Summary
In mammals, fat store levels are communicated by leptin and insulin signaling to brain centers that regulate food intake and metabolism. By using transgenic manipulation of neural activity, we report the isolation of two distinct neuronal populations in flies that perform a similar function, the c673a-Gal4 and fruitless-Gal4 neurons. When either of these neuronal groups is silenced, fat store levels increase. This change is mediated through an increase in food intake and altered metabolism in c673a-Gal4 silenced flies, while silencing fruitless-Gal4 neurons alters only metabolism. Hyperactivation of either neuronal group causes depletion of fat stores by increasing metabolic rate and decreasing fatty acid synthesis. Altering the activities of these neurons causes changes in expression of genes known to regulate fat utilization. Our results show that the fly brain measures fat store levels and can induce changes in food intake and metabolism to maintain them within normal limits.
Introduction
The relationships between caloric intake, energy expenditure, and fat deposition have been intensively studied in mammalian systems. Lipid intake, synthesis, catabolism, and storage as fat are regulated in response to the body's energy demands. This regulation involves the intestine (where dietary lipids are digested and absorbed), adipose tissue (where excess fat is stored), the liver (where de novo fatty acid synthesis occurs), and the brain (where the general metabolic state of the body is monitored and controlled). Control of caloric consumption by the brain involves hypothalamic nuclei that regulate eating and metabolism. Surgical ablation or overactivation of these nuclei alters feeding behavior and weight regulation.
The regulation of fat storage has also been studied in invertebrate model systems such as the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster. Unlike insects and vertebrates, nematodes do not have dedicated adipocytes. They store fat in droplets, and these can be visualized in live animals by staining with vital dyes such as Nile Red. These assays have been used as the basis for genetic screens that have identified many fat-regulatory genes. Some of these are conserved between worms and mammals (for review see Jones and Ashrafi, 2009). The genetic studies have also implicated serotonin and transforming growth factor β (TGF-β) as signals through which neurons might control fat storage (Greer et al. 2008, Srinivasan et. al. 2008)
In Drosophila, as in mammals, fat storage by adipocytes is regulated by interplay between the gut, fat body, and a group of cells with similar functions to the mammalian liver (oenocytes; Gutierrez et al., 2007). The coordination between these organs utilizes some molecular components whose sequences and functions are conserved between insects and mammals. These include the perilipin/Lsd-2 protein, which is required for the formation of lipid storage droplets (Miura, et al., 2002), the Brummer (Bmm) lipase, and the cytochrome P450 Cyp4g1 (Gutierrez et al., 2007, Gronke, et. al., 2007).
A major gap in our understanding of the control of fat storage and metabolism in the Drosophila system is that the brain neurons that control these processes, which might be functionally analogous to the mammalian hypothalamic feeding centers, have not been identified. To begin to fill this gap, we developed a rapid thin layer chromatography (TLC) assay to measure fat content, and used it to examine flies in which specific groups of neurons have been silenced or hyperactivated. This work has led to the identification of two sets of brain neurons that regulate Drosophila fat content, the c673a-Gal4 and fruitless-Gal4 (Fru-Gal4) neurons. Both produce increased fat content (obesity) when their synaptic activity is silenced and extreme leanness when they are hyperactivated. We have also identified behavioral and metabolic defects associated with altered fat levels in flies with abnormal c673a-Gal4 and Fru-Gal4 neural activities.
Results
c673a-Gal4 and Fru-Gal4 neurons are involved in regulation of fat storage
We developed a TLC assay specific for fat detection (see Experimental Procedures). Fly extracts produce a fat band halfway down the TLC plate. Three observations confirm that this band is indeed fat. First, the band has the same migration rate as butter, lard, or a a triglyceride standard (Supplementary Figure 1A). Second, it gradually disappears with continued starvation, which is expected of stored fat (Supplementary Figure 1B). Third, analysis by mass spectrometry confirms that this band is composed of a mixture of triglycerides (Supplementary Figure 1C).
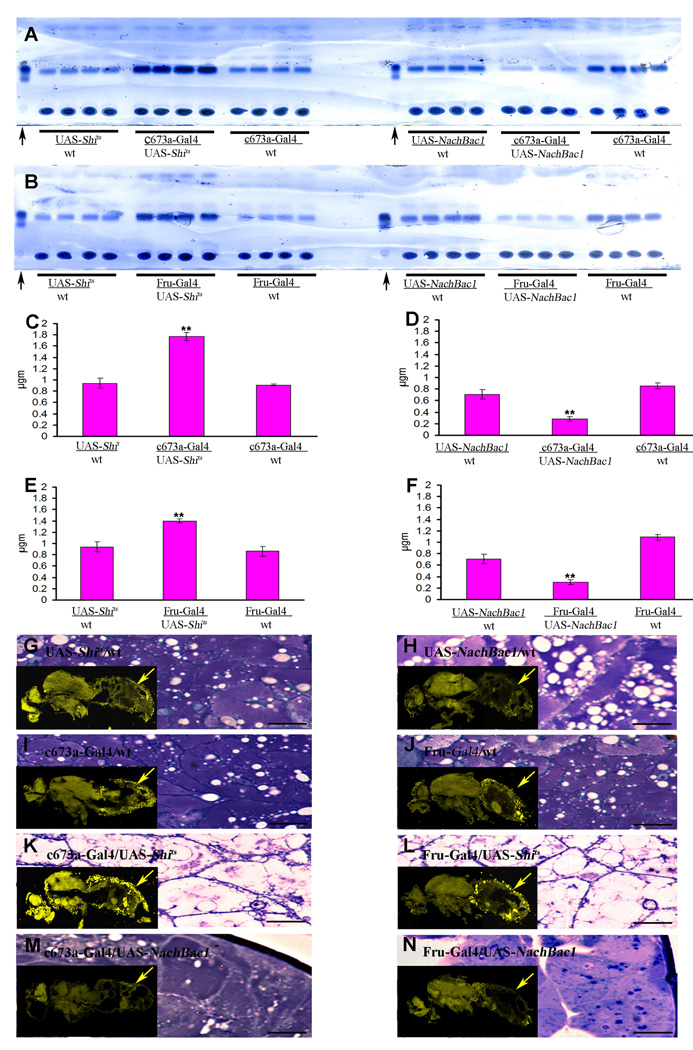
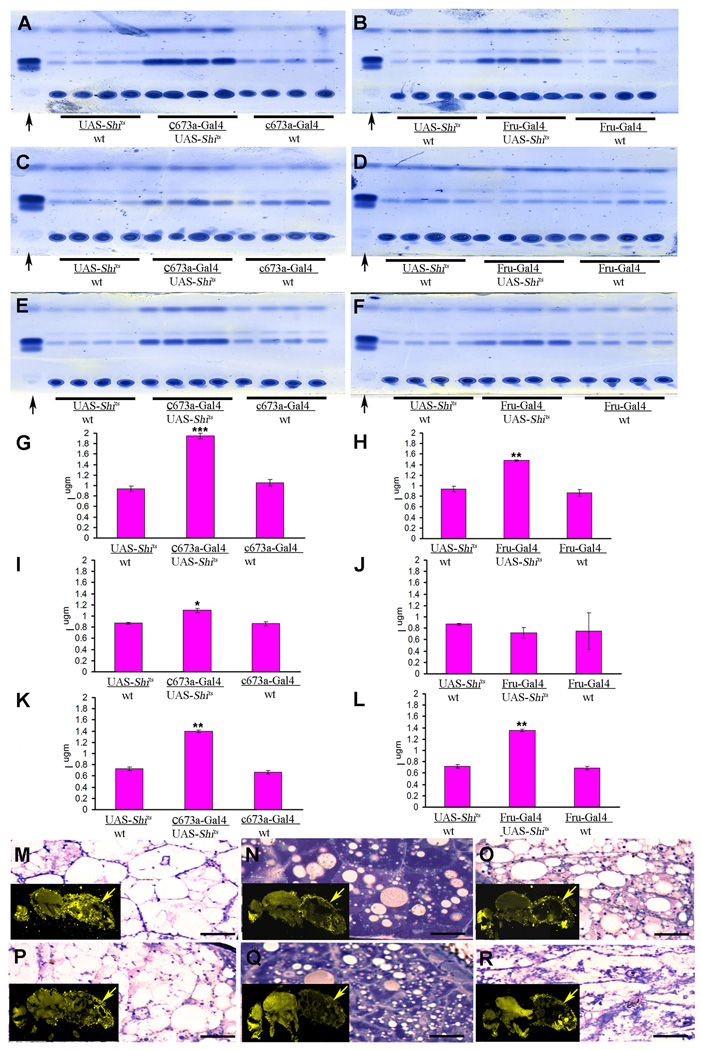
To identify Drosophila neurons that regulate fat storage, we capitalized on a collection of 350 P[Gal4] enhancer-trap lines that express the yeast transcription factor Gal4 in different subsets of neurons (Brand and Perrimon, 1993, Armstrong and Kaiser, 1997). The Gal4 was used to drive the expression of proteins that either silence or hyperactivate neurons. Neuronal silencing was achieved by driving the expression of a semi-dominant transgene containing a UAS-Temperature Sensitive Shibire allele (UAS-Shits) and incubating the resulting flies at 30 °C (Kitamoto et al., 2001), while neuronal hyperactivation was achieved by expression of a UAS- Voltage Activated Bacterial Sodium Channel transgene (UAS-NaChBac1; Navarro et al. 2001, Luan, et al., 2006). We reasoned that if a given Gal4-line is expressed in neurons that are involved in fat store regulation, hyperactivation and silencing of these neurons should produce opposite effects on fat stores. We screened the entire enhancer trap collection, and found only two lines that fulfilled this criterion: c673a-Gal4 (Armstrong and Kaiser, 1997) and fruitless-Gal4 (Fru-Gal4; Stockinger et al., 2005). Both produced major increases in the fat band when crossed to the silencing gene, and decreases in the same band when crossed to the hyperactivating channel gene (Figure 1A and B).
Figure 1.
Alteration in neural activity of the c673a-Gal4 or Fru-Gal4 neurons produces changes in fly fat content. TLC assay showing two panels with four replicates for each of three genotypes (A and B). The flanking genotypes in each panel are from control flies with single transgenes, while the central panels are from flies with both transgenes. When UAS-Shits reduce synaptic transmission in the c673a-Gal4 or Fru-Gal4 neurons, an increase in fat band is observed (central right in panel A and B respectively). Over-activation of either set of neurons with UAS-NachBac1 produces a decrease in fat band (central left in panel A and B respectively). Arrows in both panels point to butter standard lanes. The same results are observed when triglyceride levels are directly measured (C–F). Flies bearing single transgenes show some yellow fat fluorescence as examined by Nile red histological staining (G–J). The experimental double transgene flies under UAS-Shits suppression show a large amount of fat as indicated by an increase in yellow fluorescence of Nile red histological staining (K and L, arrow), while over-activation of the same neurons causes a reduction in yellow fluorescence of fat stores (M and N, arrow). The same panels also show the fat cell morphology of those flies as examined by toluene blue stained plastic section (G–J). Error bars are standard deviation of five different replicas for a given genotype. Black bar in panel G to J represent distant of 100 µm. (Asterisks denote T-test statistical significance: *; P<0.05, **; P<0.01, ***; P<0.005).
We further confirmed these results by directly measuring triglyceride levels (Figure 1C–F) and by histological staining for fat cells using Nile Red staining of cryostat sections and toluidine blue staining of plastic sections (Figure 1G–N). In control adult Drosophila, abdominal fat cells tend to have a large cytoplasm with many scattered fat droplets that surround a centrally located nucleus. Flies with silenced c673a-Gal4 or Fru-Gal4 neurons had fat cells with large fat droplets that are surrounded by thin rings of cytoplasmic material, while flies with hyperactivated c673a-Gal4 or Fru-Gal4 neurons had fat cells that either have no fat droplets or only few small ones (Figure 1C–N).
Reproductive demands on females can make it advantageous for them to accumulate more fat stores than males (Lorenz, 2003, Lorenz and Anand, 2004, ). fruitless (fru) mutations produce males with defective courtship and female-like aggressive displays, and the same behavioral defects are observed in males that express only the female-specific FRUITLESS protein isoform, or have silenced Fru-Gal4 neurons (Manoli et al., 2005; Demir et al., 2005; Stockinger et al., 2005; Villella et al., 2005). These observations would be consistent with the hypothesis that Fru-Gal4-silenced males are obese because they accumulate fat stores like females. To test this idea, we used the TLC assay to examine the fat content of males homozygous for different fru mutant alleles, or with feminized brains due to splicing of their fru transcripts in a female-specific manner. No differences in fat stores were observed in these flies as compared to wild-type flies (Supplementary Figure 2).
The c673a-Gal4 P-element is inserted 1 kbp upstream of a gene called lightoid. We have examined if mutations in this gene cause any effect on fat storage, and have found that lightoid mutants have normal fat store levels (Supplementary Figure 2C).
c673a-Gal4 and Fru-Gal4 define separate neuronal populations
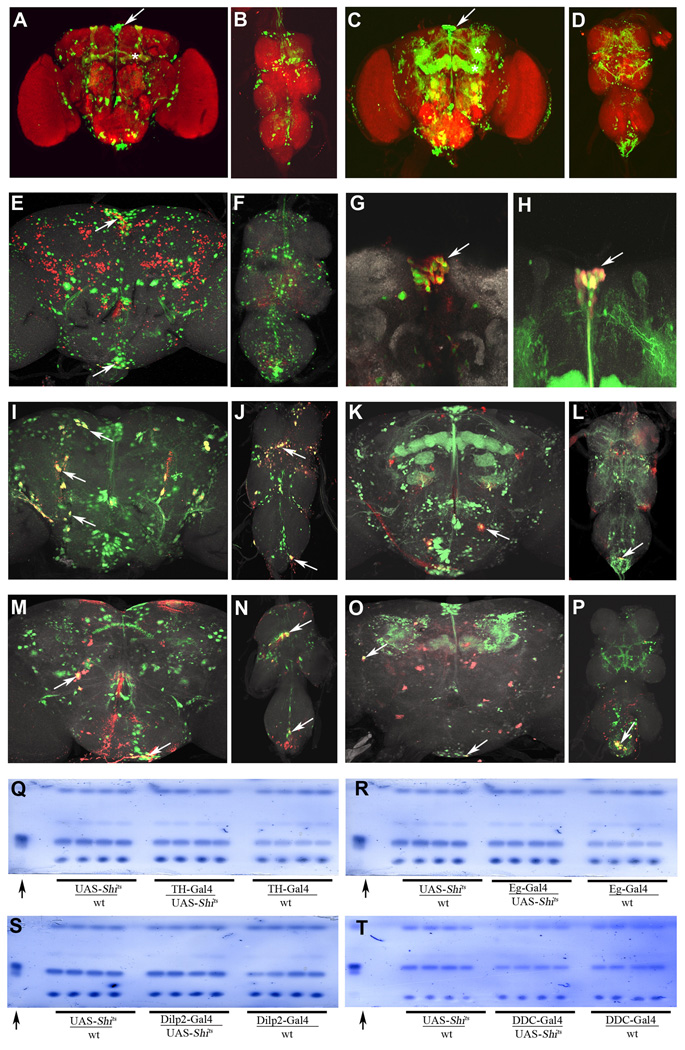
To identify the locations of c673a-Gal4 and Fru-Gal4 expressing neurons in the brain, we generated flies carrying these drivers together with two copies of a UAS-Green Fluorescent Protein transgene (2xUAS-GFP). c673a-Gal4/2xUAS-GFP neurons are scattered throughout the brain and ventral ganglion (Figures 2A and B), with a GFP-positive cluster in the neurosecretory area where Drosophila insulin-like peptide (Dilp)-expressing neurons are located (Figures 2A, arrow). There are c673a-Gal4/2xUAS-GFP clusters in the subesophageal ganglion and the ventral ganglion (B). We also observed some expression in the mushroom body (Figure 2A, asterisk). Fru-Gal4/2xUAS-GFP neurons have a much broader distribution throughout the brain, including the neurosecretory cluster (Figure 2C, arrow), the mushroom body (Figure 2C, asterisk), the subesophageal ganglion, and the ventral ganglion (Figure 2D).
Figure 2.
The c673a-Gal4 neurons overlap only minimally with the Fru-Gal4 neurons. c673a-Gal4/2xUAS-GFP brain (A) showing the neurosecretory cluster (arrow), mushroom body (asterisks), and the ventral ganglion (B). Fru-Gal4/2xUAS-GFP brain (C) showing the neurosecretory cluster (arrow), mushroom body (asterisks), and the ventral ganglion (D). Anti-FruM (red in E and F) rarely co-stained the green c673a-Gal4/UAS-nuc-GFP neurons (green in E and F), with overlap occurring in only few yellow neurons in the neurosecretory cluster and subesophageal ganglion (E and F, arrows). Anti-DILP immunostaining (red in G and H) of c673a-Gal4/2xUAS-GFP neurons (green in G) and of Fru-Gal4/2xUAS-GFP neurons (green in H) indicates that most of the insulin expressing (DILP) neurons are both c673a-Gal4 and Fru-Gal4 positive (yellow in G and H, arrows). Immunostaining with the dopaminergic marker anti-tyrosine hydroxylase (red in I–L) show that most are also c673a-Gal4/2xUAS-GFP neurons (yellow in I and J, arrows), however, only few are Fru-Gal4/2x-UAS-GFP positive neurons (yellow in K and L, arrows). Anti-serotonin immunostaining (red in M–P) shows little overlap between serotonergic neurons and c673a-Gal4/2xUAS-GFP (green in M and N) and Fru-Gal4/2xUAS-GFP neurons (green in O and P). However, few c673a-Gal4 and Fru-Gal4 neurons in the subesophageal and ventral ganglion are serotonergic (yellow in M–P, arrows). Silencing of the few neurons shared between c673a-Gal4 and Fru-Gal4 does not affect fat store level. UAS-Shits silencing of dopaminergic neurons using TH-Gal4 (Q), serotonergic neurons using Eg-Gal4 (R), insulin expressing neurons by using Dilp2-Gal4 (S), or both dopaminergic and serotonergic neurons using DDC-Gal4 (T) does not produce changes in fly fat content compared to control flies with single transgenes as examined by the TLC assay. Arrows in TLC panels point to butter standard lanes.
Changes in fat content upon altered neural activity of either c673a-Gal4 or Fru-Gal4 neurons could be due to an overlap between those two populations in a subset of neurons that are responsible for regulation of fly fat storage. To test this idea, we generated males containing c673a-Gal4 with UAS-nuclear Green Fluorescent Protein transgenes (UAS-nuc-GFP), and immunostained the resulting brains with antibodies against the FRUITLESS male-specific isoform (anti-FruM), which stains most, but not all, of the Fru-Gal4 neurons(Lee et al., 2000, Billeter et al., 2004, Stockinger et al. 2005). Only two subesophageal neurons and four to eight neurons in the neurosecretory cluster were c673a-Gal4/UAS-nuc-GFP and anti-FruM positive (Figure 2E, arrow and F),indicating little overlap between the two sets of neuron.
Due to the known roles of insulin, dopamine, and serotonin in mammalian obesity (for review see Ramos, et. al. 2005), we examined whether any of the neurons within the c673a-Gal4 and Fru-Gal4 populations belong to any of the above stated neuronal classes. To address this question, we stained c673a-Gal4/2xUAS-GFP and Fru-Gal4/2XUAS-GFP with antibodies such as anti-DILP that labels insulin expressing neurons (Cao et al., 2001), anti-tyrosine hydroxylase that marks dopaminergic neurons (Hamasaka and Nassel, 2006), and anti-serotonin that labels serotonergic neurons (Hamasaka and Nassel, 2006). The DILP-positive neurons are also c673a-Gal4 and Fru-Gal4 positive (Figure 2G and H, arrows), and can account for the overlap between the two populations within the neurosecretory cluster. While all dopaminergic neurons are c673-Gal4 positive (Figure 2I and J, arrow), only a few are Fru-Gal4 positive (Figure 2K and L, arrow). We also saw little overlap between brain serotonergic neurons and either c673-Gal4 or Fru-Gal4 neurons (Figure 2M–P, arrows). However, as reported previously, we observed that a large number of Fru-Gal4 neurons in the metathoracic neuromere of the ventral ganglion are serotonergic (Figure 2O, arrow; Billeter et al., 2004).
The fact that the two neuronal groups both contain insulin-expressing neurons, as well as some dopaminergic and serotonergic neurons, could indicate that some or all of these shared neurons are responsible for the obesity phenotype seen when either c673a-Gal4 or Fru-Gal4 circuits are silenced. If so, one might be able to produce obesity in flies by simply silencing those shared neurons. Although we do not have an enhancer line specifically expressed in c673a-Gal4/Fru-Gal4 shared neurons, we do have Gal4 lines that are only expressed in DILP-positive (Dilp2-Gal4; Ikeya et al. 2002), dopaminergic (TH-Gal4; Friggi-Grelin et al., 2003), serotonergic (Eg-Gal4; Lundell et al., 1998), and both dopaminergic and serotonergic neurons (DDC-Gal4; Li et al. 2000). When any of these neuronal sets were silenced, no change in fat content was observed (Figure 2Q–T), thus making it unlikely that the few c673a-Gal4/Fru-Gal4 shared neurons are the primary regulators of fat stores.
We also silenced both c673a-Gal4 and Fru-Gal4 neurons simultaneously, and found that the resulting flies were lethargic and died within five days after silencing. The lethargy and lethality make any conclusions about their fat store levels of questionable value.
c673a-Gal4 and Fru-Gal4 neurons affect fat storage via different behavioral, metabolic, and gene expression changes
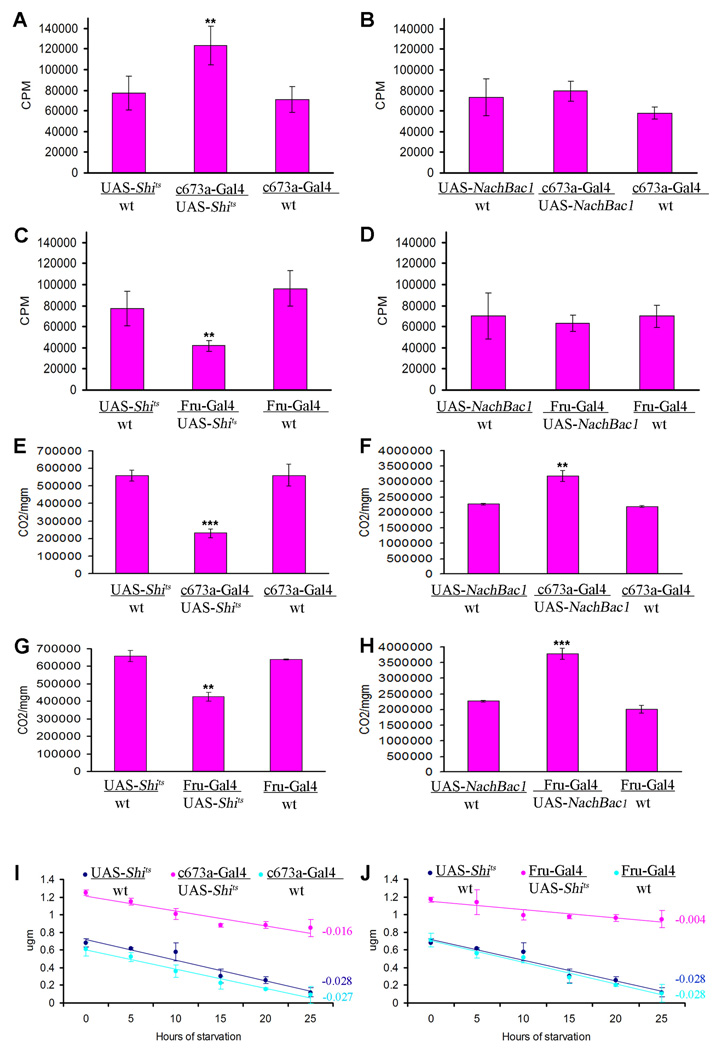
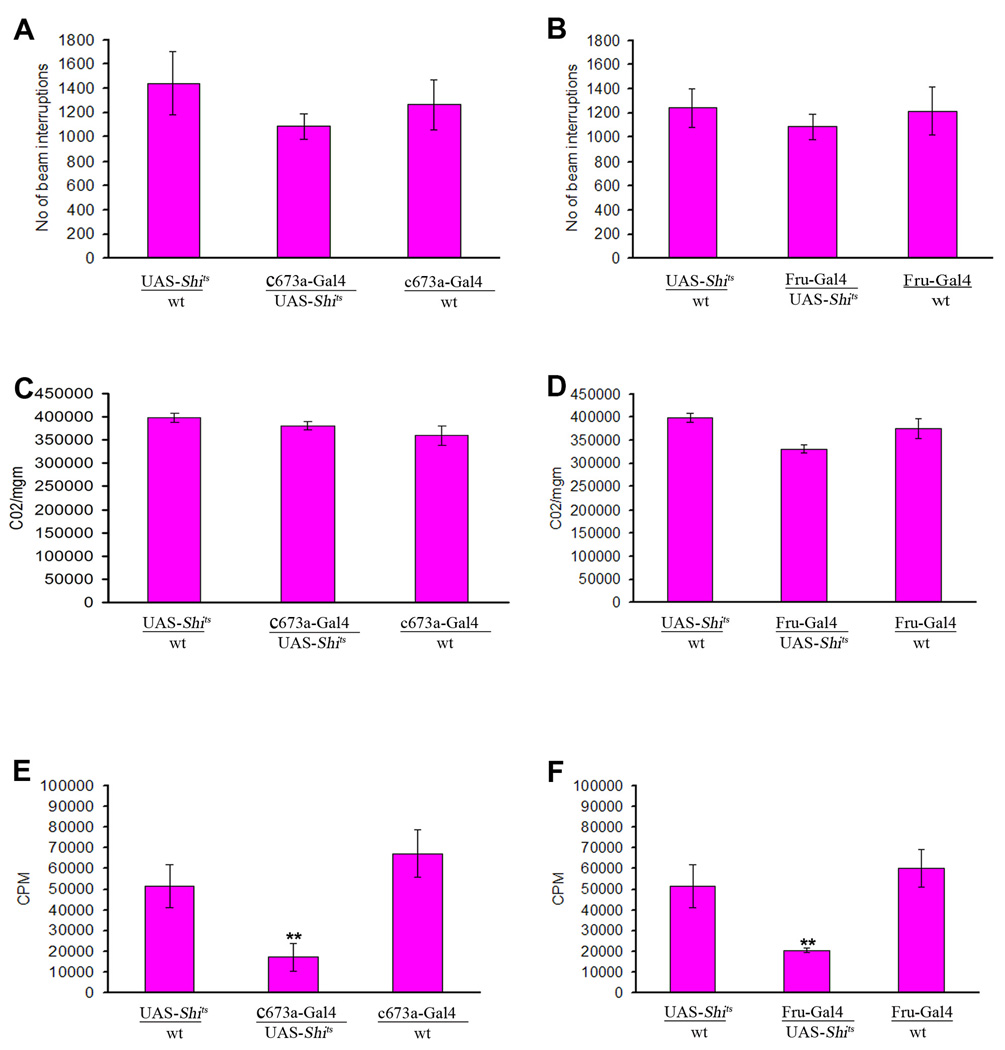
Obesity, defined here as excess fat deposition, occurs when the calories consumed far exceed the body's energy demand. This could occur as a result of reduced physical activity, an abnormal increase in food intake, a metabolic abnormality, or a combination of the above factors. To evaluate these factors, we measured the locomotor behavior of flies with suppressed or hyperactivated c673a-Gal4 or Fru-Gal4 neurons using a Trikinetics motion detector. We did not see any difference between experimental flies and controls (Supplementary Figure 3A–D). We then measured the food intake of the transgenic flies by incubating them on food mixed with 14C- labeled -leucine for 48 hours (Thompson and Reed 1987). Ingested 14C should reflect food consumed. We indeed found that flies with silenced c673a-Gal4 neurons had almost 50% more radioactivity than controls (Figure 3A). Surprisingly, though, silencing Fru-Gal4 neurons produced a marked decrease in food intake (Figure 3C). Such a decrease, without an accompanying reduction in locomotor activity, suggests a metabolic abnormality underlying the Fru-Gal4 obesity phenotype.
Figure 3.
Silencing of c673a-Gal4 neurons causes an increase in food intake when flies are exposed to 14C-labeled-leucine l food for 48 hours (A) , while silencing of Fru-Gal4 neurons produces the opposite effect as compared to controls (C). However, over-activation of either neuronal population does not produce any marked difference in radiation count as compared to control (B and D). Silencing of c673a-Gal4 and Fru-Gal4 neurons reduces metabolic rate and causes defects in fat store utilization. Reduction in CO2 emission is observed when either neuronal group is silenced (E and G) and an increase in CO2 emission is detected when they are over-activated (F and H). Measuring triglyceride levels in starved flies with 7 days silenced c673a-Gal4 or Fru-Gal4 neurons indicate a defect in their ability to utilize their fat stores as indicated by the gentler slope of their fat store depletion rate with continued starvation as compared to controls (I and J). Trendline was generated using Excel worksheet, and the slope value of a given trendline is color coded in the right side of each panel. Error bars are standard deviation of five different replicates for a given genotype. (Asterisks denote T-test statistical significance: *; P<0.05, **; P<0.01, ***; P<0.005).
We examined the metabolic status of flies with abnormal c673a-Gal4 or Fru-Gal4 neural activity by measuring the ratio of their CO2 emissions to total body weight and the levels of ATP in their tissue homogenates. We observed reduced CO2 emission when either neuronal group was silenced (Figure 3E, G.), and an increase in CO2 emission when they were over-activated (Figure 3F, H). Those changes in metabolic rate were not associated with any difference in ATP levels in all tested flies (Supplementary Figure 3E–H). Since ATP is the ultimate energy source utilized by living tissue, the normal ATP levels in flies with altered neural activity in c673a-Gal4 and Fru-Gal4 neurons can explain their ability to maintain normal activity levels despite their abnormal metabolism.
Since using fat as energy source produces more CO2 than using carbohydrate (Salway, 1994), a reduction in CO2 emission suggests a decrease in lipolysis, which would ultimately produce a net accumulation of fat stores. We examined this possibility by measuring the rate of fat store depletion in those flies when starved, and found that silencing of either c673a-Gal4 or Fru-Gal4 neurons leads to a reduced rate of fat store depletion as compared to control flies (Figure 3I, J). However, the defect is more severe in Fru-Gal4 than c673a-Gal4 silenced flies.
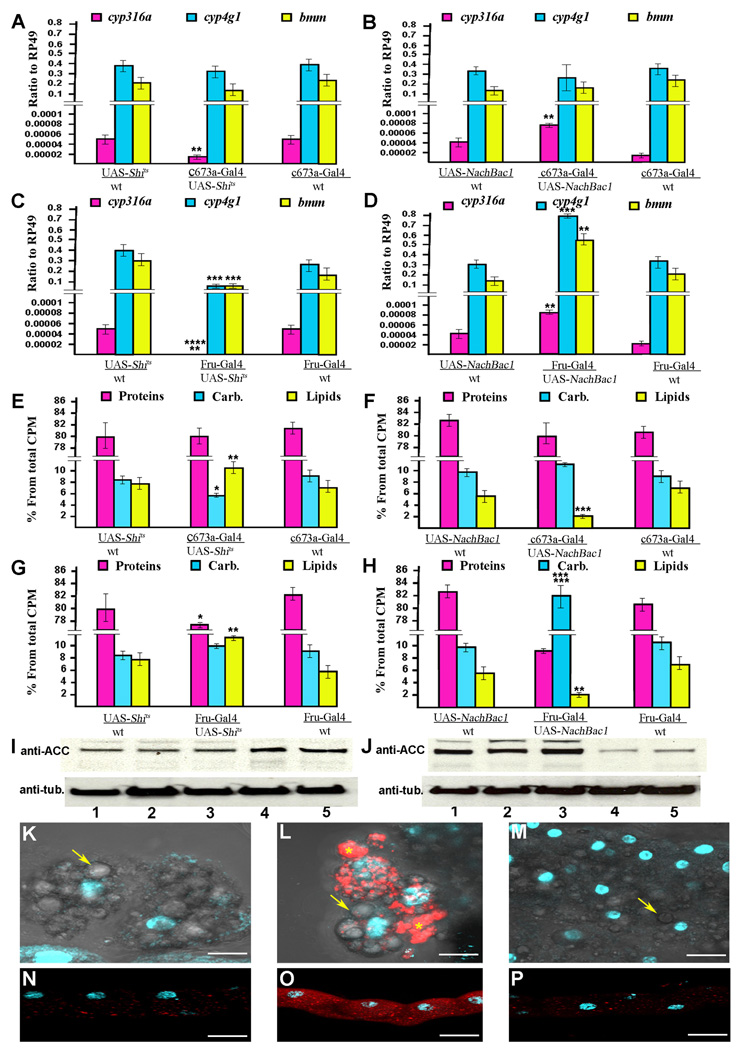
To obtain insights into the molecular mechanisms by which silencing and hyperactivation of these two neuronal groups affect metabolism, we conducted a microarray analysis of mRNA isolated from whole flies with abnormal neural activity in either c673a-Gal4 or Fru-Gal4 neurons using an Agilent Drosophila Gene Expression Microarray. This did not define a clear overall pattern of gene expression changes (data not shown). However, we did observe that the neural activity of Fru-Gal4 neurons affected the expression levels of the cytochrome P450 E-class gene cyp4g1 and the closely related cyp316a1, while altering the activity of c673a-Gal4 neurons affected the expression level of only cyp316a1. cyp4g1 is known to be an important regulator of fat storage (Gutierrez et al., 2007),
To follow up on these observations, we quantitated the changes in mRNA xpression levels for cyp4g1 and cyp316a1 using QT-PCR. We also examined xpression of mRNA encoded by the triglyceride lipase gene Brummer (bmm), another important regulator of fat storage (Gronke et al., 2007). The expression levels of these genes were measured as ratios to the levels of mRNA encoding the ribosomal protein rp49 within the same samples. In flies with silenced Fru-Gal4 neurons, a significant reduction in the expression levels of cyp4g and cyp316a1 as well as bmm was observed. Silencing c673a-Gal4 neurons caused a reduction in mRNA expression for only cyp316a1. Hyperactivation of Fru-GAL4 neurons produced increases in mRNA levels for all three genes, while hyperactivation of c673a-GAL4 neurons affected only cyp316a1 (Figure 4A–D). Thus, for both neuronal populations, silencing and hyperactivation produced changes in the expression levels of genes likely to be involved in fat store utilization.
Figure 4.
Activity of c673a-Gal4 or Fru-Gal4 neurons affect the expression levels of genes involved in fat store utilization. cyp361a1 shows a marked reduction when c673a-Gal4 or Fru-Gal4 neurons are silenced, while their hyperactivation cause the opposite (A–D). Silencing of Fru-Gal4 neurons also causes a decrease in the expression levels of cyp4g, and bmm, while their hyperactivation causes the opposite (C and D, respectively). The neural activity of c673a-Gal4 and Fru-Gal4 neurons affect the ability of flies to convert 14C-labeled-leucine in their food to different macromolecular classes.
Measurements of the percentage of the radiation signal in a given macromolecular class indicates that flies with silencing of c673a-Gal4 or Fru-Gal4 convert more 14C-labeled-leucine into fat than controls (E and G, respectively), while their hyperactivation produces the opposite (F and H, respectively). The increase in the fat radiation signal comes at the expense of the carbohydrate signal in c673a-Gal4 silenced flies (E) and protein signal in Fru-Gal4 silenced flies (G). Additionally, hyperactivation of Fru-Gal4 neurons causes a marked reduction in the protein radiation signal that is accompanied by a large increase in the carbohydrate radiation signal (H). Western blot analysis using anti-ACC indicates that these changes are associated with corresponding changes in the level of ACC enzyme; flies with silenced c673a-Gal4 or Fru-Gal4 neurons have an increase in ACC level (J4 and 5, respectively) as compared to their controls (J1, wt/UAS-Shits; 2 , c673-Gal4/wt; 3 , Fru-Gal4/wt, all were incubated in 30 °C), while flies with hyperactivated c673a-Gal4 and Fru-Gal4 neurons have lowered ACC signal (I4 and 5, respectively) as compared to their controls (I1, UAS-NachBac1/wt; 2, c673-Gal4/wt; 3 is Fru-Gal4/wt, all were incubated in room temperature). The massive shift of 14C signal into the carbohydrates observed in Fru-Gal4 hyperactivated flies is associated with an increase in autophiga in fat cells (L, asterisks designate an autophiga vesicle) and malipegian tubule cells (O). Control room temperature UAS-NachBac1/wt and Fru-Gal4/wt fat (K and M, respectively) and malipigian tubule cells (N and P, respectively) do not show such phenomena. Error bars are standard deviation of five different replicates for a given genotype. Arrows in K–M point to fat droplets, while blue color in K–P identifies nuclei stained with DAPI. White bar in panel K–M represent distant of 100 µm. anti-tub designates tubulin loading control in panel I-J.(Asterisks denote T-test statistical significance: *; P<0.05, **; P<0.01, ***; P<0.005).
Fru-GAL4 neuronal hyperactivation may produce a perceived starvation state
Fat storage can be affected by changes in the rates of conversion of non-lipid nutritional molecules into fat through the process of de novo fatty acid synthesis. To examine if flies with altered neuronal activity in either c673a-Gal4 or Fru-Gal4 neurons have any abnormality in this process, we measured their ability to convert 14C-leucine in their diet to proteins, carbohydrates, or lipids. In this assay, flies were fed 14C-leucine for two days, and their protein, lipid, and carbohydrate fractions were then separated using a modification of the method developed by Handel and Day (1988; Supplementary Figure 4). The percentage of the radiation signals in a given fraction relative to the total radiation count should reflect the ability of those flies to convert 14C-leucine to the different macromolecular classes. Even though different nutritional molecules follow different metabolic paths in order to be converted to other macromolecular classes, this method can provide an overall view of the state of anabolic processes in flies, and can allow an estimation of the relative rates of de novo fatty acid synthesis in different genotypes.
In control flies, about 80% of the ingested 14C-leucine radiation signal was found in the protein fraction, while about 9% was found in the carbohydrate fraction and about 6% in the lipid fraction (Figure 4E–H). In flies with silenced c673a-Gal4 or Fru-Gal4 neurons, an increase in the radiation signal corresponding to the lipid fraction was observed. This increase was associated with decreases in the carbohydrate signal in c673a-Gal4 silenced flies and in the protein signal in Fru-Gal4 silenced flies (Figure 4E and G, respectively). Hyperactivation of either neuronal population produced a marked reduction in lipid radiation signals (Figure 4F and H). Western blot analysis indicated that silencing and hyperactivation were associated with corresponding changes in the levels of acetyl-CoA carboxylase (ACC), the major regulatory enzyme of the de novo fatty acid synthesis pathway (Salway, 1994). Silencing of either neuronal population caused an increase in the level of ACC, while hyperactivation caused a reduction in ACC levels (Figure 4I and J). These data are consistent with a model in which silencing increases fatty acid synthesis, while hyperactivation decreases it.
Surprisingly, in flies with hyperactivated Fru-Gal4 neurons, almost 80 % of the radiation signal was observed in the carbohydrate fraction and not the protein fraction (Figure 4H), suggesting that even though those flies consumed normal amounts of food, they used amino acids to synthesize carbohydrate. This effect was not observed with c673a-GAL4 hyperactivation.
These observations suggest that Fru-GAL4 hyperactivated flies are in a state of perceived energy deficit, because they are using ingested protein precursors to make carbohydrate instead of protein. If so, these flies might also be degrading already synthesized proteins to obtain additional energy via autophagy. Cells in an autophagic state degrade their proteins and organelles to be used as an energy source. The released amino acids are then either directly used as cellular fuel or converted to more readily utilized carbohydrates. Autophagy is usually associated with an increase in lysosomal vesicles. To examine if autophagy is indeed occurring in those flies, their tissues were dissected and stained with the lysosomal marker LysoTracker Red DND-99 (Neufeld, 2008). Indeed, we observed massive autophagic vesicles in fat and malpighian tubule cells of those flies that were not detected in controls (Figure 4K–P). We also subjected flies with altered neuronal activity in c673a-Gal4 neurons to the same assay. We observed no change in LysoTracker staining in their tissues relative to wild-type (data not shown).
c673a-Gal4 and Fru-Gal4 neurons affect fat stores independently of NPY and insulin signaling
Modulation of the neuropeptide Y system (NPY) in mammals alters feeding behavior. The obesity phenotype observed in leptin-deficient mice is partially suppressed in an npy mutant background (Erickson et al., 1997). Removal of NPY/AgRP expressing hypothalamic neurons from adult mice causes weight loss and aphagia (Luquet. et. al. 2005).
Neuropeptide F (NPF) is a Drosophila NPY homolog that acts mainly through NPFR1, a Drosophila homolog of a mammalian NPY receptor (Brown et al. 1999, Shen et al., 2001, Feng et al. 2003, Wu et al., 2003, Wu et al. 2005). To test whether NPF-Gal4 and NPFR1-Gal4 neurons regulate fly fat content, we silenced them either alone or in a c673a-Gal4 or Fru-Gal4-silenced background. Neither NPF-Gal4 nor NPFR1-Gal4 silenced neurons produced any obvious change in fat content, or modified the obesity phenotype of c673a-Gal4 or Fru-Gal4-silenced flies (Supplementary Figure 5).
In mammals, injection of insulin into the brain causes a reduction in food intake, while deficiencies in neural insulin signaling cause severe obesity (Woods et al. 1979, Bruning et al. 2000). Alterations in lipid levels are observed in flies with mutations that affect insulin signaling, suggesting an evolutionarily conserved role of insulin in fat store regulation (Bohni et. al., 1999; Tatar et. al. 2001). These observations prompted us to examine the impact of manipulating insulin signaling within the c673a-Gal4 and Fru-Gal4 neurons on fly fat stores. This was achieved by overexpression in those neurons of either dominant-negative or hyperactive forms of the insulin receptor (UAS-InRDN and UAS-InRACT , respectively), or by overexpression of dominant-negative or wild-type phosphoinositide 3-kinase, the major intracellular mediator of insulin action (UAS-PI3KDN and UAS-PI3KWT , respectively; Leevers et. al. 1996; Gao et al., 1998; Barcelo et al. 2002; Wu et al., 2005). None of these manipulations had any effect on fly fat levels in TLC assays (Supplementary Figure 5G–J).
The obesity phenotype is reversible
To examine whether excess stored fat can be removed in flies, we silenced either c673a-Gal4 or Fru-Gal4 neurons for 7 days to produce the obese phenotype, and then returned those flies to room temperature for another 7 days to restore normal neural activity. Storage of excess fat was reversed in previously c673a-Gal4-silenced flies (Figure 5C, I, N) and completely reversed in previously Fru-Gal4-silenced flies (Figure. 5D, J, Q). This reversal was not due to aging, since the silencing of either circuit during the last 7 days of the 14 day incubation period did produce obesity (Figure. 5E, F, K, L).
Figure 5.
The obesity phenotype of silenced c673a-Gal4 or Fru-Gal4 neurons is reversible. Flies with c673a-Gal4 or Fru-Gal4 neurons silenced for 14 days are obese as measured by the TLC assay (A and B), triglycerides assay (G and H), and Nile red staining (M:c673a-Gal4 and P:Fru-Gal4). When neural activity of 7 day silenced c673a-Gal4 and Fru-Gal4 flies is restored for another 7 days, close to normal fat level is observed by TLC assay (C, and D), triglycerides assay (I, and J), and Nile red histological staining (N: c673a-Gal4 and Q: Fru-Gal4). These changes in fat content have nothing to do with aging since flies with normal neuronal activity in either neuronal population for the first seven days, followed by silencing in the last seven days of the incubation are capable of becoming obese as measured by the TLC assay (E and F), triglycerides assay (K and L), and by Nile red and toluene blue histological staining (O: c673a-Gal4 and R;-Fru-Gal4). Arrows on the TLC plates point to butter standard .Error bars are standard deviation of five different replicates for each genotype. Black bar in panel M–R represent distant of 100 µm (Asterisks denote T-test statistical significance: *; P<0.05, **; P<0.01, ***; P<0.005).
The reversed obesity in these flies might be due to reduced food intake, increased lipolysis, increased locomotion, or a combination of these factors. We examined these possibilities using 7 day silenced c673a-Gal4 or Fru-Gal4 flies that were shifted to room temperature for two days and were still in the process of losing fat. Using a 14C-leucine assay, we observed a severe drop in food intake in these flies as compared to controls, but no changes in CO2 emissions or general activity (Figure 6). These results indicate that loss of excess fat was achieved primarily through a reduction in food intake, and indicates the existence of a signal, possibly emanating from fly fat cells, by which the fly brain detects the general status of energy stores in the body, thus allowing it to produce changes in feeding behavior that maintain a constant level of fat storage.
Figure 6.
The reversal of the obesity phenotype in flies with previously silenced c673-Gal4 or Fru-Gal4 neurons is due to reduction in food intake. The loss of excess fat in these flies is not associated with an increase in general activity as measured by the Trikinetics motion detector (A and B) or increase in metabolism as reviled revealed by CO2 emission (C and D), but is largely due to reduction in food intake as revealed by measuring 14C -labeled Leucine food intake for 48 hours (E and F). Error bars are standard deviation of five different replicas for a given genotype. (Asterisks denote T-test statistical significance: *; P<0.05, **; P<0.01, ***; P<0.005).
Discussion
In this paper, we describe the isolation of two distinct populations of Drosophila brain neurons that regulate fat deposition. These populations, denoted as c673a-Gal4 and Fru-Gal4, were identified by using Gal4 driver lines to express neuronal silencing or hyperactivating genes. For both neuronal populations, silencing produces obesity, defined as excess fat deposition, and hyperactivation produces leanness, defined as a reduction in fat store levels (Figure 1). Silencing and hyperactivation affect the expression of genes that are likely to be regulators of fat storage (Figure 4). However, the observed phenotypes are unlikely to be mediated by signaling through receptors for NPY-like or insulin-like peptides, which are important regulators of growth, feeding and fat deposition (Supplementary Figure 5).
The two populations have only a few neurons in common (Figure 2), and our analysis suggests that the shared neurons are not responsible for the observed phenotypes. Metabolic analysis shows that the two populations affect fat deposition by different mechanisms (Figure 3). The obesity phenotype produced by silencing is reversible (Figure 5).
Metabolic, gene expression, and behavioral changes produced by perturbation of 673a-Gal4 and Fru-Gal4 neurons
Reduced use of fat stores and increases in de novo fatty acid synthesis correlate with the obesity phenotype when either Fru-Gal4 or c673a-Gal4 neurons are silenced, and c673a-Gal4-silenced animals also consume excess food. Conversely, the depletion of fat stores that occurs when either neuronal population is hyperactivated is likely to be caused by increased metabolism and decreases in de novo fatty acid synthesis. Interestingly, when Fru-Gal4 neurons, but not c673a-Gal4 neurons, are hyperactivated, the animals enter a state in which they use protein precursors to synthesize carbohydrates, and probably catabolize their own proteins via autophagy (Figure 4). This suggests that Fru-Gal4 hyperactivated animals are in a state of perceived starvation, despite the fact that they consume a normal amount of food.
Fru-Gal4 is expressed in a large number of brain neurons. Fru-Gal4 silenced flies accumulate excess fat despite consuming less food, and are less obese than c673a-Gal4 silenced flies, which consume more food. These facts suggest that driving the silencing gene with Fru-Gal4 might have two opposing effects. Neurons that are positive regulators of feeding might be silenced as well as neurons that sense fat store levels. Because of this, the flies might reduce food intake, which would decrease the severity of the obesity phenotype that would have been produced by silencing only the fat-sensing subset of the Fru-Gal4 neurons.
Alternatively (or in addition), Fru-Gal4-silenced flies might still be able to detect an increase in their fat stores, and respond to it by decreasing feeding. However, the decrease would be insufficient to prevent the accumulation of excess fat that is driven by the metabolic changes occurring when Fru-Gal4 neurons are silenced. c673a-Gal4 silenced flies probably cannot sense fat store levels at all, since they consume more food despite having an excess of energy reserves.
The different defects underlying the obesity phenotype when the two neuronal populations are silenced, and the observation that there is very little overlap between these populations, suggest that they are parts of two independent neural circuits. We speculate that c673a-Gal4 and Fru-Gal4 neurons may have different roles in the wild, regulating fat stores in response to different environmental or internal stimuli. Since silencing of c673a-GAL4 neurons increases food intake, the activity of these neurons might be turned down under unfavorable environmental conditions in order to increase the ability of the flies to accumulate additional energy stores. For Fru-Gal4 neurons, whose hyperactivation induces an autophagic state, we speculate that activity might be increased under severe starvation conditions to allow the utilization of cellular protein as an energy source.
Lipid metabolism is essential for generating much of the energy needed during periods of starvation. In Drosophila, stored fats are released from the fat body through the activity of lipases such as Bmm lipase. This is in turn causes the accumulation of fat molecules in the oenocytes, where they will be further metabolized through the activity of cytochrome P450 proteins such as Cyp4g1 (Gutierrez et al., 2007). We observed that altering the activity of Fru-Gal4 neurons affected the expression levels of the cyp316a1, cyp4g1, and bmm lipase genes, while neural activity of c673a-Gal4 only affects cyp316a1 levels. Cyp316a1 is a cytochrome P450 that is closely related to Cyp4g1, and although its role in fat metabolism has not been studied, the fact that it belongs to the same cytochrome c family as Cyp4g1 indicates that it is might have similar functions. Perturbation of both neuronal groups affects fatty acid synthesis by inducing changes in the expression of acetyl CoA-carboxylase, the main regulatory enzyme of the de novo fatty acid synthesis pathway.
Regulation of fat storage by mammalian and fly brains
In mammals, hypothalamic brain centers such as the ventromedial nuclei (VMN), paraventricular nuclei (PVN), and the lateral hypothalamic area (LHA) are informed about the status of body fat storage by the leptin and insulin pathways. These centers respond by inducing changes in food intake and metabolism that maintain constant body weight. Electrical stimulation of VMN or PVN neurons suppresses food intake, while bilateral lesions of VMN or PVN cause hyperphagia and obesity.
Leptin and insulin circulating in the bloodstream affect the activity of neurons in the arcuate nucleus of the hypothalamus (ARN). ARN is located in an area with a reduced blood-brain barrier, thus endowing it with the ability to sense leptin, insulin, and circulating nutrient levels. A subset of ARN neurons express the leptin receptor. ARN axons project to VMN, PVN, and LHA, and thereby communicate the status of fat stores to these feeding centers (for reviews see Barsh and Schwartz, 2002, Berthoud and Morrison 2008).
It is unknown whether the fly brain has feeding centers with equivalent roles to these mammalian hypothalamic nuclei. The 673a-Gal4 and Fru-Gal4 populations are dispersed throughout the brain, so the locations of neurons expressing these drivers do not indicate that any particular region of the brain is central to regulation of fat storage. However, the phenotypes produced by silencing and hyperactivation of these populations suggest that, like mammalian hypothalamic nuclei, they respond to humoral signals made by adipocytes that report on fat store levels. In particular, the fact that the obesity phenotype caused by silencing is reversible, and that previously silenced flies dramatically reduce food consumption in order to reduce fat stores back to normal levels (Figure 5 and Figure 6), suggest that adipocytes alter release of a humoral factor when their fat content changes. The levels of this humoral factor are interpreted by the c673a-Gal4 and Fru-Gal4 neurons and used to control food consumption. Thus, flies that have accumulated excess fat stores during the silencing period communicate this fact to the brain, and brain neurons respond by reducing caloric intake when the activity block is released.
The isolation of c673a-Gal4 and Fru-Gal4 neurons in Drosophila should allow the future identification of genes involved in brain/fat store communication, possibly including those encoding the putative adipocyte humoral factor(s). This might be done by examining the consequences of transgenic expression of components of candidate signaling pathways in these neurons on flies’ fat stores, or by finding transcripts selectively expressed in them. The role of such genes in regulating fat storage could then be tested by RNAi or overexpression. The expression patterns of functionally validated genes could, in turn, more precisely identify which neurons within both populations are required for regulation of fat storage and what receptors they use to detect circulating humoral regulators that convey information about fat store levels.
Experimental Procedures
Fly stock maintenance
Females from Gal4 lines obtained from various sources were crossed to males from either UAS-Shits or UAS-NachBac1 stocks at room temperature. The resulting male progeny were collected for two days in groups of 20, and kept on regular fly food (8% corn meal, 5% sucrose, 2% yeast, 1% propionic acid, 0.5% agar). Gal4-line/UAS-Shits flies and their controls were shifted to 30° C for 7–10 days, while Gal4-line/UAS-NaChBac1 flies and their control were incubated at room temperature for 7–10 days before their fat content was analyzed by TLC, triglyceride measurement, and Nile red histological staining. Unless stated otherwise, behavioral and metabolic analysis were done on Gal4/UAS-Shits flies and their controls 2 days after being shifted to 30° C as they were still in the process of becoming obese. Gal4/UAS-NaChBac1 flies and their controls were incubated 21–25 ° C for 2 days before being examined by behavioral and metabolic assays as they were still in the process of losing fat.
Fat analysis
For each TLC assay, four replicas of 10 males of a given genotype were crushed in 250 µl of a fat dissolving 2:1 chloroform methanol mixture. 2 µL of the supernatant was pipetted onto a silica TLC plate that was then run with a 4:1 mixture of hexane and ethyl ether as the mobile phase. The plate was dipped in a general oxidizing stain, ceric ammonium molybdate (CAM), prepared by dissolving 2.5 g of ammonium heptamolybdate tetrahydrate (Sigma, #431346) and1 g of cerium (IV) sulfate hydrate complex with sulfuric acid (Sigma,# 423351) in 90 ml water and 10 ml of concentrated H2SO4. After dipping, the plate was heated for 15–20 minutes in an 80° C oven.
For triglyceride measurement, five replicates of 10 males of a given genotype were crushed in 250 µl of 1XPBS with 0.1% triton-X. The mixture was then sonicated and spun down for 15 minutes at maximum speed, and 150 µl of supernatant was taken and the level of triglycerides was determined using a Stanbio LiquiColor Triglyceride Test kit (#2100-225). All samples used for triglyceride measurements had similar protein concentrations.
For Nile red staining, flies were embedded in sagittal position and cut with a cryostat at 16 µm thickness. The sections were collected on SuperFrost/Plus microscope slides (Fisher Scientific), and were defrosted at 37° C in a desiccation box for 30 minutes, followed by fixing in 4% paraformaldehyde in 1xPBS buffer for 10 minutes. The sections were washed three times for 5 minutes each in 1X PBS followed by incubation in a Nile red staining solution prepared by diluting 5mg/ml Nile red in acetone in 1:100 ratio with 1X PBS for 10–20 minutes. The stained sections were washed three times for five minutes each in 1XPBS, and were then mounted in glycerol and examined using the Cy3 fluorescence channel.
For plastic sections, incisions were made into adult flies sides. They were then incubated with 1% paraformaldhyde and 1% glutaraldehyde EM grad mixture overnight in 4°C. They were rinsed three times with 0.1 phosphate buffer for 10–15 minutes and post fixed with 1% Osmium tetroxides for 1–2 hours. The samples were rinsed with distilled water twice for 10 minutes and dehydrated in 50%, 70%, 85%, 95% ethanol, with each incubation lasting for 5 minutes. The were then incubated twice in propylene oxide for 10 minutes, and were gradually infiltrated with 1:1 propylene oxide: Epon 812 mixture for 1 hour followed by Epon 812 only solution for 1 hour. The Epon 812 was polymerized by incubation in 60 °C in vacuum oven overnight. The sample were cut to 10 µm section and collected on SuperFrost/Plus microscope slides. They were then placed on hot 40 °C plate and covered with 1% toluidine blue 1% borax mixture for 3 minutes The staining solution was then removed by rinsing in water, and the slides were left to dry on hot plate followed by mounting in paramount media..
LysoTracker staining
Adult flies were dissected in PBS followed by 3–5 minutes incubation in solution of 100 nM Lysotracker with 1 ugm/ml DAPI in PBS. The tissues were then transfer into a small drop of PBS and covered with a cover slip and viewed within 1 hour in confocal microscope. The labels were excited with a laser beam at 577 nm with capture using a 590 nm band pass filter.
Immunocytochemistry
2–4 day male brains were dissected and fixed in 4% paraformaldehyde in 1X PBS for 1 hour at room temperature followed by 5 washes of 30 minutes each in 1X PBT with 0.1% triton X-100. The brains were incubated for 1 hour in a blocking solution composed of 1% preimmune goat serum in 1X PBT. They were then incubated over night at 4° C in a 1:100 dilution of either rabbit anti-DILP, anti-serotonin (Sigma, #S5545), or antityrosine hydroxylase (Pelfreeze,# P40101-0). The brains were then washed in 1X PBT 5 times for 1 hour each followed by a 1 hour incubation in the blocking solution. They were then incubated in a 1:500 dilution of goat anti-rabbit Alexa Flour-568 (Invitrogen, #A21069) for 2 hours followed by 5 washes in PBT for 1 hour each. The brains were mounted in Vectashield and visualized with a Zeiss LSM 510 NLO confocal microscope. The GFP label was excited with a laser beam at 488nm and the images were captured with a 500–530nm bandpass filter. The Alexa Fluor 568 label was excited with a laser beam at 561nm with capture using a 575–615nm band pass filter. Autofluorescence images captured with 488nm excitation and collected with a longpass 575nm filter were used as background.
QT-PCR
Total RNA was extracted from 20 flies using Quigen RNA extraction kit (Quigen # 74104) and the amount of RNA was measured with Nanodrop spectrophotometer. Different samples were normalized to 1 µgm total RNA, and DNA contamination was removed using Dnase1 AM treatment (Sigma #048K6043). After Dnase1 deactivation, RT reaction was carried using ISCUP™ cDNA synthesis kit (BIO-RAD # 170-8891), the resulting samples were then diluted 1:3 and QT-PCR reaction was preformed using IQ-SYBRR Green kit (BIO-RAD # 170-8882). For cyp316a primer pair AGGCCAATACCTCTGAAGGACCAT and AAACAGAGAGCTGGACT were used. For cyp4g1 primer pair TTCAGGTCCACTCACTGCCAGAAA and AGAAGTCCTTGCAACGTCCCTGTA were used. For bmm primer pair GATGATGCCCTGCAATTCCTGCAT and TCGGAAACCACGAATTGTCGAACTGA were used. For house keeping gene RP49 primer pair CGATGTTGGGCATCAGTACT and CCCAAGGGTATCGACAACAG were used.
PCR reaction conditions were one cycle 95 °C for 5 minutes, followed by 35 cycles of 95 °C for 10 second and 60 °C for 30 second. Primer demerization was examined by subjecting the reaction at the end to 60 to 95 °C gradient for 65 cycles with each cycle lasting for 0.5 second. All reactions produced one primer melting peak indicating a single PCR reaction product per-tube.
Analysis of general activity level
In this assay, a single fly of a given genotype was placed in a glass tube with a food supply at one end. The flies were then allowed to adapt for 24 hours, and the tube was then placed horizontally on the Trikinetics apparatus that had a light beam passing through the tube to a detector on its other side. Fly motion was registered as the number of times in 24 hours that a fly interrupted the path of the beam to the detector as they moved back and forth. The final data was the average of readings obtained from 10 flies of each genotype.
Analysis of food intake and conversions to different macromolecular class
50 ml of hot fly food was mixed with 250 µl of 50 µCi 14C-leucine and aliquoted as 5 ml portions into regular fly food vials. A group of 20 flies of a given genotype were exposed to this food for 48 hours. They were then transferred to clean vials and allowed to groom themselves for 1 hour. For total food intake analysis, 10 flies from each genotype were transferred into scintillation vials containing 250 µl of a 1:1 mixture of perchloric acid and hydrogen peroxide and incubated at 75° C for 30 minutes, which dissolved flies to a clear fluid. The resulting mixture was mixed with 5 ml of scintillation fluid and radiation was counted using a scintillation counter..
To evaluate the rate by which those flies convert 14C-leucine into different macromolecular class, 10 flies were crushed in 250 µl of 2% sodium sulfate followed by mixing with equal volume of 2:1 chloroform methanol mixture. The samples were vortex twice for 1 minutes followed by centrifuge at maximum speed for 20 minutes. The upper, interphase, and lower layers were then separated and mixed with 250 µl of a 1:1 mixture of perchloric acid and hydrogen peroxide and incubated at 75° C for 30 minutes. The resulting mixture was mixed with 5 ml of scintillation fluid and radiation was counted using a scintillation counter. The final data was the average of readings obtained from 5 independent samples of a given genotype.
CO2 emission analysis
10 flies of a given genotype were anaesthetized with N2 to allow for quick recovery. Their weight was determined, and they were placed in a L1-COR CO2 analyzing chamber (Sable Systems, Inc.). After a 30 minute recovery period, the CO2 concentration was measured twice and the average reading was divided by the sample weight. The final data was presented as the average of five replicates for a given genotype. The DataCan V v5.4 software used was obtained from Sable Systems, Inc.
Measuring the rate of fat store depletion
c673a-Gal4 or Fru-Gal4 flies silenced for 7 days and their controls were starved in 1% agarose vials at 30° C. At different starvation time intervals, five samples of 10 flies of a given genotype were crushed 250 µl of 1XPBS with 0.1% Triton X-100. The mixture was then sonicated and spun down for 15 minutes at maximum speed, and 150 µl of supernatant was taken and the level of triglycerides was determined using a Stanbio LiquiColor Triglyceride Test kit. The results were plotted in an Excel work sheet, and the slope of the trendline was used to indicate the rate of fat store depletion (the steeper the slope the faster the rate of depletion). All samples used for measurements had similar protein concentrations.
Measuring carbohydrate content
In a 96 tissue culture plate, 20 µl of fly extract was mixed with equal volume of 8 mg/ml Rhizopus amyloglycosidase which catalysis the conversion of glycogen and trehalose into glucose. After overnight incubation in room temperature, 140 µl of Sigma Glucose Assay Reagent was added and the sample was left in room temperature of 30 minutes. The glucose concentration was measured by absorbance at 340 nm and compared to known glucose standard.
Western blot analysis
10 flies were homogenized in 200 µl of buffer composed of 1 mM DTT, 0.25 sucrose, 0.1% SDS, 1mM EDTA pH 7.4, and one pellet of protease inhibitor per 10 mL (Roche # 11873580001). The homogenate was then sonicated followed by centrifuge at maximum speed for 20 minutes. 20 µl of the supernatant was mixed with loading buffer and boiled for 5 minutes. The samples were ruin on 4–10% SDS-polyacrlyamide gel gradient and transferred onto a nitrocellulose filter using standard procedures. The E7 anti-tublin antibody used as loading control was obtained from the hyperdoma bank (Univ. of Iowa), and was used in 1:1000 dilutions. Anti-ACC was purchased from cell signaling (# 3662), and was used in 1:500 dilution.
Measuring of ATP
10 flies were homogenized in 250 µl of buffer composed of 1X PBS, 0.1% Triton X-100, 5 M Guaindine-HCl. The homogenate was then sonicated and the mixture was boiled for 10 minutes followed by centrifuge at maximum speed for 20 minutes. 20 µl of the supernatant was placed in 96 well tissue culture plate, and its ATP level was measured and compared to ATP standard using ATP Determination Kit (Invitrogen # A22066).
Mass spectrometry
Samples of fly fat were isolated by the above stated TLC assay. It was then analyzed by direct infusion electrospray ionization (ESI) mass spectrometry and on-line liquid-chromatography-mass spectrometry (LC-MS) with electrospray ionization. For direct infusion analyses, standard solutions of triglycerides tricaprin, tricaprylin, trilaurin, trimyristin, and tripalmitin were prepared to a concentration of 10 micromolar in 2:1 (v/v) choloroform:methanol, 5 mM in ammonium acetate, and analyzed by ESI in the positive ion mode. Triglycerides were detected as ammonium adducts using this solvent system. All ESI experiments were performed with an LCQ ion trap mass spectrometer (Thermo). These standards and an additional standard lipid mixture consisting of 1,3-diolein, 1,2-dioleoyl-rac-glycerol, triolein, and monoolein were used to optimize the separation of these species by reversed phase column chromatography on a 2.1 mm. i.d.× 100 mm Gemini C18 column, 5 micron particle, 110A (Phenomenex, Torrance) using a SpectraPhysics LC system. Solvent A consisted of 75:25 (v/v) acetonitrile (ACN):dichloromethane (DCM) with 4 mM ammonium acetate, and Solvent B was 25:75 (v/v) ACN:DCM saturated with ammonium acetate. The gradient started with 0% B for 4 minutes, ramped to 50%B over 34 minutes, held at 50% B for 2 minutes, to 0%B in 2 minutes and allowed to re-equilibrate for 7 minutes (total run time 45 minutes). The entire effluent (0.2 mL/min) was directed to the electrospray source, which was operated in the positive ion mode. The LCQ was programmed for automated MS/MS for ions in the mass range 350–900 Da, if they exceeded 80e4 threshold counts. Other settings for tandem MS were the usual settings of the mass spectrometer. The mass spec. peaks in our sample exhibits a fragmentation of the M+NH4 ions similar to standard triglycerides, in which results in loss of an alkyl side chain to generate a diacyl species which could be further fragmented to produce an RCO+ ion.
Supplementary Material
Acknowledgments
We would like to acknowledge M. Brown for providing the anti-DILP antibody. We would also like to acknowledge Erich Schwarz, Louise Nicholson, and Mary Lynn Formanack for their editorial assistance. This work was supported by an RO1 grant to S. Benzer, DK070154.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Armstrong JD, Kaiser K. The Study of Drosophila Brain Development. In: Houdebine LM, editor. Transgenic animals - generation and use. Harwood Academic Publishers; 1997. [Google Scholar]
- 2.Barcelo H, Stewart MJ. Altering Drosophila S6 kinase activity is consistent with a role for S6 kinase in growth. Genesis. 34:83–85. doi: 10.1002/gene.10132. [DOI] [PubMed] [Google Scholar]
- 3.Barsh G, Schwartz M. Genetic approaches to studying energy balance: perception and integration. Nature Reviews. 2002;3(8):589–600. doi: 10.1038/nrg862. [DOI] [PubMed] [Google Scholar]
- 4.Berthoud HR, Morrison C. The brain, appetite, and obesity. Annu Rev Psychol. 2008;59:55–92. doi: 10.1146/annurev.psych.59.103006.093551. [DOI] [PubMed] [Google Scholar]
- 5.Billeter J, Goodwin SF. Characterization of Drosophila fruitless-gal4 transgenes reveals expression in male-specific fruitless neurons and innervation of male reproductive structures. J. Comp. Neurol. 2004;475:270–287. doi: 10.1002/cne.20177. [DOI] [PubMed] [Google Scholar]
- 6.Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss B, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97(7):865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 7.Brown M, Crim J, Arata R, Cai H, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides. 1999;20(9):1035–1042. doi: 10.1016/s0196-9781(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 8.Brüning J, Gautam D, Burks D, Gillette J, Schubert M, Orban P, Klein R, Krone W, Müller-Wieland D, Kahn C. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289(5487):2066–2067. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- 9.Cao C, Brown M. Localization of insulin-like peptide in brains of two flies. Cell Tissue Res. 2001;304:317–321. doi: 10.1007/s004410100367. [DOI] [PubMed] [Google Scholar]
- 10.fDemir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121(5):785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 11.Erickson J, Hollopeter G, Palmiter R. Attenuation of the obesity syndrome of ob/ob mice by the loss of neuropeptide Y. Science. 1996;276(5315):1132–1133. doi: 10.1126/science.274.5293.1704. [DOI] [PubMed] [Google Scholar]
- 12.Feng G, Reale V, Chatwin H, Kennedy K, Venard R, Ericsson C, Yu K, Evans PD, Hall LM. Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Europ. J. Neurosci. 2003;18(2):227–238. doi: 10.1046/j.1460-9568.2003.02719.x. [DOI] [PubMed] [Google Scholar]
- 13.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 2003;54(4):618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- 15.Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8(2):118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronke S, Mildner A, Fellert S, Tennagel N, Stefan Petry S, Muller G, Jackle H, Kuhnlein R. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabolism. 2005;5:323–230. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez E, Wiggins D, Fielding B, Gould A. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 18.Hamasaka Y, Nassel D. Mapping of serotoinin, dopamine, and histamine in relation to different clock neurons in the brain of Drosophila. J. Comp. Neurology. 2006;494:314–330. doi: 10.1002/cne.20807. [DOI] [PubMed] [Google Scholar]
- 19.Handel E, Day J. Assay of lipids, glycogen, and sugers in individual mosquitoes: correlations with wings length in field collection Aedes vexans. J. of American Mosquito Control Assocaition. 1988;4:549–550. [PubMed] [Google Scholar]
- 20.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 2002;12(15):1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 21.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz M. Adipokinetic hormone inhibits the formation of energy stores and egg production in the cricket Gryllus bimaculatus. 2003;136(2):197–206. doi: 10.1016/s1096-4959(03)00227-6. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz M, Anand A. Changes in the biochemical composition of fat body stores during adult development of female crickets, Gryllus bimaculatus. Arch Insect Biochem Physiol. 2004;56(3):110–119. doi: 10.1002/arch.20002. [DOI] [PubMed] [Google Scholar]
- 24.Lee G, Foss M, Goodwin S, Carlo T, Taylor B, Hall J. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J. Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 25.Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- 26.Luan H, Lemon W, Peabody N, Pohl J, Zelensky P, Wang D, Nitabach M, Holmes T, White B. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J. Neurosci. 2006;26(2):573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundell MJ, Hirsh J. eagle is required for the specification of serotonin neurons and other neuroblast 7-3 progeny in the Drosophila CNS. Development. 1998;125(3):463–472. doi: 10.1242/dev.125.3.463. [DOI] [PubMed] [Google Scholar]
- 28.Luquet S, Perez F, Hnasko T, Palmiter R. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310(5748):683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 29.Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436(7049):395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 30.Navarro B, Xu H, Yue L, Shi Q, Clapham D. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 31.Neufeld T. Genetic manipulation and monitoring of autophagy in Drosophila. Methods in Enzymology. 2008;451:652–667. doi: 10.1016/S0076-6879(08)03236-9. [DOI] [PubMed] [Google Scholar]
- 32.Ramos E, Meguid M, Campos A, Coelho J. Neuropeptide Y, alpha-melanocyte-stimulating hormone, and monoamines in food intake regulation. Nutrition. 2005;21(2):269–279. doi: 10.1016/j.nut.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Salway J. Metabolism at glance. Blackwell science press; 1994. pp. 20–21.pp. 38–39. [Google Scholar]
- 34.Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J. Neurobiol. 2001;47(1):16–25. doi: 10.1002/neu.1012. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008;6:533–544. doi: 10.1016/j.cmet.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson B. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121(5):795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Thompson E, Reed B. Method for selecting exposure level for the Drosophila sex-linked recessive lethal assay. Environ. Mol. Mutagen. 1987;10(4):357–365. doi: 10.1002/em.2850100405. [DOI] [PubMed] [Google Scholar]
- 38.Tatar M, Kopelman A, Epstein D, Tu M, Yin C, Garofalo R. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 39.Villella A, Ferri SL, Krystal JD, Hall JC. Functional analysis of fruitless gene expression by transgenic manipulations of Drosophila courtship. Proc. Natl. Acad. Sci. USA. 2005;102(46):16550–16557. doi: 10.1073/pnas.0507056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods S, Lotter E, McKay L, Porte D. Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature. 1979;282(5738):503–505. doi: 10.1038/282503a0. [DOI] [PubMed] [Google Scholar]
- 41.Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39(1):147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- 42.Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. USA. 2005;102(37):13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- ad insulin-like systems. 2005;8(10):1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.