Abstract
The Drosophila Dysfusion basic-helix-loop-helix-PAS (bHLH-PAS) protein controls the transcription of genes that mediate tracheal fusion. Dysfusion is highly related to the mammalian Nxf protein that has been implicated in nervous system gene regulation. Toward the goal of understanding how Dysfusion controls fusion cell gene expression, the biochemical properties of Dysfusion were investigated using protein interaction experiments, cell culture-based transcription assays, and in vivo transgenic analyses. Dysfusion dimerizes with the Tango bHLH-PAS protein, and together they act as a DNA binding transcriptional activator. Dysfusion/Tango binds multiple NCGTG binding sites, with the following preference: TCGTG > GCGTG > ACGTG > CCGTG. This binding site promiscuity differs from the restricted binding site preferences of other bHLH-PAS/Tango heterodimers. However, it is identical to the binding site preferences of mammalian Nxf/Arnt, indicating that the specificity is evolutionarily conserved. Germ line transformation experiments using a fragment of the CG13196 Dysfusion target gene allowed identification of a fusion cell enhancer. Experiments in which NCGTG sites were mutated individually and in combination revealed that TCGTG sites were required for fusion cell expression but that the single ACGTG and GCGTG sites present were not. Finally, a reporter transgene containing four tandemly arranged TCGTG elements has strong expression in tracheal fusion cells. Transgenic misexpression of dysfusion further revealed that Dysfusion has the ability to activate transcription in multiple cell types, although it does this most effectively in tracheal cells and can only function at midembryogenesis and later.
Members within a related group of transcription factors often control expression of different gene sets despite their protein sequence conservation. This differential gene regulation can arise from a variety of mechanisms. These mechanisms include 1) different transcription factor DNA binding specificities, 2) interactions with different co-regulatory proteins, and 3) expression in different cell types that may vary in their chromatin states. The basic-helix-loop-helix-PAS (bHLH-PAS)2 proteins comprise a group of highly conserved transcription factors that control a variety of developmental and physiological events (1). The defining structural feature of this class of bHLH proteins is the presence of the PAS domain, a multifunctional interaction domain. In Drosophila there are 11 bHLH-PAS family members, and they control disparate processes, including neurogenesis, tracheal formation, tracheal fusion, dendrite morphology, retinal cell fate, circadian rhythms, hormone responsiveness, appendage identity, and the response to hypoxia. Most of these proteins have mammalian and nematode orthologs. One of the issues regarding bHLH-PAS protein function is how these related proteins regulate the different sets of genes that execute these biological phenomena.
One mechanism of differential bHLH-PAS protein gene control involves protein binding to different co-regulatory proteins. The Drosophila Single-minded (Sim) and Trachealess (Trh) bHLH-PAS proteins both dimerize with the Tango (Tgo) bHLH-PAS protein, and bind the same ACGTG sequence (2, 3). However, Trh directly interacts with the Ventral veinless (Vvl) POU-homeobox protein and activates expression of tracheal target genes that contain both Trh and Vvl binding sites (4). In contrast, Sim is unable to directly bind Vvl and, thus, unable to activate tracheal gene expression. In other cases, transcriptional specificity arises from differences in the basic region protein sequences that results in recognition of different DNA sequences. For example, the Drosophila Spineless (Ss) protein also pairs with Tgo but preferentially binds a GCGTG sequence unlike the Sim/Tgo and Trh/Tgo heterodimers that prefer ACGTG (5). Thus, similar to other transcription factor families, bHLH-PAS proteins use multiple methods to differentially regulate gene transcription in vivo.
The last Drosophila bHLH-PAS protein to be discovered was the dysfusion (dys) gene (6). This gene has a mammalian ortholog (Nxf) (7) and a nematode ortholog (C15C8.2) (8). The Dys DNA binding basic region sequence is highly conserved but not identical among the different animal species (Table 1). It is markedly divergent compared with the basic regions of other bHLH-PAS proteins. DNA binding and transient transfection studies on human Nxf revealed that Nxf dimerized with the aromatic hydrocarbon nuclear translocator (Arnt) protein, the mammalian Tgo ortholog, and bound ACGTG, GCGTG, and TCGTG sequences (7, 9, 10). Which of these sequences is utilized in vivo to control gene expression by Nxf is unknown. The Drosophila dys gene is prominently expressed in tracheal fusion cells. These cells reside at the tip of the growing tracheal tubules and mediate the fusion of adjacent tracheal branches. Elimination of dys function by mutation and RNA interference resulted in an absence of tracheal fusion (6), and four genes were identified whose fusion cell transcription was abolished or reduced in dys mutants (11) and are potential candidates to be directly regulated by Dys. The identification of these target genes allows biochemical and molecular experiments that can test the mechanistic role of dys in controlling fusion cell transcription.
TABLE 1. Basic region alignment of selected bHLH-PAS proteins.
Basic region sequences are shown along with binding site specificity for each protein. · indicates amino acid identity, and basic region numbering is at the top. Species prefixes arc: C. elegans (C), Drosophila (D), human (H), and mouse (M). Proteins are Dysfusion (Dys), Nxf, C15C8.2, Single-minded (Sim), Single-minded-1 (Sim1), Similar (Sima), Hypoxia-inducible Factor-1α (Hif-1α). HIF-like factor (Hlf). Trachealess (Trh). Spineless (Ss), and Aryl hydrocarbon receptor (Ahr).
| Protein | Basic region | Binding site | ||
|---|---|---|---|---|
| 1 | 6 | 12 | ||
| D-Dys | NKSTKGASKMRR | AC, GC, TC | ||
| H-Nxf | YR · · · · · · ·A · · | AC, GC, TC | ||
| C-C15C8.2 | QR · · R · · · ·Q · · | Unknown | ||
| D-Sim | KEKS · N · ART · · | AC | ||
| H-Sim1 | KEKS · N · ART · · | AC | ||
| D-Sima | KEKSRD · ARC · · | AC | ||
| H-Hif-1α | KEKSRD · ARS · · | AC | ||
| M-Hlf | KEKSRD · ARC · · | AC | ||
| D-Trh | KEKSRD · ARS · · | AC | ||
| D-Ss | GVTKSNP · · RH · | GC | ||
| H-Ahr | EGIKSNP · · RH · | GC | ||
In this paper we used in vitro and in vivo approaches to study how Dys regulates gene expression in tracheal fusion cells. We showed that Dys dimerizes with Tgo, resulting in nuclear translocation of the Dys/Tgo complex. The Dys/Tgo dimer then acts to activate transcription. The Dys basic region differs from mammalian Nxf at 3 amino acid sites. Yet transient transfection experiments revealed that Dys/Tgo, like Nxf/Arnt, binds to multiple NCGTG sequences with a specificity conserved between mammalian and Drosophila proteins. This is in contrast to the results of identical experiments with Sim/Tgo and Trh/Tgo which demonstrated that they are restricted to a single ACGTG binding site specificity. Drosophila transgenic approaches were employed to test how Dys/Tgo controls transcription in vivo. Reporter gene transformants containing a 1.0-kb fragment of the fusion cell-expressed CG13196 Dys target gene drives fusion cell expression. Mutation of ACGTG, GCGTG, and TCGTG sites in the CG13196 fragment along with analysis of a transgene containing TCGTG multimers revealed that TCGTG is required for expression in vivo. This indicated that Dys/Tgo uses a novel bHLH-PAS protein DNA binding specificity in vivo to control fusion cell gene expression. Further dys misexpression experiments revealed that Dys/Tgo has the ability to ectopically activate CG13196 transcription in multiple cell types but is temporally blocked from acting until mid-embryogenesis.
EXPERIMENTAL PROCEDURES
S2 Cell Transient Expression Plasmids and Assays
The dys expression plasmid, pAct-dys, was generated by cloning an EcoRV fragment of a full-length dys cDNA (6) into the EcoRV site of the pAct5CSRS (12). This pAct-dys plasmid contains the entire dys coding sequence behind an actin5C (Act) promoter. The pAct-dys-Δb plasmid has a deletion of the entire Dys basic region (NKSTKGASKMRR), which is expected to abolish DNA binding. It served as a negative control. The dys-Δb fragment was cloned into the SacI site of pAct5CSRS. The construction of pAct-sim, pAct-tgo, and pAct-trh were previously described (2). The reporter plasmids contained four tandemly linked copies of an identical 24 bp Toll CME-4 sequence (13) with a different, potential Dys/Tgo NCGTG binding site (underlined): CTAGAAATTTGTACGTGCCACAGA, CTAGAAATTTGTCCGTGCCACAGA, CTAGAAATTTGTGCGTGCCACAGA, and CTAGAAATTTGTTCGTGCCACAGA. Each fragment was cloned into pCR-Blunt II-TOPO (Invitrogen), cut with KpnI and SacI, and then cloned into the KpnI and SacI sites of the pGL-3 enhancer tester vector (Promega). pGL3 utilizes a firefly luciferase (luc) reporter gene.
The full-length Nxf cDNA coding sequence was generated by PCR from pEGFP-LE-PAS (9) using the primers 5′-GGTACCATGTACCGATCCACCAAGGGCG-3′ and 5′-GGTACCTCAAAACGTTGGTTCCCCTCCA-3′. The PCR product was cloned into pGEM-T Easy vector (Promega), digested with KpnI, and cloned into the KpnI site of pAct5CSRS, generating pAct-Nxf. The full-length human Arnt coding sequence was contained on a BamHI fragment derived from pBM5/Neo/M1-1 (14). This fragment was cloned into the BamHI site of pAct5CSRS, generating pAct-Arnt. The pAc5.1/V5-His/lacZ transfection control plasmid (Invitrogen) consists of the actin5C promoter driving β-galactosidase (lacZ) gene expression. The LE-PAS and Arnt clones were generously provided by Cam Patterson.
Transient transfection in Drosophila S2 cells was carried out using an Effectene transfection reagent protocol (Qiagen). Each transfection was performed 6 times using 1 µg of total DNA. The total DNA included 0.3 µg for each reporter and expression plasmid, 0.1 µg of pAc5.1/V5-His/lacZ internal control plasmid, and additional pAct5CSRS DNA to achieve a final DNA concentration of 1 µg. After 48 h of growth, luc expression was assayed using a luc assay kit (Promega) and a Typhoon 9400 variable mode imager (Amersham Biosciences). β-Galactosidase activity was measured with a β-galactosidase assay kit (Promega) and enzyme-linked immunosorbent assay and was used to normalize transfection efficiency.
Protein-Protein Interaction (HA Pulldown) Assays
The dys cDNA coding sequence was cloned into pAHW (T. Murphy, Carnegie Institution) in which an N-terminal hemagglutinin (HA) tag was added to dys. The dys cDNA was PCR-amplified using primers 5′-CACCATGCCAAATGCTATTGGAGCTAG-3′ and 5′-CACACTTAATACTAACCTCTATCCTC-3′. The PCR product was cloned into pENTR using the pENTR TOPO cloning kit (Invitrogen). Then the dys cDNA was recombined into pAHW using the Gateway LR Clonase enzyme mix (Invitrogen).
S2 cells were transiently transfected with 0.5 µg of pAct-tgo and 0.5 µg pAct-HA-dys individually or in combination using Effectene. pAct5CSRS DNA was added to achieve a final DNA concentration of 1 µg when necessary. After 48 h of growth, whole-cell extracts were prepared by sonication of cells in lysis buffer (20 mm Tris-HCl, pH 7.5, 0.5% Nonidet P-40, 150 mm NaCl, 3mm EDTA, 3mm EGTA, 10 µg/ml aprotinin, 10 µg/ml of leupeptin, 10 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride). Extracts were incubated with 30 µl of anti-HA-conjugated-agarose beads (Sigma) overnight at 4 °C. The extracts and HA-associated proteins were electrophoresed on 10% SDS-polyacrylamide gels followed by Western blot analysis using mAb-Tgo antibody (2).
CG13196 Transgenic Strains
The sequence from −985 to −1 in the 5′-flanking sequence of CG13196 was PCR-amplified using the primer pair GGTACCCTATAAGTATGGCAAGAGGTGGC (KpnI site is underlined) and AGATCTGATTGGGCCGCAAGTGATA (BglII site is underlined). This 1.0-kb fragment was cloned into the KpnI and BglII sites of pH-Stinger (15), which is a nuclear green fluorescent protein (GFP) P-element reporter. The CG13196 1.0-kb fragment has 3 TCGTG, 1 ACGTG, and 1 GCGTG potential Dys-Tgo binding sites. These sites were mutated individually and in combination using a QuikChange II XL site-directed mutagenesis kit (Stratagene). Each ACGTG, GCGTG, and TCGTG site was mutated to GATCC. Five CG13196 1.0-kb plasmids were generated: unmutated, all 5 (A/G/T)CGTG sites mutated, all TCGTG sites mutated, single ACGTG site mutated, and single GCGTG site mutated. Each transgene was introduced into the Drosophila germ line using standard P-element transformation techniques. Three independent lines of each transgene were analyzed for embryonic expression.
Multimerized TCGTG Transgenic Strain
The 4× 24-bp Toll CME-4 sequence that contains TCGTG, described above under “S2 Cell Transient Expression Plasmids and Assays” was used to generate a 4X-TCGTG-GFP transgenic reporter train. The 4X-TCGTG fragment was excised from the pCR-Blunt II-TOPO plasmid by cutting with KpnI and BglII and then cloned into the KpnI and BglII sites of pH-Stinger to yield P[4XTCGTG-GFP]. After introduction into the Drosophila genome, two independent lines were analyzed for embryonic and larval reporter gene expression.
Transgenic Misexpression Strains
The UAS-dys and UAS-dys-Δb transgenic lines were previously described (11). The dys mutant embryos analyzed were a trans-heterozygous combination of the dys2 and dys3 null mutants (11). Gal4 drivers included breathless (btl)-Gal4 (most tracheal cells), engrailed (en)-Gal4 (ectodermal stripes), scabrous (sca)-Gal4 (nervous system), and twist (twi)-Gal4 (mesoderm) (6, 16, 17).
Embryonic Immunostaining and in Situ Hybridization
Whole-mount embryos were immunostained and subjected to in situ hybridization using standard techniques (18). The following primary antibodies were used for immunostaining: rat anti-Dys (1:200), anti-Tgo monoclonal antibody (1:1), and rabbit anti-GFP (1:1000; AbCam). The following secondary antibodies were used: Alexafluor 488-labeled anti-rabbit IgG (1:200; Invitrogen), Alexafluor 488-labeled anti-mouse IgG (1:200; Invitrogen), and Cy3-labeled anti-rat IgG (1:200; PerkinElmer Life Sciences). The CG13196 cDNA plasmid, RE44287 (Open Biosystems), was used to generate an RNA probe for in situ hybridization. Embryos were examined using a Zeiss Pascal confocal microscope.
RESULTS
Dys Forms a Heterodimer with Tgo in Vivo
Tgo forms heterodimers in vivo with multiple bHLH-PAS proteins, including Sim, Similar (Sima), Ss, and Trh (2, 5). In the absence of a bHLH-PAS partner protein, Tgo resides at low levels in the cytoplasm. In the presence of a partner protein, the Tgo-containing heterodimer translocates and accumulates in the nucleus (16). Thus, nuclear appearance of Tgo generally indicates the occurrence of a partner bHLH-PAS protein (note, there may be exceptions to this rule; Refs. 19 and 20). It also seemed likely that Dys functions as a DNA binding heterodimer with Tgo. First, nuclear Tgo is present at sites of dys expression (6). This is particularly evident in the embryonic leading edge cells, in which no other bHLH-PAS protein besides Dys is known to be present. Second, the Nxf protein, which is the mammalian Dys ortholog, dimerizes with Arnt and Arnt2, the mammalian Tgo orthologs (7, 10). To determine whether Drosophila Dys/Tgo heterodimerization occurs, biochemical protein binding assays, in vivo genetic and misexpression experiments, and cell culture-based molecular assays were performed.
Both tgo and HA-tagged dys were cotransfected into Drosophila S2 tissue culture cells, and HA-tagged Dys protein complexes were purified with anti-HA-agarose beads. Western blot analysis of the protein complex with anti-Tgo revealed that Tgo was bound to HA-Dys (Fig. 1A), indicating a direct, biochemical association between Dys and Tgo.
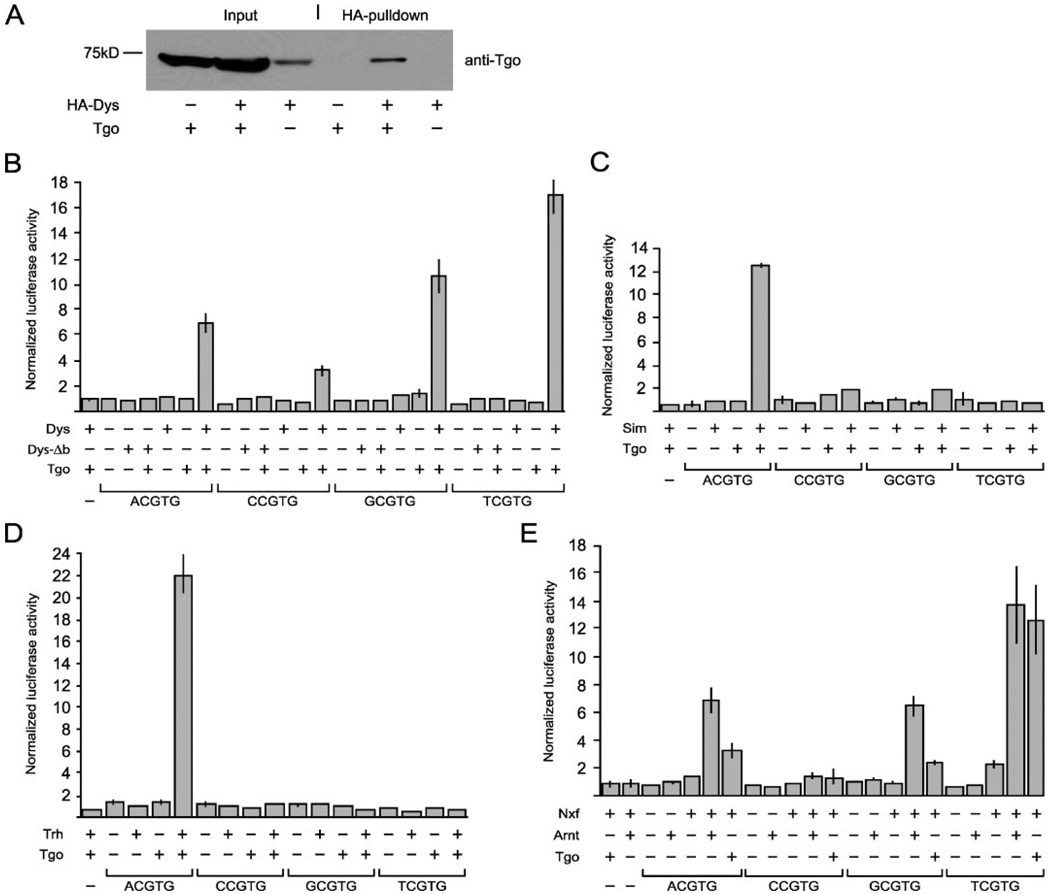
FIGURE 1. Dys binds Tgo, and transient transfection experiments with cultured Drosophila cells indicate that Dys/Tgo binds multiple NCGTG sequences but prefers TCGTG.
A, S2 cells were transfected with pAct-HA-dys (HA-Dys) and pAct-tgo (Tgo) individually or in combination. Whole-cell extracts were prepared, and proteins were immunopurified with anti-HA beads. Both cell extracts (Input) and HA pulldown complexes were subjected to 10% SDS/PAGE followed by Western blot analysis with anti-Tgo mAb. Low levels of endogenous Tgo were present in S2 cells without introduction of pAct-tgo, but the addition of pAct-Tgo greatly increased Tgo levels. Anti-Tgo recognized a protein bound to HA-Dys when cells were transfected with both pAct-HA-dys and pAct-tgo but not when either was absent. The position of the 75-kDa marker protein is shown at the left. The protein reacting with anti-Tgo was 72 kDa, identical to the predicted size of Tgo. B–E, all transfections were independently performed 6×, and the results were averaged and normalized for pAc5.1/V5-His/lacZ expression. B, S2 cells were transfected with combinations of expression plasmids (pAct-dys, pAct-dys-Δb, pAct-tgo) and reporter plasmids (4xACGTG-luc, 4xCCGTG-luc, 4xGCGTG-luc, 4xTCGTG-luc, pGL3 (−; luc negative control)). C, S2 cells were transfected with combinations of expression plasmids: pAct-sim, pAct-tgo, and the same reporters used in A. D, S2 cells were transfected with combinations of expression plasmids: pAct-trh, pAct-tgo, and the same reporters used in B. E, S2 cells were transfected with combinations of expression plasmids pAct-Nxf, pAct-Arnt, pAct-tgo, and the same reporters used in B. Vertical lines at the top of each column indicate standard deviation.
Immunostaining of wild-type embryos with anti-Dys and anti-Tgo showed strong nuclear colocalization of both proteins in the leading edge cells (Fig. 2, A–C). Immunostaining of dys mutant embryos using dys alleles predicted to generate truncated proteins terminating in the HLH regions revealed the absence of nuclear Tgo in the leading edge cells (Fig. 2,D and F), indicating that Tgo nuclear localization requires the presence of (and presumably direct interaction with) Dys. The dys gene is also prominently expressed in tracheal fusion cells, and both nuclear Dys and Tgo appear in fusion cells (Fig. 2, G–I). However, because Trh, another partner of Tgo, is also expressed in tracheal cells, including fusion cells, the appearance of nuclear Tgo cannot be unambiguously ascribed to the appearance of Dys. However, careful examination of Tgo levels in wild-type tracheal nuclei indicated that it is at higher levels in fusion cells than other tracheal cells (Fig. 2, G–I). In dys mutant embryos the levels of nuclear Tgo in fusion cells are reduced to the same levels as the surrounding tracheal cells (Fig. 2, J–L). This reduction occurs even though Trh levels are increased in fusion cells of dys mutants (6, 11). These results indicated that some of the nuclear Tgo in fusion cells is due to heterodimerization with Dys and subsequent nuclear import of the complex. It also indicated that there is a pool of cytoplasmic Tgo that normally turns over rapidly but is stabilized upon interaction with partner bHLH-PAS proteins. This pool can expand when increasing amounts of Dys are present in fusion cells.
FIGURE 2. Dys dimerizes with Tgo in vivo.
All embryos were stained with anti-Dys (magenta; A, D, G, J, M, and P) and anti-Tgo (green; B, E, H, K, N, and Q). Merge images are shown in C, F, I, L, O, and R. dys mutant embryos are dys2/dys3. Proteins derived from both alleles are predicted to lack the region that reacts with anti-Dys. Thus, mutant embryos can be identified based on the absence of Dys immunoreactivity. A–C, sagittal view of a wild-type stage 14 embryo showing overlap of Dys and Tgo in nuclei of leading edge cells (arrowheads). D–F, sagittal view of a stage 14 dys mutant embryo showing the absence of nuclear Tgo in the leading edge cells (arrowheads). G–I, sagittal view of a stage 14 wild-type embryo showing the presence of nuclear Tgo in Dys-positive tracheal fusion cells. Dorsal branch fusion cell (arrows) and dorsal trunk fusion cells (arrowheads) are shown. Note that the levels of nuclear Tgo were higher in fusion cells than adjacent tracheal cells. J–L, sagittal view of a stage 14 dys mutant embryo showing that dorsal trunk fusion cells (arrowheads) had reduced nuclear Tgo with levels comparable to adjacent tracheal cells. M–O, ventral view of a stage 11 en-Gal4 UAS-dys embryo showing the ectopic nuclear colocalization of Tgo and Dys in en stripes. P–R, ventral view of a stage 11 en-Gal4 UAS-dys-Δb embryo showing ectopic nuclear colocalization of Tgo and Dys-Δb.
Misexpression of bHLH-PAS proteins was previously employed in assays to demonstrate that these proteins dimerize with Tgo in vivo (5, 16). The UAS-dys transgene was ectopically expressed in ectodermal stripes with en-Gal4. Immunostaining with anti-Dys and anti-Tgo revealed that both proteins were localized to nuclei in en stripes (Fig. 2, M–O). This reinforces the notion that the appearance of Dys protein results in dimerization with Tgo and subsequent translocation into nuclei. Similar results were obtained with misexpression of UAS-Δb-dys (Fig. 2, P–R), indicating that DNA binding of Dys/Tgo is not required for stable localization in nuclei. This result is similar to that observed for Sim/Tgo (21). In summary, the biochemical experiments showed that Dys and Tgo dimerize. The genetic and misexpression results confirmed that this association also occurs in the embryo and further indicated that Dys/Tgo translocates into nuclei, where the dimer likely acts to bind DNA.
Dys/Tgo Activates Transcription and Binds Multiple NCGTGBinding Sites
Generally, bHLH-PAS dimers containing Tgo/Arnt bind an asymmetric E-box with the sequence NCGTG (1). The basic regions primarily contribute to DNA binding, although additional sites on the protein may contribute. Transient transfection assays using Drosophila cell culture have been successful in determining the binding site specificity of Drosophila bHLH-PAS proteins (2). Consequently, this approach was employed to study the specificity of Dys/Tgo. Tgo binds the half-site GTG (2, 3). Dys has an Arg residue at basic region position 12 (Table 1), and according to the “B-1” rule for bHLH proteins (13, 22), it will bind an NC half-site. Consequently, Dys/Tgo should bind an NCGTG sequence. This is consistent with known Nxf/Arnt/Arnt2 binding sites of ACGTG, CGGTG, GCGTG, and TCGTG (7, 9, 10). The Drosophila Dys basic region (NKSTKGASKMRR) has three amino acid substitutions compared with human Nxf (YRSTKGASKARR; differences are underlined), which could cause changes inDNAbinding specificities between the two proteins. Thus, we tested Dys/Tgo in an S2 cell transient transfection assay to test the binding specificity of Dys/Tgo as well as to determine whether Dys/Tgo was a transcriptional activator.
Reporter genes were constructed that were identical except that each had four copies of either ACGTG, CCGTG, GCGTG, or TCGTG. These constructs were cotransfected with plasmids that expressed either tgo, dys, or dys-Δb in various combinations. The dys-Δb plasmid lacks the Dys basic region and presumably is unable to bind DNA. The transfection experiments that lacked either tgo or dys or combined tgo with dys-Δb acted as negative controls. The results (Fig. 1B) showed that Dys/Tgo acts as a transcriptional activator. Strongest activation was with TCGTG (17× control) followed by GCGTG (10×). In addition ACGTG showed significant activation (7×), whereas CGGTG was significantly lower (3×). Consistent with the need for both tgo and dys, all reporters showed low levels of activation when either Tgo or Dys was absent or when Dys-Δb/Tgo heterodimers were analyzed, indicating that activation requires both Dys and Tgo proteins and is due to direct DNA binding.
Binding Site Promiscuity is Unique to Dys/Tgo
Dys/Tgo binds all four NCGTG sequences, ACGTG, CGGTG, GCGTG, and TCGTG, relatively well. Other studies have shown that Sim/Tgo preferentially binds ACGTG better than GCGTG, and Ss/Tgo preferentially binds GCGTG better than ACGTG (21). However, previous studies with Drosophila bHLH-PAS proteins did not systematically assay all four NCGTG reporters as was done here with Dys/Tgo. Thus, it is possible that other Drosophila bHLH-PAS heterodimers, besides Dys/Tgo, may also bind sequences other than their preferred sequence. To test this we utilized the same reporters assayed with Dys/Tgo in assays containing sim, tgo, and trh expression plasmids in the S2 cell transient transfection assay. The results (Fig. 1, C and D) showed that Sim/Tgo and Trh/Tgo differed from Dys/Tgo in that they were specific for ACGTG binding sites and did not significantly activate CGTGT, GCGTG, or TCGTG sites. Thus, these assays indicated that Dys/Tgo has a broader specificity than Sim/Tgo and Trh/Tgo, and Dys/Tgo also strongly binds TCGTG, a sequence unique to the Dys/Tgo/Nxf/Arnt class of bHLH-PAS proteins.
Evolutionary Conservation of Dys/Nxf Biochemical Function
Because the promiscuity of Dys/Tgo binding specificity appears unique among bHLH-PAS proteins, it is important to assess whether this feature is evolutionarily conserved. This seems likely, since the results of our analysis of Dys/Tgo DNA binding was similar to the results obtained by Ooe et al. (2004) on Nxf/Arnt using similar, but not identical, assays. In contrast, a more limited analysis demonstrated strong binding to ACGTG but not GCGTG (9). To further investigate the similarities in transcriptional specificity between Drosophila and mammalian proteins and their ability to substitute for each other, we utilized the S2 cell system and the four NCGTG reporters with human Nxf and human Arnt expression plasmids. The results shown in Fig. 1E revealed that TCGTG, GCGTG, and ACGTG were activated strongly by Nxf/Arnt, whereas CCGTG showed little activation. Essentially, the same results were obtained when Tgo was substituted for Arnt (Fig. 1E), although Tgo was slightly less active with Nxf compared with Arnt. Comparison of the results indicated that using identical assays, Dys/Tgo and Nxf/Arnt show similar promiscuous DNA binding specificities. Their specificities show a similar preference spectrum with TCGTG > GCGTG > ACGTG > CCGTG. This result confirms the analysis by Ooe et al. (2004). In addition, Dys and Nxf are both able to function with Tgo, reinforcing the strong conservation between Drosophila and mammalian proteins.
In Vivo Identification of a Tracheal Fusion Cell Enhancer
The S2 cell transient transfection results indicated that Dys can dimerize with Tgo and activate transcription of multiple NCGTG reporter genes. However, definitive insight into the role of Dys in controlling tracheal fusion cell transcription requires in vivo analysis. This is a two-step process involving transgenic identification of a fragment of DNA that can drive fusion cell transcription and then mutation and analysis of potential Dys binding sites. Genetic studies have identified four genes (CG13196, CG15252, members only (mbo), and shg (shotgun)) whose expression is abolished or reduced in dys mutants (11). The CG13196 gene was chosen for further study because (a) its embryonic expression is specific for fusion cells and, thus, its cis-regulation may be relatively uncomplicated, (b) the gene is activated in fusion cells after the appearance of Dys, consistent with direct control by Dys/Tgo, (c) its expression is ectopically expanded in all tracheal cells upon misexpression of dys (11), (d) its gene structure is relatively simple with small introns and a short 5′-flanking region, and (e) it has multiple NCGTG putative Dys/Tgo binding sites in its 5′-flanking region.
CG13196 encodes a member of the zona pellucida family of membrane proteins (23). Previously, we showed using an ectopic expression assay that CG13196 has the ability to promote tracheal fusion, suggesting a role in cell adhesion (11). The Drosophila melanogaster CG13196 gene contains four exons and lies within the large first intron of the Buffy gene. The sequence interval 5′ of CG13196 exon 1 and exon 2 of Buffy is only 985 bp. This region has 3 TCGTG, 1 ACGTG, 1 GCGTG, and no CCGTG sites (Fig. 3A). This entire fragment was cloned into pH-Stinger, which is a GFP-based enhancer tester vector (15), to yield the transgene P[1.0-CG13196-GFP]. Analysis of embryos immunostained for both GFP expression with anti-GFP and CG13196 RNA by in situ hybridization revealed that P[1.0-CG13196-GFP] is expressed in all tracheal fusion cells, identically to CG13196 (Fig. 3, B–D). Thus, the CG13196 1.0-kb upstream fragment contains a tracheal fusion cell enhancer.
FIGURE 3. 1.0-kb fragment of the CG13196 gene directs fusion cell expression.
A, schematic of 985 bp of D. melanogaster DNA 5′ to exon 1 of the CG13196 gene. The location of exon 2 of the Buffy gene is shown. Arrows indicate the direction of transcription, and scale is shown below the schematic. Five NCGTG sequences are located in this region: 3 TCGTG, 1ACGTG, and 1 GCGTG. B–D, dorsal-lateral view of a stage 16 embryo containing a P[1.0-CG13196-GFP] transgene, in which the 1.0-kb CG13196 fragment cloned into the pH-Stinger vector drives GFP expression. The embryo was hybridized to GFP (green) (B) and CG13196 (magenta) (C) probes. Merge is in D. The two probes showed complete overlap in expression in all fusion cells, including dorsal branch (arrows), dorsal trunk (white arrowheads), and lateral trunk (yellow arrowhead).
Mutational Analysis Reveals That TCGTG Is an Important Dys/Tgo Binding Site In Vivo
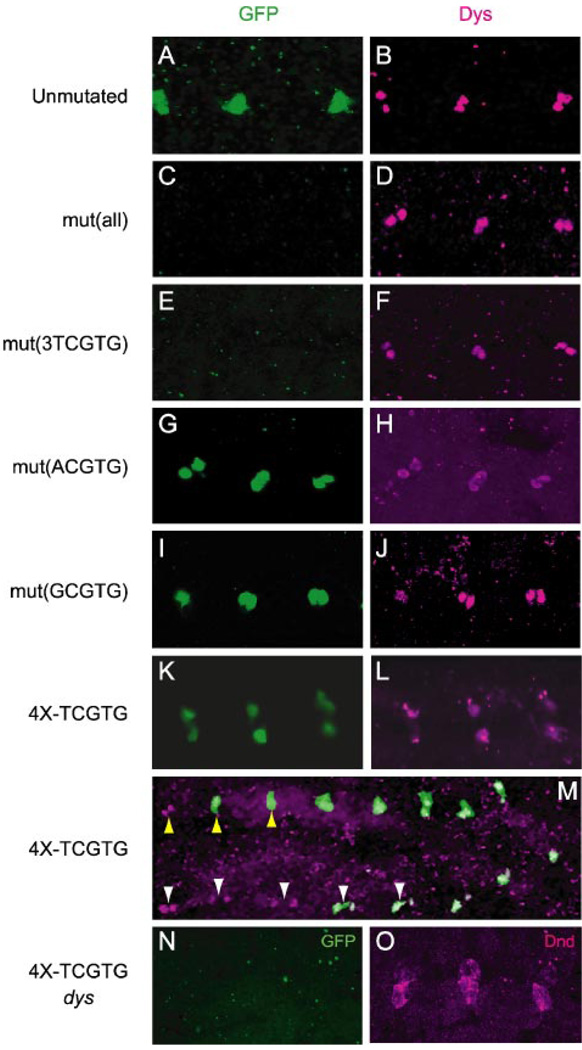
Because the 1.0-CG13196 fragment has five NCGTG sites that could act as binding sites for Dys/Tgo, these sites were mutagenized individually and in combination to the sequence GATCC, which is not expected to bind Dys/Tgo. Four variants were generated: P[1.0-mut(all)-CG13196-GFP], in which all 5 NCGTG sites were mutated, P[1.0-mut(3TCGTG)-CG13196-GFP], in which all TCGTG sites were mutated, P[1.0-mut(ACGTG)-CG13196-GFP], in which the single ACGTG was mutated, and P[1.0-mut-(GCGTG)-CG13196-GFP], in which the single GCGTG site was mutated. Three independent lines bearing each transgenic construct were analyzed for GFP expression. The results indicated that mutation of all five NCGTG sites resulted in loss of fusion cell expression (Fig. 4, C and D). Mutation of just the three TCGTG sites also resulted in loss of fusion cell expression (Fig. 4, E and F). In contrast, the mutation of just the ACGTG or GCGTG sites had no affect on fusion cell expression (Fig. 4, G–J). These results provided strong evidence that Dys/Tgo directly regulates CG13196 expression and acts as a transcriptional activator in vivo. They also demonstrated that the TCGTG sequences are required. The ACGTG and GCGTG sequences are not absolutely required for fusion cell expression, although it remains possible that they contribute to fusion cell expression in association with the TCGTG binding sites.
FIGURE 4. Fusion cell expression of CG13196 requires TCGTG sequences.
All images show sagittal views of dorsal trunk fusion cells from transgenic stage 15 embryos stained with anti-GFP (green) and anti-Dys (magenta); anterior is to the left. Dys staining acts as a marker for the presence of fusion cells. A and B, P[1.0-CG13196-GFP] strain, which has unmutated NCGTG sequences, showed GFP expression in fusion cells. C and D, P[1.0-mut(all)-CG13196-GFP] strain, in which all 5 NCGTG sites were mutated, lacked fusion cell GFP expression. E and F, P[1.0-mut(3TCGTG)-CG13196-GFP], in which all TCGTG sites were mutated, lacked fusion cell GFP expression. G and H, P[1.0-mut(ACGTG)-CG13196-GFP], in which the single ACGTG was mutated, showed fusion cell GFP expression. I and J, P[1.0-mut(GCGTG)-CG13196-GFP], in which the single GCGTG was mutated, showed fusion cell GFP expression. K and L, the P[4X-TCGTG-GFP] strain showed GFP expression in fusion cells. M, the P[4X-TCGTG-GFP] strain showed segmental differences in fusion cell GFP expression. In this embryo GFP (green) was expressed in dorsal trunk fusion cells (yellow arrowheads) in dorsal trunk units 3–9 but was absent in units 1–2 (see Manning and Krasnow (33) for a description of tracheal metameric units) even though Dys protein (magenta) was present (shown are units 2–9). Lateral trunk fusion cells (white arrowheads) showed the appearance of GFP in units 5–9, but GFP was absent in units 1–4 even though they possessed high levels of Dys (shown are units 2–9). N and O, expression of P[4X-TCGTG-GFP] was absent in dys mutant embryos. The dys mutant embryo lacked (N) GFP (green) expression in fusion cells, which were marked by staining with an antibody to the fusion cell-expressed Dead end (Dnd) protein (magenta; Jiang and Crews, unpublished information).
Tracheal Fusion Cell Expression of a Multimerized TCGTG containing Transgenic Reporter
Further evidence for the ability of Dys/Tgo to activate transcription in vivo via a TCGTG sequence was sought by analyzing a transgenic reporter, P[4XTCGTG-GFP], containing four TCGTG sequences fused to a minimal promoter. This experiment was based on the successful use of a 4× ACGTG transgenic reporter, which showed expression in CNS midline cells and trachea (2, 13) that are sites of sim and trh function, respectively. Embryonic expression of P[4X-TCGTG-GFP] was observed in two cell types, tracheal fusion cells (Fig. 4, K–M) and the salivary glands (data not shown). Recently, it was shown that the pH-Stinger vector used here is expressed in the salivary gland by itself (24), and salivary gland GFP expression was also observed in the CG13196 transgenes described above. Because dys is not expressed in the salivary gland (6), we conclude that the only sites of embryonic expression of P[4X-TCGTG-GFP] are in the fusion cells, sites of Dys/Tgo. P[4X-TCGTG-GFP] expression was absent in dys mutant embryos (Fig. 4, N and O), demonstrating that expression was dependent on dys, as expected. These results provide additional evidence that Dys/Tgo binds TCGTG in vivo.
The P[4X-TCGTG-GFP] strain showed expression in fusion cells of all four dys-positive branch types: dorsal branch, dorsal trunk, lateral trunk, and ganglionic branch. However, expression was not observed in other sites of dys expression, including brain, foregut atrium, leading edge, and anal pad (data not shown). This was reminiscent of the 4X-ACGTG transgenic strain, in which only a subset of sim and trh sites showed expression (2). Interestingly, the P[4X-TCGTG-GFP] strain showed segmental restriction of fusion cell expression in all four branch types. Expression was present in fusion cells of all posterior segments but was absent in anterior segments (Fig. 4M). Both the dorsal branch and lateral trunk failed to show GFP fusion cell expression in the four to five anterior-most tracheal segments despite the appearance of Dys protein. The dorsal trunk and ganglionic branch lacked GFP fusion cell expression in the anterior-most two to three tracheal segments. These differences in GFP expression in various Dys-positive cell types suggest that additional complexities exist regarding how Dys controls transcription in fusion cells.
dys Misexpression and Fusion Cell Transcription
In a previous paper UAS-dys was expressed throughout the entire trachea using btl-Gal4, and CG13196 was shown to be ectopically expressed throughout the trachea (11). We wondered whether dys could also drive CG13196 expression in additional cell types, since this would provide insight into the nature of other factors required for fusion cell gene expression. In these experiments, UAS-dys was misexpressed in (a) epidermal stripes using en-Gal4, (b) mesodermal cells using twi-Gal4, and (c) peripheral and central nervous system precursors using sca-Gal4. Misexpression embryos were stained with anti-Dys to gauge dys expression and hybridized to a CG13196 probe. In all cells in which dys was misexpressed, Dys protein was nuclear and appeared at high levels (Fig. 5B, E, H, K, N, and Q). This confirms results seen with other bHLH-PAS proteins that dimerization with Tgo and subsequent nuclear localization occurs in most, if not all, cell types throughout development (16).
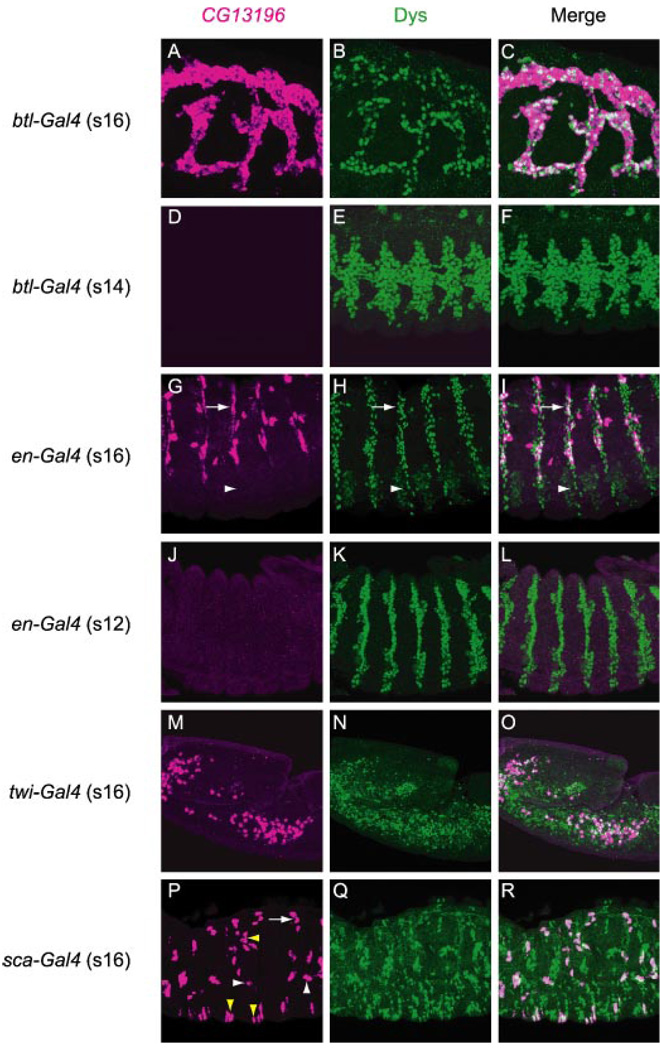
FIGURE 5. Misexpression of dys results in ectopic expression of Dys target gene CG13196 in multiple cell types.
Shown are sagittal views of embryos hybridized to a CG13196 RNA probe (magenta) and immunostained with anti-Dys (green). Merge image is shown at the right. All embryos contained the UAS-dys transgene and various Gal4 drivers. A–C, stage 16 btl-Gal4 UAS-dys embryo showed ectopic CG13196 expression in all tracheal cells. D–F, stage 14 btl-Gal4 UAS-dys embryo did not express CG13196 despite strong Dys misexpression in tracheal cells. G–I, stage 16 en-Gal4 UAS-dys embryo showed CG13196 expression in en epidermal stripes. Expression was observed in a subset of En-positive cells in the dorsal-lateral epidermis (arrows) and was absent from the ventral epidermis (arrowheads). J–L, stage 12 en-Gal4 UAS-dys embryo did not express CG13196 in epidermal stripes even though strong en expression was present. M–O, CG13196 was ectopically expressed in mesodermal cells in stage 16 twi-Gal4 UAS-dys embryos. Not all Dys-positive cells expressed CG13196. P–R, stage 16 sca-Gal4 UAS-dys embryo showed widespread dys expression in nerve cells (Q), yet CG13196 was only expressed in a small number of sca-positive cells (examples are indicated by yellow arrowheads). Many CG13196-positive cells shown are tracheal fusion cells. Shown are dorsal trunk (arrow) and longitudinal trunk (white arrowheads) fusion cells.
The ability of dys to activate CG13196 expression showed spatial and temporal differences. When misexpressed in the trachea using btl-Gal4, dys strongly activated CG13196 in most, if not all, tracheal cells when assayed at stage 16 (Fig. 5, A–C) (11). However, when assayed at stage 14, CG13196 expression was not yet induced in btl-Gal4 UAS-dys embryos even though Dys protein was present at high levels (Fig. 5, D–F). This indicated an early developmental block to Dys/Tgo activation. When expressed in en epidermal stripes, CG13196 expression was observed in some cells but not others when assayed at stage 16 (Fig. 5, G–I). Expression is absent from the ventral epidermis but present in the dorsal epidermis, indicating a regional difference. However, in the dorsal epidermis expression was still patchy, with no clear pattern. Thus, equivalent cells in different segments showed varying levels of CG13196 expression, indicating that ectopic CG13196 expression in epidermal cells is sporadic and not restricted to specific epidermal cell types. Relative levels of Dys appeared high in both cells that were CG13196-positive and also CG13196-negative, indicating that high level dys expression is not an obvious reason for CG13196 expression differences. Similar to activation of CG13196 by tracheal dys, CG13196 was not activated in en stripes when assayed at earlier stages (Fig. 5, J–L). Similar results to en misexpression were observed in the mesoderm. Dys could induce CG13196 in some mesodermal cells, but not all, when induced by twi-Gal4 (Fig. 5, M–O). The misexpression of dys can induce developmental defects. For example, the twi-Gal4 UAS-dys embryos fail to germ-band retract (Fig. 5, M–O). Ectopic expression of CG13196 in neural precursors and their progeny by sca-Gal4 was relatively rare (Fig. 5, P and R). Thus, ectopic expression of dys can activate CG13196 transcription ectopically, but the robustness of activation is dependent on both temporally and spatially controlled factors.
DISCUSSION
The Drosophila Dys bHLH-PAS protein is an important regulator of tracheal fusion cell gene expression. This paper mechanistically deals with how it regulates transcription. The results demonstrate that Dys dimerizes with Tgo, and together they activate transcription of target genes. The target specificity of Dys/Tgo was analyzed systematically in cell culture transient transfection assays and showed promiscuity in binding site specificity by binding multiple NCGTG sequences. The quantitative preference was TCGTG > GCGTG > ACGTG > CGGTG. Using the same assay it was shown that this promiscuity is biochemically distinct from the actions of the Sim/Tgo and Trh/Tgo bHLH-PAS proteins, which significantly bind only ACGTG. This broad specificity of Dys/Tgo is identical to that for mammalian Nxf/Arnt, suggesting that the broad specificity has functional significance. Expression of the CG13196 gene in tracheal fusion cells requires dys function, which suggested that it might be a direct Dys/Tgo target gene. Using germ line transformation, we identified a 1.0-kb fragment of CG13196 that drives expression of a reporter gene in fusion cells identically to the endogenous gene. This fragment had ACGTG, GCGTG, and TCGTG sites, and these sites were mutated and tested for their potential role in regulating fusion cell transcription. Mutation of the TCGTG sites abolished transcription, whereas mutation of either ACGTG or GCGTG did not. Additional evidence that Dys/Tgo binds TCGTG in vivo emerged from results showing that a multimerized 4X-TCGTG transgene was expressed exclusively in tracheal fusion cells in a dys-dependent manner. Consequently, we conclude that Dys/Tgo binds TCGTG sequences in vivo to regulate CG13196 gene expression. All 12 sequenced Drosophila species have at least 2 TCGTG sequences in the homologous intergenic regions between CG13196 and Buffy exon 2. Interestingly, all 12 species have a GCGTG sequence adjacent to the TCGTG sequences, whereas ACGTG sequences are often absent. Thus, although TCGTG has been shown to play an in vivo role as a Dys/Tgo binding site and GCGTG is not required, the sequence conservation suggests that GCGTG sequences could still play an accessory role in CG13196 regulation.
Binding Site Specificities of bHLH-PAS Proteins
To a partial extent, the ability of bHLH and bHLH-PAS proteins to regulate different target genes depends on their DNA binding site specificities. Considerable insight into this issue has been gained from mutational and structural analyses of bHLH proteins. However, no mutational or structural data exists for Dys/Tgo or Nxf/Arnt, so correlating the unique DNA binding specificity of Dys and Nxf to specific amino acid residues within the basic region is only speculative at this point. In addition, it is possible that protein sequences outside the basic regions, such as the HLH and PAS domains, may influence DNA binding specificity (25). Nevertheless, there are several features of the Dys basic region worth noting.
Table 1 shows the basic regions of selected bHLH-PAS proteins whose DNA binding specificities are known. The Arg-12 residue, which is the only residue conserved in all the basic regions shown, dictates the C at position −1 of the binding site (−2NC|GTG3), which is also a conserved element of all of the half-sites (22). The Dys and Nxf binding regions are identical at 9/10 residues from residues 3 to 12 and are quite divergent from the Sim/Hif-1α/Trh and Ss/Ahr subgroups. The Caenorhabditis elegans C15C8.2 Dys/Nxf ortholog also shares 8/10 residues from residues 3–12. This Dys/Nxf sequence conservation likely contributes to their binding to TC, unique among bHLH-PAS proteins and their similar affinities for AC and GC. Some of these conserved amino acid residues are shared with other bHLH-PAS proteins. The Sim/Hif-1α/Trh subgroup proteins all show identity to Dys/Nxf at Ala-7 and Arg-11 in addition to Arg-12. This identity could contribute to the Dys/Nxf AC specificity. It has been shown that both Ala-7 and Arg-11 amino acids are required for the binding of Hif-like factor-Arnt to ACGTG (26). The Ss/Ahr basic regions share identity with Dys/Nxf at Ser-8 and Lys-9 in addition to Arg-12. This could contribute to the GC binding specificity of Dys/Nxf, and all three residues are required for Ahr DNA binding (27). Thus, the Dys/Nxf basic region may represent a hybrid protein structure that combines recognition elements both unique and similar to those of other basic regions to bind a variety of DNA sequences.
Dys Regulation of Fusion Cell Transcription
The dys gene is expressed in all tracheal fusion cells and is required for proper branch fusion in all branches except the dorsal trunk. Genetic analysis has shown that dys function is required for transcription of four genes (CG13196, CG15252, mbo, and shg) and down-regulates levels of Trh protein but not trh RNA (6, 11). Other fusion-expressed genes are not regulated by dys, although some of these, including dys, are regulated by the Escargot zinc finger protein (28, 29). The work described in this paper indicates that Dys/Tgo acts as a transcriptional activator and directly regulates CG13196 expression. Thus, it is also possible that Dys/Tgo directly regulates CG15252, mbo, and shg. Analysis of the sequence of these genes indicates that all three have multiple TCGTG elements that could bind Dys/Tgo, although the expansive intergenic sequences flanking CG15252 and shg make bioinformatic identification of relevant fusion cell enhancers challenging. More promising for future analysis is the D. melanogaster mbo gene, which has no introns and is closely wedged between Cyp313a4 and CG6188. It has two TCGTG sequences and a single ACGTG in its 409-bp 3′-flanking region. Closely related species of the D. melanogaster group have at least one of the TCGTG sequences present, often accompanied by an ACGTG sequence, consistent with a role of TCGTG in Dys-mediated transcription. However, the more distantly related Drosophila pseudoobscura and Drosophila persimilis species have no TCGTG sequences in either the 5′ or 3′ intergenic regions. Future transgenic and mutational work identifying mbo fusion cell enhancers in D. melanogaster and the more distantly related D. pseudoobscura and D. persimilis species will be necessary to understand how this gene is regulated by Dys.
In contrast, the negative regulation of trh is unlikely to be direct, since only Trh protein levels, but not RNA levels, are reduced in dys mutants. Thus, Dys/Tgo is proposed to activate transcription of a gene(s) that encodes a protein involved in translation or decay of Trh protein. Consequently, Dys/Tgo may only be able to activate transcription, similar to Sim/Tgo (21). Genetic evidence also suggests that Ss/Tgo, Sima/Tgo, and Trh/Tgo are generally, if not exclusively, transcriptional activators (2, 5). The dys target genes include two proteins involved in cell adhesion (shg, CG13196), one that is in nuclear protein export (mbo) and another that is possibly a cytoskeletal component (CG15252). In addition, dys may regulate expression of a gene that controls Trh levels post-transcriptionally. This argues that Dys target genes constitute a diverse family of genes, consistent with multiple roles in tracheal migration and morphogenesis (11). It will be important to identify additional Dys/Tgo target genes and determine whether they function in related cellular processes.
The results of dys misexpression data indicate that additional factors can influence Dys/Tgo gene activation and also reveal a temporal aspect of fusion cell gene expression. Two aspects of dys expression and function during embryogenesis stand out. One is that dys is expressed in a relatively small, but diverse group of embryonic cell types (6). These include tracheal fusion cells, anal pad, foregut atrium, brain subset, and leading edge cells. Despite their diversity, one feature of dys expression in these cell types is that its expression appears rather late in embryogenesis, beginning at stage 12. Thus, dys activation of target gene expression, although widespread spatially, is restricted temporally. When dys is expressed ectopically in most tracheal cells, it can activate CG13196 expression in all of these cells at stage 15 or later but not earlier at stages 11–14. Ectopic expression of dys in epidermal stripes and mesoderm also results in widespread, but not uniform, expression at stages 15 and later but not earlier. These temporal and spatial restrictions are unlikely to be due to restricted function of Tgo, since tgo is ubiquitously expressed, and experiments with other bHLH-PAS proteins have demonstrated that Tgo functions similarly in most, if not all, cell types throughout development (5, 16). Thus, there may be factors in tracheal cells that are absent or at lower levels in other cell types that account for the enhanced ability of Dys/Tgo to activate transcription of CG13196 in trachea and additional factors or chromatin states that allow Dys/Tgo to activate transcription throughout the embryo at later stages of development but not earlier stages.
Another unusual result is the occurrence of GFP expression in only posterior fusion cells of P[4X-TCGTG-GFP] embryos. GFP was not detected in anterior fusion cell units or other Dys-positive embryonic cell types. Segmentally different Dys protein levels are unlikely to explain the GFP differences. Levels of Dys protein are generally higher in fusion cells than in leading edge, brain, and atrial foregut cells, but levels in the anal pad are comparable to fusion cells (6). In addition, levels of Dys in anterior fusion cells are comparable to posterior fusion cells as are levels of dys target gene expression (e.g. CG13196). Differences are also unlikely to be due to the timing of detectable GFP accumulation, since anterior GFP expression was still absent even in first instar larvae. Anterior repression is unlikely to be due to silencing by the pH-Stinger vector, since the P[1.0-CG13196-GFP] pH-Stinger-based transgenes were expressed in all fusion cells. Nor is it likely that the non-TCGTG sequences present in the 96-bp 4×-TCGTG fragment mediates segmental differences, since the same fragment containing ACGTG showed CNS midline and tracheal expression in all segments (2). Consequently, anterior expression of Dys target genes may require a DNA binding factor in addition to Dys/Tgo, whose binding site is absent from the P[4X-TCGTG-GFP] reporter transgene.
The Dys and Nxf proteins have unique basic region sequences. Not surprisingly, they also show unique DNA binding specificities. Using identical assays, we showed that Sim/Tgo and Trh/Tgo only bind ACGTG in transient transfection experiments (and also probably in vivo), but Dys/Tgo and Nxf/Arnt strongly bind TCGTG, ACGTG, and GCGTG. One important question concerns the functional significance of this binding site promiscuity. Our data currently leave this issue unresolved. Because identical binding specificities are conserved between Drosophila Dys/Tgo and mammalian Nxf/Arnt, this argues that the promiscuity is biologically important. However, our in vivo mutational analysis of CG13196 provided evidence for a requirement of TCGTG for fusion cell gene expression but not other NCGTG sequences. However, these results demonstrated that ACGTG and GCGTG were not required by themselves (i.e. sufficient); they did not rule out that either site might contribute to gene activation with each other or with TCGTG sequences. The ACGTG and GCGTG sites could also play a more prominent role in regulating other fusion cell target genes of Dys/Tgo or controlling expression in other cell types.
Could the different binding sites for Dys/Tgo or Nxf/Arnt allow competitive or synergistic interactions with other bHLH-PAS or bHLH proteins that use ACGTG or GCGTG binding sites? This is an attractive possibility, but no evidence currently exists to support it. In Drosophila, Dys is expressed in several non-tracheal embryonic sites, including the leading edge, anal pad, foregut atrium, and brain cells. Yet, the presence in most of these cell types does not obviously overlap with other bHLH-PAS proteins. The exception is the anal pad, in which sim is also expressed (30). Dys and Trh are both expressed in tracheal fusion cells, but Dys levels increase during development, whereas Trh levels decrease. It is possible that Dys could regulate expression of some Trh target genes, as Trh becomes increasingly unable to do so, via the ACGTG sites in these genes. Another possibility is that Dys and Sima could influence the tracheal transcriptional output of each other under conditions of hypoxia, since Sima influences tracheal gene expression under hypoxic conditions (31, 32). Similarly, Nxf and Sim2 have been proposed to be co-expressed in subsets of mouse hippocampal cells (7). However, in this case, it is proposed that Nxf levels are negatively regulated by direct repression by Sim2, not that they act together. Although the conserved binding site specificities of Dys/Tgo and Nxf/Arnt are intriguing, functional significance awaits direct in vivo tests and a greater appreciation of the biological functions of these proteins.
Acknowledgments
We thank Cam Patterson for providing human LE-PAS (Nxf) and Arnt clones and Joe Kearney for helpful comments on the manuscript.
Footnotes
This project was funded by a grants from the National Science Foundation (Developmental Mechanisms) and the National Center for Research Resources, National Institutes of Health (to S. T. C.).
The abbreviations used are: bHLH-PAS, basic-helix-loop-helix-PAS; Act, actin5C; Arnt, aromatic hydrocarbon nuclear translocator; btl, breathless; dys, dysfusion; en, engrailed; GFP, green fluorescent protein; HA, hemagglutinin; Hif-1α, hypoxia-inducible factor-1α; lacZ, β-galactosidase; luc, luciferase; mbo, members only; sca, scabrous; shg, shotgun; sim, single-minded; sima, similar; ss, spineless; tgo, tango; trh, trachealess; twi, twist; vvl, ventral veinless; kb, kilobase; Ahr, aryl hydrocarbon receptor.
REFERENCES
- 1.Crews ST, editor. PAS Proteins: Regulators and Sensors of Development and Physiology. Norwell, MA: Kluwer Academic Publishers; 2003. [Google Scholar]
- 2.Sonnenfeld M, Ward M, Nystrom G, Mosher J, Stahl S, Crews S. Development. 1997;124:4583–4594. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- 3.Ohshiro T, Saigo K. Development. 1997;124:3975–3986. doi: 10.1242/dev.124.20.3975. [DOI] [PubMed] [Google Scholar]
- 4.Zelzer E, Shilo B. Mech. Dev. 2000;19:163–173. doi: 10.1016/s0925-4773(99)00295-6. [DOI] [PubMed] [Google Scholar]
- 5.Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, Crews ST. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L, Crews ST. Mol. Cell. Biol. 2003;23:5625–5637. doi: 10.1128/MCB.23.16.5625-5637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Mol. Cell. Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell-Coffman JA. In: PAS Proteins: Regulators and Sensors of Development and Physiology. Crews ST, editor. Norwell, MA: Kluwer Academic Publishers; 2003. pp. 51–68. [Google Scholar]
- 9.Moser M, Knoth R, Bode C, Patterson C. Brain Res. Mol. Brain Res. 2004;128:141–149. doi: 10.1016/j.molbrainres.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Hester I, McKee S, Pelletier P, Thompson C, Storbeck C, Mears A, Schulz JB, Hakim AA, Sabourin LA. Brain Res. 2007;1135:1–11. doi: 10.1016/j.brainres.2006.11.083. [DOI] [PubMed] [Google Scholar]
- 11.Jiang L, Crews ST. Mol. Cell. Biol. 2006;26:6547–6556. doi: 10.1128/MCB.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han K, Levine MS, Manley JL. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 13.Wharton JKA, Franks RG, Kasai Y, Crews ST. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman EC, Reyes H, Chu F, Sander F, Conley LH, Brooks BA, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 15.Barolo S, Carver LA, Posakony JW. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- 16.Ward MP, Mosher JT, Crews ST. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- 17.Manning L, Doe CQ. Development. 1999;126:2063–2071. doi: 10.1242/dev.126.10.2063. [DOI] [PubMed] [Google Scholar]
- 18.Patel NH, Snow PM, Goodman CS. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- 19.Kim MD, Jan LY, Jan YN. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crews ST, Brenman JE. Genes Dev. 2006;20:2773–2778. doi: 10.1101/gad.1487706. [DOI] [PubMed] [Google Scholar]
- 21.Estes P, Mosher J, Crews ST. Dev. Biol. 2001;232:157–175. doi: 10.1006/dbio.2001.0174. [DOI] [PubMed] [Google Scholar]
- 22.Dang CV, Dolde C, Gillison ML, Kato GJ. Proc. Natl. Acad. Sci. U. S. A. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jazwinska A, Affolter M. Gene Expr. Patterns. 2004;4:413–421. doi: 10.1016/j.modgep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Q, Halfon M. Fly. 2007;1:55–56. doi: 10.4161/fly.3892. [DOI] [PubMed] [Google Scholar]
- 25.Chapman-Smith A, Whitelaw ML. J. Biol. Chem. 2006;281:12535–12545. doi: 10.1074/jbc.M512145200. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita K, Kikuchi Y, Sasakura Y, Suzuki M, Fujii-Kuriyama Y, Sogawa K. Nucleic Acids Res. 2004;32:3169–3179. doi: 10.1093/nar/gkh637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong L, Ma Q, Whitlock JP., Jr J. Biol. Chem. 1996;271:7942–7948. doi: 10.1074/jbc.271.14.7942. [DOI] [PubMed] [Google Scholar]
- 28.Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Development. 1996;122:3531–3536. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Development. 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- 30.Pielage J, Steffes G, Lau DC, Parente BA, Crews ST, Strauss R, Klambt C. Dev. Biol. 2002;249:283–299. doi: 10.1006/dbio.2002.0770. [DOI] [PubMed] [Google Scholar]
- 31.Lavista-Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, Bocca SN, Muzzopappa M, Ratcliffe PJ, Wappner P. Mol. Cell. Biol. 2002;22:6842–6853. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorr TA, Gassmann M, Wappner P. J. Insect Physiol. 2006;52:349–364. doi: 10.1016/j.jinsphys.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Manning G, Krasnow MA. In: The Development of D. melanogaster. Bate M, Martinez Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 609–685. [Google Scholar]