far have raised two questions: first, why does nanocomposite G result in the higher efficiency of gene transfer by DNA being endocytosed compared to nanocomposites derived from other formulations? Second, why do most of the cells exhibit the highest gene transfer efficiency when nanocomposite G was used? Clearly, the efficiency of gene transfer by endocytosed DNA correlates with both properties of the nanocomposites and that of cells. The intracellular fate of DNA is closely associated with the phagosomal pH and pH responsiveness of delivery systems[4] [16]. Therefore, we set out to determine the correlation between the efficiency of gene transfer by endocytosed DNA, phagosomal pH of cells and pH responsiveness of nanocomposites.
Figure 5. Correlation of gene transfer efficiency with DNA uptake and pH50 of nanocomposites.
a, The gene transfer efficiency normalized by the cellular uptake of DNA by cells. b, The pH responsiveness, pH50, of nanocomposites. pH50 is the pH at which 50% of calcium ions were released (MIN: mineralization). c, The early phagosomal pH of cells at 15 minutes post-phagocytosis of 100 nm beads. d, Correlation of the normalized gene transfer efficiency with the phagosomal pH of cells and the pH50 values of nanocomposites. Red: the relative normalized gene transfer efficiency ≥ 0.9; green: 0.9> the relative normalized gene transfer efficiency ≥ 0.5; blue: the relative normalized gene transfer efficiency < 0.5. The relative normalized gene transfer efficiency is the normalized gene transfer efficiency divided by highest normalized gene transfer efficiency of that cell type among different mineral formulations. The dashed line stands for equal pH. Values shown are the mean of the triplicates with a standard deviation less than 5% of the mean.
We first examined the pH responsiveness of DNA-doped nanocomposites. The pH at which 50% of the total calcium was released from nanocomposites, denoted as pH50, was used as an indicator of pH responsiveness. As illustrated in Fig. 5b, nanocomposite A, B, H, and G-Sr, had pH50 values ranging from 7.0–7.3 for the duration of mineralization examined. In contrast, nanocomposite G and G-F were more resistant to acidification, particularly as the mineralization duration was prolonged. For nanocomposite G, as the duration of mineralization increased from 8 h to 48 h, the pH50 decreased from 6.8 to 5.6, respectively; for formulation G-F, the pH50 decreased from 7.1 to 4.7 and finally to 4.2 as the mineralization time increased from 8h, 24h, to 48h, respectively. Nanocomposites derived from the initial library of mineral solutions demonstrate appreciably different pH responsiveness.
We subsequently measured the phagosomal pH of cells at 15 min post exposure to 100 nm-polystyrene beads (Fig. 5c) by a flow cytometry-based method[17]. Different cell types exhibited significantly different early phagosomal pH. Although the phagosomal pH of the cell types we examined has not been extensively characterized, the value we obtained for the J774A.1 macrophage cell line was consistent with those reported in literature[18].
We then correlated the normalized gene delivery efficiency with the deviation of pH50 of nanocomposites from the phagosomal pH of cells (Fig. 5d). For eight out nine cell types, nanocomposites with a pH50 that displayed the slightest deviation from the early phagosomal pH of cells yielded the highest gene transfer efficiency per endocytosed DNA. As the pH50 of nanocomposites deviated from the phagosomal pH of cells, the gene transfer efficiency correspondingly de creased. This may indicate that both the pH sensitivity of nanocomposites and phagosomal pH of cells are critical parameters that determine the optimal release of DNA from nanocomposites to mediate gene delivery to the cell nucleus. For nanocomposites that possess a pH50 that is above the phagosomal pH of cells, the DNA may not be completely released as it is trafficked in the phagosomes, thus leading to inefficient gene transfer. Nanocomposites with a pH50 that match the phagosomal pH of cells would enable the optimal release of DNA. Further studies are required to provide detailed intracellular mechanisms in which DNA nanocomposites facilitate the efficient transport of DNA to the cell nucleus.
Our results so far suggest that the gene transfer efficiency medicated by DNA-doped nanocomposites strongly correlates with the cellular uptake of DNA and the deviation of pH50 of nanocomposites from phagosomal pH of individual cell types. This correlation will enable us to customize the gene delivery system with respect to each individual cell type.
3.7. Optimization of gene transfer in B35 and EMT6 cells
Based on our preliminary analysis on how cellular uptake and deviation of pH50 of nanocomposites from phagosomal pH affected gene transfer efficiency, we attempted to improve gene transfer in both B35 and EMT6 cells. The cellular uptake in both cells was very low compared to other cell types; therefore, we initially focused on improving cellular uptake. It has been shown that the cellular uptake can be facilitated by concentrating high levels of DNA on cell substrates[9, 19]. Therefore, we aimed to control the spatial distribution of DNA to achieve higher local concentrations of DNA molecules per cell.
DOTAP, a cationic lipid able to condense DNA[20], has been widely used as a non-viral gene delivery vector. We first complexed DNA with DOTAP at two different ratios, followed by subsequent mineralization to form DNA/DOTAP nanocomposites. Fluorescent microscopy images of fluorescent DNA in modified nanocomposites revealed large, punctuate structures, suggesting a high local concentration of DNA (Supplemental Fig. 2); a higher DNA: DOTAP ratio (Supplemental Fig. 2c) resulted in more localized distributions compared to a lower ratio (Supplemental Fig. 2b). In contrast, DNA in un-modified nanocomposites showed a more diffuse distribution of DNA, suggesting lower local concentration of DNA (Supplemental Fig. 2a). The inclusion of DOTAP slightly affected the pH responsiveness of nanocomposites, but not the DNA precipitation efficiency (Supplemental Fig. 3).
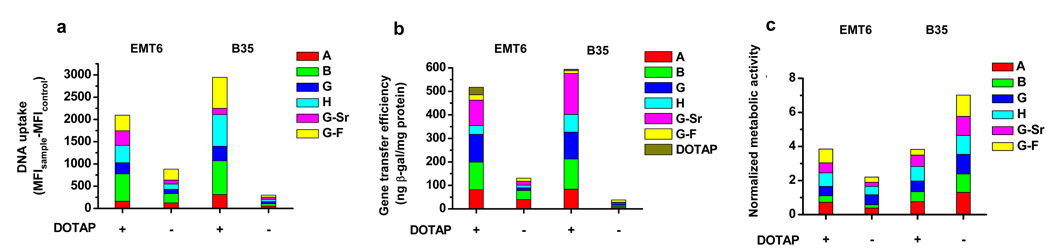
DOTAP-nanocomposites greatly enhanced the uptake of DNA compared to un-modified ones for both EMT-6 and B35 cell lines, respectively (Fig. 6a). Consequently, the gene transfer efficiency was markedly enhanced, with the greatest improvement shown by formulations B, G, and G-Sr (Fig. 6b). The gene transfer efficiency directly correlated with the enhanced uptake of DNA for all mineral formulations. Modified nanocomposites enhanced the viability of EMT6 cells, yet slightly reduced the viability of B35 cells compared to un-modified nanocomposites (Fig. 6c). Currently, we are carrying out more detailed experiments to examine the mechanisms by which DOTAP enhances the cellular uptake of DNA incorporated into nanocomposites.
Figure 6. The gene transfer in EMT6 and B35 cells on DOTAP/DNA nanocomposite-coated substrates.
a, Cellular uptake of DNA. Fluorescein-labeled DNA was complexed with DOTAP at a DNA:DOTAP ratio (w/w) of 1:6 for EMT6 and 1:12 for B35 cells, respectively. DNA uptake was quantified by flow cytometry and expressed as the MFI of cells cultured on nanocomposites subtracted the MFI of control cells without DNA. The uptake for cells on un-modified nanocomposites is shown for comparison. b, The gene transfer efficiencies. The LacZ reporter gene was complexed with DOTAP at a DNA:DOTAP ratio (w/w) of 1:6 for EMT6 and 1:12 for B35 cells, respectively. The formed complex was mineralized with different mineral formulations for 8h and the gene transfer efficiency was evaluated. c, The metabolic activity of cells. Values shown are the mean of the triplicates with a standard deviation less than 5% of the mean.
4. Conclusions
The survey of gene transfer in nine cell types suggest that surface-induced nanocomposites provide a flexible platform to examine the efficiency of nanocomposites derived from a wide variety of mineral formulations in various cell types in a high throughput fashion. The initial success in correlating the gene transfer efficiency with cellular uptake, pH responsiveness of surface-induced nanocomposites and phagosomal pH of cells suggest that this platform potentially allows us develop a design principle, which can be applied to optimize and design gene delivery systems for cell type of interest in silica. Further studies are under way to translate this platform to different substrates for the applications in both systematic and surface-mediated gene delivery.
Supplementary Material
Acknowledgement
This work was supported by the grant R21EB007494-01 from the National Institutes of Health and by the National Science Foundation CAREER Award awarded to H. Shen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tripathy SK, Black HB, Goldwasser E, Leiden JM. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2(5):545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 2.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11:S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 3.Wickham TJ. Targeting adenovirus. Gene Ther. 2000;7(2):110–114. doi: 10.1038/sj.gt.3301115. [DOI] [PubMed] [Google Scholar]
- 4.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18(1):33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 5.Niidome T, Huang L. Gene therapy progress and prospects: Nonviral vectors. Gene Ther. 2002;9(24):1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 6.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem-Int Edit. 2003;42(27):3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 7.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyane A, Tsurushima H, Ito A. Novel gene-transferring scaffolds having a cell adhesion molecule-DNA-apatite nanocomposite surface. Gene Ther. 2007;14(24):1750–1753. doi: 10.1038/sj.gt.3303041. [DOI] [PubMed] [Google Scholar]
- 9.Shen H, Tan J, Saltzman WM. Surface-mediated gene transfer from nanocomposites of controlled texture. Nat Mater. 2004;3(8):569–574. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- 10.Tan J, Saltzman WM. Biomaterials with hierarchically defined micro- and nanoscale structure. Biomaterials. 2004;25(17):3593–3601. doi: 10.1016/j.biomaterials.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 11.Morin LG. Direct Colorimetric Determination of Serum-Calcium with Ortho Cresolphthalein Complexon. Am J Clin Pathol. 1974;61(1):114–117. doi: 10.1093/ajcp/61.1.114. [DOI] [PubMed] [Google Scholar]
- 12.Brash DE, Reddel RR, Quanrud M, Yang K, Farrell MP, Harris CC. Strontium Phosphate Transfection of Human-Cells in Primary Culture - Stable Expression of the Simian-Virus 40 Large-T-Antigen Gene in Primary Human Bronchial Epithelial-Cells. Mol Cell Biol. 1987;7(5):2031–2034. doi: 10.1128/mcb.7.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driessen Fc. Relation between Apatite Solubility and Anti-Cariogenic Effect of Fluoride. Nature. 1973;243(5407):420–421. doi: 10.1038/243420a0. [DOI] [PubMed] [Google Scholar]
- 14.Nancollas GH, Wu WJ. Biomineralization mechanisms: a kinetics and interfacial energy approach. J Cryst Growth. 2000;211(1–4):137–142. [Google Scholar]
- 15.Mayr-Wohlfart U, Fiedler J, Gunther KP, Puhl W, Kessler S. Proliferation and differentiation rates of a human osteoblast-like cell line (Saos-2) in contact with different bone substitute materials. J Biomed Mater Res. 2001;57(1):132–139. doi: 10.1002/1097-4636(200110)57:1<132::aid-jbm1152>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 16.Jeong B, Gutowska A. Lessons from nature: stimuli-responsive polymers and their biomedical applications. Trends Biotechnol. 2002;20(7):305–311. doi: 10.1016/s0167-7799(02)01962-5. [DOI] [PubMed] [Google Scholar]
- 17.Tran KK, Shen H. The role of phagosomal pH on the size-dependent efficiency of cross-presentation by dendritic cells. Biomaterials. 2009;30(7):1356–1362. doi: 10.1016/j.biomaterials.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haggie PM, Verkman AS. Cystic fibrosis transmembrane conductance regulator-independent phagosomal acidification in macrophages. J Biol Chem. 2007;282(43):31422–31428. doi: 10.1074/jbc.M705296200. [DOI] [PubMed] [Google Scholar]
- 19.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjugate Chem. 2002;13(3):621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 20.Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN. Improved DNA: Liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol. 1997;15(7):647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.