Abstract
Rationale: Prenatal exposure to tobacco smoke increases the risk for diseases later in the child's life that may be mediated through alterations in DNA methylation.
Objectives: To demonstrate that differences in DNA methylation patterns occur in children exposed to tobacco smoke and that variation in detoxification genes may alter these associations.
Methods: Methylation of DNA repetitive elements, LINE1 and AluYb8, was measured using bisulfite conversion and pyrosequencing in buccal cells of 348 children participating in the Children's Health Study. Gene-specific CpG methylation differences associated with smoke exposure were screened in 272 participants in the Children's Health Study children using an Illumina GoldenGate panel. CpG loci that demonstrated a statistically significant difference in methylation were validated by pyrosequencing. Estimates were standardized across loci using a Z score to enable cross-comparison of results.
Measurements and Main Results: DNA methylation patterns were associated with in utero exposure to maternal smoking. Exposed children had significantly lower methylation of AluYb8 (β, −0.31; P = 0.03). Differences in smoking-related effects on LINE1 methylation were observed in children with the common GSTM1 null genotype. Differential methylation of CpG loci in eight genes was identified through the screen. Two genes, AXL and PTPRO, were validated by pyrosequencing and showed significant increases in methylation of 0.37 (P = 0.005) and 0.34 (P = 0.02) in exposed children. The associations with maternal smoking varied by a common GSTP1 haplotype.
Conclusions: Life-long effects of in utero exposures may be mediated through alterations in DNA methylation. Variants in detoxification genes may modulate the effects of in utero exposure through epigenetic mechanisms.
Keywords: DNA methylation, epigenetics, prenatal, smoke
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Exposure to prenatal tobacco smoke is associated with several childhood diseases, but the mechanism for this association is not well understood.
What This Study Adds to the Field
Life-long effects of in utero tobacco smoke exposure may be mediated through alterations in DNA methylation. Children exposed to maternal smoking have differences in global and gene-specific DNA methylation.
Scientific evidence increasingly suggests that exposures during the intrauterine period can increase the risk for developing diseases in later life (1, 2). Prenatal exposure to maternal tobacco smoking is associated with lower pulmonary function and increased asthmatic symptoms in childhood (3, 4). Moreover, maternal and grand maternal smoking during pregnancy are associated with increased risk of childhood asthma, suggesting a persistent heritable effect (5). The clinical and public health implications of life-long or transgenerational effects after preventable exposures indicate an urgent need to understand better the potential mechanisms that mediate these effects.
Alterations to the epigenome are one mechanism by which prenatal exposures affect disease risk later in life. DNA methylation is the most studied type of epigenetic mark. In mammals, there are at least two developmental periods, in germ cells and in preimplantation embryos, in which methylation patterns are reprogrammed genome wide, generating cells with a broad developmental potential (6). Generally, this period involves demethylation and later remethylation in a cell or tissue specific manner. Thus, prenatal tobacco smoke exposure in early pregnancy may have important and lasting effects on DNA methylation and consequently influence gene expression and disease phenotypes across the life course.
Genetic variants can modulate the effects of prenatal tobacco smoke exposure on DNA methylation patterns. For example, risk of childhood asthma associated with prenatal tobacco smoke exposure differs by GSTM1 genotype. Children carrying the common null genotype whose mothers smoked during pregnancy are at greater risk for developing asthma (7, 8). Children who are null for GSTM1 and exposed to second hand smoke (SHS) have a reduced peak expiratory flow rate (9). Haplotypes in GSTP1 and maternal smoking jointly increase the odds of childhood asthma (10). Thus, GSTP1 and GSTM1, along with tobacco, smoke may jointly contribute to alteration of epigenetic marks, which may affect later disease onset.
In this study, we sought to provide the first evidence that differences in methylation patterns occur in children exposed to tobacco smoke and that these differences can be detected in DNA from buccal cells, an aero-digestive tissue cell type. We investigated two types of DNA methylation that have been associated with disease phenotypes: global methylation and promoter CpG island methylation. We evaluated the effect of prenatal tobacco smoke exposure on two DNA repetitive elements that are surrogate markers of global DNA methylation, long interspersed nucleotide element (LINE1) and a short interspersed nucleotide element (AluYb8). To assess promoter CpG island methylation, we studied methylation at 1,505 specific CpG loci in 807 gene promoters using the Illumina GoldenGate Bead Array DNA Methylation assay. To investigate the effects of common genetic variants associated with susceptibility to tobacco smoke, we assessed the effects of variants in GSTP1 and GSTM1 genes on susceptibility to prenatal tobacco smoke–related changes in DNA methylation.
METHODS
Study Population
This study was nested in the ongoing Children's Health Study (11). We sampled children from the 5,341 kindergarten and first graders who were enrolled in 2002. For the purposes of this study, a subset of 348 children with DNA and genotype information was selected for global DNA methylation analysis. A separate subset of 272 children was selected for analysis using the Illumina Platform. Subjects were chosen to ensure a balanced distribution of the exposure and ethnicity. Approximately 180 of the children selected for the global methylation were also selected for the Illumina methylation panel.
The institutional review board for human studies at the University of Southern California approved the study protocol, and parents or legal guardians consented for all study subjects.
Exposure Information
Information on maternal smoking and current second hand tobacco smoke exposure was collected annually using a structured questionnaire (see online supplement). Personal and family covariates and housing characteristics were also collected.
Genotyping
Buccal scrapes were collected using standard protocols, and genomic DNA was isolated using a PURGENE DNA isolation Kit (Gentra Systems, Minneapolis, MN). The GSTM1 deletion was chosen because of its functional relevance to respiratory disease (7). Four GSTP1 tagging single-nucleotide polymorphisms (SNPs) were chosen to represent the GSTP1 locus (rs1695, rs6591255, rs4147581, and rs749174) and accounted for greater than 80% of the haplotype variation (12). Greater details of SNP selection, haplotype creation, and haplotype frequencies are provided in the online supplement and in Table E1 in the online supplement. The SNPs were identified using allele-specific MGB probes on an ABI PRISM 7700 Sequence Detector (Applied Biosystems, Foster City, CA). Ten percent of the samples in each batch were repeated to verify inter- and intraplate validity.
DNA Methylation
Laboratory personnel performing DNA methylation analysis were blinded to study subject information. Bisulfite modification of genomic DNA was performed by using the EZ-96 DNA Methylation-Gold Kit (Zymo Research, Orange, CA). Methylation analyses of LINE1 and AluYb8 were performed by bisulfite-polymerase chain reaction (PCR) pyrosequencing assay using the HotMaster Mix (Eppendorf, Hamburg, Germany) and the PSQ HS 96 Pyrosequencing System (Biotage AB, Uppsala, Sweden) (13). See the online supplement for additional details.
The quality control steps we implemented are described in detail in the online supplement. To minimize scanner-to-scanner variation effects, timed mean normalization was used to normalize β values across plates (14). An additional 411 loci on the panel that contained SNPs or repetitive elements that could interfere with measurement of DNA methylation (Hyang-Min Byun, personal communication) were removed. Sixty-three loci had fewer than 30 values across all samples in at least one exposure stratum and were also removed from our analyses. In total, 1,031 CpG loci remained for analysis after all quality control steps were applied.
Statistical Analysis
We used a stepped-discovery approach for exposure-related CpG methylation that included a broad initial screen followed by quantitative validation. Descriptive analyses were first conducted to examine the distribution of methylation by subject characteristics and exposure groups. For each methylation endpoint, we applied a Z score transform to standardize the effects of exposure across the range of methylation to enable cross-comparison of results. Thus, the estimates obtained are reported as a difference in standard deviation (SD) units.
To assess the effects of tobacco smoke exposure on global methylation, we fitted linear regression models adjusted for gender, race, and Hispanic ethnicity. For CpG gene–specific methylation analysis, the Illumina GoldenGate Cancer methylation panel I was used (Illumina, San Diego, CA). This panel spans 1,505 CpG loci selected from 807 genes. Two independent runs of the Illumina methylation panel were performed by the USC Epigenome Center. A total of 182 samples were used for the first run, and 90 unique samples and 94 replicate samples were included in the second run. Each run was screened to generate a list of loci that demonstrated a change in methylation ‘Beta’ value of 0.05 or more comparing children exposed to prenatal tobacco smoke with unexposed children. The two lists were compared, and loci were retained if they were statistically significant at a P value of 0.05 on at least one list. Loci identified through the two separate Illumina runs were analyzed in a pooled analysis to improve power.
To assess differences in CpG methylation, we used nonparametric Wilcoxon rank-sum tests to screen the CpG loci from the Illumina panel. Linear regression models were then used to estimate the effects of tobacco smoke exposure adjusted for important covariates, including gender, race, Hispanic ethnicity, and Illumina plate. The methylation status of each CpG locus quantified by the Illumina Beta value was the dependent variable, and prenatal tobacco smoke exposure was the independent variable.
These genes were then validated by demonstrating consistency of effect across the two sets of unique samples (182 samples from the first Illumina run and 90 unique samples from the second Illumina run, excluding the replicate samples common to both groups) and then by using pyrosequencing as a second methylation assay. All confirmation pyrosequencing assays were designed to cover the same CpG sites interrogated by Illumina (Table E2). Linear regression models were used to analyze the pyrosequencing data for genes selected for validation.
To evaluate the joint effects of variants in GSTM1 and GSTP1 and prenatal smoke exposure, a recessive genetic model was used for GSTM1 null, and an additive model for haplotypes was used for GSTP1. The statistical interaction between SNP or haplotype and smoke exposure was evaluated by adding the corresponding product terms to the regression model and using a likelihood ratio test. All tests assumed a two-sided alternative hypothesis, assumed a 0.05 significance level, and were conducted using SAS/STAT software version 9.1.
RESULTS
Samples from 348 and 272 children were selected for global DNA methylation and Illumina analyses, respectively. The groups of children were similar (Table 1), with approximately 50% of the population being male and of Hispanic ethnicity. For global DNA methylation, 29% of the population was exposed to prenatal tobacco smoke, whereas for Illumina methylation, 49% was exposed. Mean methylation levels for selected Illumina assays and Pyrosequencing assays are shown in Table 2.
TABLE 1.
DEMOGRAPHIC CHARACTERISTICS FOR PARTICIPANTS IN THE CHILDREN'S HEALTH STUDY SELECTED FOR GLOBAL METHYLATION AND CpG-SITE SPECIFIC METHYLATION ASSAYS
| Global DNA Methylation (N = 348)
|
Illumina Methylation (N = 272)
|
|||
|---|---|---|---|---|
| n* | % | n* | % | |
| Gender | ||||
| Female | 183 | 53 | 144 | 53 |
| Male | 165 | 47 | 128 | 47 |
| In utero smoke | ||||
| No | 239 | 69 | 138 | 51 |
| Yes | 102 | 29 | 134 | 49 |
| Secondhand smoke | ||||
| No | 265 | 76 | 176 | 65 |
| Yes | 83 | 24 | 96 | 35 |
| Hispanic ethnicity | ||||
| No | 170 | 49 | 138 | 51 |
| Yes | 175 | 50 | 128 | 47 |
Differences in the number of subjects reflect available data for each item.
TABLE 2.
DISTRIBUTION OF METHYLATION VALUES FOR GENE-SPECIFIC PROMOTER CpGs AND GLOBAL DNA METHYLATION FOR ILLUMINA AND PYROSEQUENCING ASSAYS
| Illumina*
|
PSQ†
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Probe | Mean (SD) | n | Mean (SD) | n | ||||
| Gene-specific methylation | ||||||||
| AXL | 0.30 (0.17) | 231 | 7.94 (4.32) | 262 | ||||
| KLK11 | 0.67 (0.16) | 226 | 55.61 (9.25) | 246 | ||||
| MET | 0.37 (0.16) | 230 | 14.43 (4.50) | 265 | ||||
| NBL1 | 0.42 (0.18) | 211 | n/a | n/a | ||||
| PTPRO | 0.21 (0.14) | 209 | 15.70 (5.57) | 220 | ||||
| SNCG | 0.66 (0.17) | 203 | 46.66 (11.62) | 266 | ||||
| SPDEF | 0.38 (0.17) | 204 | 21.78 (6.46) | 266 | ||||
| TGFB3 | 0.59 (0.16) | 272 | 47.74 (13.40) | 267 | ||||
| Global methylation | ||||||||
| LINE1 | n/a | 75.4 (2.6) | 322 | |||||
| AluYb8 | n/a | 90.7 (0.87) | 306 | |||||
Methylation is expressed in Illumina ‘beta’ values.
Methylation is expressed as % methylation.
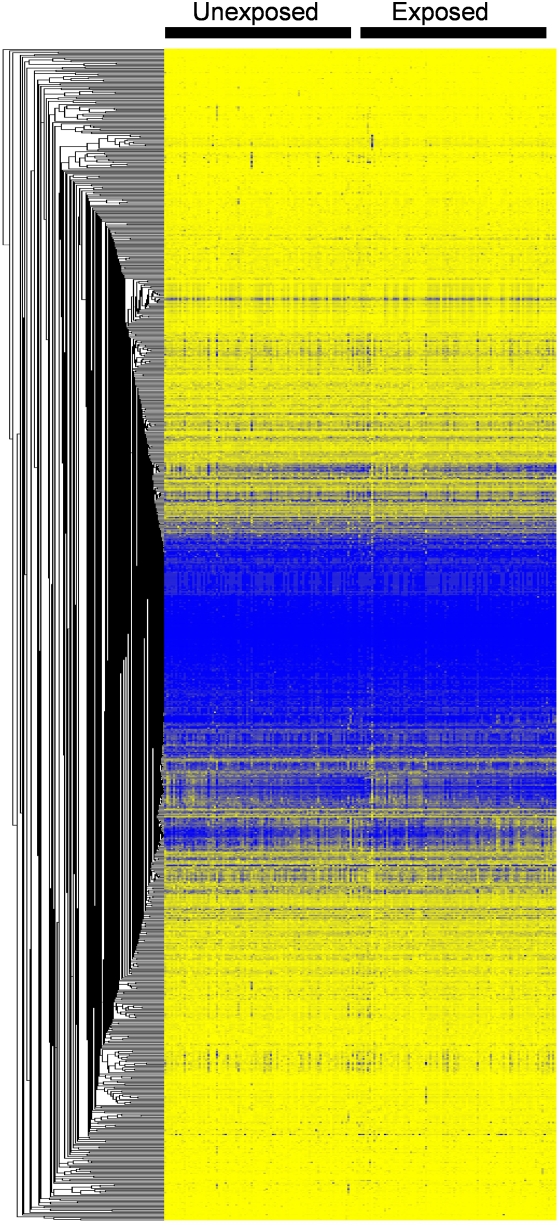
Prenatal tobacco smoke exposure was associated with detectable changes in DNA methylation patterns. Exposed children had a significantly lower level of methylation for AluYb8 (β, −0.31; P = 0.03) but not for LINE1 (β, 0.13; P = 0.32) compared with unexposed children (Table 3). Visual representation of methylation levels at the 1,031 gene-specific CpG loci is shown in Figure 1. There was no strong clustering of methylation patterns by prenatal smoke exposure.
TABLE 3.
THE EFFECT OF PRENATAL EXPOSURE TO MATERNAL SMOKING ON TWO MEASURES OF GLOBAL DNA METHYLATION
Z score transformed estimate is interpreted as a change in SD units. All models were adjusted for gender, race, and Hispanic ethnicity.
Figure 1.
One-way clustering of 1,031 Illumina CpG loci by prenatal tobacco smoke exposure.
Nine candidate CpG loci whose methylation levels differed significantly depending on the child's exposure to prenatal tobacco smoke were identified from the two separate runs of the Illumina panel (Table E3). Adjustment for important covariates eliminated HOXA5 from our candidate list. In all cases, methylation of the CpG sites was greater for exposed than unexposed children.
To confirm the observed differences in the loci methylation on the Illumina screen, pyrosequencing was performed for each of the eight genes in two independent sets of samples. One of the probes, NBL1, could not be pyrosequenced for technical reasons. Of the seven remaining probes, AXL and PTPRO showed effect estimates for tobacco smoke that were consistent across independent replication groups and were statistically significant in pooled analyses using the pyrosequencing assay (Table 4). AXL was most strongly affected by prenatal tobacco smoke, showing an increase in methylation of 0.37 (P = 0.005) in exposed children. PTPRO methylation increased by 0.34 (P = 0.02) in exposed children.
TABLE 4.
THE EFFECT OF PRENATAL EXPOSURE TO MATERNAL SMOKING ON METHYLATION OF CpG IN PROMOTERS FOR EIGHT GENES BY REPLICATION GROUP AND POOLED, USING PYROSEQUENCING
| Group 1‡ (n = 182)
|
Group 2‡ (n = 90)
|
Pooled (n = 272)
|
||||
|---|---|---|---|---|---|---|
| Gene* | β† | P Value | β† | P Value | β† | P Value |
| AXL | 0.51 | 0.001 | 0.45 | 0.09 | 0.37 | 0.005 |
| PTPRO | 0.38 | 0.03 | 0.35 | 0.23 | 0.34 | 0.02 |
| KLK11 | 0.48 | 0.004 | −0.12 | 0.67 | 0.29 | 0.04 |
| TGFB3 | 0.32 | 0.04 | 0.07 | 0.80 | 0.22 | 0.09 |
| MET | 0.26 | 0.09 | −0.02 | 0.93 | 0.16 | 0.21 |
| SPDEF | 0.24 | 0.12 | −0.07 | 0.79 | 0.12 | 0.37 |
| SNCG | −0.003 | 0.98 | −0.19 | 0.49 | −0.04 | 0.76 |
| NBL1 | n/a | n/a | n/a | n/a | n/a | n/a |
CpG loci interrogated by Illumina were located at the following positions relative to the transcription start site: AXL(−223), PTPRO(−371), KLK11(−103), TGFB3(+58), MET(+333), SPDEF(−6), SNCG(+119), and NBL1(−24).
Z score–transformed estimate is interpreted as a change in SD units. All models were adjusted for gender, race, and Hispanic ethnicity.
Groups 1 and 2 represent unique sets of individuals, all of whom were initially screened using the Illumina platform.
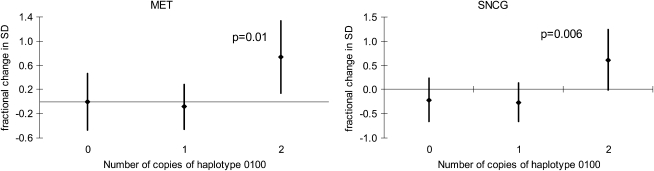
Functional variants in GSTM1 and GSTP1 genes altered susceptibility to prenatal tobacco smoke exposure on global DNA methylation. Prenatal exposure was associated with lower LINE1 methylation in the GSTM1-null children but higher methylation in the GSTM1-present children (Pint = 0.03) (Table 5). No interactions with any of the GSTP1 haplotypes were observed for LINE1 or AluYb8 or with GSTM1 and AluYb8. The association between prenatal tobacco smoke exposure and methylation of CpG loci varied among children with the most common GSTP1 haplotype, “0100,” which carries the variant allele for SNP rs4147581 (Figure 2). Prenatal tobacco smoke exposure increased methylation of loci in MET and SNCG genes to a greater extent in children who had two copies of this haplotype compared with those with fewer copies. No interactions with the GSTP1 haplotypes were observed for the other methylation loci.
TABLE 5.
THE EFFECTS PRENATAL EXPOSURE TO MATERNAL SMOKING ON LINE1 AND AluYb8 METHYLATION AMONG CHILDREN WITH AND WITHOUT GSTM1
| Prenatal Smoke Exposure | AluYb8
|
LINE1*
|
|||
|---|---|---|---|---|---|
| GSTM1 | β† | P Value | β† | P Value | |
| Present | No | ref | ref | ||
| Present | Yes | −0.31 | 0.14 | 0.41 | 0.04 |
| Null | No | ref | ref | ||
| Null | Yes | −0.39 | 0.07 | −0.14 | 0.47 |
P value for the interaction was 0.03.
Z score–transformed estimate is interpreted as a change in SD units.
Figure 2.
Effects of prenatal tobacco smoke exposure on methylation of CpG loci in MET and SNCG by number of copies of GSTP1 haplotype “0100.” 1 is the minor allele, and 0 is the common allele, with SNPs in the following order: rs6591255, rs4147581, rs1695, and rs749174.
Because SHS might also affect methylation and can be correlated with prenatal tobacco smoke exposure, we conducted a sensitivity analysis to evaluate the effect of SHS on the estimates for prenatal exposure. A sensitivity analysis of adjustment for season of buccal cell collection was also conducted because one study showed that season could confound their results (15). Adjustment for SHS or season did not appreciably change our results.
DISCUSSION
Prenatal tobacco smoke exposure was associated with small yet significant changes in global and CpG-specific DNA methylation. Exposed children had lower mean methylation for AluYb8 repetitive elements compared with unexposed children. Moreover, a preliminary scan of specific CpG loci suggested that eight genes were hypermethylated in exposed children, of which two genes were validated by pyrosequencing. Genetic susceptibility appeared to play a role in smoking-related effects based on differences in LINE1 methylation in children with the common GSTM1 null genotype and in CpG-specific methylation in children with a common GSTP1 haplotype. These findings provide some of the first evidence that the life-long effects of in utero exposures may be mediated through alterations in DNA methylation patterns.
Although changes in methylation have been evaluated extensively in relation to cancer, research into whether prenatal environmental exposures can alter methylation in humans is nascent. Nutritional differences during pregnancy affect methylation in humans and animals (16, 17). Aberrant promoter methylation in smoking-related lung tumors has been found for several genes (18, 19). Moreover, tobacco smoke generates oxidative stress, which can cause a wide range of DNA lesions. Such DNA lesions can interfere with the binding of DNA methyltransferases to DNA, resulting in global hypomethylation (20).
Only one study in humans has documented an association between prenatal tobacco smoke exposure and global methylation (21). Terry and colleagues reported that prenatal exposure resulted in higher levels of DNA methylation. In contrast, we found prenatal exposure results in decreased global methylation. These inconsistent findings may be related to differences in the assays and tissues used to measure methylation in the two studies. Terry and colleagues measured methylation in isolated mononuclear cells from whole blood, whereas in our study methylation was measured in buccal cells. The [3H]-methyl acceptance assay used by Terry and colleagues is limited by high day-to-day variability and inaccurate measurements of DNA concentration (22, 23). In contrast, pyrosequencing is a highly sensitive, quantitative, and high-throughput technique for DNA methylation detection (24). Moreover, the methyl acceptance assay infers global methylation by measuring the ability of DNA to accept radiolabeled methyl groups. It is unclear how much this assay should correlate with specific methylation of AluYb8, which has a high level of intrinsic methylation.
Two genes demonstrated consistent differential methylation dependent upon prenatal smoke exposure status. AXL, a receptor tyrosine kinase, promotes antiapoptosis, mitogenesis, invasion, and cell survival (25). The CpG site in AXL was located in the promoter at −223 base pairs upstream of the transcription start site in part of the core promoter region located −556 to −182, which is a known Sp1/Sp3 transcription factor binding site (26). PTPRO, a protein tyrosine phosphatase receptor, has been linked to differentiation and axonogenesis of central and peripheral nervous system neurons during gestation (27). The CpG site in PTPRO was located in the promoter at −371 base pairs upstream of the transcription start site. Gene expression in the promoters of AXL and PTPRO has previously been correlated with DNA methylation levels (26, 28). Although we were not directly able to evaluate correlations between smoking-induced methylation changes and subsequent changes in gene expression in our population, future research in this area is warranted. Given that these genes are susceptible to changes in methylation, exposure to tobacco smoke during gestation could alter expression of these genes, the consequences of which are unknown.
Genes involved in metabolism of tobacco smoke, particularly GSTM1 and GSTP1, can alter risks of childhood diseases that are associated with tobacco smoke (7, 8). We hypothesized that functional variants or haplotypes in these genes alter the associations between tobacco smoke exposure and methylation levels. Consistent with this hypothesis, we observed that the effects of maternal smoking on LINE1 methylation varied among children with and without the common GSTM1 null genotype. Smoke-related hypermethylation of CpG loci in MET and SNCG was also increased more in children who had two copies of the GSTP1 “0100” haplotype compared with those with fewer copies. We previously found this haplotype to protect against asthma in the larger CHS population, and variants in GSTM1 and GSTP1 are involved in asthma susceptibly from exposure to maternal smoking (10).
In this study, we observed that LINE1 was hypomethylated, whereas single CpG loci in eight genes were hypermethylated, in response to prenatal tobacco smoke exposure. Though not fully understood, this trend of global hypomethylation with region-specific hypermethylation has been observed previously in cancers (29). Global hypomethylation is believed to result in chromosomal instability and increased mutation events, whereas promoter hypermethylation can affect gene expression. Global hypomethylation could result from smoke-related ROS DNA damage during gestation if it prevents maintenance methyltransferases from binding (20). Tobacco smoke exposure could also affect de novo methylation in specific gene promoters, perhaps by incomplete erasure during methylation reprogramming that occurs in the embryo after fertilization.
Although the results of this study show that differences in methylation can be detected in children exposed to prenatal tobacco smoke, the differences were small for the endpoints evaluated. Therefore, the potential for bias needs to be considered in interpreting our findings. The modest differences in methylation level may result from having investigated CpG methylation in only 807 genes from a cancer panel in which some genes may have been less pertinent to the exposure evaluated. Examining a larger number of genes, using a genome-wide approach, or using a custom-targeted panel might identify additional genes with larger differences in methylation levels.
An association between prenatal tobacco smoke exposure and global methylation was observed only for AluYb8 but not LINE1; however, these are different repetitive elements with different residence times in the human genome and are not expected to respond to environmental stimuli in the same manner. The two repetitive elements are controlled through different mechanisms and have different transcription patterns in response to cellular stressors (30). Moreover, other modifying factors likely exist that we have failed to adequately capture, such as any interaction between genotypes and exposure on DNA methylation. Our analyses of GSTM1 suggest this may be the case.
The association with prenatal tobacco smoke may be explained by other factors not addressed in this study. Women who smoke during pregnancy may have a less healthy lifestyle in general than women who do not, which could include differences in diet, alcohol consumption, or the use of illegal drugs. Smoking has also been associated with reduced folate levels, and folate-deficient diets have been associated with global hypomethylation and gene-specific hypermethylation (31).
Given the limitations of collecting airway cells in children, we chose to use buccal mucosa cells, an aero-digestive tract epithelium, as an easily measurable surrogate for respiratory tract epithelium. Although the ideal tissue to use for assessing epigenetic changes in studies of respiratory health has not been defined, previous studies have demonstrated that DNA methylation changes of the oral epithelium can serve as a surrogate tissue for measuring DNA methylation changes of the lung (32).
Our data support the utility of Illumina as an epigenome-wide screening platform but also demonstrate the need to use more quantitative DNA methylation assays, such as pyrosequencing, to confirm gene-specific analysis. We used a two-step approach—an Illumina methylation panel followed by pyrosequencing—to identify CpG loci associated with prenatal tobacco smoke exposure. This approach may become the paradigm for how these studies are performed. The purpose of the Illumina screen was to identify the most promising candidates for further follow-up and replication, based first on the magnitude of associations detected and then by statistical significance as a way to further narrow the list. Therefore, we did not adjust for multiple comparisons.
In this study, we provide some of the first evidence that differences in methylation patterns occur in children exposed in utero to tobacco smoke and that these differences can be detected in DNA from buccal cells, an aerodigestive tissue cell type. Exposure to maternal smoking affected two types of DNA methylation that have been associated with disease phenotypes: global methylation and promoter CpG island methylation. The effects of smoke exposure differed among children with common variants in genes involved in the detoxification of tobacco smoke. Our findings support the urgent need for research to understand better the potential mechanisms for life-long or transgenerational effects after preventable environmental exposures.
Supplementary Material
Supported by NIEHS grants 5P01 ES009581, 5P01 ES011627, and 5P30 ES007048; by the Hastings Foundation; and by a American Society of Clinical Oncology Career Development Award in Geriatric Oncology (A.Y.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200901-0135OC on June 4, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Reamon-Buettner SM, Borlak J. A new paradigm in toxicology and teratology: altering gene activity in the absence of DNA sequence variation. Reprod Toxicol 2007;24:20–30. [DOI] [PubMed] [Google Scholar]
- 2.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 2007;8:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lodrup Carlsen KC, Carlsen KH. Effects of maternal and early tobacco exposure on the development of asthma and airway hyperreactivity. Curr Opin Allergy Clin Immunol 2001;1:139–143. [DOI] [PubMed] [Google Scholar]
- 4.Carlsen KH, Lodrup Carlsen KC. Parental smoking and childhood asthma: clinical implications. Treat Respir Med 2005;4:337–346. [DOI] [PubMed] [Google Scholar]
- 5.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 2005;127:1232–1241. [DOI] [PubMed] [Google Scholar]
- 6.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001;293:1089–1093. [DOI] [PubMed] [Google Scholar]
- 7.Gilliland FD, Li YF, Dubeau L, Berhane K, Avol E, McConnell R, Gauderman WJ, Peters JM. Effects of glutathione s-transferase m1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med 2002;166:457–463. [DOI] [PubMed] [Google Scholar]
- 8.Kabesch M, Hoefler C, Carr D, Leupold W, Weiland SK, von Mutius E. Glutathione s transferase deficiency and passive smoking increase childhood asthma. Thorax 2004;59:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer CN, Doney AS, Lee SP, Murrie I, Ismail T, Macgregor DF, Mukhopadhyay S. Glutathione s-transferase m1 and p1 genotype, passive smoking, and peak expiratory flow in asthma. Pediatrics 2006;118:710–716. [DOI] [PubMed] [Google Scholar]
- 10.Li YF, Gauderman WJ, Conti DV, Lin PC, Avol E, Gilliland FD. Glutathione s-transferase p1, maternal smoking, and asthma in children: a haplotype-based analysis. Environ Health Perspect 2008;116:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell R, Berhane K, Yao L, Jerrett M, Lurmann F, Gilliland F, Kunzli N, Gauderman J, Avol E, Thomas D, et al. Traffic, susceptibility, and childhood asthma. Environ Health Perspect 2006;114:766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam T, Berhane K, McConnell R, Gauderman WJ, Avol E, Peters JM, Gilliland FD. Glutathione-S-transferase (GST) P1, GSTM1, exercise, ozone and asthma incidence in school children. Thorax 2009;64:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite pcr of repetitive DNA elements. Nucleic Acids Res 2004;32:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroll TC, Wolfl S. Ranking: a closer look on globalisation methods for normalisation of gene expression arrays. Nucleic Acids Res 2002;30:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baccarelli A, Wright R, Bollati V, Tarantini L, Litonjua A, Suh H, Zanobetti A, Sparrow D, Vokonas P, Schwartz J. Genomic DNA methylation, cardiovascular disease, and short-term exposure to traffic air pollution [abstract]. ISEE Conference 2008; Pasadena, CA, October 2008.
- 16.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res 2006;600:46–57. [DOI] [PubMed] [Google Scholar]
- 17.Mathers JC. Early nutrition: impact on epigenetics. Forum Nutr 2007;60:42–48. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of dnmt3b. Oncogene 2007;26:5900–5910. [DOI] [PubMed] [Google Scholar]
- 19.Pulling LC, Vuillemenot BR, Hutt JA, Devereux TR, Belinsky SA. Aberrant promoter hypermethylation of the death-associated protein kinase gene is early and frequent in murine lung tumors induced by cigarette smoke and tobacco carcinogens. Cancer Res 2004;64:3844–3848. [DOI] [PubMed] [Google Scholar]
- 20.Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 2008;266:6–11. [DOI] [PubMed] [Google Scholar]
- 21.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, Gamble MV, Susser E. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev 2008;17:2306–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology 2003;4:233–250. [DOI] [PubMed] [Google Scholar]
- 23.Nephew KP, Balch C, Skalnik DG. Methyl group acceptance assay for the determination of global DNA methylation levels. Methods Mol Biol 2009;507:35–41. [DOI] [PubMed] [Google Scholar]
- 24.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of cpg sites. Biotechniques 2003;35:146–150. [DOI] [PubMed] [Google Scholar]
- 25.Shankar SL, O'Guin K, Cammer M, McMorris FA, Stitt TN, Basch RS, Varnum B, Shafit-Zagardo B. The growth arrest-specific gene product gas6 promotes the survival of human oligodendrocytes via a phosphatidylinositol 3-kinase-dependent pathway. J Neurosci 2003;23:4208–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mudduluru G, Allgayer H. The human receptor tyrosine kinase axl gene: promoter characterization and regulation of constitutive expression by sp1, sp3 and cpg methylation. Biosci Rep 2008;28:161–176. [DOI] [PubMed] [Google Scholar]
- 27.Stepanek L, Stoker AW, Stoeckli E, Bixby JL. Receptor tyrosine phosphatases guide vertebrate motor axons during development. J Neurosci 2005;25:3813–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob ST, Motiwala T. Epigenetic regulation of protein tyrosine phosphatases: potential molecular targets for cancer therapy. Cancer Gene Ther 2005;12:665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000;21:461–467. [DOI] [PubMed] [Google Scholar]
- 30.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009;130:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nafee TM, Farrell WE, Carroll WD, Fryer AA, Ismail KM. Epigenetic control of fetal gene expression. BJOG 2008;115:158–168. [DOI] [PubMed] [Google Scholar]
- 32.Bhutani M, Pathak AK, Fan YH, Liu DD, Lee JJ, Tang H, Kurie JM, Morice RC, Kim ES, Hong WK, et al. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prev Res 2008;1:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.