Abstract
In vitro, high-risk human papillomavirus E6 proteins have been shown, in conjunction with E6-associated protein (E6AP), to mediate ubiquitination of p53 and its degradation by the 26S proteasome by a pathway that is thought to be analogous to Mdm2-mediated p53 degradation. However, differences in the requirements of E6/E6AP and Mdm2 to promote the degradation of p53, both in vivo and in vitro, suggest that these two E3 ligases may promote p53 degradation by distinct pathways. Using tools that disrupt ubiquitination and degradation, clear differences between E6- and Mdm2-mediated p53 degradation are presented. The consistent failure to fully protect p53 protein from E6-mediated degradation by disrupting the ubiquitin-degradation pathway provides the first evidence of an E6-dependent, ubiquitin-independent, p53 degradation pathway in vivo.
Keywords: ubiquitination, p53, human papillomavirus, E6, degradation, proteasome
Introduction
p53 is a tumor suppressor protein that is activated by a wide variety of cellular stresses. During periods of cellular stress wild-type full-length p53 acts as a transcription factor, involved in both transactivation and transrepression of target genes. This leads to either cell cycle arrest or apoptosis preventing any genomic aberrations from being passed on to daughter cells which could ultimately promote cancer, reviewed by Harris and Levine (2005).
During periods where there are no stressful stimuli, the transcriptional activity of p53 is latent and the cell is free to progress through its normal cycle (Hupp et al., 1992). The levels of p53 are tightly controlled at this time primarily through degradation by the ubiquitin-proteasome system. The principal endogenous E3 ligase involved was identified as Mdm2 (Haupt et al., 1997; Honda et al., 1997).
The ubiquitin–proteasome pathway is involved in many diverse functions, including cell cycle regulation, signal transduction and transcription, but was originally described for its role in the degradation of cellular proteins. Although ubiquitination is not required for the degradation of all proteins by the 26S proteasome (Li and Coffino, 1996b), the ubiquitin-proteasome system is the usual pathway.
Ubiquitination is a multistep process that initially involves the formation of a thioester bond between a cysteine residue of the E1 ubiquitin-activating enzyme and the terminal glycine residue of ubiquitin. The ubiquitin moiety is then transferred to a ubiquitin-conjugating E2 enzyme which in turn covalently links the ubiquitin molecule to the α-amino group of a lysine residue in the target protein. In most cases, the final linkage requires the activity of an E3 ligase that is required for substrate specificity. Successive rounds of ubiquitination allow the formation of polyubiquitin chains which target the protein for degradation by the 26S proteasome (reviewed by Glickman and Ciechanover, 2002).
Human papillomavirus (HPV) infection is a major risk factor in the development of anogenital cancers with HPV DNA present in more than 90% of cervical cancers, the second most common cancer in women worldwide. ‘High risk’ HPV types include HPV-18, HPV-16, HPV-45, HPV-33 and HPV-56, and are strongly associated with cervical cancer, whereas ‘low risk’ types are generally associated with genital warts (for a review see zur Hausen, 2000). As the viral DNA integrates into the host cell genome, the expression of two viral proteins, E6 and E7, is enhanced disturbing the normal terminal differentiation process of cervical cells. E7 binds the retinoblastoma gene product, whereas E6 mediates the degradation of p53, among other functions. As a result, crucial cell cycle checkpoints are compromised an leading to transformation of the host cells (zur Hausen, 2000).
E6 binds to two distinct sites on p53 (Lechner and Laimins, 1994; Li and Coffino, 1996a): one in the C-terminus (residues 376-384) and one in the DNA-binding domain (residues 66-326) (Li and Coffino, 1996a). The binding of E6 to the DNA-binding domain of p53 requires the co-interaction of E6-associated protein (E6AP), an endogenous cellular E3 ligase that has no effect on p53 in the absence of E6. This E6AP-dependent interaction is thought to mediate proteasome-dependent p53 degradation by promoting the ubiquitination of p53 (Scheffner et al., 1990, 1993). The interaction between E6 and the C-terminus of p53 does not involve E6AP and the outcome of this interaction is currently unknown.
The pathway by which E6 mediates the degradation of p53 has been fully dissected in vitro and it has been clearly demonstrated that E6, E6AP, ubiquitin, E1 and E2 enzymes are all required (Huibregtse et al., 1993; Scheffner et al., 1993, 1994). There is no doubt that the ubiquitination cascade is of primary importance in E6-mediated p53 degradation in vitro. However, such in-depth analysis of the degradation pathway has not been performed in vivo.
Here we show that in addition to the previously established ubiquitin-dependent pathway, E6 mediates the degradation of p53 by a pathway that is independent of ubiquitination in vivo. Combined, the two pathways lead to extremely efficient degradation of p53.
Results
E6 promotes the ubiquitination of p53 in vitro
Previous work demonstrating that E6 promotes the degradation of p53 through the ubiquitination pathway has been carried out using proteins in vitro translated in rabbit reticulocyte lysate (Huibregtse et al., 1993; Scheffner et al., 1993, 1994). To re-confirm these findings, we performed an in vitro translation experiment in which 35S labelled p53, in the absence or presence HPV-16 E6, was incubated in the presence of an increasing concentration of purified methylated ubiquitin. This ubiquitin is methylated on all lysine residues so that it is unable to form poly-ubiquitin chains. The addition of purified methylated ubiquitin to the reticulocyte lysate, therefore, promotes the premature termination of chains of endogenous wild-type (WT) ubiquitin leading to a bias towards lower molecular weight ubiquitinated p53 species. In the absence of any additional ectopic proteins involved in the ubiquitination pathway, E6 efficiently promotes the degradation of p53 in rabbit reticulocyte lysate with ubiquitination visible as a faint smear of high molecular weight species at the top of the gel (Figure 1, lanes 1 and 2). This pattern is consistent with poly-ubiquitination, which is the type of ubiquitination E6 has been reported to promote (Scheffner et al., 1993). In the presence of increasing concentrations of purified methylated ubiquitin, a ladder of multiple mono-ubiquitinated p53 species is detected as the methylated ubiquitin is incorporated into the chains (Figure 1, lanes 3-6). In parallel with the shift from poly- to multiple mono-ubiquitination, there is a concurrent increase in the levels of unmodified p53. This confirms that E6 does promote the ubiquitination of p53 in vitro and also suggests that the ubiquitination is required for E6-dependent p53 degradation, in complete agreement with the literature. The lower band of p53 detected corresponds to a previously described isoform of p53 produced by internal initiation of translation from Codon 40 (termed p47 or DeltaN-p53) (Courtois et al., 2002; Yin et al., 2002; Ghosh et al., 2004). E6 protein is visible and is not ubiquitinated in this assay.
Figure 1.
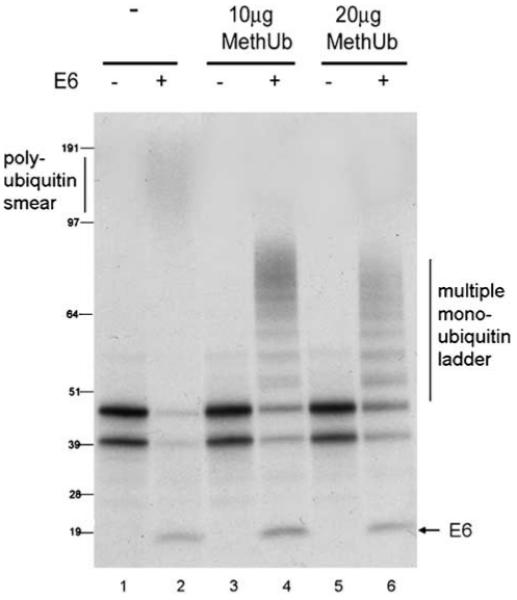
E6 mediates ubiquitination of p53 in vitro. p53, E6 or empty vector control (all under a T7 promoter) were translated separately in rabbit reticulocyte lysate in the presence of [35S]methionine. Translated p53 and pcDNA3 (lanes 1, 3 and 5) or E6 (lanes 2, 4 and 6) were mixed in the absence (lanes 1 and 2) or presence of 10 μg (lanes 3 and 4) or 20 μg (lanes 5and 6) purified methylated ubiquitin (MethUb) and further incubated at 25°C for 90 min.
Unlike the situation in vitro, we have previously shown that, in contrast to Mdm2, E6-mediated ubiquitination of p53 is extremely difficult to detect in vivo (Camus et al., 2003). An explanation for this was recently provided by Stewart et al. (2005), who demonstrated that E6-mediated ubiquitinated p53 species could only be detected in nuclear fractions. The overall ubiquitinated p53 levels detected in whole-cell extracts were significantly lower in the presence of E6 compared with Mdm2. However, in nuclear extracts ubiquitinated p53 levels were approximately equivalent in the presence of E6 compared with Mdm2 (Stewart et al., 2005). The authors provide the first definitive evidence that E6-mediated ubiquitination of p53 does occur in vivo as well as a possible explanation for why it had previously been difficult to detect. The reason why the level of total cellular ubiquitinated p53 is much lower in the presence of E6 compared with Mdm2, however, still remains unclear.
To compare E6- and Mdm2-mediated p53 degradation in vivo, it was first necessary to determine the ratio of E6:p53 that promotes efficient and approximately equivalent levels of p53 degradation to that seen with Mdm2. Supplementary Figure 1 shows a titration of E6 or Mdm2 against a constant level of p53 transfected in HPV-negative H1299 cells. All subsequent experiments were performed at a p53:E6/Mdm2 ratio of 1:2 (Supplementary Figure S1, lanes 4 and 9). Importantly, the E6-mediated degradation of p53 is not saturated at this ratio and therefore E6AP is not a limiting factor.
Ubiquitin that is unable to form chains partially protects p53 from E6-mediated degradation in vivo
A similar approach to using methylated ubiquitin in vitro was employed to investigate the importance of ubiquitination in the in vivo pathway. A vector expressing His6-tagged ubiquitin with each of its lysine residues mutated to arginine, 7KR-ubiquitin, was titrated against a single concentration of p53 in the presence or absence of either E6 or Mdm2 (Figure 2). As 7KR-ubiquitin has no lysine residues, it is unable to support chain formation and therefore initiates premature termination of ubiquitin chains analogous to the methylated ubiquitin used in vitro in Figure 1. All of the proteins were ectopically expressed in H1299 cells, ubiquitinated species were purified through the His-tag using Nickel chelated beads and p53 was detected using anti-p53 antibody DO-1. It is clear that, at the highest concentration of 7KR-ubiquitin transfected, p53 is almost completely protected from Mdm2-mediated degradation (Figure 2a, lanes 3-6). However, at the same concentration, p53 is only partially protected from E6-mediated degradation (Figure 2b, lanes 3-6). No overall increase in p53 protein levels is observed in the absence of an ectopic E3 ligase with any concentration of 7KR-ubiquitin (Figure 2c, lanes 3-5).
Figure 2.
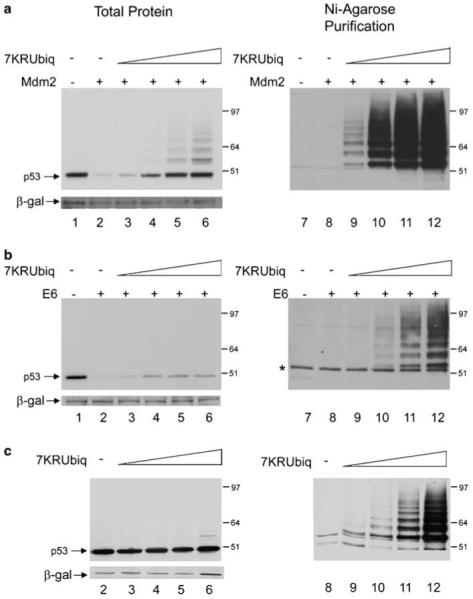
7KR-ubiquitin only partially protects p53 from E6-mediated degradation. (a) H1299 cells were transfected with 1 μg p53 together with 2 μg Mdm2 (lanes 2-6 and 8-12) or control pcDNA3 vector (lanes 1 and 7). Co-expression of a titration of 5 μg (lanes 3 and 9), 10 μg (lanes 4 and 10), 15 μg (lanes 5 and 11) or 20 μg (lanes 6 and 12) His6-tagged 7KR-ubiquitin was performed. An Ni-agarose purification of His6-tagged 7KR-ubiquitinated species is shown in the right panel. (b) As in (a) but with E6 instead of Mdm2. (c) As in (a) (minus lanes 1 and 7) but with pcDNA3 instead of Mdm2. DNA concentrations are maintained with empty pcDNA3 control vector and β-gal is used as a loading and transfection-efficiency control throughout. p53 is detected with DO-1 antibody.
The density of the p53 bands in Figure 2 was measured using the computer software QuantityOne. The difference in the level of protection of p53 from degradation provided by 7KR-ubiquitin in the presence of E6 compared with Mdm2 is represented on a graph of the percentage protection of p53 levels against concentration of 7KR-ubiquitin transfected. At the highest concentration of 7KR-ubiquitin, the p53 levels were 100% protected from degradation (compared with the control, no-ectopic E3 ligase, levels), whereas at the same concentration of 7KR-ubiquitin only just over 40% of protection from E6-mediated degradation was detected (Supplementary Figure S2, graph A).
Importantly, at the highest concentration of 7KR-ubiquitin, there is a substantial increase in the detection of ubiquitinated p53 species in the presence of E6 (Figure 2, lane 12). At this concentration, in the presence of E6, the ubiquitinated p53 species detected are of a higher molecular weight than those detected in the presence of Mdm2 (Figure 2, compare a and b, lane 12). This would support that E6 promotes high molecular weight poly-ubiquitination of p53, and that these high molecular weight species are shifted down to form a ubiquitination ladder only in the presence of high levels of 7KR-ubiquitin. The difference in ubiquitin pattern promoted by E6 compared with Mdm2 indicates that the ubiquitination detected in the presence of E6 is E6-dependent, confirming previous reports that E6/E6AP can utilize this ubiquitin mutant as a substrate (Glockzin et al., 2003) and that the ubiquitinated p53 species detected are not promoted by the endogenous Mdm2 present in these cells. To further ensure that Mdm2 is not responsible for any of the effects seen, the experiments were repeated in p53 and Mdm2 double knockout cells, in which the same ubiquitination pattern and protection of p53 protein levels was observed (data not shown).
The protection of p53 from Mdm2-mediated degradation by 7KR-ubiquitin reaffirms that poly-ubiquitination is necessary for this process as blocking the formation of poly-ubiquitin chains reduces the efficiency of degradation. The fact that p53 was partially protected from E6-mediated degradation by high concentrations of 7KR-ubiquitin indicates that ubiquitination also plays at least a partial role in this process. To ensure that the short ubiquitin-chains produced in the presence of high concentrations of 7KR-ubiquitin could not be utilized as a substrate for degradation by E6/E6AP, the highest concentration of 7KR-ubiquitin was co-transfected with p53 and E6 or Mdm2 in the presence and absence of proteasome inhibitor MG132. As expected, MG132 caused no further increase in the levels of p53 in the presence of Mdm2 and high levels of 7KR-ubiquitin as the ubiquitin mutant alone protects p53 completely from Mdm2-mediated degradation (Figure 3a). However, in the presence of E6 MG132 brings the total p53 protein levels back up to the control (no ectopic E3-ligase) level (Figure 3b, left panel). Importantly, despite the increase in total p53 protein levels, MG132 does not lead to the further accumulation of E6-mediated ubiquitinated p53 species (Figure 3b, right panel), indicating that these species are not subject to degradation by the 26S proteasome. The density of the bands was measured, as before, and a graph comparing the level of protection of p53 levels from E6- and Mdm2-mediated degradation under each condition is shown (Supplementary Figure S2, graph B).
Figure 3.
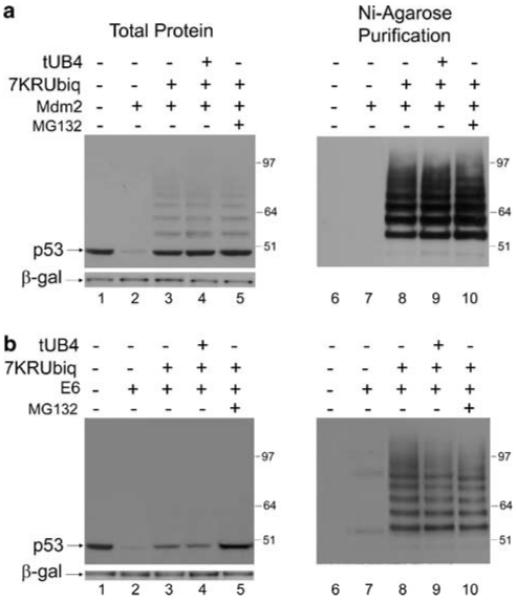
Effects of 7KR-ubiquitin and tUB4 on E6- and Mdm2-mediated degradation of p53. (a) H1299 cells were transfected with 0.5 μg p53 together with 1 μg Mdm2 (lanes 2-6), or control pcDNA3 vector (lane 1), as above. Twenty micrograms 7KR-ubiquitin was co-expressed in lane 3 and 20 μg 7KR-ubiquitin plus 20 μg tUB4 was co-expressed in the absence (lane 4) or presence (lane 5) of MG132, incubated for 3 h before harvesting. DNA concentrations were maintained with empty pcDNA3 control vector. (b) As in (a) but with E6 instead of Mdm2.
Blocking ubiquitin-recognition by the proteasome leads to a partial protection of p53 from E6-mediated degradation
A possible complication in the interpretation of the results obtained by overexpression of the 7KR-ubiquitin arises because ubiquitination is a reversible process. In order for the protection of p53 from degradation to be comparable in the presence of E6 versus Mdm2, it must be assumed that any de-ubiquitination of 7KR-ubiquitin from p53 occurs with a similar efficiency in the presence of each E3 ligase. Although Blattner and co-workers show that both E6- and Mdm2-mediated ubiquitinated p53 species can be substrates for de-ubiquitination (Glockzin et al., 2003) there is not enough evidence provided (either by us or in the literature) to show that these pathways are comparable. It was therefore important to disrupt the ubiquitin-dependent degradation pathway at different stages in order to substantiate the reliability of the interpretations drawn. Thus, the impact of blocking ubiquitin-recognition by the proteasome on p53 degradation was investigated. A vector expressing a tandem uncleavable chain of four ubiquitin molecules (tUB4), joined together head-to-tail, was used to analyse its effect on Mdm2- and E6-mediated p53 degradation. The tandem ubiquitin chain cannot itself be conjugated to substrates because the final glycine residue has been mutated to a valine residue and therefore does not affect the efficiencies of either the ubiquitination or the de-ubiquitination pathways. tUB4 has been shown to inhibit the degradation of ubiquitinated proteins by binding to the 19S subunit of the proteasome and blocking the access of ubiquitinated substrates (Saeki et al., 2004), thereby effectively acting as a proteasome inhibitor that is specific for ubiquitin-dependent degradation pathways. The catalytic activity of the 20S core subunit is not affected by the chain, allowing substrates that do not require ubiquitination to be degraded normally (Saeki et al., 2004). Like 7KR-ubiquitin, tUB4 was also found to completely inhibit Mdm2-mediated degradation but found to only partially inhibit E6-mediated degradation of p53.
A titration of tUB4 in the presence of a constant concentration of p53 and either E6 or Mdm2 was performed in H1299 cells. Similar to the results obtained with the 7KR-ubiquitin titrations, in the presence of Mdm2 p53 protein levels were completely recovered by the expression of high levels of tUB4 (Figure 4a, lanes 3-6). In contrast, the highest concentration of tUB4 transfected only partially recovered p53 levels in the presence of E6 (Figure 4b, lanes 3-6). No increase in p53 levels was observed in the absence of an ectopic E3 ligase (Figure 4c). Measurement of the band density was also performed and, similar to the results with 7KR-ubiquitin, protection of p53 levels from Mdm2-mediated degradation by high levels of tUB4 was complete, whereas protection from E6-mediated degradation was only approximately 50% (Supplementary Figure S2, graph C). As the tUB4 chains have been shown to bind to, and block, ubiquitin recognition motifs on the proteasome, thereby inhibiting degradation of poly-ubiquitinated substrates (Saeki et al., 2004), the partial protection of p53 from E6-mediated degradation obtained upon expression of tUB4 suggests at least a partial involvement of ubiquitin-recognition (and therefore ubiquitination) in E6-mediated p53 degradation.
Figure 4.
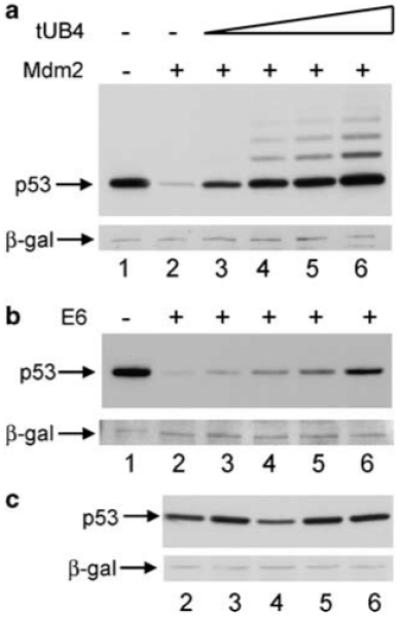
Tandem chains of four ubiquitin moieties only partially protect p53 from E6-mediated degradation. (a) H1299 cells were transfected with 0.5 μg p53 together with 1 μg Mdm2 (lanes 2-6) or control pcDNA3 vector (lane 1), as above. Co-expression of a titration of 5 μg (lane 3), 10 μg (lane 4), 15 μg (lane 5) or 20 μg (lane 6) pcDNA3 tUB4 was performed. (b) As in (a) but with E6 instead of Mdm2. (c), As in (a) (minus lane 1) but with pcDNA3 instead of Mdm2.
As with 7KR-ubiquitin, this result indicates either that ubiquitination only plays a partial role in E6-mediated degradation of p53 or that the pathway by which E6 promotes the degradation of p53 is too efficient to be disrupted by the tools used. It is unlikely that the latter is true as concentrations of E6 and Mdm2 were chosen that promote the degradation of p53 with approximately equal efficiency (Supplementary Figure S1) and the Mdm2-mediated p53 degradation pathway is severely disrupted by each tool employed. To further address this, the effect of co-transfecting the maximum concentration of 7KR-ubiquitin along with the maximum concentration of tUB4 chains was also investigated. Under these conditions, there is no further increase in the level of p53 in the presence of either E6 or Mdm2 compared with expression of 7KR-ubiquitin alone (Figure 3a and b, lane 4 and Supplementary Figure S2, graph B). This indicates that the level of protection conferred by 7KR-ubiquitin alone is already at the point of saturation and no additional protection of p53 from degradation can be achieved by further blocking the ubiquitination pathway.
However, as p53 and E6 or Mdm2 are overexpressed in these experiments, it is unclear whether the super-physiological levels of these proteins could affect the outcome through, for example, saturation of components of the ubiquitin-proteasome pathway. As there are no antibodies available that can detect E6 protein levels, we carried out real-time polymerase chain reaction to compare the mRNA levels of p53 and E6 in our transfection system with the levels of p53 and E6 mRNA found in HPV-16 positive cell lines. A titration of p53 and E6, transfected in H1299 cells, was performed maintaining the 1:2 ratio of p53:E6 used in our experiments. At the lowest level transfected (0.25 μg p53:0.5 μg E6), the mRNA levels of E6 are within the range of those detected in the HPV-positive cells and the p53 mRNA levels are approximately 1.5-2.7 times higher than those found in SiHa and Caski cells, respectively (Supplementary Figure S3, graph A). In addition, the p53:E6 ratio of 1:2 used in our transfection conditions is within the range of the p53:E6 mRNA ratio found in Caski and SiHa cells (Supplementary Figure S3, graph B), suggesting that the ratio chosen is physiologically relevant. Because the level of p53 mRNA was higher than the physiological levels detected in HPV-positive cells, the 7KR-ubiquitin and tUB4 experiments shown were repeated using a transfection concentration of p53 that was 10 times lower than previously used (a new concentration of 0.1 μg p53:0.2 μg E6), resulting in the same outcome. Figure 5a shows the level of protection that co-transfection of the maximum level of tUB4 (20 μg) has on these low levels of p53 from E6- or Mdm2-mediated degradation. As these data show, when physiological levels of p53 and E6 are used in these experiments, the level of protection from degradation conferred by high levels of tUB4 is still only approximately 50% from E6-compared with total protection from Mdm2-mediated degradation, suggesting that no saturation of the components of the ubiquitin-degradation pathway occurs. The same is true for 7KR-ubiquitin (data not shown). In addition, the transfected H1299 samples were run alongside an equal total protein concentration of HPV-positive HeLa cells and the p53 protein levels detected were comparable to the p53 protein levels detected in the presence of E6 (and Mdm2) in the transfected H1299 cells (Figure 5b), indicating that this concentration of p53 DNA translates into a physiological level of p53 protein.
Figure 5.

The non-canonical pathway of E6-mediated p53 degradation occurs at physiologically relevant levels of p53 and E6 expression. (a) H1299 cells were transfected with 0.1 μg p53 together with 0.2 μg E6 (lanes 2 and 4), Mdm2 (lanes 3 and 5) or control pcDNA3 vector (lane 1). (20 μg) tUB4 was co-transfected with E6 (lane 4) or Mdm2 (lane 5) as indicated. (b) H1299 cells transfected with 0.1 μg p53 in the absence (lane 2) or presence of 0.2 μg E6 (lane 3) or Mdm2 (lane 4) and samples were run beside an extract of the same total protein concentration of HeLa cell lysate (lane 1).
Although these results suggest that the system is robust at physiological levels of p53 and E6 proteins, they do not directly demonstrate that the ubiquitin-independent pathway is actually active in HPV-infected cells. In order to investigate this, a titration of tUB4 was performed in HPV-positive HeLa cells with an MG132 treatment used as a ‘negative’ control in place of the no-ectopic E3 ligase control used in H1299 cells (data not shown). Concurrent with the results observed in transfected H1299 cells, the protection from degradation by high levels of tUB4 was approximately 50% compared with the protection obtained by MG132 treatment, indicating that it is likely that both the ubiquitin-dependent and the ubiquitin-independent pathways are active in HPV-positive cell lines.
E6-mediated p53 degradation is only partially inhibited by inactivation of E1
The consistent failure to protect p53 levels completely from E6-mediated degradation using tools that disrupt ubiquitination or ubiquitin recognition suggests that an alternative degradation pathway may exist in vivo. To further explore this, a murine cell-line expressing a temperature-sensitive E1 enzyme was employed. The cell line has previously been used to demonstrate that the normal cellular turnover of p53 is ubiquitin dependent (Chowdary et al., 1994). This cell line completely inhibits the ubiquitination pathway at the non-permissive temperature, presenting a robust system for analysing the involvement of ubiquitination in p53 degradation that is not subject to any complications that may arise from differences in the efficiencies of the de-ubiquitination pathways that may be active in the presence of Mdm2 and E6. Two sets of temperature-sensitive cells (TS20b) were transiently transfected with p53 in the presence or absence of E6 or Mdm2 and incubated at the permissive temperature (35°C) until 20 h before harvesting. At this point, one set was moved to the non-permissive temperature (39°C). Three hours before harvesting, cells were treated with proteasome inhibitor MG132 or left untreated, as indicated. The parental cell line (H38-5), which expresses the functional E1 enzyme at both temperatures, was transfected in the same way and also moved to 39°C 20 h before harvesting.
In the case of Mdm2, it is clear that p53 is degraded efficiently, with inhibition by MG132, in the TS20b cell line at the permissive temperature (Figure 6a, lanes 5 and 6) and at both temperatures in the parental cell line, which has an active E1 enzyme at both temperatures (Figure 6c), as expected. However, at the non-permissive temperature in the TS20b cell line Mdm2 fails to mediate the degradation of p53 (Figure 6a, lanes 11 and 12), indicating that Mdm2-mediated p53 degradation is ubiquitin dependent. In the case of E6, there is also degradation of p53 in the TS20b cell line at the permissive temperature (Figure 6a, lanes 1 and 2) and in the parental cell line at the non-permissive temperature (Figure 6c, lanes 7 and 8). However, in contrast to Mdm2, at the non-permissive temperature, E6-dependent degradation of p53 is observed (with concurrent inhibition by MG132) in the TS20b cell line, but at a reduced level compared with the level observed at the permissive temperature (Figure 6a, compare lanes 7 and 8 with lanes 1 and 2). Figure 6b shows alternative exposure lengths of the same membrane at each temperature.
Figure 6.
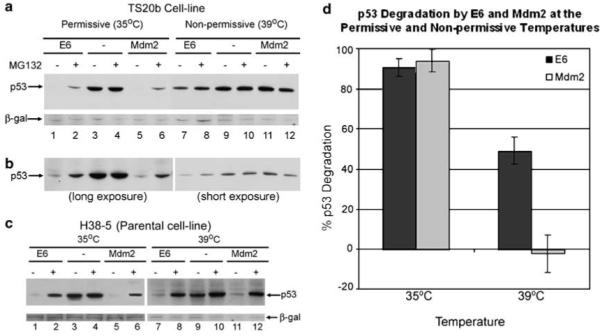
E6-mediated p53 degradation is only partially inhibited by inactivation of E1. (a) TS20b cells were transfected with p53 together with Mdm2 (lanes 5, 6, 11 and 12), E6 (lanes 1, 2, 7 and 8) or pcDNA3 (lanes 3, 4, 9and 10) and left at 35°C (lanes 1-6) or moved to the non-permissive temperature of 39°C (lanes 7-12) 20 h before harvesting. Cells were incubated for 3 h before harvesting with 20 μM MG132 as indicated (lanes 2, 4, 6, 8, 10 and 12). (b) As above but a longer exposure of the 35°C samples (lanes 1-6) and a shorter exposure of the 39°C samples (lanes 7-12). (c) Parental H38-5cells were transfected as in (a) and moved to 39°C 20 h before harvesting. (d) Graph showing degradation of p53 mediated by E6 (dark shaded bars) and Mdm2 (light shaded bars) at the permissive temperature (35°C) and the non-permissive temperature (39°C).
In order to evaluate the difference in p53-degradation mediated by the two E3-ligases, the density of the p53 bands (detected by Western blot) was quantitated by densitometry as used before. The difference between the band intensity in the presence and absence of either E6 or Mdm2 (with no MG132) was determined. A graph was then plotted showing the percentage degradation of p53 in the presence of E6 compared with Mdm2 at 35°C and 39°C (Figure 6d). Whereas Mdm2-mediated p53 degradation is approximately equivalent to the degradation induced by E6 at the permissive temperature, at the non-permissive temperature Mdm2-mediated p53 degradation is completely inhibited. Interestingly, the E6-mediated p53 degradation observed at the non-permissive temperature is approximately half of the degradation observed at the permissive temperature. This decrease in p53 degradation is analogous to the decrease in degradation detected at high concentrations of 7KR-ubiquitin or tUB4 when similarly measured.
There have been previous reports that the temperature sensitive cell line used may have leaky E1 expression after a 16 h incubation at the non-permissive temperature (Salvat et al., 2000). To address this, the Western blot membrane was re-probed with an antibody against ubiquitin (Supplementary Figure S4). Total cellular poly-ubiquitin levels are detected in every lane at the permissive temperature (Supplementary Figure S4, lanes 1-6), whereas at the non-permissive temperature no poly-ubiquitination is detected (Supplementary Figure S4, lanes 7-12). This indicates that at the incubation time used (20 h) there was no detectable ubiquitination at 39°C, suggesting that there was no leaky expression of E1 at the non-permissive temperature. This, combined with the other data presented, strongly advocates the existence of both a ubiquitin-dependent and a ubiquitin-independent E6-mediated p53 degradation pathway.
The C-terminus of p53 is important for the ubiquitin-independent degradation pathway
The function of the E6AP-independent E6-binding site in the C-terminus of p53 is unknown, but it is not required for E6-dependent p53 degradation in vitro (Li and Coffino, 1996a). Removal of the last 30 amino acids of p53 creates a p53 deletion mutant (p53CΔ30) that no longer contains this second E6-binding site. As the C-terminus of p53 has been shown to be largely dispensable for E6-mediated degradation in vivo (Kubbutat et al., 1998) and E6 has been shown to be insensitive to the loss of the six C-terminal lysine residues (also lost in this deletion mutant) (Nakamura et al., 2002; Camus et al., 2003), it was expected that E6 would still be able to degrade this mutant. This is in fact the case, as the levels of p53CΔ30 are reduced in the presence of E6 compared with the empty vector control (Figure 7, compare lanes 1 and 2). To determine whether the C-terminal binding site is important for the ubiquitin-independent degradation of p53, the formation of polyubiquitin chains and ubiquitin recognition were disrupted as before (7KR-ubiquitin and tUB4, respectively). In contrast to WTp53, overexpression of high levels of both 7KR-ubiquitin (Figure 7a) and tUB4 (Figure 7b) completely inhibits E6-mediated degradation of p53CΔ30. The level of inhibition of E6-mediated degradation of p53CΔ30 by high levels of 7KR-ubiquitin and tUB4 is analogous to that observed with WTp53 in the presence of Mdm2. Neither 7KR-ubiquitin nor tUB4 had a significant effect on the overall level of p53CΔ30 in the absence of E6 (data not shown). A graph of the protection of p53CΔ30 levels by tUB4 and 7KR-ubiquitin, determined by densitometry of the Western blot bands, shows a protection of over 97% of the control p53 levels in the presence of high levels of both tUB4 and 7KR-ubiquitin (Supplementary Figure S2, graph D).
Figure 7.
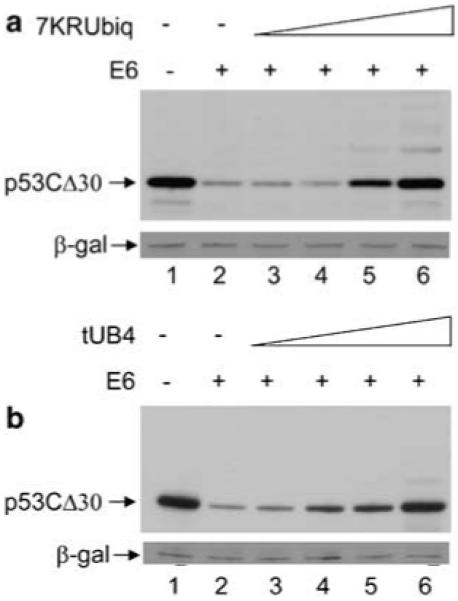
The C-terminus of p53 is important for ubiquitin independent degradation. (a) H1299 cells were transfected with 0.5 μg p53CΔ30 together with 1 μg E6 (lanes 2-6), or control pcDNA3 vector (lane 1). Co-expression of a titration of 5 μg (lane 3), 10 μg (lane 4), 15 μg (lane 5) or 20 μg (lane 6) pcDNA3 7KR-ubiquitin was performed with DNA concentrations maintained with empty pcDNA3 control vector. (b) As in (a) but with tUB4 instead of 7KR-ubiquitin.
The ability of these proteins to completely disrupt E6-mediated degradation of p53CΔ30 but only partially inhibit E6-mediated degradation of WTp53 suggests that the C-terminus of p53 is required for the ubiquitin-independent degradation pathway.
Discussion
Dissection of the E6-mediated p53 degradation pathway in vitro has demonstrated its dependence on ubiquitination. However, a number of differences in the requirements for degradation has been reported between E6-mediated p53 degradation in vitro compared with in vivo. Vousden and co-workers (Crook et al., 1996) have observed that E6-mediated degradation of p53 is sensitive to mutation of certain p53 lysine residues in in vitro degradation assays but that no lysine residue, or group of lysine residues tested, is critical to the in vivo degradation pathway. The existence of an additional ubiquitin-independent degradation pathway in vivo would explain the insensitivity of the E6-mediated degradation pathway to loss of p53 lysine residues. Additionally, E6 mutants have been described that are unable to promote the degradation of p53 in vitro, but that retain the ability in vivo. For example, amino acids 118-122 have been shown to be required for efficient degradation of p53 in vitro, but deletion of these residues has little affect on p53 degradation in vivo (Foster et al., 1994). Furthermore, mutation of E6 residues Cys 63 to serine and Trp 132 to arginine renders E6 defective for p53 degradation in vitro but still able to promote rapid p53 turnover in in vivo degradation assays, prompting the authors to suggest that E6 may mediate p53 loss in vivo through a pathway that is distinct from the pathway utilized in rabbit reticulocyte lysate (Dalal et al., 1996). There are also p53 mutants that show markedly different susceptibilities to E6-induced degradation in vitro compared with in vivo (Gardiol and Banks, 1998). Such discrepancies (highlighted in Table 1) indicate that the requirement for ubiquitination apparent in the in vitro pathway does not necessarily mean that ubiquitination is required for the in vivo pathway.
Table 1.
Differential effects of mutation of p53 and HPV E6 on E6 dependent degradation of p53 in vivo and in vitro
| Mutation |
Degradation of p53
|
References | |
|---|---|---|---|
| In vivo | In vitro | ||
| p53 I381/382/386 (C-terminal lysine mutant) | Yes | Very weak | Crook et al. (1996) |
| E6 Δ118-122 | Yes | Very weak | Foster et al. (1994) |
| E6 F125V | Yes | Very weak | Liu et al. (1999) |
| G134V | Yes | Very weak | |
| E6 Cys-63 Ser | Yesa | No | Dalal et al. (1996) |
| Trp-132 Arg | Yesa | No | |
| E6 Δ94-97 | Yes | Reduced | Gardiol and Banks (1998) |
| Δ126-130 | Yes | Reduced | |
| 10S/11G | Yes | Reduced | |
Abbreviations: HPV, human papillomavirus. Table showing mutated regions/residues of E6 and p53 that confer different susceptibilities of p53 to degradation by E6 under in vitro compared to in vivo conditions.
In immortalized MEC cells but not normal MECs.
Here, we present evidence that disruption of various stages of the ubiquitin-proteasome pathway, either through the inhibition of poly-ubiquitination (7KR-ubiquitin) or interruption of ubiquitin recognition (tUB4 chains) has the ability to protect p53 almost entirely from Mdm2-mediated degradation yet only partially disrupts E6-mediated degradation of p53. This observation is supported by Blattner and co-workers (Glockzin et al., 2003), who reported that overexpression hHR23 (which was suggested to inhibit the access of ubiquitinated p53 to the proteasome) protected p53 from Mdm2-mediated degradation in vivo but that E6-induced p53 degradation was reproducibly less affected, despite hHR23 protecting p53 from E6-mediated degradation completely in vitro.
To establish definitively whether ubiquitination is required for E6-mediated p53 degradation in vivo, an E1 temperature-sensitive cell line was employed. At the non-permissive temperature (39°C) the E1 enzyme is inactive, preventing the ubiquitination cascade from occurring (Chowdary et al., 1994). As no ubiquitination is detectable at the non-permissive temperature, the observation that there is functional, but reduced, E6-mediated p53 degradation in these cells at 39°C indicates that E6 promotes the degradation of p53 through a ubiquitin-independent pathway. This provides clear evidence that E6-mediated p53 degradation occurs through both an ubiquitin-dependent pathway, which is inhibited in these cells at the non-permissive temperature, and an additional ubiquitin-independent pathway, which is unaffected by the inactivation of the E1 enzyme at the non-permissive temperature. Collectively, the results presented here consistently show that E6 can degrade p53 by a pathway that is independent of ubiquitination in vivo. This does not appear to be the case in vitro, where extensive data have shown the requirement for an active ubiquitination pathway for E6-mediated p53 degradation (Scheffner, 1998).
Recently a number of proteins have been reported to be degraded in a proteasome-dependent, ubiquitin-independent manner, as summarized in Table 2. This has prompted some groups to develop assays to test the dependency of a specific protein’s degradation on ubiquitination, generally through adaptations to the pathway by which antizyme mediates the degradation of ornithine decarboxylase (ODC) (Hoyt et al., 2005; Kahana and Reiss, 2005). The ODC degradation pathway has been characterized in detail and has been shown to be independent of any post-translational modifications to ODC, despite being dependent on the 26S proteasome (Rosenberg-Hasson et al., 1989; Bercovich and Kahana, 1993). The growing list of ubiquitin-independent degradation substrates being discovered suggests that alternative routes to the proteasome are becoming an important field of research.
Table 2.
Previously reported ubiquitin-independent substrates of proteasome-dependent degradation
| Substrate protein | Reference |
|---|---|
| Ornithine decarboxylase | Rosenberg-Hasson et al. (1989); Coffino (2001) |
| pRB | Sdek et al. (2005) |
| pRB family proteinsa | Kalejta and Shenk (2003) |
| HMG-CoA | Ness and Holland (2005) |
| Thymidylate synthase | Pena et al. (2005) |
| α-catenin | Hwang et al. (2005) |
| P21 | Jin et al. (2003); Chen et al. (2004) |
| RPN4 | Ju and Xie (2004) |
| IκBα | Kretz-Remy and Arrigo (2003) |
| TCRα | Kretz-Remy and Arrigo (2003) |
| Calmodulin | Benaroudj et al. (2001) |
| Troponin C | Benaroudj et al. (2001) |
| α-synuclein | Tofaris et al. (2001) |
| SRC-3/AIB1 coactivator | Li et al. (2006) |
Abbreviation: SRC, spleen repopulating cells. Table showing the variety of substrates that have been reported in the literature to be degraded by the proteasome in a ubiquitin-independent manner.
Degraded by a viral protein.
The additional in vivo ubiquitin-independent p53 degradation pathway also offers an explanation for why antisense ablation of E1, E6AP or the E2 enzyme involved in the (ubiquitin-dependent) E6-mediated p53 degradation pathway failed to inhibit p53 degradation by more than 20-30% in a study by Rolfe et al. (1995). It also explains the E6 mutants that show reduced binding to E6AP and fail to degrade p53 in vitro, but retain the ability to degrade p53 in vivo (Liu et al., 1999), and the antibodies that inhibit E6 binding to E6AP but have little effect on p53 degradation (Lagrange et al., 2005). However, in contrast to these findings, Kelley et al. (2005) show an approximately equivalent increase in p53 protein levels by small interfering RNA (siRNA) knockdown of E6 compared to E6AP in HPV-positive cell-lines, suggesting E6AP does play an important role in E6-mediated p53 degradation. Although the knockdown of E6AP and E6 in these experiments was not equivalent, these results suggest a degradation pathway that involves E6AP. This could be supported by Nomine et al. (2006), who, in addition to describing E6 mutants that fail to degrade p53 in vitro while retaining normal degradative function in vivo, also show that recruitment of an E6AP mutant that is dead for E3 ligase activity into the E6/E6AP/p53 tertiary complex still allows E6-dependent p53 degradation These authors hypothesize that E6AP is only required to mediate the interaction between E6 and p53 and that an additional E3 ligase may provide the enzymatic activity. However, the ubiquitin-independent degradation pathway may also provide an explanation for this result. Scheffner and co-workders (Hengstermann et al., 2005) have also performed a study in which E6 and E6AP mRNAs were individually ablated using siRNA techniques. Although in their data the knockdown of E6 appears to be more effective than E6AP at increasing p53 protein in cell-staining data, no direct comparison is made between the increases in p53 levels in the presence of E6 siRNA compared with E6AP siRNA. In light of the controversial data discussed here, such information would be interesting to give further insight into the role of E6AP in p53 degradation in HPV-infected cells. The possibility that E6AP could offer dual roles, one function being its known enzymatic E3 ligase activity and another mediating the interaction between E6 and p53, or possibly merely stabilizing this interaction, offers an interesting avenue of further study.
Although, the role of the E6AP in the ubiquitin-independent pathway has yet to be formally ascertained, the requirement for the C-terminus of p53 in the ubiquitin-independent pathway suggests that this pathway may require the second (E6AP-independent) binding site on p53. The original investigations into the function of the second E6-binding site in the C-terminus of p53 concluded that this region of p53 was not necessary for degradation (Lechner and Laimins, 1994; Li and Coffino, 1996a). These experiments were performed in vitro where the ubiquitin-dependent pathway appears to be the predominant degradation pathway. No function for the second binding site has been found. Here we show that the C-terminus of p53 is important for ubiquitin-independent, but not ubiquitin-dependent, degradation of p53, providing a possible function for the E6AP-independent-binding site.
Combined, the results presented here provide the first evidence that E6 mediates the degradation of p53 by a pathway that is independent of ubiquitination in vivo. The combination of a ubiquitin-independent pathway, along with the previously described ubiquitin-dependent pathway, allows E6 to mediate the degradation of p53 extremely efficiently. This discovery has important implications on future research into p53 degradation, particularly with regard to the use of E6 as a control for ubiquitination studies in vivo, as well as on research into possible therapeutic targets for HPV-positive tumors. Additionally, the discovery of a novel p53 degradation pathway opens up new avenues of investigation into the pathways utilized by other tumor viruses that promote the degradation of p53 as well as other substrates.
Materials and methods
Cells, antibodies and reagents
H1299 cells were cultured in Rosewell Park Memorial Institute medium supplemented with 10% fetal calf serum (FCS) and gentamicin at 37°C, 5% CO2 in a humidified atmosphere. Mouse TS20b and H38-5cells were a kind gift from Harvey Ozer and were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS and gentamicin. Human p53 was detected using the mouse monoclonal antibody DO-1. Anti-β-galactosidase (β-gal) mouse monoclonal antibodies were obtained from Oncogene. Proteasome inhibitor MG132 was obtained from Calbiochem (Merck, Singapore, Singapore). Ni2-NTA agarose beads were obtained from Qiagen (Research Biolabs, Singapore). Purified methylated ubiquitin was purchased from Affinity (Biomol International, Exeter, UK).
Plasmids
Expression from constructs pcDNA3 E6, pCOC-X2mdm2, pcDNA3 β-gal, pcDNA3 His6 WT ubiquitin, pcDNA3 His6 7KR-ubiquitin, pcDNA3 tUB4, pcDNA3 p53 and pcDNA3 p53CΔ30 was under the control of the CMV promoter. pcDNA3 tUB4 was cloned from a vector generously supplied by Akio Toh-e.
Transfection of cells and Western blotting
Cells were transfected using the calcium-phosphate method as described in (Xirodimas et al., 2001). In all experiments 1 μg β-gal encoding plasmid was co-transfected as a transfection control. Forty-eight hours post-transfection, cells were lysed in Novex loading buffer supplemented with 0.1 m dithiothreitol and proteins were separated on 4-12% Novex polyacrylamide gels, transferred to nitrocellulose membrane and developed with the relevant antibodies as described previously in Xirodimas et al. (2001).
In vitro translation degradation assays
One microgram of the relevant DNA was in vitro transcribed and translated in the presence of 0.56 MBq. [35S]methionine (or 0.5 μg unlabelled methionine for the pcDNA3 empty vector controls) in 50 μl of a transcription/translation coupled with rabbit reticulocyte lysate system (Promega UK, Southampton, UK), according to the manufacturers instructions. One microliter of extract containing translated p53 protein was mixed with 3 μl extract containing translated E6 protein or pcDNA3 control, as indicated, and incubated for a further 90 min at 25°C to allow degradation. Methylated ubiquitin was added to the relevant samples before degradation step. Reactions were stopped by adding 2 × sodium dodecyl sulfate loading buffer. Proteins were separated on 4-12% polyacrylamide gels and analysed by direct autoradiography.
Purification of His-tagged ubiquitin conjugates
Purification of His-tagged ubiquitin-conjugated proteins was as described in (Xirodimas et al., 2001). Briefly, cells were lysed in 6 m guanidinium/HCl pH 8 and His-tagged conjugates were isolated by incubation with Ni2-NTA agarose beads. His-ubiquitin-tagged proteins were washed, eluted and then analyzed by Western blot with DO-1 antibody against p53.
Acknowledgements
This work was supported by Cancer Research UK and by the generosity of Reverend and Mrs Cameron through the Kitty Cameron Fund. We thank D Coomber for help with the manuscript preparation and M Scheffner for advice and reagents at the project outset.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- Benaroudj N, Tarcsa E, Cascio P, Goldberg AL. The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie. 2001;83:311–318. doi: 10.1016/s0300-9084(01)01244-5. [DOI] [PubMed] [Google Scholar]
- Bercovich Z, Kahana C. Involvement of the 20S proteasome in the degradation of ornithine decarboxylase. Eur J Biochem. 1993;213:205–210. doi: 10.1111/j.1432-1033.1993.tb17749.x. [DOI] [PubMed] [Google Scholar]
- Camus S, Higgins M, Lane DP, Lain S. Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett. 2003;536:220–224. doi: 10.1016/s0014-5793(03)00054-1. [DOI] [PubMed] [Google Scholar]
- Chen X, Chi Y, Bloecher A, Aebersold R, Clurman BE, Roberts JM. N-acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1) Mol Cell. 2004;16:839–847. doi: 10.1016/j.molcel.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffino P. Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie. 2001;83:319–323. doi: 10.1016/s0300-9084(01)01252-4. [DOI] [PubMed] [Google Scholar]
- Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, et al. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–6728. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- Crook T, Ludwig RL, Marston NJ, Willkomm D, Vousden KH. Sensitivity of p53 lysine mutants to ubiquitin-directed degradation targeted by human papillomavirus E6. Virology. 1996;217:285–292. doi: 10.1006/viro.1996.0115. [DOI] [PubMed] [Google Scholar]
- Dalal S, Gao Q, Androphy EJ, Band V. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J Virol. 1996;70:683–688. doi: 10.1128/jvi.70.2.683-688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Demers GW, Etscheid BG, Galloway DA. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiol D, Banks L. Comparison of human papillomavirus type 18 (HPV-18) E6-mediated degradation of p53 in vitro and in vivo reveals significant differences based on p53 structure and cell type but little difference with respect to mutants of HPV-18 E6. J Gen Virol. 1998;79:1963–1970. doi: 10.1099/0022-1317-79-8-1963. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Stewart D, Matlashewski G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol. 2004;24:7987–7997. doi: 10.1128/MCB.24.18.7987-7997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol Cell Biol. 2003;23:8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hengstermann A, D’silva MA, Kuballa P, Butz K, HoppeSeyler F, Scheffner M. Growth suppression induced by downregulation of E6-AP expression in human papillomavirus-positive cancer cell lines depends on p53. J Virol. 2005;79:9296–9300. doi: 10.1128/JVI.79.14.9296-9300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Zhang M, Coffino P. Probing the ubiquitin/proteasome system with ornithine decarboxylase, a ubiquitin-independent substrate. Methods Enzymol. 2005;398:399–413. doi: 10.1016/S0076-6879(05)98033-6. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Yu SS, Ryu JH, Jeon HB, Yoo YJ, Eom SH, et al. Regulation of beta-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of alpha-catenin. J Biol Chem. 2005;280:12758–12765. doi: 10.1074/jbc.M413367200. [DOI] [PubMed] [Google Scholar]
- Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju D, Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J Biol Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- Kahana C, Reiss Y. Cell-free assay for ubiquitin-independent proteasomal protein degradation. Methods Mol Biol. 2005;301:83–96. doi: 10.1385/1-59259-895-1:083. [DOI] [PubMed] [Google Scholar]
- Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc Natl Acad Sci USA. 2003;100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley ML, Keiger KE, Lee CJ, Huibregtse JM. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J Virol. 2005;79:3737–3747. doi: 10.1128/JVI.79.6.3737-3747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz-Remy C, Arrigo AP. Modulation of the chymotrypsin-like activity of the 20S proteasome by intracellular redox status: effects of glutathione peroxidase-1 overexpression and antioxidant drugs. Biol Chem. 2003;384:589–595. doi: 10.1515/BC.2003.066. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Ludwig RL, Ashcroft M, Vousden KH. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange M, Charbonnier S, Orfanoudakis G, Robinson P, Zanier K, Masson M, et al. Binding of human papillomavirus 16 E6 to p53 and E6AP is impaired by monoclonal antibodies directed against the second zinc-binding domain of E6. J Gen Virol. 2005;86:1001–1007. doi: 10.1099/vir.0.80607-0. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Laimins LA. Inhibition of p53 DNA binding by human papillomavirus E6 proteins. J Virol. 1994;68:4262–4273. doi: 10.1128/jvi.68.7.4262-4273.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Coffino P. High-risk human papillomavirus E6 protein has two distinct binding sites within p53, of which only one determines degradation. J Virol. 1996a;70:4509–4516. doi: 10.1128/jvi.70.7.4509-4516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Coffino P. Identification of a region of p53 that confers lability. J Biol Chem. 1996b;271:4447–4451. doi: 10.1074/jbc.271.8.4447. [DOI] [PubMed] [Google Scholar]
- Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen JJ, Gao Q, Dalal S, Hong Y, Mansur CP, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminus of p53 make it resistant to degradation mediated by MDM2 but not by human papillomavirus E6 and induce growth inhibition in MDM2-overexpressing cells. Oncogene. 2002;21:2605–2610. doi: 10.1038/sj.onc.1205343. [DOI] [PubMed] [Google Scholar]
- Ness GC, Holland RC. Degradation of HMG-CoA reductase in rat liver is cholesterol and ubiquitin independent. FEBS Lett. 2005;579:3126–3130. doi: 10.1016/j.febslet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, et al. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Pena MM, Xing YY, Koli S, Berger FG. Role of N-terminal residues in the ubiquitin-independent degradation of human thymidylate synthase. Biochem J. 2005;394:355–363. doi: 10.1042/BJ20051479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe M, Beer-Romero P, Glass S, Eckstein J, Berdo I, Theodoras A, et al. Reconstitution of p53-ubiquitinylation reactions from purified components: the role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP) Proc Natl Acad Sci USA. 1995;92:3264–3268. doi: 10.1073/pnas.92.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem. 1989;185:469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Isono E, Oguchi T, Shimada M, Sone T, Kawahara H, et al. Intracellularly inducible, ubiquitin hydrolase-insensitive tandem ubiquitins inhibit the 26S proteasome activity and cell division. Genes Genet System. 2004;79:77–86. doi: 10.1266/ggs.79.77. [DOI] [PubMed] [Google Scholar]
- Salvat C, Acquaviva C, Scheffner M, Robbins I, Piechaczyk M, Jariel-Encontre I. Molecular characterization of the thermosensitive E1 ubiquitin-activating enzyme cell mutant A31N-ts20Requirements upon different levels of E1 for the ubiquitination/degradation of the various protein substrates in vivo. Eur J Biochem. 2000;267:3712–3722. doi: 10.1046/j.1432-1327.2000.01404.x. [DOI] [PubMed] [Google Scholar]
- Scheffner M. Ubiquitin, E6-AP, and their role in p53 inactivation. Pharmacol Ther. 1998;78:129–139. doi: 10.1016/s0163-7258(98)00003-5. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Howley PM. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Sdek P, Ying H, Chang DL, Qiu W, Zheng H, Touitou R, et al. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Stewart D, Ghosh A, Matlashewski G. Involvement of nuclear export in human papillomavirus type 18 E6-mediated ubiquitination and degradation of p53. J Virol. 2005;79:8773–8783. doi: 10.1128/JVI.79.14.8773-8783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofaris GK, Layfield R, Spillantini MG. Alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509:22–26. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–467. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]


