Abstract
Chronic obstructive pulmonary disease (COPD) is a heterogeneous syndrome, including emphysema and airway disease. Phenotypes defined on the basis of chest computed tomography (CT) may decrease disease heterogeneity and aid in the identification of candidate genes for COPD subtypes. To identify these genes, we performed genome-wide linkage analysis in extended pedigrees from the Boston Early-Onset COPD Study, stratified by emphysema status (defined by chest CT scans) of the probands, followed by genetic association analysis of positional candidate genes. A region on chromosome 1p showed strong evidence of linkage to lung function traits in families of emphysema-predominant probands in the stratified analysis (LOD score = 2.99 in families of emphysema-predominant probands versus 1.98 in all families). Association analysis in 949 individuals from 127 early-onset COPD pedigrees revealed association for COPD-related traits with an intronic single-nucleotide polymorphism (SNP) in transforming growth factor-β receptor-3 (TGFBR3) (P = 0.005). This SNP was significantly associated with COPD affection status comparing 389 cases from the National Emphysema Treatment Trial to 472 control smokers (P = 0.04), and with FEV1 (P = 0.004) and CT emphysema (P = 0.05) in 3,117 subjects from the International COPD Genetics Network. Gene-level replication of association with lung function was seen in 427 patients with COPD from the Lung Health Study. In conclusion, stratified linkage analysis followed by association testing identified TGFBR3 (betaglycan) as a potential susceptibility gene for COPD. Published human microarray and murine linkage studies have also demonstrated the importance of TGFBR3 in emphysema and lung function, and our group and others have previously found association of COPD-related traits with TGFB1, a ligand for TGFBR3.
Keywords: betaglycan, chronic obstructive pulmonary disease, computed tomography, linkage, single nucleotide polymorphism
CLINICAL RELEVANCE
In this article we have identified transforming growth factor-β receptor 3 as a novel candidate gene for chronic obstructive pulmonary disease (COPD) susceptibility, in a relevant COPD pathway. We also demonstrate how more precise phenotypic characterization may aid in the identification of genes for a heterogeneous complex disease.
Genetic linkage and association studies have made significant strides in uncovering the genes for monogenic lung diseases, such as familial pulmonary hypertension (1) and autosomal dominant familial spontaneous pneumothorax (2). However, when these same techniques have been applied to common, complex respiratory disorders such as asthma and chronic obstructive pulmonary disease (COPD), the results have been less definitive; often the initial positive results were inconsistently replicated in follow-up studies (3–5). Multiple explanations have been proposed for the lack of replication, including small sample sizes, genotyping error, and population stratification (6). Differences in disease phenotypes between study populations may be a reason for nonreplication that is particularly relevant for COPD, a heterogeneous condition that includes both emphysema and airway disease.
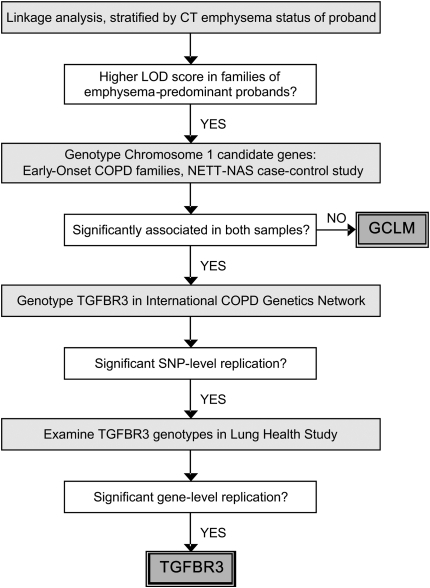
Measurement of lung function using spirometry, though essential for the diagnosis of COPD (7), fails to distinguish between emphysema and airway disease. On the other hand, computed tomography (CT) scans of the chest can be used to confirm the presence and to grade the severity of emphysema. Therefore, we hypothesized that using chest CT scans to identify subjects with emphysema-predominant COPD would yield a more precise phenotype that would allow for the more efficient identification of candidate genes for COPD. We tested this hypothesis by performing stratified genome-wide linkage analysis in families of probands with CT-defined emphysema from the Boston Early-Onset COPD Study (8) to identify candidate genomic regions (Figure 1). Candidate genes from the linkage regions identified in the emphysema-stratified analysis were then tested for association with airflow obstruction in the Boston Early-Onset COPD Study families and in a case-control study comparing patients with emphysema from the National Emphysema Treatment Trial (NETT) (9) with community control subjects from the Normative Aging Study (NAS) (10). The results were replicated in the family-based International COPD Genetics Network (ICGN) study (11, 12) and in a cohort of patients with COPD from the Lung Health Study (LHS) (13). Results from this study have been previously presented as an abstract (14).
Figure 1.
Study design incorporating stratified linkage analysis in the Boston Early-Onset COPD Study followed by genetic association studies in the Boston Early-Onset COPD Study, the National Emphysema Treatment Trial (NETT)–Normative Aging Study (NAS) case-control study, the International COPD Genetics Network, and the Lung Health Study.
MATERIALS AND METHODS
Study Subjects
The Boston Early-Onset COPD Study enrolled extended pedigrees recruited through probands with physician-diagnosed COPD (age < 53 yr and FEV1 < 40% predicted) (15). The linkage data set consisted of 585 individuals in 72 families (16); these individuals plus additional enrolled subjects constituted the association dataset of 949 individuals in 127 families (Table 1) (4). Phenotyping involved a questionnaire (17) and spirometry before and after inhaled bronchodilator (18). Chest CT scans were not performed as part of the study protocol, but chest CTs were available from the clinical records of 44 of the probands in the linkage sample and 75 probands in the association sample. Chest CTs were scored by three to four readers, using a modification of the scoring system used in NETT (19, 20). Probands in the upper three quartiles of emphysema severity were defined as “emphysema-predominant” (8).
TABLE 1.
CHARACTERISTICS OF SUBJECTS IN THE BOSTON EARLY-ONSET COPD (EOCOPD) STUDY, THE NATIONAL EMPHYSEMA TREATMENT TRIAL (NETT) GENETICS ANCILLARY STUDY, THE INTERNATIONAL COPD GENETICS NETWORK (ICGN), AND THE LUNG HEALTH STUDY (LHS)
| Characteristic | EOCOPD Probands | NETT | ICGN Probands | ICGN Relatives | LHS |
|---|---|---|---|---|---|
| Subjects | 127 | 389 | 1,132 | 1,985 | 427 |
| Age | 48.1 (± 4.7) | 67.4 (± 5.8) | 58.3 (± 5.4) | 57.9 (± 9.4) | 48.7 (± 6.9) |
| Sex | |||||
| Male | 32 (25.2%) | 250 (64.3%) | 679 (60.0%) | 1,025 (51.6%) | 280 (65.5%) |
| Female | 95 (74.8%) | 139 (35.7%) | 453 (40.0%) | 960 (48.4%) | 147 (34.5%) |
| Pack-years of smoking | 38.9 (± 21.9) | 66.4 (± 30.4) | 52.2 (± 29.1) | 39.1 (± 25.0) | 40.3 (± 19.1) |
| FEV1% predicted | 21.9 (± 8.4)* | 28.0 (± 7.4) | 36.1 (± 12.9)* | 83.1 (± 26.1)* | 79.2 (± 6.2) |
| FEV1/FVC Ratio | 0.31 (± 0.10)* | 0.32 (± 0.06) | 0.37 (± 0.12)* | 0.64 (± 0.14)* | 0.65 (± 0.06) |
| Subjects with chest CT scans | 75 | 389 | 527 | 640 | – |
| Emphysema score† | 13.2 (± 5.1) | 16.3 (± 4.0) | 2.8 (± 1.3) | 1.3 (± 1.1) | – |
| Subjects with quantitative CT analysis | – | 358 | 409 | 578 | – |
| CT percent emphysema, −950 Hounsfield Units | – | 17.0 (± 10.9) | 26.3 (± 14.8) | 18.8 (± 11.6) | – |
Values are shown as N (%) or mean (± SD)
Subjects with post-bronchodilator (BD) values: EOCOPD N = 118; ICGN probands N = 1,092; ICGN relatives N = 1,918.
EOCOPD and NETT: range, 0–24; ICGN: range, 0–5.
In the case-control study, emphysema cases were from NETT, a multicenter clinical trial of lung volume reduction surgery versus medical management for emphysema (9). Subjects had FEV1 less than or equal to 45% predicted, hyperinflated lung volumes, and bilateral emphysema on chest CT scan (19). Participants in the NETT Genetics Ancillary Study provided a blood sample for genetic analysis. Control subjects with normal spirometry at the last study visit and with a smoking history of at least 10 pack-years (packs of cigarettes smoked per day multiplied by number of years smoking) were from the Normative Aging Study, a longitudinal cohort study of healthy male veterans in the Greater Boston area (10).
Subjects in the International COPD Genetics Network (ICGN) were recruited from ten centers in North America and Europe. Probands were aged 45 to 65 years, with post-bronchodilator FEV1 less than 60% predicted, FEV1/VC less than 90% predicted, a smoking history of at least 5 pack-years, and at least one sibling with a smoking history of 5 pack-years or more. Phenotype measurements included questionnaires, spirometry, and chest CT scans in a subset of the participants. Density mask analysis was used to quantify emphysema at a threshold of −950 HU; detailed chest CT methods have been described previously (11).
The Lung Health Study (LHS) was a 10-center randomized clinical trial, sponsored by the National Heart, Lung, and Blood Institute, to determine whether a program of smoking intervention and use of an inhaled bronchodilator could slow the rate of decline in pulmonary function in patients with COPD over a 5-year follow-up period (13). This analysis included 427 European American participants with mild-to-moderate COPD who were randomly selected from the LHS. At baseline, phenotyping involved questionnaires and spirometry before and after inhaled bronchodilator.
All studies were approved by the relevant institutional review boards, and study subjects provided written informed consent.
Linkage Analysis
A genome-wide panel of 377 autosomal short tandem repeat (STR) markers had been previously genotyped in the Boston Early-Onset COPD Study (16, 21). Linkage analyses for FEV1 and FEV1/FVC ratio (both pre- and post-bronchodilator) were performed in the 44 families with available proband chest CT scans and in the subset of 34 families with emphysema-predominant probands. Variance component linkage models were calculated in SOLAR (22), adjusted for age, sex, height, race, smoking status (ever versus never), pack-years of smoking, and quadratic terms where appropriate. Covariates significant at P < 0.05 were retained in the linkage models. Statistical significance for the subgroup analysis was determined by randomly permuting the emphysema-predominant proband status over 10,000 simulations.
Association Analysis
The initial association analyses were performed in the Boston Early-Onset COPD Study families and in the NETT-NAS case-control study. In TGFBR3, we initially genotyped six haplotype tagging (ht) single-nucleotide polymorphisms (SNPs), based on publicly available DNA sequence data from 23 European-American subjects in the Innate Immunity Program in Genomic Applications (IIPGA) (23); sequencing primers and results are available on the IIPGA website (www.innateimmunity.net). An additional 31 SNPs were genotyped to complete linkage disequilibrium (LD) tagging (r2 > 0.9, minor allele frequency [MAF] ≥ 0.1), based on the sequencing data. In GCLM, we selected five LD-tagging SNPs based on data from white subjects (CEU) in the International HapMap Project (24). SNPs were genotyped using Taqman (Applied Biosystems, Foster City, CA) or Sequenom (San Diego, CA) platforms.
In the Boston Early-Onset COPD Study, SNPs were analyzed using the Family Based Association Test (25) implemented in PBAT software (26). Analyses of FEV1 were adjusted for age, sex, height, smoking status, and pack-years, under additive genetic models, unless otherwise noted. SNPs were tested under the null hypothesis of linkage, but no association. SNPs in the NETT-NAS case-control study were analyzed in SAS/Genetics (SAS Institute, Cary, NC), using the Armitage trend test (additive model), unless otherwise noted. LD was calculated in Haploview (27).
In the NETT-NAS case-control study, an additional panel of 195 common intergenic SNPs throughout the genome, excluding regions linked to COPD, was genotyped using the Illumina (San Diego, CA) GoldenGate assay in the NETT cases and in a majority of the NAS control subjects (n = 409). Based on this panel, there was no evidence of population stratification (χ2195 df = 211.9, P = 0.19) (28).
In the ICGN family-based study, the screening panel of 6 ht-SNPs in TGFBR3 plus two additional SNPs that had been significant in both the Boston Early-Onset COPD Study and in NETT-NAS were genotyped using Sequenom assays. Data were analyzed using PBAT, under additive genetic models. Quantitative traits were adjusted for age, sex, pack-years of smoking, height (for spirometry), or weight (for CT densitometry measurement), and study center.
In the LHS, SNP selection and genotyping were conducted independently from the other three study samples. A total of 48 SNPs from TGFBR3 were selected from the dbSNP database (http://www.ncbi.nlm.nih.gov). Criteria for SNP selection included the following, in order of priority: (1) regulatory and coding SNPs; (2) SNPs with known frequency, preferably MAF greater than or equal to 0.10 and no less than 0.05; (3) validated SNPs; (4) SNPs in intron/exon boundaries; and (5) SNPs in promoter and 3′UTR regions. SNPs were genotyped at the SNP Center at the Johns Hopkins Genetic Resources Core Facility (http://snpcenter.grcf.jhmi.edu/), with the Illumina BeadStation 500G GoldenGate genotyping platform. SNPs with MAF less than 0.01 (n = 13) were excluded from analyses. All SNPs were in Hardy-Weinberg equilibrium. The primary outcome was post-bronchodilator FEV1, expressed as a percent of predicted (29). Linear regression models were used to adjust for potential confounders, including age, sex, and pack-years of smoking. Age and pack-years were significantly associated with FEV1 (% predicted) in separate bivariate analyses (P < 0.05) and were included in the final model. The residuals from the linear regression model for FEV1 (% predicted) adjusted for age and pack-years were then analyzed in an additive genetic model for each SNP. Analyses were performed with StataSE, version 8.0 (Stata Corp, College Station, TX) and PLINK (30).
RESULTS
Stratified Linkage Analysis
In the Boston Early-Onset COPD Study probands, chest CT scans were obtained for clinical indications and not as part of the study protocol. Since CTs were available for many of the probands but not their relatives, we could not perform stratified linkage analyses for CT emphysema scores. However, we were able to perform linkage analyses for spirometric measures of lung function (FEV1 and FEV1/FVC ratio, both before and after bronchodilator administration) in families of 34 probands with emphysema-predominant COPD, defined on the basis of the proband CT scans. The stratified linkage analyses for lung function in emphysema-predominant proband families were considered surrogate phenotypes for emphysema. We compared the results of the stratified linkage analyses to the linkage analyses in the full set of 44 families in which the proband had a CT scan available. Chromosomal regions that showed increased evidence for linkage in the subgroup analysis were presumed to be linked to the emphysema subtype of COPD.
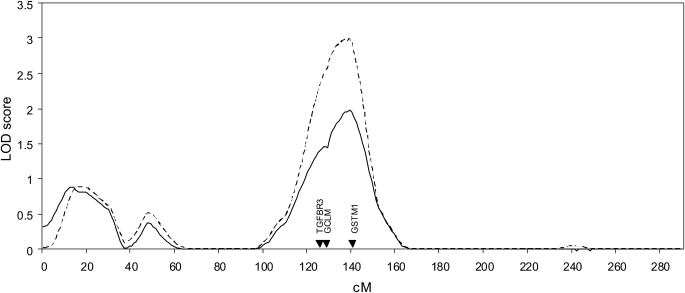
For FEV1, a region on chromosome 1p showed stronger linkage evidence in the stratified analysis (Table 2 and Figure 2). For post-bronchodilator values of FEV1, the LOD (logarithm of the odds of linkage) score of 1.98 in all families increased to 2.99 in the emphysema-predominant proband families; similar results were seen for pre-bronchodilator values. The increases in LOD scores were statistically significant (P ≤ 0.01). A region on chromosome 8p also had an increase in LOD score in the stratified analysis of post-bronchodilator FEV1. By genotyping additional STR markers on chromosome 8p, we had previously shown an attenuation of LOD score in this region, in an analysis that did not account for CT subtypes (31). The LOD score was also attenuated in the current analyses when we included the additional markers; therefore, we did not pursue further study of this chromosome 8p region.
TABLE 2.
GENOME-WIDE LINKAGE ANALYSIS FOR FEV1 IN THE BOSTON EARLY-ONSET COPD STUDY
| Pre-Bronchodilator
|
Post-Bronchodilator
|
||||
|---|---|---|---|---|---|
| Chromosome | cM | Emphysema* | All† | Emphysema* | All† |
| 1 | 138 | 2.89 (0.0006) | 1.85 (0.0039) | 2.99 (0.001) | 1.98 (0.0044) |
| 2 | 227 | 1.75 (0.006) | 1.75 (0.0049) | 1.50 (0.016) | 1.53 (0.011) |
| 3 | 207 | 1.45 (0.018) | 1.57 (0.010) | ||
| 6 | 89 | 2.17 (0.0047) | 0.98 (0.040) | ||
| 6 | 155 | 1.58 (0.014) | 1.32 (0.019) | ||
| 8 | 6 | 1.68 (0.0074) | 1.68 (0.0058) | 3.64 (0.0005) | 3.09 (0.0005) |
| 11 | 85 | 2.12 (0.0055) | 1.11 (0.029) | ||
| 14 | 135 | 1.78 (0.0097) | 0.93 (0.045) | ||
Chromosomal regions with LOD score > 1.5 (with empirical P values) in analyses of either families of emphysema-predominant probands or all families are shown.
Significant covariates (P < 0.05) included age, sex, height, and pack-years of smoking.
Significant covariates (P < 0.05) included age, sex, height, pack-years of smoking, and (pack-years)2.
Figure 2.
Linkage analysis on chromosome 1 for forced expiratory volume in 1 s (FEV1, post-bronchodilator) in all subjects (solid line) and in families of probands with emphysema-predominant COPD (dashed line) in the Boston Early-Onset COPD Study.
For FEV1/FVC ratio, chromosome 1p demonstrated a slight increase in LOD scores in the stratified analyses (see Table E1 in the online supplement). The most significant results in any of the linkage analyses were found for FEV1/FVC on chromosome 2q, with higher LOD scores in all subjects than in the emphysema subgroup. We have previously demonstrated significant linkage for FEV1/FVC on chromosome 2q (16, 32), and our group and others have found replicated association for SNPs in SERPINE2, a candidate gene located in this linked region (12, 33).
Genetic Association Analysis
Three potential candidate genes with relevance to COPD reported in the literature were located under the linkage peak on chromosome 1p (Figure 2). Several authors have shown the null deletion of glutathione S-transferase M1 (GSTM1) to be associated with COPD (34–36). In a previous article, we had found no association between the GSTM1 null variant and COPD in the NETT-NAS case-control sample (4); with this negative result, consistent replication would not have been possible, so we did not examine GSTM1 further in the present study. Two studies have examined variants in glutamate-cysteine ligase, modifier subunit (GCLM), though no association was found with COPD (37) or lung function decline (38). The gene for vascular cell adhesion molecule-1 (VCAM1) is also located in the region. However, this gene may be more relevant for airway disease than for emphysema (39), so we did not genotype VCAM1. We did examine transforming growth factor β receptor-3 (TGFBR3), given the prior associations between TGF-β1 variants and COPD (40, 41).
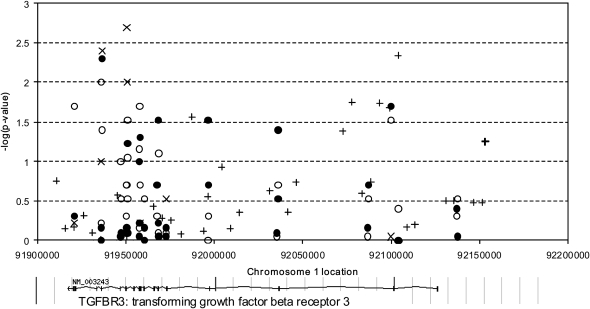
In the first stage, we genotyped six ht-SNPs in TGFBR3 and five LD-tagging SNPs in GCLM; subsequently, an additional 31 LD-tagging SNPs in TGFBR3 were typed (Figure 3). Over both stages, there were average genotype completion rates greater than 95% with very few pedigree inconsistencies in the family-based data. All SNPs were in Hardy-Weinberg equilibrium (P > 0.01) in the founder individuals from the Boston Early-Onset COPD Study families and in the NAS control subjects.
Figure 3.
Association results for transforming growth factor-β receptor 3 single-nucleotide polymorphisms in the Boston Early-Onset COPD Study families (post-bronchodilator FEV1, solid circles), the National Emphysema Treatment Trial (NETT)–Normative Aging Study (NAS) case-control study (open circles), the International COPD Genetics Network (ICGN) families (post-bronchodilator FEV1, symbol “X”) and the Lung Health Study cohort (post-bronchodilator FEV1 percent predicted, symbol “+”).
To improve statistical power, SNPs were tested for association in 949 individuals in 127 families from the Boston Early-Onset COPD Study rather than using only the families with proband CT scans. Results for the 6 ht-SNPs, plus two additional significant SNPs, in TGFBR3 in the Boston Early-Onset COPD Study, the NETT-NAS case-control study, and the ICGN family study are shown in Table 3. One intronic SNP, rs2296621, was significantly associated in all three cohorts (P = 0.005 in Boston Early-Onset COPD Study; P = 0.04 in NETT-NAS; P = 0.004 in ICGN), and another intronic SNP, rs2291477, was significantly associated in the NETT-NAS (P = 0.03) and ICGN (P = 0.01) studies and showed a trend for association in the Boston Early-Onset COPD Study (P = 0.06). These two SNPs are in LD with each other (r2 = 0.91 in Boston Early-Onset COPD Study; r2 = 0.87 in NETT-NAS and in ICGN).
TABLE 3.
ASSOCIATION ANALYSIS OF 8 SNPS (INCLUDING 6 HAPLOTYPE-TAGGING SNPS) IN TGFBR3 IN THE BOSTON EARLY-ONSET COPD STUDY (EOCOPD) FAMILIES, THE NATIONAL EMPHYSEMA TREATMENT TRIAL (NETT)–NORMATIVE AGING STUDY (NAS) CASE-CONTROL STUDY, AND THE INTERNATIONAL COPD GENETICS NETWORK (ICGN) FAMILY-BASED STUDY
| EOCOPD
|
NETT-NAS
|
ICGN
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chrom. 1 Location* | Role | MAF | P Value FEV1† | MAF Cases | MAF Controls | Odds Ratio (95% CI) | P (trend) | MAF | P Value FEV1† |
| rs902 | 91920815 | 3′ UTR | 0.27 | 0.5 | 0.22 | 0.27 | 0.75 (0.60, 0.95) | 0.02 | 0.24 | 0.6 |
| rs2765889 | 91935931 | Intron | 0.23 | 1.0 | 0.19 | 0.25 | 0.73 (0.58, 0.93) | 0.01 | 0.24 | 0.1 |
| rs2296621 | 91936374 | Intron | 0.20 | 0.005 | 0.22 | 0.18 | 1.29 (1.01, 1.64) | 0.04 | 0.19 | 0.004 |
| rs1805113 | 91950526 | Coding, synonymous | 0.49 | 0.7 | 0.47 | 0.43 | 1.16 (0.95, 1.42) | 0.2 | 0.45 | 0.002 |
| rs2291477 | 91950923 | Intron | 0.22 | 0.06 | 0.24 | 0.19 | 1.30 (1.02, 1.64) | 0.03 | 0.21 | 0.01 |
| rs1805112 | 91958245 | Coding, synonymous | 0.40 | 0.046 | 0.42 | 0.39 | 1.15 (0.94, 1.42) | 0.2 | 0.41 | 0.6 |
| rs2306886 | 91972699 | Intron | 0.45 | 0.9 | 0.42 | 0.42 | 1.03 (0.84, 1.25) | 0.8 | 0.45 | 0.3 |
| rs1805110 | 92099633 | Coding, Ser15Phe | 0.065 | 0.02‡ | 0.095 | 0.069 | 1.51 (1.04, 2.20)‡ | 0.03‡ | 0.086 | 0.9‡ |
Definition of abbreviations: MAF, minor allele frequency; SNP, single-nucleotide polymorphism; TGFBR3, transforming growth factor-β receptor 3.
March 2006 human genome reference sequence (NCBI Build 36).
P value for association with post-bronchodilator FEV1, using the Family-Based Association Test, implemented in PBAT software.
Due to low minor allele frequency, rs1805110 was analyzed using a dominant model.
In the ICGN study, these two SNPs were also significantly associated with an increased risk of severe COPD, defined by GOLD stages 3 or 4 (rs2296621, P = 0.001; rs2291477, P = 0.002), consistent with the increased odds ratios for COPD in the NETT-NAS case-control study. In the ICGN, the two SNPs showed trends for association with increased emphysema on quantitative image analysis of chest CT scans at −950 HU (rs2296621, P = 0.05; rs2291477, P = 0.06). Chest CT scans were available on a subset of NETT subjects, but the two TGFBR3 intronic SNPs and the Ser15Phe coding SNPs were not associated with percent emphysema at −950 HU in NETT. However, power was limited for this analysis, due to the reduced sample size of NETT subjects with quantitative CT scan data (Table 1).
Given the low minor allele frequency, the Ser15Phe (rs1805110) coding SNP in TGFBR3 was analyzed in dominant models, with significant associations for both pre-bronchodilator (P = 0.01) and post-bronchodilator FEV1 (P = 0.02) in the Boston Early-Onset COPD Study and a significantly increased risk of COPD in the NETT-NAS case-control study (odds ratio, 1.51; 95% confidence interval, 1.04–2.20; P = 0.03). In the Boston Early-Onset COPD Study, TGFBR3 Ser15Phe was associated with GOLD stage 2 or greater COPD (P = 0.03) and was significantly associated with FEV1 in an analysis restricted to 34 emphysema-predominant families (pre-bronchodilator, P = 0.03; post-bronchodilator, P = 0.045). However, Ser15Phe was not associated with FEV1 or COPD in the ICGN study. Ser15Phe is not in LD with either of the two replicated intronic SNPs (r2 < 0.05). Association results in the Boston Early-Onset COPD Study and in the NETT-NAS case-control study for the full set of 37 LD-tagging SNPs in TGFBR3 are shown in Table E2. Although several SNPs had P values < 0.05 in each cohort, no other SNPs showed replicated associations.
In the Lung Health Study (LHS), six SNPs in TGFBR3 were significantly associated with postbronchodilator values for FEV1, expressed as a percent of predicted (Table 4). Neither rs2296621 nor rs2291477 nor any SNPs in tight LD (r2 > 0.8) with these two SNPs were genotyped in the LHS. SNP rs1805110 (Ser15Phe) was not genotyped in the LHS, but two significant intronic SNPs, rs2634028 (P = 0.02) and rs2046737 (P = 0.02), are in complete LD (r2 = 1) with rs1805110 in the white subjects (CEU) from the International HapMap Project (24). Another significantly associated intronic SNP, rs2129972 (P = 0.02), is also in LD with rs1805110 in the HapMap (r2 = 0.81).
TABLE 4.
ASSOCIATION ANALYSIS OF TGFBR3 SNPS AND POST-BRONCHODILATOR FEV1 (% PREDICTED) IN THE LUNG HEALTH STUDY
| SNP | Chrom. 1 Location* | β | P Value |
|---|---|---|---|
| rs12026758 | 91910754 | 1.9 | 0.2 |
| rs2810894 | 91915822 | −0.17 | 0.7 |
| rs1804506 | 91920601 | 0.24 | 0.7 |
| rs6604050 | 91926079 | −0.30 | 0.5 |
| rs2765887 | 91931035 | 0.11 | 0.8 |
| rs2765889 | 91935931 | −0.01 | 1.0 |
| rs6660484 | 91945196 | 0.47 | 0.3 |
| rs1805113 | 91950526 | 0.06 | 0.9 |
| rs7532589 | 91965267 | 0.46 | 0.4 |
| rs11578802 | 91970265 | 0.29 | 0.5 |
| rs923378 | 91975471 | −0.38 | 0.6 |
| rs4658112 | 91981254 | 0.09 | 0.8 |
| rs284170 | 91987216 | 1.3 | 0.03 |
| rs7529421 | 91994013 | −0.18 | 0.8 |
| rs284176 | 91996564 | 0.49 | 0.3 |
| rs12044500 | 92003913 | −0.81 | 0.1 |
| rs6700529 | 92008930 | 0.26 | 0.7 |
| rs1473486 | 92014164 | −0.32 | 0.4 |
| rs4658270 | 92031370 | −0.52 | 0.2 |
| rs284165 | 92041549 | −1.5 | 0.4 |
| rs10493859 | 92046232 | −0.59 | 0.2 |
| rs10874996 | 92072672 | 1.0 | 0.04 |
| rs2129972 | 92077838 | −2.0 | 0.02 |
| rs6686126 | 92083462 | 0.92 | 0.3 |
| rs17575497 | 92088484 | 2.0 | 0.2 |
| rs2634028 | 92093441 | −2.0 | 0.02 |
| rs2046737 | 92099224 | −2.0 | 0.02 |
| rs12727153 | 92104106 | 4.9 | 0.005 |
| rs6656018 | 92108672 | 0.18 | 0.7 |
| rs1192524 | 92113272 | 0.22 | 0.6 |
| rs6680463 | 92130774 | −0.43 | 0.3 |
| rs12566180 | 92135390 | −0.43 | 0.3 |
| rs12141128 | 92146335 | −0.42 | 0.3 |
| rs2770186 | 92151431 | −0.42 | 0.3 |
| rs883873 | 92152890 | −1.6 | 0.06 |
Definition of abbreviations: SNP, single-nucleotide polymorphism; TGFBR3, transforming growth factor-β receptor 3.
March 2006 human genome reference sequence (NCBI Build 36).
SNP associations with P < 0.05 are shown in boldface type.
A SNP 3′ to the GCLM gene (rs2235970) was significantly associated with FEV1 in the Boston Early-Onset COPD study, but no association with COPD susceptibility was seen in the NETT-NAS study (Table E3). No other SNPs in GCLM were significantly associated in either sample, and therefore were not followed up in the ICGN.
DISCUSSION
Using clinical chest CT scans, we have previously identified emphysema-predominant probands in the Boston Early-Onset COPD Study (8). Without CT scans on all family members, we could not perform genome-wide linkage analysis of CT emphysema; stratified linkage analysis of airflow obstruction phenotypes in families of emphysema-predominant probands served as a surrogate phenotype. We identified a region on chromosome 1p with significantly increased evidence for linkage in the subset of families of the emphysema-predominant probands. This linkage peak from the stratified analysis pointed to a chromosomal region that may be important in emphysema, in addition to the linkage peaks for spirometric traits previously identified in all subjects (16). In a previous publication, we found no association with GSTM1, a potential candidate gene (4). In the current study, we have tested two additional candidate genes for emphysema located in the linked region, finding SNP-level replication of the association between two intronic SNPs in TGFBR3 (also known as betaglycan) and COPD phenotypes in three study samples (Boston Early-Onset COPD Study, NETT-NAS and ICGN), with gene-level replication in a fourth study (LHS). A coding SNP (Ser15Phe), or proxy SNPs for this coding SNP, was associated in three of the four populations. In the Boston Early-Onset COPD Study, the ICGN, and the LHS, the main associated phenotype was FEV1; subjects in NETT-NAS were ascertained based on FEV1 levels, among other criteria. The two intronic SNPs also showed a trend toward association with quantitative emphysema measurements in the subset of ICGN participants who had chest CT scans. In NETT, all subjects had emphysema on chest CT, and in the Boston Early-Onset COPD Study, the association remained significant in the families of emphysema-predominant probands, also pointing to the potential relevance of TGFBR3 as a candidate gene for the emphysema component of COPD. Chest CT scans were not available in the Lung Health Study.
The only reported genome-wide linkage analyses for COPD-related traits have been performed in the Boston Early-Onset COPD Study (16, 21). However, a genome-wide linkage analysis in 144 families from the Collaborative Study for the Genetics of Asthma found evidence of linkage to asthma on chromosome 1p only in subjects exposed to environmental tobacco smoke (42), pointing to the potential relevance of genes on chromosome 1p in smoking-related lung diseases. Our study is the first published report of an association between polymorphisms in TGFBR3 and COPD.
Other human and murine genetics studies demonstrate the potential importance of TGFBR3 in COPD and emphysema. In a microarray study of human lung tissue, Golpon and colleagues found reduced expression of TGFBR3 in lungs from five patients with emphysema compared with five normal lungs (43); this difference was validated by RT-PCR. In a genome-wide linkage analysis in mice, Reinhard and coworkers found significant linkage to lung function on murine chromosome 5, where the orthologous murine gene tgfbr3 is located (44). Sequencing of the tgfbr3 gene found three nonsynonymous coding variants between the two parental strains. However, Reinhard and colleagues did not describe a SNP corresponding to the human Ser15Phe variant, and a similar mouse variant is not found in the dbSNP database.
The functional effects of the two associated intronic SNPs are unknown. These SNPs may have functional consequences, or more likely, they are in LD with the causal variant(s). The Ser15Phe variant is located in the signal peptide of TGFBR3; the functional consequence of this amino acid change is not known, but it is predicted to be possibly damaging, according to PolyPhen analysis (45). We demonstrated associations with several SNPs in this large gene (223.5 kb), suggesting that multiple TGFBR3 variants may affect emphysema susceptibility. It also possible that the causal variant or variants are not located in the TGFBR3 gene, and that the associations we found are due to LD with other chromosome 1 genes. However, we did not find associations with two other COPD candidate genes in the region, and TGFBR3 is known to be expressed in human emphysematous lung (43). Several other lines of evidence have pointed to the importance of the TGF-β pathway in COPD (40, 46, 47).
The linkage and association analyses in the Boston Early-Onset COPD Study are limited by the availability of chest CT scans only on the probands; since CT scans were performed for clinical indications at the discretion of the local physician, and were not part of the study protocol, CTs were not available in the family members. Linkage and association analyses for qualitative or quantitative CT traits could not be performed, though we have previously demonstrated that emphysema predominance on CT can be used to define clinically relevant subgroups (8). We did not find association with emphysema measurements in NETT, though the sample size in NETT with quantitative CT data was limited. In the larger sample from the ICGN study, we were able to find trends for association with quantitative emphysema measurements.
Spurious association due to multiple testing is a concern in genetic association studies (6). To guard against multiple testing, we used a staged genotyping and replication approach (Figure 1). First, we screened a limited number of SNPs in TGFBR3 in the Boston Early-Onset COPD Study families and in the NETT-NAS case-control study. Only after identifying a replicated association did we test additional SNPs in that gene. The screening set of ht-SNPs plus additional significant SNPs were then tested in the large ICGN family study. We then compared the results to independent genotyping performed in a separate cohort study of patients with COPD from the LHS. Consistent replication of the results across multiple patient samples, using different study designs, offers strong protection against false positive results (5). The use of family-based study designs in the Boston Early-Onset COPD Study and the ICGN and the testing of a panel of unlinked SNPs in the NETT-NAS case-control study protect against spurious association due to population stratification.
To refine the heterogeneous syndrome of COPD, we used chest CT scans to define a set of early-onset COPD probands with emphysema-predominant COPD. Genome-wide linkage analysis followed by association testing identified variants in TGFBR3, a biologically plausible candidate gene, to be associated with COPD and with FEV1, an important intermediate phenotype. SNP-level replication was confirmed in follow-up studies comparing emphysema cases from NETT with control smokers and in a large family-based study from the ICGN. The same SNPs were not tested in the LHS, but gene-level replication was found for TGFBR3. COPD is, by definition, a complex disease. There are likely to be some genes relevant for COPD in general and other genes relevant only for COPD subtypes, such as emphysema or airway disease. Genetic studies in patients with COPD defined only on the basis of airflow obstruction may be sufficient to identify the former set of genes. However, more precise characterization of subjects with COPD, using chest CT scans, will be required to find distinct genes for emphysema and airway disease.
Supplementary Material
Acknowledgments
The authors thank Jody Sylvia and Lisa Catalano for their assistance with genotyping and sample management; Laura Kaufman for her assistance in cataloging the CT films; and Scott Weiss, Frank Speizer, Jeffrey Drazen, Hal Chapman, Leo Ginns, and Steven Mentzer for their roles in developing the Boston Early-Onset COPD Study. Co-investigators in the NETT Genetics Ancillary Study include Joshua Benditt, Gerard Criner, Malcolm DeCamp, Philip Diaz, Mark Ginsburg, Larry Kaiser, Marcia Katz, Mark Krasna, Neil MacIntyre, Barry J. Make, Rob McKenna, Fernando Martinez, Zab Mosenifar, Andrew Ries, Paul Scanlon, Frank Sciurba, and James Utz. International COPD Genetics Network (ICGN) Investigators (in addition to Drs. Lomas and Silverman): Alvar Agusti, Son Dureta Hospital and Fundación Caubet-Cimera, Palma de Mallorca, Spain; Peter M. A. Calverley, University of Liverpool, Liverpool, UK; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, BC, Canada; Barry J. Make, National Jewish Health, Denver, CO; Peter D. Paré, University of British Columbia, Vancouver, BC, Canada; Stephen I. Rennard, University of Nebraska, Omaha, NE; Jørgen Vestbo, Department of Cardiology and Respiratory Medicine, Hvidovre Hospital, Copenhagen, Denmark; Emiel F. M. Wouters, University Hospital Maastricht, The Netherlands. Co-investigators in the LHS genotyping project include Terri Beaty, Audrey Grant, Li Gao, Monica Campbell, Cassandra Foster, Kathy Farnell, and Helen Voelker. The authors thank the NHLBI and the LHS investigators, project coordinators, safety and data monitoring board members, and morbidity and mortality review board members. The principal investigators of the clinical and coordinating centers of the Lung Health Study are as follows. Case Western Reserve University, Cleveland, OH: M. D. Altose, M.D. (Principal Investigator); Henry Ford Hospital, Detroit, MI: M. S. Eichenhorn, M.D. (Principal Investigator); Johns Hopkins University School of Medicine, Baltimore, MD: R. A. Wise, M.D. (Principal Investigator), C. S. Rand, Ph.D. (Co-Principal Investigator); Mayo Clinic, Rochester, MN: P. D. Scanlon, M.D. (Principal Investigator); Oregon Health Sciences University, Portland, OR: A. S. Buist, M.D. (Principal Investigator), L. R. Johnson, Ph.D. (LHS Pulmonary Function Coordinator); University of Alabama at Birmingham, Birmingham, AL: W. C. Bailey, M.D. (Principal Investigator); University of California, Los Angeles, CA: D. P. Tashkin, M.D. (Principal Investigator); University of Manitoba, Winnipeg, MB, Canada: N. R. Anthonisen, M.D. (Principal Investigator, Steering Committee Chair), J. Manfreda, M.D. (Co-Principal Investigator), R. P. Murray, Ph.D. (Co-Principal Investigator); University of Minnesota Coordinating Center, Minneapolis, MN: J. E. Connett, Ph.D. (Principal Investigator), P. L. Enright, M.D., P. G. Lindgren, M.S., P. O'Hara, Ph.D., (LHS Intervention Coordinator), M. A. Skeans, M.S., H. T. Voelker; University of Pittsburgh, Pittsburgh, PA: R. M. Rogers, M.D. (Principal Investigator); University of Utah, Salt Lake City, UT: R. E. Kanner, M.D. (Principal Investigator).
This work was supported by National Institutes of Health grants HL080242, HL71393, HL075478, U01 HL065899, P01 HL083069, HL076322, U01 HL 066583; the Alpha-1 Foundation; the Flight Attendant Medical Research Institute. The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119), Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), Boston, MA. The Lung Health Study was supported by National Heart, Lung, and Blood Institute N01HR46002. K.C.B. was supported in part by the Mary Beryl Patch Turnbull Scholar Program. Genotyping services on the Lung Health Study samples were provided by the Johns Hopkins University under U.S. Federal Government contract number N01-HV-48195 from the National Heart, Lung, and Blood Institute. The International COPD Genetics Network was supported by GlaxoSmithKline.
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2008-0427OC on January 8, 2009
Conflict of Interest Statement: S.G.P. is a full time employee of GlaxoSmithKline (GSK). H.O.C. received $11,000 in 2005 and $4,800 in 2006 and 2007 for serving on an advisory board for GSK. In addition, H.O.C. is the co-investigator on two multi-center studies sponsored by GSK and has received travel expenses to attend meetings related to the project. H.O.C. has three contract service agreements with GSK to quantify the CT scans in subjects with COPD and a service agreement with Spiration, Inc. to measure changes in lung volume in subjects with severe emphysema. A percentage of H.O.C.'s salary between 2003 and 2006 ($15,000/year) derives from contract funds provided to a colleague (Peter D. Pare) by GSK for the development of validated methods to measure emphysema and airway disease using computed tomography. H.O.C. is the co-investigator (principal investigator: D. Sin) on a Canadian Institute of Health–Industry (Wyeth) partnership grant. There is no financial relationship between any industry and the current study. E.K.S. received an honorarium for a talk on COPD genetics in 2006, and grant support and consulting fees from GSK for two studies of COPD genetics. E.K.S. received an honorarium from Wyeth for a talk on COPD genetics in 2004. E.K.S. received an honorarium from Bayer for a symposium at the ERS Meeting in 2005. E.K.S. received an honorarium for a talk at the Lund Symposium in 2007 and consulting fees in 2008 from AstraZeneca. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA III, Loyd JE, Nichols WC, Trembath RC. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. The International PPH Consortium. Nat Genet 2000;26:81–84. [DOI] [PubMed] [Google Scholar]
- 2.Painter JN, Tapanainen H, Somer M, Tukiainen P, Aittomaki K. A 4-bp deletion in the Birt-Hogg-Dube gene (FLCN) causes dominantly inherited spontaneous pneumothorax. Am J Hum Genet 2005;76:522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet 2001;29:306–309. [DOI] [PubMed] [Google Scholar]
- 4.Hersh CP, Demeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, et al. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol 2005;33:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hersh CP, Raby BA, Soto-Quiros ME, Murphy AJ, Avila L, Lasky-Su J, Sylvia JS, Klanderman BJ, Lange C, Weiss ST, et al. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med 2007;176:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet 2003;361:865–872. [DOI] [PubMed] [Google Scholar]
- 7.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 2001;163:1256–1276. [DOI] [PubMed] [Google Scholar]
- 8.Hersh CP, Jacobson FL, Gill R, Silverman EK. Computed tomography phenotypes in severe, early-onset chronic obstructive pulmonary disease. COPD 2007;4:331–337. [DOI] [PubMed] [Google Scholar]
- 9.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med 2003;348:2059–2073. [DOI] [PubMed] [Google Scholar]
- 10.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging & Human Development 1972;3:5–17. [Google Scholar]
- 11.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, Donner CF, Make BJ, Muller NL, Rennard SI, Vestbo J, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008;178:500–505. [DOI] [PubMed] [Google Scholar]
- 12.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med 2007;176:167–173. [DOI] [PubMed] [Google Scholar]
- 13.Anthonisen NR, Connett JE, Kiley JP, Altose MD, Bailey WC, Buist AS, Conway WA Jr, Enright PL, Kanner RE, O'Hara P, et al. Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. JAMA 1994;272:1497–1505. [PubMed] [Google Scholar]
- 14.Hersh CP, Jacobson FL, Gill R, Litonjua AA, Reilly JJ, Silverman EK. Using disease subtypes to reduce phenotypic heterogeneity: the example of TGFBR3 and pulmonary emphysema [abstract]. 2007. American Society of Human Genetics 57th Annual Meeting, San Diego, CA.
- 15.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O'Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med 1998;157:1770–1778. [DOI] [PubMed] [Google Scholar]
- 16.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002;70:1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferris BG. Epidemiology Standardization Project (American Thoracic Society). Am Rev Respir Dis 1978;118:1–120. [PubMed] [Google Scholar]
- 18.American Thoracic Society. Standardization of Spirometry, 1994 Update. Am J Respir Crit Care Med 1995;152:1107–1136. [DOI] [PubMed] [Google Scholar]
- 19.The National Emphysema Treatment Trial Research Group. Rationale and design of The National Emphysema Treatment Trial: a prospective randomized trial of lung volume reduction surgery. Chest 1999;116:1750–1761. [DOI] [PubMed] [Google Scholar]
- 20.Hersh CP, Washko GR, Jacobson FL, Gill R, Estepar RS, Reilly JJ, Silverman EK. Interobserver variability in the determination of upper lobe-predominant emphysema. Chest 2007;131:424–431. [DOI] [PubMed] [Google Scholar]
- 21.Silverman EK, Mosley JD, Palmer LJ, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum Mol Genet 2002;11:623–632. [DOI] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998;62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarus R, Vercelli D, Palmer LJ, Klimecki WJ, Silverman EK, Richter B, Riva A, Ramoni M, Martinez FD, Weiss ST, et al. Single nucleotide polymorphisms in innate immunity genes: abundant variation and potential role in complex human disease. Immunol Rev 2002;190:9–25. [DOI] [PubMed] [Google Scholar]
- 24.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 2000;50:211–223. [DOI] [PubMed] [Google Scholar]
- 26.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 1999;65:220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crapo RO, Morris AH, Gardner RM. Reference spirometric values using techniques and equipment that meet ATS recommendations. Am Rev Respir Dis 1981;123:659–664. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hersh CP, DeMeo DL, Raby BA, Litonjua AA, Sylvia JS, Sparrow D, Reilly JJ, Silverman EK. Genetic linkage and association analysis of COPD-related traits on chromosome 8p. COPD 2006;3:189–194. [DOI] [PubMed] [Google Scholar]
- 32.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2004;170:1294–1301. [DOI] [PubMed] [Google Scholar]
- 33.Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison DJ, Cantlay AM, Rae F, Lamb D, Smith CA. Frequency of glutathione S-transferase M1 deletion in smokers with emphysema and lung cancer. Hum Exp Toxicol 1997;16:356–360. [DOI] [PubMed] [Google Scholar]
- 35.Baranova H, Perriot J, Albuisson E, Ivaschenko T, Baranov VS, Hemery B, Mouraire P, Riol N, Malet P. Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum Genet 1997;99:822–826. [DOI] [PubMed] [Google Scholar]
- 36.Cheng SL, Yu CJ, Chen CJ, Yang PC. Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur Respir J 2004;23:818–824. [DOI] [PubMed] [Google Scholar]
- 37.Chappell S, Daly L, Morgan K, Guetta-Baranes T, Roca J, Rabinovich R, Lotya J, Millar AB, Donnelly SC, Keatings V, et al. Genetic variants of microsomal epoxide hydrolase and glutamate-cysteine ligase in COPD. Eur Respir J 2008;32:931–937. [DOI] [PubMed] [Google Scholar]
- 38.Siedlinski M, Postma DS, van Diemen CC, Blokstra A, Smit HA, Boezen HM. Lung function loss, smoking, vitamin C intake, and polymorphisms of the glutamate-cysteine ligase genes. Am J Respir Crit Care Med 2008;178:13–19. [DOI] [PubMed] [Google Scholar]
- 39.Riise GC, Larsson S, Lowhagen O, Andersson BA. Circulating leukocyte adhesion molecules in stable asthma and nonobstructive chronic bronchitis. Allergy 1995;50:693–698. [DOI] [PubMed] [Google Scholar]
- 40.Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-{beta}1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649–1656. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 2004;59:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colilla S, Nicolae D, Pluzhnikov A, Blumenthal MN, Beaty TH, Bleecker ER, Lange EM, Rich SS, Meyers DA, Ober C, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol 2003;111:840–846. [DOI] [PubMed] [Google Scholar]
- 43.Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol 2004;31:595–600. [DOI] [PubMed] [Google Scholar]
- 44.Reinhard C, Meyer B, Fuchs H, Stoeger T, Eder G, Ruschendorf F, Heyder J, Nurnberg P, de Angelis MH, Schulz H. Genomewide linkage analysis identifies novel genetic loci for lung function in mice. Am J Respir Crit Care Med 2005;171:880–888. [DOI] [PubMed] [Google Scholar]
- 45.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002;30:3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Boer WI, van Schadewijk A, Sont JK, Sharma HS, Stolk J, Hiemstra PS, van Krieken JH. Transforming growth factor beta1 and recruitment of macrophages and mast cells in airways in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1998;158:1951–1957. [DOI] [PubMed] [Google Scholar]
- 47.Morris DG, Huang X, Kaminski N, Wang Y, Shapiro SD, Dolganov G, Glick A, Sheppard D. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes MMP12-dependent emphysema. Nature 2003;422:169–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.