Abstract
The claustrum is a prominent but ill-defined forebrain structure that has been suggested to integrate multisensory information and perhaps transform percepts into consciousness. The claustrum's shape and vague borders have hampered experimental assessment of its functions. We used matrix-assisted laser desorption ionization–imaging mass spectrometry to reveal a novel protein marker, G-protein gamma2 subunit (Gng2), which is enriched in the claustrum but not adjacent structures of the rat forebrain. The spatial pattern of Gng2 expression suggests key differences from commonly held views of the claustrum's structure. Using anatomical methods, we found that the rat claustrum is present only at striatal levels of the telencephalon and does not extend to frontal cortical territories. Moreover, the claustrum is surrounded on all sides by layer VI insular cortex cells in both the rat and primate. Using these defining characteristics of the claustrum, we found that the claustrum projects to cortical but not to subcortical sites. The definition of the claustrum as a cortical site is considered. The identification of a claustrum-specific protein opens the door to selective molecular lesions and the subsequent evaluation of the role of the claustrum in cognition.
Keywords: cognition, cortex, G protein, matrix-assisted laser desorption ionization–imaging mass spectrometry, primate, rat
Introduction
The structure and function of the claustrum are enigmatic, consistent with the derivation of its name (a hidden place). The claustrum is present in the forebrain of arguably all mammals (Kowiański et al. 1999; Ashwell et al. 2004), but the exact structural boundaries of the claustrum have long puzzled investigators. Meynert (1885), Brodmann (1994), and Rae (1954) all commented on the claustrum's lack of clear structural boundaries. Indeed, the claustrum was considered by Brodmann (1994) to represent layer VIII of cortex (layer VII being the extreme capsule). These early reports were part of an ongoing debate concerning claustral structural boundaries, hodology, and function that is largely unresolved.
Over the last century, it became widely accepted that the claustrum is sandwiched between the external capsule (EC) and insular cortex (IC) in species lacking an extreme capsule, including rodents, and embedded between the EC and the extreme capsule in higher order species, including primates. Using this structural definition, contemporary studies of the claustrum have concentrated on hodology, showing the claustrum to be reciprocally connected with virtually all cortical sites (Olson and Graybiel 1980; Levay and Sherk 1981; Pearson et al. 1982; Carey and Neal 1985; Li et al. 1986) as well as projecting to a number of subcortical nuclei (Amaral and Cowan 1980; Sloniewski et al. 1986; Volz et al. 1990; Dinopoulos et al. 1992; Peyron et al. 1998; Erickson et al. 2004; Yoshida et al. 2006).
The claustrum's shape and proximity to white matter bundles and the cortex preclude selective lesions of the structure, thus hampering evaluation of its function. Limited electrophysiological and functional imaging studies suggest that the claustrum is involved in processing of multimodal sensory information (Segundo and Machne 1956; Spector et al. 1974; Hadjikhani and Roland 1998). Based on these and other data, Crick and Koch (2005) posited that the claustrum binds sensory input both within and across sensory modalities to generate conscious percepts. Crick and Koch further called for the identification of proteins expressed selectively in the claustrum, with the idea that this information could be exploited to define and molecularly lesion the claustrum and thus permit a detailed analysis of its role in cognition.
The rodent claustrum is generally viewed as being medially contiguous with the EC at striatal levels and located in the incurvature of the forceps minor at more anterior (frontal) levels. Attempts to define the claustrum on cytoarchitectonic grounds have been controversial and led to varying descriptions of claustral boundaries (Druga et al. 1993; Swanson 2004; Paxinos and Watson 2007). Because the shape and proximity of the claustrum to other areas renders accurate placement of anatomical tracers restricted to the claustrum almost impossible, current information on the connections of the claustrum is largely based on retrograde tract–tracing studies. However, a prerequisite of such studies is clearly defined anatomical boundaries.
In order to delineate the anatomical boundaries of the claustrum, we used proteomic methods to reveal proteins enriched in the claustrum but not adjacent structures. We then used classical anatomical methods to validate the picture revealed by the proteomic studies and elucidate the claustrum's connections with other brain areas.
Materials and Methods
Subjects
Adult male Sprague-Dawley rats (Harlan, Indianapolis, IN) served as subjects. In addition, archival forebrain tissue from vervet monkey (Chlorocebus sabaeus) was used. All studies were performed in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals.
Proteomic Analyses
In order to obtain a protein signature of the claustrum with appropriate anatomical resolution, we used matrix-assisted laser desorption ionization–imaging mass spectrometry (MALDI IMS), a method that applies matrix-assisted laser desorption ionization (MALDI) to tissue sections and allows mapping the distribution of proteins with relatively high spatial resolution (Stoeckli et al. 2001; Andersson et al. 2008). Coronal 12-μm sections through the rat forebrain were cut on a cryostat and thaw mounted on gold MALDI target plates. The methods used generally followed our previously described approach (Andersson et al. 2008). After quick rinses in ethanol, sections were dried in a vacuum desiccator before the application of the MALDI matrix sinapinic acid (SA; 20 mg/mL SA in 50% acetonitrile [ACN] and 0.3% trifluoroacetic acid [TFA] in water) to the section as 100-pL drops in a Cartesian array, using an acoustic picoliter droplet ejector (Portrait 630; Labcyte, Sunnyvale, CA). Matrix-applied sections were analyzed on a MALDI time-of-flight instrument operating in linear mode (Autoflex; Bruker Daltonics Inc., Billerica, MA). The size of the laser spot was 50 μm, and the laser was rastered across the brain section every 50 μm (on dead center). Data were analyzed for anatomical localization of protein signals with an imaging software tool (BioMap; http://www.maldi-msi.org).
Protein Identification
In order to identify the protein marked by the IMS peak of mass-to-charge ratio (m/z) 7725, microdissected samples enriched in rat claustrum were homogenized and fractionated, followed by tryptic digestion of a gel-isolated band and tandem mass spectrometry (MS)/MS peptide sequencing, with subsequent database comparisons (see Andersson et al. 2008).
Briefly, the claustrum was sonicated in 50-mg wet weight tissue/1-mL T-PER protein extraction buffer (Cartagen Inc., San Carlos, CA) and then centrifuged at 14 000 × g for 10 min at 4 °C in order to pellet cell/tissue debris. The supernatant was further purified and proteins in 200 μL of supernatant were separated by reverse phase high performance liquid chromatography (HPLC) on a C8 column. Fractions were collected at 1-min intervals, and aliquots (∼0.5 μL) of each fraction were robotically spotted onto the MALDI target on top of prespotted (∼0.5 μL) SA matrix (20 mg/mL SA in 40:60 ACN:H2O with 0.1% TFA [v/v]), using a SymBiot XVI (Applied Biosystems, Foster City, CA). MALDI MS analyses were performed using a Bruker Daltonics Autoflex III L200 mass spectrometer in positive-ion linear acquisition mode under delayed extraction conditions. A mixture of protein standards (bovine insulin, equine heart cytochrome C, equine apomyoglobin, and bovine pancreatic trypsinogen) covering a mass range of ∼5734–23 981 and the SA matrix were spotted onto the MALDI target for external mass calibration. To achieve better mass accuracy (∼200 ppm) by internal mass calibration, the standard protein mixture was comixed with matrix and HPLC fractions of interest. Spectra were evaluated using flexAnalysis software (Bruker Daltonics).
Reconstituted HPLC fractions of interest from the MALDI analyses were separated by gel electrophoresis on a 10–20% gradient Tricine gel (Invitrogen, Carlsbad, CA). Gels were fixed and then stained overnight with Colloidal Blue. Stained protein bands were excised, washed, and equilibrated with 150 μL of 100 mM ammonium bicarbonate. The disulfide bonds of proteins in the gel were reduced with 10 μL of 100 mM dithiothreitol at 50 °C for 15 min. The samples were allowed to cool and cysteine thiols alkylated at room temperature in the dark for 15 min by adding 10 μL of 500 mM iodoacetamide. The gel pieces were equilibrated twice with 1:1 ACN (100 μL):ammonium bicarbonate (50 mM) for 15 min. Gel pieces were then dehydrated in 100 μL ACN for 10 min and dried under vacuum. The reduced and alkylated proteins in the gel were digested overnight at 37 °C with ∼20 μL of 25 mM ammonium bicarbonate containing 0.01 μg/μL sequencing grade trypsin.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of peptides resulting from enzymatic digestion was performed using a Bruker Daltonics HCTultra PTM Discovery System ion-trap mass spectrometer equipped with a FAMOS 920 autosampler (LC Packings-A Dionex Company, Sunnyvale, CA). Peptides were separated on a C18 column. Peptides eluted from the capillary tip were introduced into the nanoelectrospray source in positive-ion mode with an ion-transfer capillary voltage of approximately 1.5 kV. MS/MS spectra of peptides were acquired using data-dependent scanning whereby one full MS spectrum (375–1200 mu) was followed by 4 MS/MS spectra of the 4 most intense ions from the full scan.
Peptide sequences and protein coverage of the MS/MS data were determined using a Sequest algorithm and the Trans-Proteomic Pipeline, which utilizes PeptideProphet and ProteinProphet (Seattle Proteome Center, http://tools.proteomecenter.org/TPP.php). The Trans-Proteomic Pipeline protein results were filtered by a ProteinProphet probability score of ≥0.8 and protein matches with less than 2 peptides identified were eliminated.
Histochemistry and Immunohistochemistry
Rats were transcardially perfused with 0.1 M sodium phosphate buffer, pH 7.3, followed by ice-cold 4% paraformaldehyde in phosphate buffer. After cryoprotection, frozen coronal sections through the forebrain were cut at 40 μm.
Primary antibodies used included rabbit anti–G-protein gamma2 subunit (Gng2) (1:100 immunofluorescence [IF]; Sigma-Aldrich, Inc., St. Louis, MO), mouse anti-parvalbumin (1:1000 IF; 1:3000 immunoperoxidase [IP]; Sigma-Aldrich, Inc.), rabbit anti-parvalbumin (1:2000 IF; SWANT, Bellinzona, Switzerland), rabbit anti-FluoroGold (1:3000 IF; Chemicon Inc., Temecula, CA), goat anti-cholera toxin B (1:5000 IF; List Biological, Campbell, CA), mouse anti-mu crystallin (Crym; 1:150 IF; Novus Biologicals, Littleton, CO), and mouse anti-NeuN (1:1000 IP; Chemicon Inc.). Acetylcholinesterase (AChE) histochemistry was performed according to the method of Tago et al. (1986) without iso-OMPA preincubation. Cytochrome oxidase (CO) histochemistry was performed following the procedure of Wong-Riley and Welt (1980). Primate (C. sabaeus) tissue was obtained from archival samples. Immunohistochemical protocols on the primate tissue follow our previously described methods (Deutch et al. 1986).
Neuronal Tract Tracing
Retrograde tract tracing was performed in rats using FluoroGold (FG; Fluorochrome, Englewood, CO) or cholera toxin B (List Biological Laboratories, Campbell, CA). FG in 0.1 M cacodylate was iontophoretically deposited (+2.5 μA, 7 s on/7s off for 10 min) through glass micropipettes with 15- to 20-μm tip diameters into a variety of cortical areas, including the prelimbic (area 32) aspect of the medial prefrontal cortex (N = 8), the pregenual anterior cingulate cortex (area 24b; N = 6), primary somatosensory cortex (N = 6), M1 motor cortex (N = 4), lateral parietal association cortex (N = 5), and visual cortices (areas 17–19) (N = 3). In addition, FG deposits were placed in the mediodorsal thalamus (N = 8). In 7 animals, 50 nL of a 1% cholera toxin B solution in 0.l M phosphate was pressure injected into the lateral hypothalamus.
Dual immunohistochemical methods followed our previously described protocol (Bubser et al. 2000). Injection sites and location of retrogradely labeled cells were charted using NeuroLucida (MicroBrightfield, Williston, VT).
Results
MALDI IMS Identification of Gng2 as a Claustral Marker
MALDI IMS revealed ∼50 peaks in the region of the claustrum with a signal:noise ratio of greater than 3:1. A specific assessment of these peaks revealed only one (m/z 7725) that was enriched in the claustrum but not adjacent areas (see Fig. 1). The same species was seen in much lower abundance in restricted aspects of the cingulate and insular cortices (Fig. 1); little to no expression was seen in the striatum and white matter. Identification of the m/z 7725 protein by HPLC fractionation of dissected samples of the claustrum followed by trypsinization of the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)–isolated protein and subsequent LC-MS/MS mass fingerprint analysis revealed that the peak is Gng2 (accession EDL86207) (see Fig. 2).
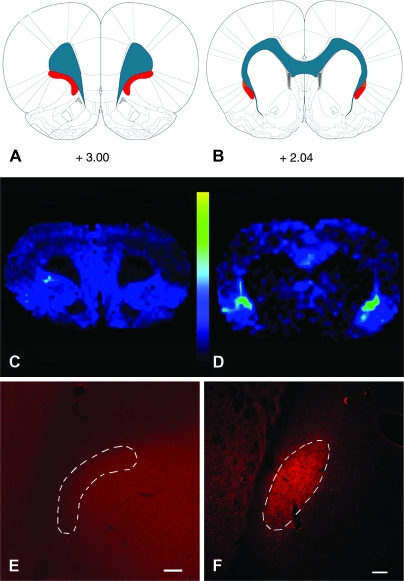
Figure 1.
MALDI IMS reveals a protein enriched in the claustrum. (A, B) Images acquired at 2 different rostrocaudal levels are depicted on the rat brain atlas plates from Paxinos and Watson (2007); values represent distance in millimeters relative to bregma. Blue represents white matter tracts, whereas red depicts the claustrum. (D) At striatal levels, a m/z 7725 protein peak can be seen to be enriched in the claustrum relative to surrounding structures. This peak is not seen at frontal levels (panel C). (E, F) Immunohistochemical localization of Gng2 validates the MALDI IMS data: Gng2-ir is enriched in the neuropil of the claustrum only at striatal levels (F) and is not seen at frontal (E) levels. The outlined areas depict the current definitions of claustral structural boundaries. Scale bars: (E), 200 μm; (F), 100 μm.
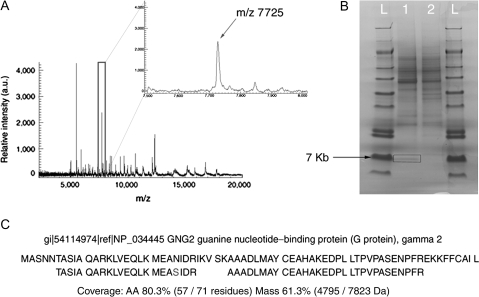
Figure 2.
Mass spectrum of proteins and peptides in the rat claustrum as revealed by MALDI IMS. (A) A representative spectrum obtained from IMS analysis of the claustrum is shown, with an expanded view of the spectrum outline in the box shown at the upper right. The peak corresponding to m/z 7725 was enriched in the claustrum. (B) SDS-PAGE isolation of the m/z 7725 protein species. Following fractionation of the claustral homogenate by HPLC, 2 fractions containing the m/z 7725 species were identified by MALDI MS. These fractions were separated by gel electrophoresis. A single band of ∼7 kDA was seen in the first fraction (lane 1) but was not present in the next fraction (lane 2); the lanes marked L refers are molecular weight ladders. The band from lane 1 was excised (box) for subsequent in-gel trypsinization. (C) Alignment of peptides derived from trypsinization of the m/z 7725 protein with the predicted trypsinized mouse full-length Gng2. The full-length rat Gng2 protein was not available in the following searchable databases: NCBI, ExPASy, EBI, PIR, MIPs, and SGD. In total, peptides of the m/z 7725 protein had a mass coverage of 61.3%, matching 57 of 72 amino acid residues. The rat protein contains a serine (S) at residue 24.
The spatial distribution of basolateral forebrain Gng2 that was revealed by MALDI IMS roughly corresponded to current definitions of the claustrum at striatal levels, with the exception that the protein signal did not appear to be contiguous with the white matter of the EC but rotated slightly away from the EC (see Fig. 1). The Gng2 IMS signal was not present dorsal to the claustrum, in the overlying IC, or ventral to the claustrum in the endopiriform nucleus. There was no detectable Gng2 signal in the area ventrolateral to the forceps minor in the frontal cortex, suggesting that this frontal territory differs substantially from the claustrum. Consistent with the picture revealed by MALDI IMS, immunohistochemical examination of the distribution of Gng2 in the forebrain revealed a strong plexus of Gng2-immunoreactivity (-ir) in the claustrum at striatal levels but no specific staining at frontal levels (Fig. 1)
The Pattern of Gng2 Expression and Anatomical Boundaries of the Rat Claustrum
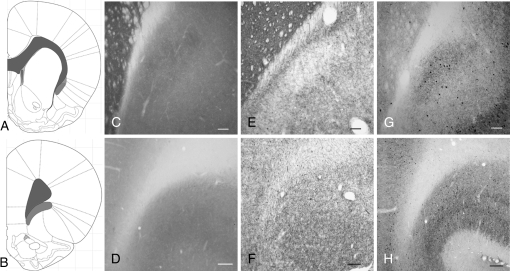
We validated the definition of claustral anatomy obtained from the MALDI IMS data in a series of experiments using classical neuroanatomical methods. CO histochemistry revealed moderately dense staining in the claustrum, with low staining intensity in a thin zone between the claustrum and EC (Fig. 3); we could not distinguish a well-defined CO-positive structure in the frontal territory usually termed the rostral claustrum (Fig. 3). AChE staining revealed a paucity of AChE fibers in the claustrum at striatal levels (Fig. 3) but illuminated a band of densely stained AChE fibers running dorsoventrally between the claustrum and the EC. Again, AChE staining did not reveal a distinct “claustrum” in regions rostral to the striatum. Finally, a dense plexus of parvalbumin (PV)-ir processes and perikarya defined the claustrum at striatal levels, as has been previously reported (Druga et al. 1993); this dense PV-ir staining was not present between the claustrum and the white matter of the EC (Fig. 3). Thus, a dense plexus of PV-ir is in register with the Gng2-defined claustrum.
Figure 3.
The claustrum is present at striatal but not at more anterior levels. The atlas depiction seen in panel (A) represents the rostrocaudal level for panels (C, E, and G); panel (B) represents the rostrocaudal level for panels (D, F, and H). (C, D) CO histochemistry reveals the structural boundaries of the claustrum only at striatal levels (panel C), where a distinct body of relatively dense staining can be seen that is rotated slightly lateral to the EC. In contrast, there is not a zone of dense CO staining that marks the inferior concavity of the forceps minor at frontal levels (D). (E, F) AChE histochemistry and (G, H) PV-ir reveal relatively dense staining in the claustrum at striatal (panels E and G) but not frontal levels (panels F and H). Scale bars: (C, E, and G), 200 μm; (D, F, and H), 100 μm.
The Claustrum Is Surrounded by Cortex in Rodent and Primate Species
Our histochemical observations suggest that cortical cells are interspersed between the EC and the medial aspect of the claustrum, in addition to being present along the lateral border of the claustrum in the deep layers of the IC. The intervening region between the claustrum and EC that we observed with histochemical stains was typically between 20 and 100 μm in mediolateral extent, being greater more anteriorly. This territory interspersed between the claustrum and EC contained neuronal cell bodies, as identified by the neuronal marker NeuN (Fig. 4).
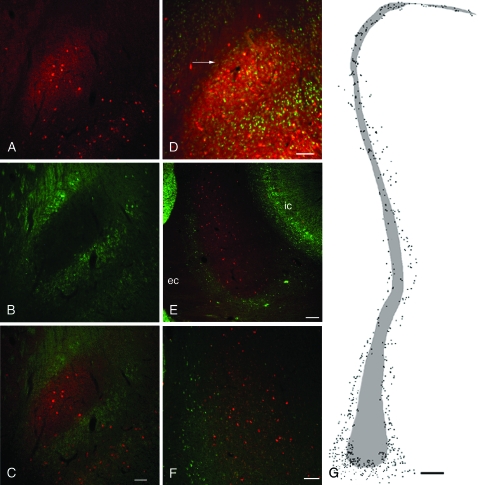
Figure 4.
The claustrum of the rat and primate is embedded in layer VI of IC. (A) A dense PV-ir neuropil (red) marks the claustrum. (B) In contrast, immunohistochemical staining for Crym (green), a protein whose expression is restricted to deep layers of cortex, avoids the claustrum and instead surrounds it (see merged image in panel C). (D) In the rat claustrum, NeuN-ir neurons (green) can be seen to surround the dense PV-ir plexus marking the claustrum (red), including in the zone between the EC and the claustrum (arrow). (E) The primate claustrum, marked by PV-ir (red), is also surrounded by Crym-ir (IC) cells (green). (F) In the middorsal aspect of the primate claustrum many Crym-ir cortical neurons (green) are seen at the border of the claustrum (red). (G) A charting of the distribution of Crym-ir cells (dots) surrounding and admixed with the PV-ir claustrum (shaded area) in the primate. Scale bar shown in (C) (100 μm) applies to panels (A, B, and C). (D), 100 μm; (E), 200 μm; (F), 100 μm; and (G), 500 μm.
To examine further this anatomical organization, we used dual-label immunohistochemistry for PV and Crym, a marker of deep layer (V/VI) cortical neurons (Arlotta et al. 2005). The PV-ir dense cluster defining the rat claustrum was surrounded on all sides by Crym-ir cortical neurons, including Crym neurons interposed between the claustrum and EC (Fig. 4). Crym-ir cells were not present in the body of the claustrum.
The claustrum in primate species is thought to be completely surrounded by the white matter of the EC medially and the extreme capsule laterally. As such, one would not expect IC cells to surround the primate claustrum. However, we observed that Crym-ir neurons were indeed present along the periphery of the claustrum of an old world primate; in the dorsal third of the primate claustrum Crym-ir and PV-ir neurons were completely admixed (Fig. 4). Thus, the claustrum of primates as well as that of rats is surrounded by a zone of deep layer IC neurons.
The Claustrum Projects to Cortical but Not Diencephalic Sites
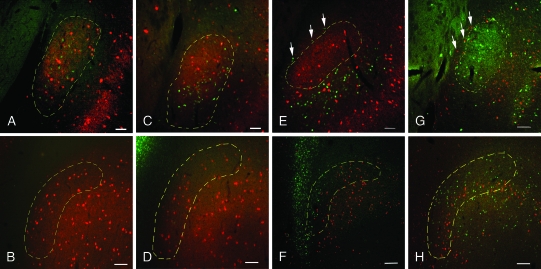
The MALDI IMS data as well as classical anatomical methods showed that the claustrum of the rat is present only at striatal levels, where it is rotated away from the white matter of the EC, and that the claustrum is enveloped by deep layer insular cells. However, it was not clear if the connections of this IC territory medial to the claustrum or of the frontal region commonly designated rostral claustrum were similar to those of the body of the claustrum as defined by Gng2 and PV. We therefore examined the projection targets of these regions in the rat in a series of retrograde tracer studies. We focused most of our attention on the primary somatosensory cortex and the prelimbic (area 32) and pregenual anterior cingulate (area 24b) cortices in the medial prefrontal area (see Supplemental Fig. S1) and examined a smaller number of cases involving cortices of other sensory modalities or motor function. In all cases involving FG deposits into cortical sites (N = 32), retrogradely labeled cells were confined to the claustrum, as defined by PV-ir (Fig. 5). Notable was the lack of retrograde labeling in the region rostral to the striatum, in the area previously designated as the anterior portion of the rat claustrum (see Fig. 5).
Figure 5.
Retrograde labeling of claustrum from cortical and subcortical sites. Injections of retrograde tracers into various cortical sites revealed retrograde labeling in the claustrum at striatal but not frontal levels. Conversely, tracer deposits into subcortical sites did not label cells in the claustrum at striatal levels but did label cells interposed between the claustrum and EC. (A) FG deposits of the anterior cingulate cortex retrogradely labeled cells (green) in the PV-rich claustrum (red) at striatal levels but not at frontal levels (B). (C, D) Injections of FG into the medial prefrontal cortex show a similar picture, with labeled cells in the claustrum at caudal (panel C) but not rostral (panel D) levels. (E) Following FG injections into the mediodorsal thalamus, FG-positive cell bodies (green) at striatal levels surround the claustrum (red), including in the zone between the claustrum and EC (arrows), but are not present in the claustrum. (F) What was previously designated anterior claustrum contains retrogradely labeled cells (green) from the thalamus. (G, H) Following injection of cholera toxin B into the lateral hypothalamus, retrogradely labeled cells (shown in red in panels G and H) are seen only at frontal levels (panel H) but are not seen in the claustrum at striatal levels (G). At the level of the striatum, retrogradely labeled cells from the hypothalamus instead surround the claustrum, including the territory between the claustrum and EC (arrows) Scale bars: (A, C, E, and G), 100 μm; (B, D, F, and H), 200 μm.
We also deposited FG or another retrograde tracer, cholera toxin B, in subcortical regions, focusing on the mediodorsal thalamus (including the mediodorsal, paraventricular, and intermediodorsal nuclei) and the lateral hypothalamus (see Supplemental Fig. S1), both regions identified in earlier studies as receiving afferents from the claustrum. In contrast to cortical deposits, thalamic FG injections retrogradely labeled IC neurons between the claustrum and the EC (Fig. 5) but did not retrogradely label cells in the claustrum. At frontal levels, FG-positive cells were densely packed in the concavity of the forceps minor, that is, in the territory previously identified as the anterior claustrum. A similar picture emerged with retrograde tracer deposits into the lateral hypothalamus (Fig. 5). In addition, we examined a number of injections into various striatal territories and did not observe any retrogradely labeled cells in the claustrum (data not shown).
Discussion
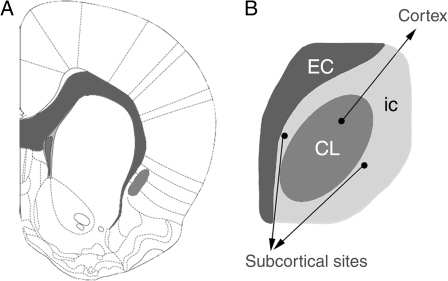
Our MALDI IMS analysis and the anatomical verification of the proteomic data indicate that the structural boundaries of the rat and the nonhuman primate claustrum differ markedly from current definitions. Using these new structural data on the organization of the claustrum, we showed that the claustrum projects to cortical sites only and that subcortical projections previously ascribed to the claustrum arise from IC neurons that surround the claustrum (see Fig. 6).
Figure 6.
Organization of the claustrum. A revised view of the structural boundaries and organization of efferent projections of the rat claustrum. White matter structures are shaded dark gray and the claustrum is depicted in a lighter shade of gray. Current data do not support the presence of an anterior claustrum, ventrolateral to the forceps minor at frontal cortical levels; this region appears to be deep layers of IC. However, at striatal levels (panel A) the claustrum is present, where it is surrounded on all sides by deep layer IC cells (light gray in panel B). These insular cells project to subcortical but not cortical targets. In contrast, the claustrum (CL, darker gray) projects solely to cortical regions. Image in panel (A) is modified from Paxinos and Watson (2007).
Identification of Gng2 as a Claustral Marker
Our definition of claustrum anatomy was driven by the discovery of Gng2 as a protein marker that is specific to the claustrum. The identification of the IMS m/z 7725 peak as Gng2 is supported by several lines of evidence, including the isolation of a single band by SDS-PAGE that contained the peak. Peptide mass fingerprint analysis of this gel-isolated protein species revealed a single database hit. Immunohistochemical localization of Gng2 confirmed the claustral distribution of the protein revealed by IMS. Finally, Gng2 mRNA is present in the claustrum but not the IC or anterior cingulate cortex, with a few scattered cells seen in the striatum (Allen Brain Atlas, http://www.brain-map.org), consistent with Gng2 being expressed by claustral neurons and not by afferents to the claustrum. As such, the low-level Gng2 signal seen in cingulate cortex and IC using MALDI IMS or immunohistochemistry suggests that the extraclaustral Gng2 IMS signal reflects the presence of the protein in claustral axons innervating these cortices. The presence of Gng2 protein in the claustrum, with small amounts detectable in cingulate and IC, also suggests that Gng2 is transported rapidly to dendrites or axons of claustral neurons, where the protein may be part of autoreceptor or heteroreceptor G-protein receptor complexes.
Revised Anatomical Boundaries and Connections of the Claustrum
Our anatomical data indicate that current structural definitions of the claustrum need to be revised, with 3 major changes.
First, the claustrum is not present at frontal levels but is confined to an anteroposterior position corresponding to that of the striatum. As noted earlier, it has proven difficult to define the claustrum based on cytoarchitectonics, particularly at frontal levels. Based on protein expression patterns as well as connectivity, our revised definition of the claustrum sets a more limited anteroposterior domain of the rat claustrum than previously held. Interestingly, this revised definition of the anterior extent of the claustrum is consistent with the anterior border of the claustrum in primates, where the claustrum has not been considered to extend to frontal cortical territories. Finally, it is of interest to note that our data indicate that the projections of the frontal territory in the rat previously identified as claustrum are similar to those of the IC but differ significantly from the connections of the claustrum present at striatal levels. These observations further bolster the argument that the frontal territory previously called anterior claustrum in the rat should be characterized on hodological grounds as IC.
Second, deep layer IC cells surround the claustrum, thereby separating the claustrum from nearby white matter bundles. This observation echoes the early comments of Rae (1954), who noted that the perimeter of the claustrum harbored fusiform somata not unlike those seen in the adjacent IC. We were able to define these cells as cortical neurons on the basis of Crym expression, which is a marker of layer V/VI cortical neurons (Arlotta et al. 2005). This arrangement of cortical cells enveloping the claustrum was also seen in the primate brain, with Crym-ir cells surrounding the claustrum.
Third, the claustrum projects to cortical but not diencephalic sites. Consistent with previous studies (Olson and Graybiel 1980; Pearson et al. 1982; Carey and Neal 1985; Li et al. 1986), we observed retrogradely labeled cells in the claustrum after tracer deposits into all cortical areas examined. However, previous reports also indicated that the claustrum has extensive projections to subcortical targets, including the mediodorsal thalamus and lateral hypothalamus (Dinopoulos et al. 1992; Erickson et al. 2004; Yoshida et al. 2006). Our data indicate that these diencephalic projections do not arise from the claustrum but instead from the IC cells that surround the claustrum; this conclusion is consistent with known IC projections to the thalamus and hypothalamus (Saper 1982). For example, Erickson et al. (2004) commented on labeling of the claustrum after retrograde tracer injections into the mediodorsal thalamus. However, the retrogradely labeled neurons in the claustrum were all located at the periphery of the nucleus, whereas more dorsally in the claustrum, retrogradely labeled neurons were scattered throughout the nucleus. This pattern is strikingly similar to what we observed in sections stained for Crym-ir neurons adjacent to and within the primate claustrum.
We examined diencephalic sites that were selected on the basis of earlier studies indicating that these regions received claustral afferents. Although we cannot comment directly on other subcortical sites that are supposed to receive claustral projections (Amaral and Cowan 1980; Volz et al. 1990; Peyron et al. 1998), a more extensive characterization of the precise origin of these projections is required. It is nonetheless interesting to speculate that the claustrum may lack any direct subcortical projections, placing it in a unique position for executive function.
Our observations in an Old World primate suggest that the revised structural boundaries of the claustrum that we outlined also apply to the human brain; a comprehensive comparative anatomical study of the claustrum is clearly warranted. This revised view of the structure of the claustrum and its relationship to the IC has important implications for research and clinical applications of in vivo imaging, particularly as magnet strength and resolution of the imaging instruments increase. Future studies examining claustrum or insular function using functional imaging approaches will need to closely attend to our anatomical findings.
The Claustrum as a Cortical or Subcortical Site
Historically, there has been a debate over whether the claustrum should be considered a cortical or subcortical structure or a hybrid structure with elements of both cortex and subcortical nuclei (Edelstein and Denaro 2004). Ariens Kappers et al. (1936) tentatively suggested that the claustrum was a part of the cortex. However, the claustrum lacks a laminar organization, a defining characteristic of the cortex. Moreover, the connections of the claustrum differ from those of cortex. For instance, the claustrum does not send projections to the striatum, in contrast to cortical regions (see Graybiel and Ragsdale 1979). Moreover, there is no evidence for claustral projections to the thalamic reticular nucleus. For example, retrograde labeling after tracer injections encompassing virtually the entire thalamus (including reticular nucleus as well as medial thalamic sites) of the hedgehog was interpreted by Dinopoulos et al. (1992) as evidence for claustral projections to the thalamus, but inspection of their photomicrographs reveals a ring of cells that appear to surround—but not be located within—the claustrum.
There are also data supporting a cortical affiliation of the claustrum. Primary among these is the lateral pallial derivation of the claustrum (Puelles et al. 2000). Rae (1954) also suggested that the claustrum is cortical, noting fusiform somata with a morphology similar to IC neurons at the perimeter of the claustrum. However, it now seems likely that these fusiform cells are the IC cells that surround the claustrum. Golgi studies of the claustrum have revealed prominent densely spinous pyramidal cells that resemble those seen in other cortical areas, including long axons that exit the claustrum (Brand 1981; Braak H and Braak E 1982). Adding to this, our data indicates that the claustrum resides within the cortical mantle.
One reason that the claustrum has been considered by some to be a subcortical structure is because in species with an extreme capsule, the claustrum is located beneath the cortical mantle. If one considers that through evolution the extreme capsule formed laterally around the claustrum, our data suggests that the IC was bifurcated by this process, leaving some IC cells medial to the extreme capsule, where they remained surrounding the claustrum and the rest laterally in the place typically recognized as the insula. These considerations suggest that the definition of “subcortical” structures may be better phrased as those internal or beneath the EC rather than the cortical mantle.
Taken together, it appears that the claustrum is a nuclear (nonlayered) structure that is neither cortical nor subcortical but may be best described as “noncortical.” As such, the claustrum represents a novel reconsideration of the definition of “cortex” or at least a blatant exception to the rule.
Conclusions
MALDI IMS is a powerful unbiased approach that can be used to define tissue structures on the basis of protein expression. We used IMS to identify Gng2 as a novel marker of the claustrum. The use of MALDI IMS should open new doors to unraveling defining characteristics of discrete anatomical regions on the basis of protein expression. Although we uncovered a single protein that marked the claustrum, more complex patterns of multiple proteins or peptides may define other regions.
Our anatomical studies confirmed and extended the MALDI IMS data and suggest that previous views of the anatomical organization and connectivity of the claustrum are in error and need to be revised, arguably in all therian species, including human. As such, our MALDI IMS data revealed a solution to a long-standing puzzle in mammalian neuroanatomy. The identification of Gng2 as a marker of the claustrum paves the way to generating animals in which the claustrum can be specifically lesioned without incurring incidental damage to adjacent areas, using the expression of various toxins or their receptors under the control of a Gng2 promoter. This would allow one to test the Crick and Koch (2005) prediction that the claustrum functions as a generator of conscious percepts or determine if the claustrum is more simply functioning as a relay/processing center subserving a distributed network of cortical sites involved in attentional allocation.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxford journals.org/
Funding
National Institutes of Health (T32 MH64913 to B.N.M. and R01 GM58008 to R.M.C.); the National Parkinson Foundation Center of Excellence (A.Y.D.).
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Mental Health or General Medicine or the National Institutes of Health or the National Parkinson Foundation. The authors gratefully thank Lisa Manier for assistance in protein identification. We appreciate the assistance of Drs David L. Tabb, David B. Friedman, and Amy-Joan L. Ham for assistance with database searching and enjoyed helpful discussions with Drs Michael Bubser and Jon H. Kaas on the anatomy of the claustrum and surrounding cortical areas. Conflict of Interest: None declared.
References
- Amaral DG, Cowan WM. Subcortical afferents to the hippocampal formation in the monkey. J Comp Neurol. 1980;189:573–591. doi: 10.1002/cne.901890402. [DOI] [PubMed] [Google Scholar]
- Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- Ariens Kappers CU, Huber GC, Crosby EC. The comparative anatomy of the nervous system of vertebrates, including man. New York: Macmillan; 1936. 1845 p. [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Ashwell KW, Hardman C, Paxinos G. The claustrum is not missing from all monotreme brains. Brain Behav Evol. 2004;64:223–241. doi: 10.1159/000080243. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuronal types in the claustrum of man. Anat Embryol (Berl) 1982;163:447–460. doi: 10.1007/BF00305558. [DOI] [PubMed] [Google Scholar]
- Brand S. A serial section Golgi analysis of the primate claustrum. Anat Embryol (Berl) 1981;162:475–488. doi: 10.1007/BF00301872. [DOI] [PubMed] [Google Scholar]
- Brodmann K. In: Localisation in the cerebral cortex. Garey L, translator. Garey L, translator. Leipzig (Germany): J.A. Barth; 1994. [Google Scholar]
- Bubser M, Scruggs JL, Young CD, Deutch AY. The distribution and origin of the calretinin-containing innervation of the nucleus accumbens of the rat. Eur J Neurosci. 2000;12:1591–1598. doi: 10.1046/j.1460-9568.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- Carey RG, Neal TL. The rat claustrum: afferent and efferent connections with visual cortex. Brain Res. 1985;329:185–193. doi: 10.1016/0006-8993(85)90524-4. [DOI] [PubMed] [Google Scholar]
- Crick FC, Koch C. What is the function of the claustrum? Philos Trans R Soc Lond B Biol Sci. 2005;360:1271–1279. doi: 10.1098/rstb.2005.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutch AY, Lewis DA, Iadarola MJ, Redmond DE, Jr, Roth RH. Effects of D2 dopamine receptor antagonists on Fos protein expression in the striatal complex and entorhinal cortex of the nonhuman primate. Synapse. 1986;23:182–191. doi: 10.1002/(SICI)1098-2396(199607)23:3<182::AID-SYN7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Dinopoulos A, Papadopoulos GC, Michaloudi H, Parnavelas JG, Uylings HB, Karamanlidis AN. Claustrum in the hedgehog (Erinaceus europaeus) brain: cytoarchitecture and connections with cortical and subcortical structures. J Comp Neurol. 1992;316:187–205. doi: 10.1002/cne.903160205. [DOI] [PubMed] [Google Scholar]
- Druga R, Chen S, Bentivoglio M. Parvalbumin and calbindin in the rat claustrum: an immunocytochemical study combined with retrograde tracing frontoparietal cortex. J Chem Neuroanat. 1993;6:399–406. doi: 10.1016/0891-0618(93)90014-u. [DOI] [PubMed] [Google Scholar]
- Edelstein LR, Denaro FJ. The claustrum: a historical review of its anatomy, physiology, cytochemistry and functional significance. Cell Mol Biol (Noisy-le-grand) 2004;50:675–702. [PubMed] [Google Scholar]
- Erickson SL, Melchitzky DS, Lewis DA. Subcortical afferents to the lateral mediodorsal thalamus in cynomolgus monkeys. Neurosci. 2004;129:675–690. doi: 10.1016/j.neuroscience.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Ragsdale CW., Jr Fiber connections of the basal ganglia. Prog Brain Res. 1979;51:239–283. [PubMed] [Google Scholar]
- Hadjikhani N, Roland PE. Cross-modal transfer of information between the tactile and the visual representations in the human brain: a positron emission tomographic study. J Neurosci. 1998;18:1072–1084. doi: 10.1523/JNEUROSCI.18-03-01072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowiański P, Dziewiatkowski J, Kowiański J, Moryś J. Comparative anatomy of the claustrum in selected species: a morphometric analysis. Brain Behav Evol. 1999;53:44–54. doi: 10.1159/000006581. [DOI] [PubMed] [Google Scholar]
- LeVay S, Sherk H. The visual claustrum of the cat. I. Structure and connections. J Neurosci. 1981;9:956–980. doi: 10.1523/JNEUROSCI.01-09-00956.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Takada M, Hattori T. Topographic organization and collateralization of claustrocortical projections in the rat. Brain Res Bull. 1986;17:529–532. doi: 10.1016/0361-9230(86)90220-0. [DOI] [PubMed] [Google Scholar]
- Meynert T. In: Psychiatry: a clinical treatise on diseases of the forebrain. Based upon a study of its structure, functions, and nutrition. Part 1: the anatomy, physiology, and chemistry of the brain. Sachs B, translator. New York: Putnam; 1885. 285 p. [Google Scholar]
- Olson CR, Graybiel AM. Sensory maps in the claustrum of the cat. Nature. 1980;288:479–481. doi: 10.1038/288479a0. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 6thed. London: Academic Press; 2007. 462 p. [Google Scholar]
- Pearson RC, Brodal P, Gatter KC, Powell TP. The organization of the connections between the cortex and the claustrum in the monkey. Brain Res. 1982;234:435–441. doi: 10.1016/0006-8993(82)90883-6. [DOI] [PubMed] [Google Scholar]
- Peyron C, Petit JM, Rampon C, Jouvet M, Luppi PH. Forebrain afferents to the rat dorsal raphe nucleus demonstrated by retrograde and anterograde tracing methods. Neurosci. 1998;82:443–468. doi: 10.1016/s0306-4522(97)00268-6. [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Rae AS. The form and structure of the human claustrum. J Comp Neurol. 1954;100:15–39. doi: 10.1002/cne.901000103. [DOI] [PubMed] [Google Scholar]
- Saper CB. Convergence of autonomic and limbic connections in the insular cortex of the rat. J Comp Neurol. 1982;210:163–173. doi: 10.1002/cne.902100207. [DOI] [PubMed] [Google Scholar]
- Segundo JP, Machne X. Unitary responses to afferent volleys in lenticular nucleus and claustrum. J Neurophysiol. 1956;19:325–339. doi: 10.1152/jn.1956.19.4.325. [DOI] [PubMed] [Google Scholar]
- Sloniewski P, Usunoff KG, Pilgrim C. Diencephalic and mesencephalic afferents of the rat claustrum. Anat Embryol (Berl) 1986;173:401–411. doi: 10.1007/BF00318925. [DOI] [PubMed] [Google Scholar]
- Spector I, Hassmannova J, Albe-Fessard D. Sensory properties of single neurons of cat's claustrum. Brain Res. 1974;66:39–65. [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain maps: structure of the rat brain. 3rd ed. San Diego: Academic Press; 2004. 215 p. [Google Scholar]
- Tago H, Kimura H, Maeda T. Visualization of detailed acetylcholinesterase fiber and neuron staining in rat brain by a sensitive histochemical procedure. J Histochem Cytochem. 1986;34:1431–1438. doi: 10.1177/34.11.2430009. [DOI] [PubMed] [Google Scholar]
- Volz HP, Rehbein G, Triepel J, Knuepfer MM, Stumpf H, Stock G. Afferent connections of the nucleus centralis amygdalae. A horseradish peroxidase study and literature survey. Anat Embryol (Berl) 1990;181:177–194. doi: 10.1007/BF00198957. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Welt C. Histochemical changes in cytochrome oxidase of cortical barrels after vibrissal removal in neonatal and adult mice. Proc Natl Acad Sci USA. 1980;77:2333–2337. doi: 10.1073/pnas.77.4.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, McCormack S, España RA, Crocker A, Scammell TE. Afferents to the orexin neurons of the rat brain. J Comp Neurol. 2006;494:845–861. doi: 10.1002/cne.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.