Abstract
Rationale: T-helper type 2 (Th2) inflammation, mediated by IL-4, IL-5, and IL-13, is considered the central molecular mechanism underlying asthma, and Th2 cytokines are emerging therapeutic targets. However, clinical studies increasingly suggest that asthma is heterogeneous.
Objectives: To determine whether this clinical heterogeneity reflects heterogeneity in underlying molecular mechanisms related to Th2 inflammation.
Methods: Using microarray and polymerase chain reaction analyses of airway epithelial brushings from 42 patients with mild-to-moderate asthma and 28 healthy control subjects, we classified subjects with asthma based on high or low expression of IL-13–inducible genes. We then validated this classification and investigated its clinical implications through analyses of cytokine expression in bronchial biopsies, markers of inflammation and remodeling, responsiveness to inhaled corticosteroids, and reproducibility on repeat examination.
Measurements and Main Results: Gene expression analyses identified two evenly sized and distinct subgroups, “Th2-high” and “Th2-low” asthma (the latter indistinguishable from control subjects). These subgroups differed significantly in expression of IL-5 and IL-13 in bronchial biopsies and in airway hyperresponsiveness, serum IgE, blood and airway eosinophilia, subepithelial fibrosis, and airway mucin gene expression (all P < 0.03). The lung function improvements expected with inhaled corticosteroids were restricted to Th2-high asthma, and Th2 markers were reproducible on repeat evaluation.
Conclusions: Asthma can be divided into at least two distinct molecular phenotypes defined by degree of Th2 inflammation. Th2 cytokines are likely to be a relevant therapeutic target in only a subset of patients with asthma. Furthermore, current models do not adequately explain non–Th2-driven asthma, which represents a significant proportion of patients and responds poorly to current therapies.
Keywords: asthma, phenotypes, inflammation, airway remodeling
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Clinical studies increasingly suggest that asthma is heterogeneous, but the molecular basis for this heterogeneity is uncertain.
What This Study Adds to the Field
This study suggests that asthma can be divided into at least two distinct molecular phenotypes defined by degree of Th2 inflammation. Therapies targeting Th2 cytokines may be effective in only a subset of patients with asthma. Non–Th2-driven asthma represents a significant proportion of patients and responds poorly to current therapies.
Asthma is traditionally thought to result from aeroallergen-induced inflammation driven by T-helper type 2 (Th2) responses and mediated by cytokines including IL-4, IL-5, and IL-13. IL-13 is produced by activated T cells, basophils, eosinophils, and mast cells and is thought to be a central mediator of inflammation in asthma based on animal models (1, 2) and on findings of elevated levels of IL-13 in the airways of patients with asthma (3). Consequently, much of the ongoing basic research in asthma is directed at understanding how Th2 cytokines cause asthma-like pathology and physiology (4), and inhibitors of Th2 cytokines are under development as novel asthma therapies (5). However, there is increasing evidence that a significant proportion of human asthma may be driven by alternative forms of inflammation. In particular, studies of the cellular components of airway inflammation in asthma provide evidence for distinct eosinophilic and noneosinophilic phenotypes of asthma (6–8). Whether the molecular mechanisms underlying these clinical and cellular phenotypes of asthma differ is unknown. The identification and development of biomarkers for distinct molecular phenotypes of asthma would guide the direction of basic research and the clinical application of emerging asthma therapies that specifically target Th2 responses in the lung.
In a previous study, we identified periostin (POSTN), chloride channel regulator 1 (CLCA1), and serpin peptidase inhibitor, clade B, member 2 (SERPINB2) as epithelial genes that were specifically induced in asthma (as compared with healthy control subjects and smokers with mild-to-moderate chronic obstructive pulmonary disease) and directly regulated by IL-13 in vitro (9). Thus, epithelial expression of POSTN, CLCA1, and SERPINB2 may serve as a surrogate marker of Th2-driven inflammation in the lung. We hypothesized that this gene expression signature could be used to identify subsets of patients with asthma who differ with respect to the molecular mechanism underlying their airway inflammation and that these subsets would constitute distinct inflammatory, pathological, and clinical phenotypes. Some of the results of these studies have been previously reported in the form of abstracts (10–12).
METHODS
Airway Tissue Bank at the University of California, San Francisco
We studied samples stored in the University of California, San Francisco (UCSF) Airway Tissue Bank that had been collected during research bronchoscopy in healthy volunteers and volunteers with asthma. Bronchoscopy included collection of epithelial brushings, bronchoalveolar lavage (BAL), and bronchial biopsies using methods previously described (9, 13). BAL cell counts and differentials had been performed and databased, and macrophages had been sorted from BAL fluid using flow cytometry (14). Four to six bronchial biopsies had been obtained from second- through fifth-order carinae (contralateral to the brushing site), formalin fixed, and then paraffin embedded in isotropic uniform random orientation (15) to enable quantitative measures of remodeling using design-based stereology (16). An additional two bronchial biopsies had been homogenized using a rotor-stator homogenizer (Power Gen 35 H-01; Fisher Scientific, Pittsburgh, PA) for 20 seconds in RLT lysis buffer (Qiagen Inc., Valencia, CA) and processed for RNA using the Qiagen RNeasy minikit (Qiagen Inc.). RNA extracted from epithelial brushings, homogenates of bronchial biopsies, and lavage macrophages had been quality assured and aliquoted for future microrray- and PCR-based gene profiling. All studies were approved by the UCSF Committee on Human Research, written informed consent was obtained from all subjects, and all studies were performed in accordance with the principles expressed in the Declaration of Helsinki. The Airway Tissue Bank procedures were also reviewed and approved by UCSF's Committee on Human Research. Samples of epithelial brushings and macrophages from this tissue bank have been used in previously reported studies (9, 13, 14, 17, 18). For this study, we conducted new analyses on our previously generated epithelial microarray data and generated new data using samples from the tissue bank. In each of these new analyses, we used samples from all subjects in the initial epithelial array analyses for whom the respective samples were available (9), including gene expression profiles in biopsy homogenates and alveolar macrophages, quantitative measures of fibrosis and airway epithelial mucin stores in bronchial biopsies, and total and differential cell counts in BAL.
Study Population
Subjects with asthma (n = 42) had a prior physician diagnosis of asthma, symptoms consistent with asthma confirmed by a study physician, airway hyperresponsiveness, defined as a drop in FEV1 of 20% or greater with inhalation of less than 8 mg/mL of methacholine (PC20 methacholine) and either symptoms on 2 or more days per week, β-agonist use on 2 or more days per week, or an FEV1 less than 85% predicted. They did not take inhaled or oral corticosteroids for 4 weeks before enrollment. Healthy control subjects (n = 28) had no history of lung disease and lacked airway hyperresponsiveness (PC20 methacholine >16 mg/mL). Exclusion criteria for all subjects included a history of smoking (excluded for ≥10 pack-years total or any smoking in the past year); upper respiratory tract infection in the previous 4 weeks; asthma exacerbation within 6 weeks; and current use of salmeterol, astemizole, nedocromil sodium, sodium cromoglycate, methlyxanthines, montelukast, or zafirlukast. Healthy control subjects were excluded for a history of allergic rhinitis. Subjects underwent baseline evaluation by study physicians, including spirometry and methacholine challenge testing as described previously (9) and repeat spirometry before and after four puffs of albuterol at the second visit to measure bronchodilator reversibility. Subjects underwent allergen skin prick testing with a panel of 12 aeroallergens, a positive control, and a negative control (see Table E1 in the online supplement). Thirty-two of the subjects with asthma were also randomized to receive inhaled fluticasone (500 μg BID) or matched placebo in a blinded manner (ClinicalTrials.gov Identifier: NCT00187499). Subjects in the clinical trial underwent baseline bronchoscopy and were then randomized to study medication before undergoing repeat bronchoscopy 1 week later. They continued study medication for a total of 8 weeks. Spirometry was repeated after 4 and 8 weeks on study medication and after a 1-week run-out. These subjects were block randomized to yield a greater number of subjects on fluticasone as compared with placebo to maximize the power of analyses performed within the inhaled-steroid treated group.
Gene Expression Analyses
Microarray analyses on epithelial brushings had been performed as described previously (9). These data are available in MIAME-compliant format at GEO (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE4302). Additional real-time PCR (qPCR) analyses were performed using RNA from homogenized bronchial biopsies (34 patients with asthma and 14 healthy control subjects) and from lavage macrophages (14 patients with asthma and 15 healthy control subjects). Real-time PCR was performed using methods described previously (9) and with the primers and probes listed in Table E2.
Quantitative Morphometry
Reticular basement membrane thickness, a measure of subepithelial fibrosis, was measured in trichrome–stained, 3-μm sections using the orthogonal intercept method (15). Mucin stores were measured using Alcian blue/Periodic acid Schiff 3-μm sections using the line and point intersect counting method (13).
Statistical Methods
Microarray data analyses were performed using Bioconductor (19) in the R statistical environment. Unsupervised hierarchical clustering was performed using the Euclidean metric with complete linkage. All other analyses were performed using the JMP software package (SAS Institute, Cary, NC). Values are presented as mean ± SD or median (range) unless otherwise specified. Correlation was performed using Spearman's rank order correlation. For significance testing of PC20 and serum IgE levels, data were log transformed for normality. A P value less than 0.05 was taken as statistically significant, and sidak correction for multiple comparisons was used after initial three-group comparisons by analysis of variance.
RESULTS
Subject Characteristics
Characteristics of the 42 subjects with asthma and 28 healthy control subjects are presented in Table 1. These groups differed with respect to ethnicity (with a greater number of African Americans and Hispanics in the asthma group). Subjects with asthma also had FEV1, greater reversibility to albuterol and responsiveness to methacholine, higher serum IgE levels, and higher blood eosinophil numbers. Only two subjects with asthma and one healthy control subjects had a history of smoking (5, 1, and 4 pack-years, respectively), and none had smoked within the past year.
TABLE 1.
OVERALL SUBJECT CHARACTERISTICS*
| Healthy Control Subjects | Subjects with Asthma | P Value | |
|---|---|---|---|
| Sample size | 28 | 42 | — |
| Age, years | 36 ± 9 | 36 ± 11 | 0.98 |
| Gender, M:F (% F) | 12:16 (56) | 17:25 (60) | 0.84 |
| Ethnicity | |||
| White | 20 | 18 | 0.012 |
| African American | 0 | 8 | |
| Hispanic | 3 | 11 | |
| Asian/Pacific Islander | 5 | 5 | |
| FEV1, % predicted | 107 ± 13% | 87 ± 12% | <0.0001 |
| ΔFEV1 with albuterol (% of baseline) | 2.7 ± 3.4% | 11.1 ± 8.7% | <0.0001 |
| Methacholine PC20 | 64 (22–64) | 0.43 (0.05–7.3) | <0.0001 |
| IgE, IU/ml | 27 (3–287) (n = 26) | 214 (19–2627) | <0.0001 |
| Blood eosinophils, ×109/L | 0.10 ± 0.07 | 0.31 ± 0.22 | <0.0001 |
Definition of abbreviation: PC20 = provocative concentration required to cause a 20% decline in FEV1.
Values are presented as mean ± SD or median (range) unless otherwise specified. For significance testing of PC20 and IgE, data were log transformed for normality. In case of missing data, number of subjects for whom data exist are noted.
Asthma Phenotyping Based on Epithelial Markers
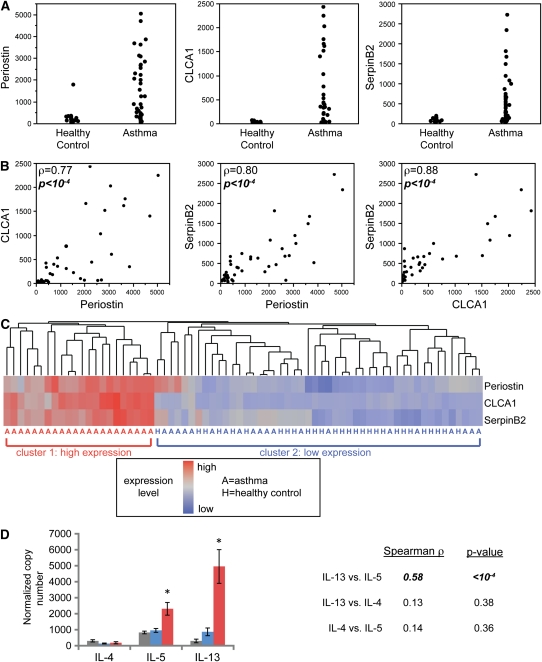
In previous gene expression microarray and qPCR studies, we identified three genes as highly induced in airway epithelial cells from subjects with asthma: POSTN, CLCA1, and SERPINB2 (9). We showed that IL-13 directly induces the expression of these three genes in epithelial cells in vitro (9); POSTN (20) and CLCA1 (21) can also be induced in vitro by IL-4. Because expression levels of IL-4 and IL-13 were low in epithelial cells and could not be used in profiling, these three genes served as surrogate markers of the effects of IL-13 and/or IL-4 on epithelial cells in the human airway. In the present study, we reanalyzed the previously published data on epithelial gene expression of POSTN, CLCA1, and SERPINB2 and observed that each of these three genes is overexpressed in some, but not all, of the 42 subjects with asthma (Figure 1A). Furthermore, expression levels of these three genes are highly correlated within individual subjects (Figure 1B). Together, these data suggest that markers of Th2 inflammation are overexpressed in a specific subset of patients with asthma. To formally define this subset, we performed unsupervised hierarchical clustering of all 70 subjects (42 subjects with asthma and 28 healthy control subjects) based on the microarray expression levels of POSTN, CLCA1, and SERPINB2 (Figure 1C). Approximately half of subjects with asthma (n = 22) showed consistently high expression of Th2 cytokine–induced genes and grouped together in one major branch of the dendrogram (cluster 1). Although POSTN, CLCA1, and SERPINB2 were significantly overexpressed when comparing all 42 patients with asthma to all 28 healthy control subjects (9), nearly half of the patients with asthma examined (n = 20) were indistinguishable from healthy control subjects on the basis of expression of these three genes. This subset of patients with asthma and all the healthy control subjects are grouped together in the second major branch of the dendrogram in Figure 1C (cluster 2).
Figure 1.
Expression levels of three IL-13 induced genes in the airway define two subgroups of patients with asthma. (A) Relative expression levels (normalized fluorescence units) of periostin (POSTN), chloride channel regulator 1 (CLCA1), and serpin peptidase inhibitor, clade B, member 2 (SERPINB2) in healthy control subjects (n = 28) and in patients with (n = 42). (B) Two-way comparisons of expression levels of POSTN, CLCA1, and SERPINB2 in the 42 patients with asthma. Spearman's rank order correlation (ρ) and P values are indicated. (C) Heatmap depicting unsupervised hierarchical clustering (Euclidean complete) of POSTN, CLCA1, and SERPINB2 expression levels in bronchial epithelium across all subjects at baseline. (D) Mean (±SEM) expression levels of IL-4, IL-5, and IL-13 in bronchial biopsy homogenates obtained contemporaneously with bronchial brushings from a subset of subjects depicted in A through C (cluster 1: 18 patients with T-helper type 2 [Th2]-high asthma, red bars; cluster 2: 14 healthy control subjects, gray bars; and 16 patients with Th2-low asthma, blue bars). Two-way correlations across all subjects between IL-4, IL-5, and IL-13 indicated at right (ρ).
Validation Using Bronchial Biopsies
To confirm the validity of subsetting using these previously published epithelial cell gene expression data, we performed new analyses of the expression levels of Th2 cytokines in bronchial biopsies obtained contemporaneously from 48 of the subjects (14 healthy control subjects, 18 cluster 1 patients with asthma, and 16 cluster 2 patients with asthma). Using qPCR, we found that IL-13, IL-5 and IL-4 expression was detectable in bronchial biopsy homogenates. IL-13 and IL-5 expression were significantly higher (*P < 0.002) in cluster 1 patients with asthma than in cluster 2 patients with asthma or healthy control subjects, but IL-4 expression was not (Figure 1D). Possible interpretations of this finding are that IL-4 functions predominantly to skew T-lymphocyte function rather than to exert effects in the peripheral tissues, that very low levels of IL-4 are sufficient to exert effects locally, that IL-4 mRNA is comparatively unstable in vivo, or that the role played by IL-4 in asthma is small compared with that played by IL-13 or IL-5. By contrast, cluster 2 patients with asthma did not differ from healthy control subjects in expression of IL-4, IL-5, or IL-13. In addition, we found that expression levels of IL-13 and IL-5 were highly correlated across all of the subjects with asthma (r = 0.58; P < 0.0001; Figure 1D), so that subjects with high-level of expression of IL-13 also had high-level expression of IL-5. Based on these results, we designated subjects in cluster 1 as having “Th2-high” asthma and patients with asthma in cluster 2 as having “Th2-low” asthma. To further investigate pathways contributing to Th2-low asthma, we measured the expression of Th1, Th17, and Treg cytokines including IL-12A, IFN-γ, TNF-α, IL-17A, IL-17F, and IL-10 in bronchial biopsies. We did not observe differential expression of TNF-α, IL-10, IL-17A, or IL-17F between healthy control subjects and either group of patients with asthma (data not shown). However, although we observed no significant differences in Th1 cytokine expression (IL-12A and IFN-γ) between patients with Th2-low asthma and healthy control subjects, patients with Th2-high asthma showed reduced expression of IL-12A and IFN-γ, as would be predicted based on the expected reciprocal balance of high Th2- and low Th1-driven inflammation in this group (Figure E1). To further explore this reciprocal balance, we calculated the ratio (log difference) for each pair of Th2 (IL-13, IL-5) and Th1 (IL-12A, IFN-γ) cytokines (i.e., IL-13/IL-12A, IL-13/IFN-γ, IL-5/IL-12A, and IL-5/IFN-γ). We found in each instance that the ratio of Th2 to Th1 was higher in the Th2-high group than in healthy control subjects or in subjects in the the Th2-low group (all P < 0.012), and in each comparison the Th2-low group did not differ significantly from healthy control subjects.
Clinical and Inflammatory Features of Th2-high Asthma
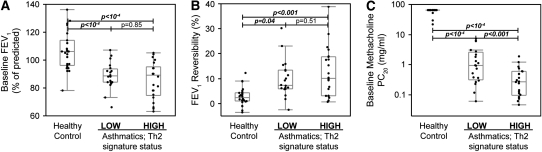
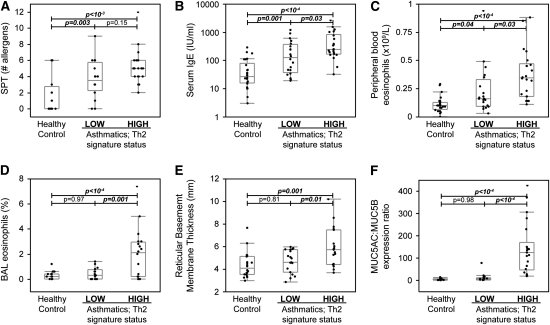
Although subjects with Th2-high asthma were indistinguishable from those with Th2-low asthma based on demographic characteristics, severity (lung function), or bronchodilator responsiveness (Table 2; Figures 2A and 2B), the degree of airway hyperresponsiveness (PC20 to methacholine) in Th2-high asthma was significantly worse than in Th2-low asthma (Figure 2C). With regard to measures of allergic inflammation, Th2-high and Th2-low subgroups had increased allergen skin prick test reactivity as compared with healthy control subjects (Figure 3A), although the Th2-low subgroup tended to have fewer positive skin tests than the Th2-high subgroup and tended to be sensitized less frequently to aeroallergens such as dog and house dust mite (Figure E2a). Subjects with Th2-high asthma had higher serum IgE levels and peripheral blood eosinophil counts than subjects with Th2-low asthma, even subjects with though Th2-low asthma differed from healthy control subjects (Figures 3B and 3C). In addition, eosinophil numbers in the BAL were significantly elevated (on average) in subjects with Th2-high asthma (Figure 3D), although five subjects with Th2-high asthma had BAL eosinophil numbers that overlapped with healthy control subjects. BAL eosinophil numbers were similar to healthy control subjects in Th2-low asthma. These data demonstrate enrichment for airway hyperresponsiveness, IgE levels, and eosinophilic inflammation in Th2-high asthma but that allergen skin test reactivity was not restricted to this subgroup.
TABLE 2.
SUBJECT CHARACTERISTICS AND BRONCHOSCOPIC FEATURES BY ASTHMA PHENOTYPE*
| Subjects with Asthma |
||||
|---|---|---|---|---|
| Healthy Control Subjects | Th2 Signature Low | Th2 Signature High | P Value, Low vs. High | |
| Sample size | 28 | 20 | 22 | — |
| Age, years | 36 ± 9 | 36 ± 11 | 37 ± 12 | 0.98 |
| Gender, M:F (% F) | 12:16 (56) | 6:14 (70) | 11:11 (50) | 0.19 |
| Ethnicity | ||||
| White | 20 | 9 | 9 | 0.98 |
| African American | 0 | 4 | 4 | |
| Hispanic | 3 | 5 | 6 | |
| Asian or Pacific Islander | 5 | 2 | 3 | |
| FEV1, % predicted | 107 (13) | 89 (10) | 85 (13) | 0.85 |
| ΔFEV1 with albuterol (% of baseline) | 2.7 ± 3.4 | 9.7 ± 7.4 | 12.5 ± 9.8 | 0.51 |
| Methacholine PC20 | 64 (22–64) | 0.93 (0.06–7.3) | 0.27 (0.05–1.9) | <0.001 |
| IgE, IU/ml | 27 (3–287); n = 26 | 125 (19–1,194) | 244 (32–2,627) | 0.031 |
| Blood eosinophils, ×109/L | 0.10 ± 0.07 | 0.23 ± 0.21 | 0.37 ± 0.22 | 0.027 |
| BAL eosinophil % | 0.26 ± 0.29; n = 22 | 0.42 ± 0.46; n = 16 | 1.9 ± 1.9; n = 20 | 0.001 |
| RBM thickness, μm | 4.34 ± 1.11; n = 22 | 4.67 ± 0.99; n = 19 | 5.91 ± 1.72; n = 19 | 0.014 |
| ΔFEV1 with fluticasone at 4 wk, L | N/A | 0.03 ± 0.12; (−0.12 to 0.21); n = 6 | 0.35 ± 0.2; (−0.02 to 0.73); n = 10 | 0.004 |
| ΔFEV1 with fluticasone at 8 wk, L | N/A | 0.04 ± 0.12; (−0.11 to 0.26); n = 6 | 0.25 ± 0.23;(−0.18 to 0.52); n = 10 | 0.05 |
Definition of abbreviations: BAL = bronchoalveolar lavage; PC20 = provocative concentration required to cause a 20% decline in FEV1; RBM = reticular basement membrane.
Values are presented as mean ± SD or median (range) unless otherwise specified. P values are sidak corrected for multiple testing (across the three groups). For significance testing of PC20 and IgE, data were log transformed for normality. In case of missing data, the number of subjects for whom data exist is noted. P values relative to healthy control subjects are also depicted in Figures 2 and 3.
Figure 2.
Clinical features of asthma are present in patients with Th2-high and Th2-low asthma. (A) FEV1, a measure of airway obstruction. (B) Improvement in FEV1 after four puffs (360 μg) of albuterol (bronchodilator reversibility testing). (C) Provocative concentration of methacholine required to induce a 20% decline in FEV1 (PC20), a measure of airway hyperresponsiveness.
Figure 3.
Markers of allergy, eosinophilic inflammation, and airway remodeling are increased in Th2-high asthma. (A) Skin prick test (SPT) results using a panel of 12 aeroallergens. (B) Serum IgE. (C) Peripheral blood eosinophil count. (D) Eosinophils as a percentage of total cells in bronchoalveolar lavage (BAL) fluid. (E) Stereologic measurement of reticular basement membrane thickness on bronchial biopsy, a measure of subepithelial fibrosis. (F) Ratio of MUC5AC to MUC5B expression in epithelial brushings as determined by real-time PCR.
Subepithelial Fibrosis and Epithelial Mucins in Th2-high Asthma
To assess the relationship between Th2 phenotype and structural changes in the airway, we performed new analyses of long-term pathological changes known as “airway remodeling” in bronchial biopsies from the same subjects. We specifically analyzed reticular basement membrane thickness (a measure of subepithelial fibrosis) and epithelial mucin stores.
We found that reticular basement membrane thickness was increased in subjects with Th2-high asthma but normal in subjects with Th2-low asthma (Figure 3E). In addition, we found that epithelial mucin stores were increased in both groups of subjects with asthma, but this increase was significant only in subjects with Th2-high asthma (Figure E3A). Although these differences in total mucin stores were modest, qPCR revealed a striking difference in expression of the major gel-forming mucins in airway epithelial cells in Th2-high asthma as compared with patients with Th2-low asthma and healthy control subjects (Figures E3B–E3D). Specifically, we found induction of MUC5AC and MUC2 and repression of MUC5B expression that was specific to Th2-high asthma. This alteration in the expression of specific mucin genes in Th2-high asthma is most evident in the ratio of MUC5AC to MUC5B expression (Figure 3F).
Alveolar Macrophage Gene Expression
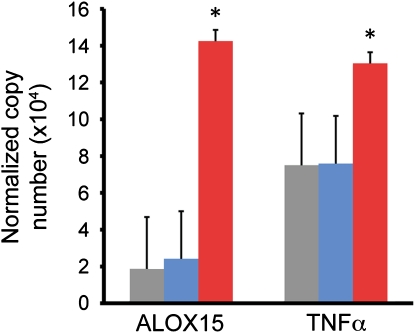
Alveolar macrophages may modulate allergic airway inflammation in asthma as a source of IL-13 (22) and leukotrienes or eicosanoid lipids (23, 24) or through “alternative activation” under the influence of IL-13 (25). To determine whether alveolar macrophages from subjects with Th2-high asthma manifest any of these findings, we performed new analyses of the expression levels of relevant genes using qPCR in 14 subjects with asthma and 15 healthy control subjects (Table E3). We found no evidence for induction of Th2 cytokines or of usual alternative activation markers in asthma generally or in the Th2-high subgroup specifically (data not shown). Levels of expression of IL-13 were below the limit of detection (cycle threshold >40) in 26 of 29 subjects, and IL-4 was below the limit of detection in 20 of the 29 subjects (no differences between the three groups for either cytokine; all P > 0.35). All other genes were within the limit of detection across samples. However, we did find increased expression of 15-lipoxygenase in Th2-high asthma (Figure 4; Table E4), consistent with prior findings of increased 15-lipoxygenase products in the airways in severe eosinophilic asthma (24). We also found an increase in expression of TNF-α that was limited to the Th2-high subgroup (Figure 4; Table E4).
Figure 4.
Alveolar macrophage gene expression in Th2-high and Th2-low asthma. Mean (+SEM) expression levels of 15-lipoxygenase (ALOX15) and TNF-α as determined by real-time PCR (*P < 0.03 for subjects with Th2-high asthma vs. healthy control subjects). n = 15 healthy control subjects (gray bars); n = 5 patients with Th2-low asthma (blue bars); and n = 9 patients with Th2-high asthma (red bars).
Reproducibility and Responsiveness to Inhaled Corticosteroids
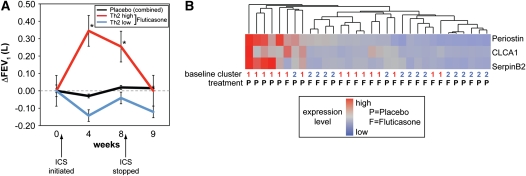
To identify gene expression markers of corticosteroid responsiveness, we measured FEV1 during the 8-week randomized controlled trial of inhaled fluticasone versus matched placebo, as previously described (9). When we reanalyzed these previously described data while stratifying subjects by Th2 status, we found that improvements in FEV1 were limited to those in the Th2-high subgroup. Specifically, the subjects with Th2-high asthma who were treated with inhaled fluticasone had statistically significant improvements in FEV1 at 4 and 8 weeks compared with subjects treated with placebo, whereas subjects with Th2-low asthma did not (Figure 5A). At 4 weeks, only 1 of 6 subjects with Th2-low asthma had a greater than 150-ml improvement in FEV1, whereas 9 of 10 subjects with Th2-high asthma had this degree of improvement (P < 0.003 by Fishers exact test). At 8 weeks, only 1 of 6 subjects with Th2-low asthma had greater than 150-ml improvement, as compared with 7 of 10 patients with Th2-high asthma (P = 0.038). These improvements in FEV1 in the Th2-high group were lost after a 1-week run-out period off drug. To determine the reproducibility of the Th2-high phenotype and the responsiveness of Th2 markers to steroid therapy, we analyzed data from the second bronchoscopy performed 1 week after the initiation of treatment. While stratifying subjects by Th2 status, subjects with Th2-high asthma at baseline continued to exhibit a strong Th2 signature after 1 week of placebo. However, after 1 week of fluticasone, subjects with Th2-high asthma clustered with subjects who were Th2-low at baseline. Finally, subjects who were Th2-low at baseline remained Th2-low on repeat evaluation, whether they were treated with fluticasone or with placebo (Figure 5B).
Figure 5.
Responsiveness of Th2-high asthma to inhaled steroids and reproducibility of phenotypic markers after placebo in a randomized controlled trial. (A) FEV1 measured at baseline (week 0), after 4 and 8 weeks on daily fluticasone (500 μg BID), and 1 week after the cessation of fluticasone (week 9). *See Table 2 for number of subjects and P values. There was no significant change in FEV1 in response to placebo at any time point in either group. (B) Heatmap depicting unsupervised hierarchical clustering of POSTN, CLCA1, and SERPINB2 as in Figure 1C in bronchial epithelium of patients with asthma 1 week after the initiation of fluticasone or placebo treatment (n = 19 receiving fluticasone; 13 receiving placebo). Cluster at baseline for individual subjects in Figure 1C and treatment are indicated below the heatmap.
DISCUSSION
Basic research has established IL-13 and related Th2 cytokines as central regulators of allergic inflammation and many of the pathophysiologic changes associated with asthma (1, 2). Our work challenges these current concepts of asthma pathogenesis by showing that a gene signature for Th2-driven inflammation in airway epithelial cells is prominent in only half of patients with asthma; non–Th2-driven mechanisms therefore operate in the remaining half. This finding leads us to propose that asthma can be divided broadly into Th2-high and Th2-low molecular phenotypes. We validate this classification scheme through confirmatory analyses of gene expression in another tissue compartment (bronchial biopsies); through analysis of reproducibility on repeat examination; and through comprehensive characterization of the distinct clinical, inflammatory, pathological, and treatment-related characteristics of these two molecular phenotypes of asthma. These findings are timely in that they provide a mechanistic framework for the emerging clinical observation that asthma is a complex and heterogeneous disease (26).
Molecular phenotyping of asthma based on Th2 inflammation has important therapeutic implications. First, airway obstruction improves with inhaled steroids in the Th2-high sub-group, whereas it does not in the Th2-low subgroup. These findings identify subjects with Th2-low asthma as a subgroup with a clinical need that is poorly met by current therapies. Second, blockade of IL-13 and related Th2 cytokines is under active clinical development as a therapeutic strategy in asthma (5). Our data suggest that clinical response to these therapies may be limited to the specific subgroup of patients with Th2-high asthma. It has been noted that patient-specific treatments for asthma depend on the development of discriminatory markers of distinct asthma subtypes (27), and the markers of Th2-high asthma identified here should have direct application in patient selection for clinical trials. Whether patient heterogeneity has influenced recently completed clinical trials of IL-13 blockade (5) is uncertain, but these data provide a basis for investigating that possibility.
Our data suggest that Th2-driven inflammation is the molecular mechanism underlying the cellular phenotype of asthma known as “eosinophilic asthma” (28) because of the airway eosinophilia that we demonstrate in Th2-high asthma and because eosinophilic asthma and Th2-high asthma are characterized by subepithelial fibrosis (8, 29), ALOX15 production by alveolar macrophages (24), and lung function responses to inhaled corticosteroids (30, 31). In addition to these recognized features of eosinophilic asthma, we have identified further clinical features of Th2-high asthma, including increased mucin stores, which occur because of increases in MUC2 and MUC5AC and despite a marked decrease in MUC5B expression. We found increased expression of TNF-α in macrophages in Th2-high asthma. TNF-α is not considered a Th2 cytokine, but it has been previously associated with severe asthma (32). We predict that all of these features will also be found in eosinophilic asthma because it is likely that IL-5 is a major contributor to the airway and systemic eosinophilia in Th2-high asthma, and we found that IL-5 expression is significantly co-regulated with IL-13 expression and is increased in Th2-high asthma (see Figure 1D). IL-5 is a major stimulus of eosinophil differentiation, recruitment, activation, and survival (33). Two recent clinical trials of mepolizumab in patients with relatively severe asthma highlight the importance of IL-5 and eosinophilia in asthma exacerbations (34, 35). Despite reducing the frequency of asthma exacerbations, these studies did not show a reduction in airway hyperresponsiveness, suggesting that IL-5 blockade may leave residual tissue inflammation, that other Th2 cytokines (e.g., IL-13) drive airway hyperresponsiveness, or that airway hyperresponsiveness is an epiphenomenon. Other studies indicate that IL-13 can strongly induce the expression of eosinophil chemoattractants such as CCL11, CCL24, and CCL26 in the airway (36) and thus represents an IL-5–independent pathway that promotes eosinophil infiltration, activation, and survival in the airways. Residual IL-13 activity may therefore explain the incomplete tissue depletion of eosinophils observed in previous clinical trials of IL-5 blockade in asthma (37, 38).
Molecular phenotyping of asthma has some advantages over phenotyping by sputum cell counts or measures of airway pathology. First, underlying molecular mechanisms likely drive the inflammatory and pathological processes and thus represent proximal targets for directed therapy. Second, phenotyping by gene expression identifies the genes that may contribute to this complex disease based on DNA sequence variation and will help reduce phenotypic heterogeneity in genetic studies. Third, these gene expression array data facilitate the identification of secreted proteins that mark the phenotype and may be developed into diagnostic tests.
One strength of our approach to molecular phenotyping is the use of multiple sample types to provide a broad view of gene expression in the airway. Our preparations of epithelial cells and alveolar macrophages are valuable for gene expression profiling because they represent relatively homogeneous populations of cells. On the other hand, bronchial biopsies are valuable because the mix of cell types they contain (including inflammatory cells) permits direct measurement of Th1 and Th2 cytokines. However, because of the cell type heterogeneity inherent to bronchial biopsies, the cellular source of these cytokines remains uncertain.
Finally, these data reveal that a significant percentage of patients with asthma have a Th2-low phenotype that manifests clinical features of asthma, airway obstruction, airway hyperresponsiveness, and bronchodilator reversibility despite a paucity of Th2-driven inflammation. The causes of Th2-low asthma remain obscure, but possibilities include neutrophilic inflammation, IL-17–driven inflammation, intrinsic defects in barrier function and chronic subclinical infection by viruses, and atypical intracellular bacteria. Whatever the mechanism, our data suggest that it does not relate to uncontrolled allergic inflammation but rather to alternative mechanisms that are not steroid responsive. Future work should be directed at identifying noninvasive biomarkers that correlate with these molecular phenotypes in the airway and at developing a deeper understanding of the pathophysiologic mechanisms underlying asthma that is not driven by Th2 inflammation.
There are limitations inherent to our study design. First, our subjects were not treated with inhaled or oral steroids for at least 4 weeks before enrollment, and the use of leukotriene antagonists was excluded. Although these steps facilitated identification of this Th2 signature in the absence of the confounding effects of antiinflammatory medications, these inclusion/exclusion criteria limit the generalizability of the study to patients with more severe asthma. Whether these markers can be used to detect Th2-driven inflammation in patients who are poorly controlled despite corticosteroid treatment remains to be determined. Second, repeat bronchoscopy was performed 1 week after initiation of treatment, which was continued for a total of 8 weeks. Thus, the timing of the second bronchoscopy may underestimate the extent to which inhaled steroids influence inflammation and our molecular signature. Third, we excluded healthy control subjects who had a history of allergic rhinitis. Thus, it is possible that our Th2 signature may also be detectable in the upper or lower airway of atopic nonasthmatic subjects. Finally, our study design does not definitively rule out the possibility that the Th2-low phenotype in part represents patients with well-controlled asthma. However, the majority of our subjects had not been using inhaled steroids for a long period before enrollment (i.e., we did not stop the use of inhaled steroids in recruitment of our subjects but rather sought patients who were not using inhaled steroids chronically of their own accord), minimizing the effect of prior steroid therapy in our analyses. Furthermore, patients with Th2-high and Th2-low asthma did not significantly differ with regard to airway obstruction or reversibility at baseline in this study, which lends further credence to the notion that the severity of their disease was comparable and not due to differences in asthma control.
Supplementary Material
Acknowledgments
Inhaled fluticasone and matching placebo were kindly provided as a gift from GlaxoSmithKline. The authors thank members of the UCSF Airway Clinical Research Center, including Homer Boushey, Stephen Lazarus, Cindy Nguyen, Peggy Cadbury, Hofer Wong, Roderick Carter, Steven Hays, and Anh Innes.
Supported by National Institutes of Health grants HL56385, HL080414, and HL66564 (J.V.F.); RR17002 and HL095372 (P.G.W.); by the Sandler Asthma Basic Research Center (J.V.F.) from the General Clinical Research Centers of Moffitt Hospital (RR-00079) and San Francisco General Hospital (RR-00083); and by a grant from Genentech Inc.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200903-0392OC on May 29, 2009
Conflict of Interest Statement: P.G.W. received more than $100,001 research grant to study biomarkers in asthma from Genentech and is the co-inventor on a patent for the development of biomarkers in asthma. B.M. received more than $100,001 in salary from employment by Genentech and has $10,001 to $50,000 in stock owernship or options from Genentech. D.F.C. is an employee of Genentech. G.J. is a full-time employee of Roche/Genentech Inc. A.R.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.E. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.L.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.R.A. received more than $100,001 in salary from employment by Genentech and holds a patent for methods for treating and diagnosing asthma from Genentech. J.V.F. received $1,001 to $5,000 from Amira, up to $1,000 from Gilead, $1,001 to $5,000 from Merck, $1,001 to $5,000 from Roche, up to $1,000 from Aerovance in consultancy fees, $5,001 to $10,000 as a member on a scientific advisory board from Cytokinetics, more than $100,001 in grants for research in asthma and cystic fibrosis from Genentech, $50,001 to $100,000 grant for research in asthma from Roche, and more than $100,001 grant for clinical trial in COPD from Boehringer Ingelheim. J.V.F. is the coinventor on a patent for the development of biomarkers of asthma.
References
- 1.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science 1998;282:2261–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science 1998;282:2258–2261. [DOI] [PubMed] [Google Scholar]
- 3.Humbert M, Durham SR, Kimmitt P, Powell N, Assoufi B, Pfister R, Menz G, Kay AB, Corrigan CJ. Elevated expression of messenger ribonucleic acid encoding IL-13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J Allergy Clin Immunol 1997;99:657–665. [DOI] [PubMed] [Google Scholar]
- 4.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest 2008;118:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet 2007;370:1422–1431. [DOI] [PubMed] [Google Scholar]
- 6.Haldar P, Pavord ID. Noneosinophilic asthma: a distinct clinical and pathologic phenotype. J Allergy Clin Immunol 2007;119:1043–1052. (quiz 1053−1054). [DOI] [PubMed] [Google Scholar]
- 7.Douwes J, Gibson P, Pekkanen J, Pearce N. Non-eosinophilic asthma: importance and possible mechanisms. Thorax 2002;57:643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzel SE. Asthma: defining of the persistent adult phenotypes. Lancet 2006;368:804–813. [DOI] [PubMed] [Google Scholar]
- 9.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA 2007;104:15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modrek B, Woodruff PG, Fahy JV, Arron JR. Gene expression signatures in bronchial epithelium define distinct molecular subtypes of asthma. Am J Respir Crit Care Med 2008;177:A814. [Google Scholar]
- 11.Koth LL, Hou L, Woodruff PG. Absence of alternative activation markers but induction of ALOX15 and TNFα in alveolar macrophages in “IL-13 high” asthma [abstract]. Am J Respir Crit Care Med 2009;179:A3723. [Google Scholar]
- 12.Arron JR, Jia G, Modrek B, Choy DF, Abbas AR, Woodruff PG, Fahy JV. Peripheral biomarkers of an IL-13 induced bronchial epithelial gene signature in asthma. J Allergy Clin Immunol 2009;123:S74. [Google Scholar]
- 13.Innes AL, Woodruff PG, Ferrando RE, Donnelly S, Dolganov GM, Lazarus SC, Fahy JV. Epithelial mucin stores are increased in the large airways of smokers with airflow obstruction. Chest 2006;130:1102–1108. [DOI] [PubMed] [Google Scholar]
- 14.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med 2005;172:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrando RE, Nyengaard JR, Hays SR, Fahy JV, Woodruff PG. Applying stereology to measure thickness of the basement membrane zone in bronchial biopsy specimens. J Allergy Clin Immunol 2003;112:1243–1245. [DOI] [PubMed] [Google Scholar]
- 16.Weibel ER, Hsia CC, Ochs M. How much is there really? Why stereology is essential in lung morphometry. J Appl Physiol 2007;102:459–467. [DOI] [PubMed] [Google Scholar]
- 17.Seibold MA, Donnelly S, Solon M, Innes A, Woodruff PG, Boot RG, Burchard EG, Fahy JV. Chitotriosidase is the primary active chitinase in the human lung and is modulated by genotype and smoking habit. J Allergy Clin Immunol 2008;122:944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol 2005;116:305–311. [DOI] [PubMed] [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, McKenzie AN, Nagai H, Hotokebuchi T, Izuhara K. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J Allergy Clin Immunol 2006;118:98–104. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Dong Q, Louahed J, Dragwa C, Savio D, Huang M, Weiss C, Tomer Y, McLane MP, Nicolaides NC, et al. Characterization of a calcium-activated chloride channel as a shared target of Th2 cytokine pathways and its potential involvement in asthma. Am J Respir Cell Mol Biol 2001;25:486–491. [DOI] [PubMed] [Google Scholar]
- 22.Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, Benoit LA, Byers DE, Alevy Y, Tucker J, et al. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med 2008;14:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters-Golden M. The alveolar macrophage: the forgotten cell in asthma. Am J Respir Cell Mol Biol 2004;31:3–7. [DOI] [PubMed] [Google Scholar]
- 24.Chu HW, Balzar S, Westcott JY, Trudeau JB, Sun Y, Conrad DJ, Wenzel SE. Expression and activation of 15-lipoxygenase pathway in severe asthma: relationship to eosinophilic phenotype and collagen deposition. Clin Exp Allergy 2002;32:1558–1565. [DOI] [PubMed] [Google Scholar]
- 25.Gordon S. Alternative activation of macrophages. Nat Rev Immunol 2003;3:23–35. [DOI] [PubMed] [Google Scholar]
- 26.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet 2008;372:1107–1119. [DOI] [PubMed] [Google Scholar]
- 27.Adcock IM, Caramori G, Chung KF. New targets for drug development in asthma. Lancet 2008;372:1073–1087. [DOI] [PubMed] [Google Scholar]
- 28.Simpson JL, Scott R, Boyle MJ, Gibson PG. Inflammatory subtypes in asthma: assessment and identification using induced sputum. Respirology 2006;11:54–61. [DOI] [PubMed] [Google Scholar]
- 29.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med 1999;160:1001–1008. [DOI] [PubMed] [Google Scholar]
- 30.Berry M, Morgan A, Shaw DE, Parker D, Green R, Brightling C, Bradding P, Wardlaw AJ, Pavord ID. Pathological features and inhaled corticosteroid response of eosinophilic and non-eosinophilic asthma. Thorax 2007;62:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavord ID, Brightling CE, Woltmann G, Wardlaw AJ. Non-eosinophilic corticosteroid unresponsive asthma. Lancet 1999;353:2213–2214. [DOI] [PubMed] [Google Scholar]
- 32.Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, Bradding P, Brightling CE, Wardlaw AJ, Pavord ID. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med 2006;354:697–708. [DOI] [PubMed] [Google Scholar]
- 33.Takatsu K, Nakajima H. IL-5 and eosinophilia. Curr Opin Immunol 2008;20:288–294. [DOI] [PubMed] [Google Scholar]
- 34.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med 2009;360:973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O'Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med 2009;360:985–993. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann N, Hershey GK, Foster PS, Rothenberg ME. Chemokines in asthma: cooperative interaction between chemokines and IL-13. J Allergy Clin Immunol 2003;111:227–242. (quiz 243). [DOI] [PubMed] [Google Scholar]
- 37.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med 2003;167:199–204. [DOI] [PubMed] [Google Scholar]
- 38.Flood-Page P, Swenson C, Faiferman I, Matthews J, Williams M, Brannick L, Robinson D, Wenzel S, Busse W, Hansel TT, et al. A study to evaluate safety and efficacy of mepolizumab in patients with moderate persistent asthma. Am J Respir Crit Care Med 2007;176:1062–1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.