Abstract
The transcription factor Runx2 is highly expressed in breast cancer cells compared to mammary epithelial cells and contributes to metastasis. Here we directly show that Runx2 expression promotes a tumor cell phenotype of mammary acini in three dimensional culture. Human mammary epithelial cells (MCF-10A) form polarized, growth-arrested acini-like structures with glandular architecture. The ectopic expression of Runx2 disrupts acini formation, and electron microscopic ultrastructural analysis revealed the absence of lumens. Characterization of the disrupted acini structures showed increased cell proliferation (Ki-67 positive cells), decreased apoptosis (Bcl-2 induction) and loss of basement membrane formation (absence of β4-integrin expression). In complementary experiments, inhibition of Runx2 function in metastatic MDA-MB-231 breast cancer cells by stable expression of either shRNA-Runx2 or a mutant Runx2 deficient in subnuclear targeting resulted in reversion of acini to more normal structures and reduced tumor growth in vivo. These novel findings provide direct mechanistic evidence for the biological activity of Runx2, dependent on its subnuclear localization, in promoting early events of breast cancer progression and suggest a molecular therapeutic target.
Keywords: Runx2, acini, mammary epithelial cells, breast cancer, 3D culture
Introduction
Loss of cell polarization and luminal filling of mammary glands are crucial structural alterations in breast cancer (1, 2). The expression of cancer related genes in breast epithelial cells can lead to disorganization of mammary gland structure in in vitro models and cause a pathology similar to epithelial tumors, indicating a critical requirement for the fidelity of tissue and cellular organization of acini (2–4). Molecular events leading to disruption of the glandular structure include expression of anti-apoptotic genes, reduced expression of pro-apoptotic factors, inactivation of pRB and ectopic expression of ERBB2 and TIMPs (1, 4, 5). However, the role of transcription factors in regulating the expression of genes implicated in formation of the luminal space and in cell polarization during mammary epithelial gland formation is not well defined.
The Runx transcription factors (Runx1, Runx2, and Runx3) are essential for organogenesis, and mutations in these genes have been linked to several types of cancer (6, 7). Runx1 and Runx3 mutations promote leukemia and gastric cancer, respectively (8, 9). Runx2 is a key factor for bone formation (10, 11) and deregulation of Runx2 is associated with osteosarcoma (12, 13). In normal mammary epithelial cells Runx2 is expressed at low levels, but it is expressed at high levels in metastatic cancer cells (14–16) and promotes bone metastatic properties of breast and prostate cancer cells (17–21). The consequences of aberrant Runx2 expression in breast cancer cells have been well studied (20). Runx2 directly increases expression of metastatic genes (IHH, MMP9, MMP13, TGFβR, VEGF and others) and increases activity of the osteolytic cycle in metastatic bone disease (20). However, the consequences of induced expression of Runx2 in normal breast epithelial cells in contributing to a cancer cell phenotype are unknown and constitute the focus of this study. Understanding the relationship between deregulation of transcription factors and associated molecular and architectural abnormalities that result in histological phenotypes observed in tumors, is critical for developing novel therapeutic targets for early stages of cancer progression
Normal MCF-10A mammary epithelial cells cultured on a basement membrane form polarized, growth-arrested acini-like spheroids, recapitulating several aspects of glandular architecture in vivo (1, 22). In these studies, we took advantage of the 3D culture model to examine the contribution of Runx2 to the tumorigenic phenotype of mammary acini. Ectopic expression of Runx2 in MCF-10A cells results in disrupted acini structures that resemble cancer phenotype. Importantly, complementary experiments in metastatic breast cancer MDA-MB-231 cells, which do not form an organized structure in basement membrane, show that knockdown of endogenous Runx2 or expression of mutant Runx2 proteins results in reversion to more normal acini-like structures. Thus, our results directly demonstrate that induced expression of Runx2 in mammary epithelial cells to levels found in metastatic breast cancer cell lines, contributes to early events in development of tumor cell properties and may play a critical role in the loss of cellular organization of mammary acini tissue.
Materials and Methods
Cell Culture, viral constructs and treatments
Mammary epithelial MCF-10A cells and the metastatic MDA-MB-231 human breast cancer cell line were cultured in DMEM containing 10% fetal bovine serum (FBS) (Invitrogen Inc., Carlsbad, CA). The construction and transduction procedure for adenovirus expressing Runx2 and control are reported previously (23). Three-dimensional 3D cultures were set up according to Debnath et al. (1, 24); briefly, single cell suspensions in 2% Matrigel (BD Biosciences, San Jose, CA) were overlaid on a thin layer of Matrigel, cultures were fed every second day with 2% Matrigel in DMEM medium. Generation of stable MDA-MB-231-shRNA- control or Runx2 cells by lentiviral gene delivery are reported previously (21).
Western blot analysis
Runx2 protein in normal and breast cancer cells was detected by western blot analysis as described previously (18). Whole cell lysates or nuclear lysates were mixed with direct lysis buffer and separated in a 10% SDS–PAGE. Proteins were transferred to PVDF membrane and incubated with mouse monoclonal Runx2 antibody (R&D) followed by incubation with HRP-conjugated secondary antibodies (Santa Cruz, CA). Immunoreactive proteins were detected using an enhanced chemiluminescence kit (Pierce, Rockford, IL).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (18). Briefly, formaldehyde crosslinking was performed followed by sonication to obtain DNA fragment with average size of 0.3 kb. Protein-DNA complexes were immunoprecipitated using Runx2 antibody (M-70, Santa Cruz) or IgG as a control. Purified DNA was subjected to real time PCR amplification with Sybr Green dye on an ABI real time thermocycler. IL8 and Bcl-2 promoter fragments containing Runx elements were amplified using forward primer: IL8: 5’ GCC GAA TGG GAC GTA AAT AA −3’ and reverse primer: 5’ TAA ATG GGC TTA GGC GGA AA-3’. Bcl-2: forward primer: 5’GGG GGA GAA CTT CGT AGC CAG-3’ and reverse primer: 5’CCG AAA AGC TGC TGG ATA AA-3’.
Real time RT-PCR analysis
Expression levels of IL8, Bcl-2, Runx2, and GAPDH in MCF-10A cells were analyzed after adenovirus transduction. Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s specification. Purified RNA was oligo dT primed and cDNA synthesized at 42°C with SuperScript II RNA polymerase (Invitrogen). For PCR amplification, the following primers were used: IL8 forward 5’GTG CAG TTT TGC CAA GGA GT, reverse 5’ CTC TGC ACC CAG TTT TCC CTT-3’, Runx2 forward 5’- CGG CCC TCC CTG AAC TCT −3’, reverse 5’- TGC CTG CCT GGG GTC TGT A −3’, GAPDH forward 5’- ATG TTC GTC ATG GGT GTG AA −3’ and reverse 5’- TGT GGT CAT GAG TCC TTC CA −3’.
Electrophoretic mobility shift analysis
Nuclear extracts were prepared from normal mammary epithelial MCF-10A or breast cancer MDA-MB-231 cells as previously described (18). Complementary oligonucleotides representing the consensus Runx binding element were synthesized: 5’- ACC CTG AGT TCT GTG GTT GTT TCC TGT GGG TCT. The plus strand (10 pmol) was labeled with γ[-32P]ATP for 1 h at 37°C with T4 polynucleotide kinase (New England Biolabs, Beverly, MA). For immunoshift analysis, 200 ng of Runx2 antibody was incubated with nuclear extract at 22°C for 30 min prior to the addition of probe. The samples were electrophoresed at 200 V for 3 h at 4°C. Gels were dried and subjected to autoradiography at 70°C.
Electron microscopy
For electron microscopy of MCF-10A acini, tissue structures were harvested by suspending the cultures in ice cold serum free DMEM/F12 and centrifuging at 600 g for 5 min, followed by three washes in cold DMEM/F12 to remove the Matrigel. Acini were fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate, pH 7.4 at 48ºC for 1–2 h, and then washed in 0.1M sodium cacodylate. Samples were post-fixed in 1% osmium in 0.1 M sodium cacodylate at 4ºC for 30 min, washed again, and dehydrated in graded ethanols. Samples were embedded in Epon and the transitional solvent was propylene oxide. Sections were stained with 1.4% uranyl acetate in 40% ethanol and then with lead citrate. Sections were imaged with a Philips CM10. Negatives were scanned and processed digitally. Size measurements were made from the digitized images.
Immunofluorescence studies
For confocal microscopy, cells were cultured in Reduced Growth Factor Matrigel on 4-well chamber slides (Thermo Scientific Nalgene And Nunc, Rochester, NY), before fixation and staining as described (24) and then imaged with a Leica SP1 confocal microscope. DNA was stained with DRAQ5 after the second antibody wash.
Mammary fat pad injections and bioluminescence imaging
Anesthetized mice were inoculated with 1×106 MDA-MB-231 cells with matrigel under mammary fat pads. Bioluminescence images were acquired by using the IVIS Imaging System (Xenogen) 2–15 min after injection 150 mg/kg of D-Luciferin (Gold BioTech, St. Louis, MO) in PBS.
Results
Ectopic Runx2 expression disrupts normal acini structure
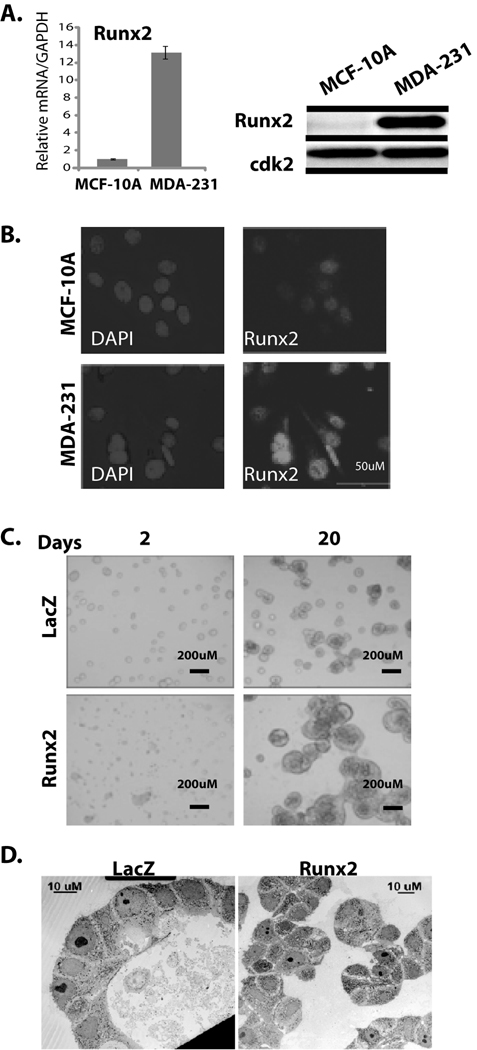
To investigate the consequences of Runx2 expression in normal epithelial cells, we first characterized Runx2 levels in the MCF-10A cells which form normal acini structures in 3D cultures. Analysis of total RNA and nuclear extracts from MCF-10A cells shows the presence of low levels of Runx2 compared to metastatic MDA-MB-231 cells by real time PCR analysis and western blotting (Fig. 1A left and right panel). The presence of Runx2 DNA binding activity in MCF-10A cells was further confirmed by antibody supershift studies (see Supplementary Fig. S1). Examination of subnuclear organization using in situ immunofluorescence microscopy shows that Runx2 is localized in nuclear foci in MCF-10A cells, albeit at lower levels compared to MDA-MB-231 cells (Fig. 1B). The observed low levels of Runx2 in MCF-10A cells make this cell line a suitable model to study the effect of Runx2 expression on mammary acini formation and the potential induction of a phenotype related to cancer progression.
Figure 1. Ectopic Runx2 expression disrupts normal acini structure.
(A) left panel: Runx2 mRNA levels were examined by qRT-PCR using total RNA (5µg) from MCF-10A and MDA-MB-231 cells. Right panel: Whole cell lysate from MCF-10A and MDA-MB-231 was subjected to western blot analysis to detect Runx2 protein by mouse monoclonal antibody (1:2000). Expression of cdk2 is shown as internal loading control. (B) Immuno-localization of endogenous Runx2 protein in MDA-MB-231 and MCF10-A cells shows punctate nuclear signal. Nuclei are revealed by 4, 6-diamidino-2-phenylindole staining (DAPI). (C) MCF-10A cells transduced with adenovirus expressing either Runx2 or β-galactosidase as control were allowed to grow in 3D culture system for 20 days after transduction. Acini structures examined by light microscopy are shown. AdRunx2 treated cells form disorganized and proliferating cell masses compared to acini structures in control AdLacZ treated cells. (D) Ultrastructure of acini (day 20) of LacZ control or Runx2 expressing MCF-10A cells by electron microscopy.
MCF-10A cells were transduced with adenovirus expressing either Runx2 or β-gal as control. Increased levels of Runx2 were confirmed by western blot analysis (Supplementary Fig. S2) and cells were grown in 3D culture for 20 days after transduction. We find that ectopic expression of Runx2 causes a significant disruption of normal acini structure as examined by light microscopy at days 2 and 20 (Fig. 1C). Runx2 treated cells form disorganized cell masses compared to the organized acini structures formed by control cells (infected with AdLacZ). Runx2 expressing cells exhibited structures with a 2 fold increased diameter compared to controls (AdRunx2: 176.6µM ± 29.5 vs AdLacZ control: 91.43µM ± 16.32). We examined the ultrastructure of acini by electron microscopy which revealed that the disorganized structures from Runx2 expressing cells lack a hollow lumen and apoptotic cells. Control cells formed polarized acini with hollow lumens (Fig. 1D). These results demonstrate that ectopic expression of Runx2 in normal mammary epithelial cells results in disorganization of mammary acini architecture.
Runx2 expression disrupts acinus formation by increasing proliferation and decreasing basement membrane formation
Formation of acini structure is a result of stringent control of cell proliferation, polarization, apoptosis and formation of basement membrane (1). Our findings (Fig. 1) suggest that ectopic Runx2 expression results in disruption of normal acini structure in 3D culture of MCF-10A cells, due to either continuous cell proliferation or inability of the MCF-10A cells to undergo apoptosis. We find a significant increase in proliferation of cells transduced with Runx2 adenovirus compared to control cells at day 20 of differentiation as shown by Ki-67 staining (Fig. 2A). The low levels of Ki-67 staining in control cells (AdLacZ) indicate that MCF-10A cells are primarily non-proliferative. These results suggest that continued proliferation of Runx2 expressing cells may contribute to disorganization of acini structure.
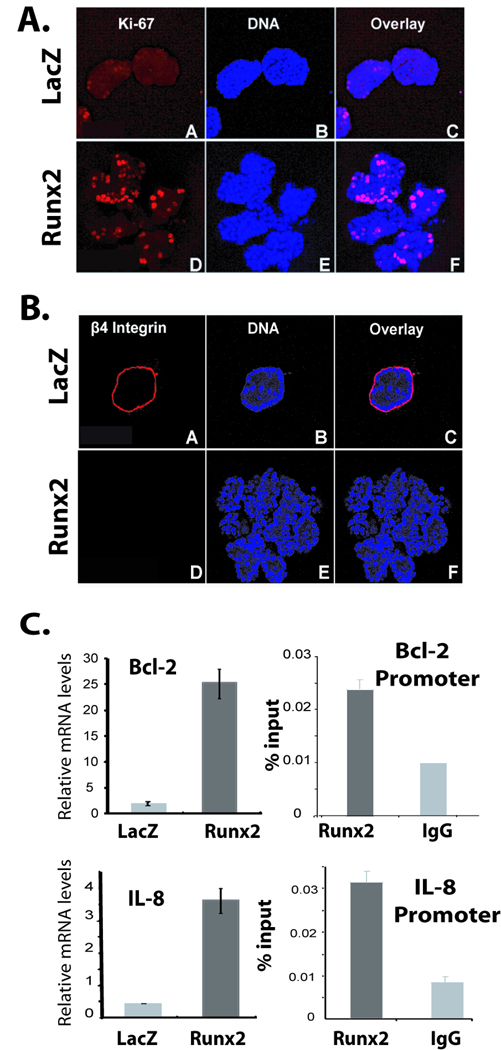
Figure 2. Runx2 expression causes increased proliferation and decreases β4 integrin levels.
3D cultures of MCF-10A cells infected with AdLacZ control or AdRunx2 were subjected to Ki-67 (A) or β4 integrin (B) staining at day 20. Ki-67 and β4 integrin are shown in the red channel of the single confocal section of the upper part. Nuclei shown in the blue channel were stained with Draq5. (C) Left panel: Expression levels of IL-8 and Bcl-2 in MCF-10A cells transduced with either LacZ or Runx2 adenovirus as determined by qRT-PCR. Right panels: MCF-10A cells were subjected to chromatin immunoprecipitation assay with Runx2 antibody. Immunoprecipitated DNA was further amplified by real-time qPCR with primer pairs for proximal promoters of Bcl-2 and IL-8.
Acini formation is a highly regulated process in which cells communicate with each other to form a polarized structure. These structures also show the presence of basement membrane around well formed acini. We examined the MCF-10A 3D cultures treated with Runx2 or control adenovirus for β4 integrin as a marker for basement membrane. MCF-10A cells infected with control virus exhibit well formed basement membrane as shown by β4 integrin staining after day 20 of culture (Fig. 2B), while ectopic expression of Runx2 prevents basement membrane formation in culture. These findings demonstrate that Runx2 overexpression in MCF-10A cells causes loss of polarization as these cells continue to proliferate in disorganized acini. Moreover, Runx2 expressing cellular aggregates accumulate characteristics similar to architectural features of breast cancer.
Runx2 activates cancer-related Bcl-2 and IL-8 genes in MCF-10A cells
The organizational changes in MCF-10A acini with Runx2 ectopic expression might be due to alteration of Runx2 target gene expression. To gain further insight into molecular alterations leading to disorganization of acini architecture due to ectopic expression of Runx2, we performed a focused cDNA expression profile of cancer related genes. Total RNA from MCF-10A cells transduced with either control LacZ or Runx2 adenovirus was labeled and hybridized with membranes containing probes for 96 cancer related genes (supplementary Fig. S3). Our results show a significant increase in expression of Bcl-2 and IL-8 genes in cells treated with Runx2 adenovirus compared to controls. We validated the increased expression of these genes by qRT-PCR analysis (Fig. 2C, left panels). We further confirmed the in vivo recruitment of Runx2 on Bcl-2 and IL-8 gene promoters by chromatin immunoprecipitation assay in MCF-10A cells (Fig. 2C, right panels). These studies identified Bcl-2 and IL-8 as novel Runx2 responsive genes. Increased Bcl-2 expression in response to Runx2 overexpression might contribute to luminal filling by reducing apoptosis. These results suggest that aberrant Runx2 expression in mammary epithelial cells leads to activation of downstream cancer-related genes and disrupts the balance between cell growth and apoptosis.
Loss of functions associated with the Runx2 C-terminus prevents cell growth and tumor cell like phenotype
To understand the molecular mechanism of Runx2-mediated regulation of cellular processes required for maintaining highly organized epithelial glandular structures, we utilized mutants of Runx2. A point mutant (RY: Y428A &R398A) and one deletion mutant (ΔC: Runx2 1–361) of Runx2 protein are known to abrogate subnuclear targeting to the nuclear matrix and to disrupt organization of Runx2 regulatory complexes with co-regulatory proteins involved in the TGFβ and c-Src signaling pathways (7, 25, 26). Both these pathways are linked to tumor progression (27). Furthermore, the mutants failed to promote in vivo bone formation (11) and prevented tumor growth when expressed in MDA-MB-231 cells (15, 17).
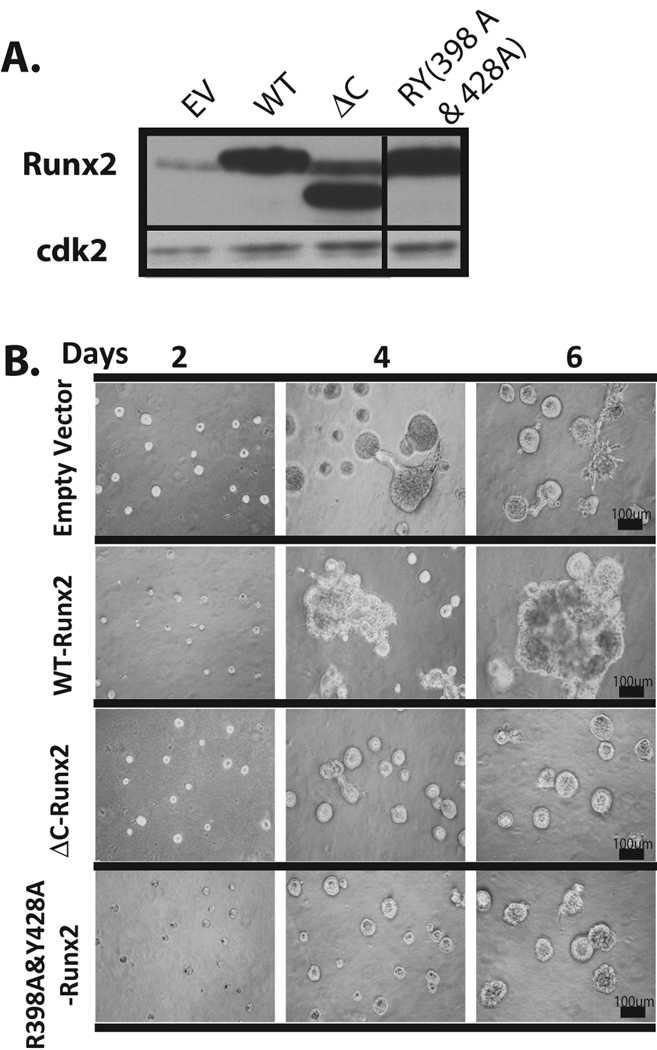
MDA-MB-231 cells stably expressing the Runx2 mutants were generated by lentiviral mediated gene delivery, and protein levels were confirmed by western blot analysis (Fig. 3A). We examined the functional activity of these Runx2 mutants on endogenous levels of cancer related markers and found that both mutations reduce activation of Runx2 target genes (MMP2, MMP9, VEGF and Osteopontin) compared to wild type Runx2 (supplementary Fig. S4). The morphology of 3D cultures of MDA-MB-231 cells expressing the RY mutant Runx2 protein showed significantly smaller acini compared to empty vector control, and this difference was more striking on days 2 and 4 (Fig. 3B). Cells expressing wild type Runx2 form large and disorganized structures compared to empty vector control cells, while functionally deficient ΔC-Runx2 and Runx2-RY expressing cells show reversion to acini structures compared to the empty vector control. Taken together, our results demonstrate that Runx2 interactions with the nuclear matrix and its co-regulatory proteins via the C-terminus are critical for mediating disorganization of mammary acini.
Figure 3. Loss of functions associated with the Runx2 C-terminus prevents cell growth and tumor cell like phenotype.
(A) Expression levels of ectopic wild type or mutant Runx2 proteins in MDA-MB-231 cells (ΔC: C-terminal deletion; double point mutant RY: Y428A &R398A) by lentiviral mediated gene delivery is shown by western blot analysis. (B) MDA-MB-231 cells stably expressing Runx2 mutants show significantly smaller acini in mutant cell lines compared to empty vector control. Cells expressing wild type Runx2 formed large and disorganized structures compared to empty vector control cells.
Depletion of Runx2 in metastatic cancer cells partially restores organized acini structure and inhibits tumor growth in vivo
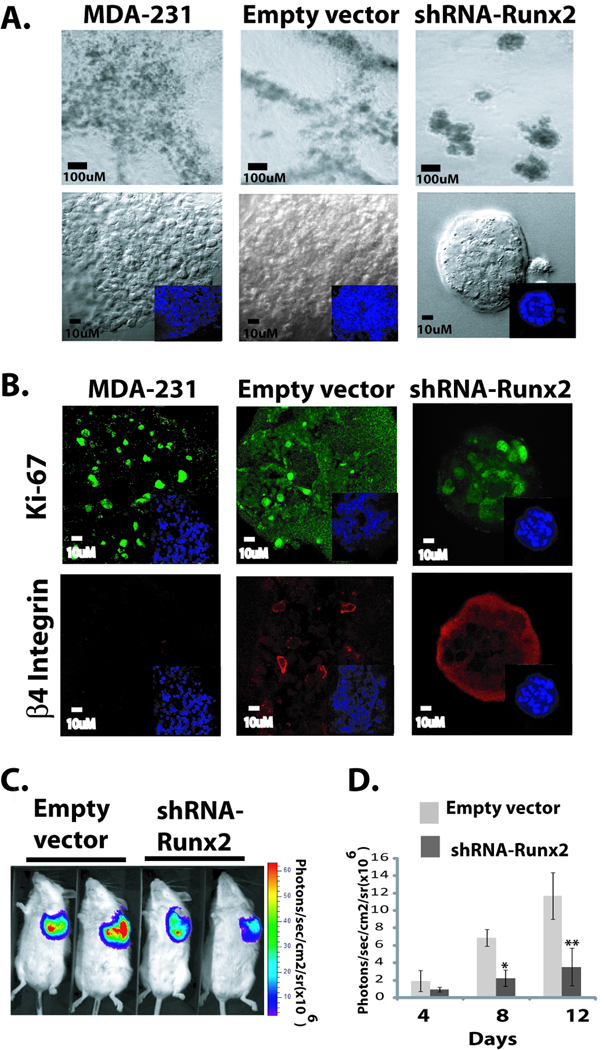
The MDA-MB-231 metastatic breast cells do not form acini-like structures but instead make highly branched masses when grown in 3D matrigel culture (Fig. 4A upper panels). We tested the hypothesis that abnormal levels of expressed Runx2 (see Fig.1) are contributing to the unregulated cell growth that prevents acini formation in the 3D culture model. The experimental design examined whether depleting Runx2 in MDA-MB-231 cells would rescue normal acini structure. We generated MDA-MB-231 cells stably expressing an shRNA for Runx2 and achieved knockdown of more than 80% of endogenous Runx2, as shown by western blot analysis (supplementary Fig. S5). The 3D culture of shRNA-Runx2 expressing cells for 12 days showed significant reversion, in that the structures which formed more closely resembled MCF-10A acini than the structures formed by the parental cells or empty vector controls (Fig. 4A lower panels). The morphology and diameter of acini formed by shRNA-Runx2 cells (110 µm at day 12 of culture) were comparable to normal mammary epithelial cell acini. Microscopic characterization of the reverted structures revealed reduced proliferation compared to controls, as examined by Ki-67 immunostaining (Fig. 4B, upper panels), and significantly higher staining of integrin β4 around these structures, indicating the formation of basement membrane (Fig. 4B, lower panels).
Figure 4. Knock down of Runx2 in metastatic cancer cells partially restores organized acini structure and inhibits tumor growth in vivo.
(A). The parental MDA-MB-231 cells, empty vector or shRNA-Runx2 expressing cells were grown in 3D culture for 12 days. Parental and empty vector cells show highly branched masses when grown in 3D matrigel culture. The 3D culture of shRNA-Runx2 expressing cells showed significant reversion to normal acini like structures compared to empty vector or parental cells. (B) Immunostaining of Ki-67 (upper panel) and integrin β4 (lower panel) of acini structures revealed reduced proliferation and formation of basement membrane in MDA-MB-231 cells expressing shRNA-Runx2 compared to controls. (C) MDA-MB-231 cells stably expressing firefly luciferase and shRNA-Runx2 or empty vector were injected in SCID mice under mammary fat pads. Anesthetized mice (Empty vector-MDA-MB-231 group (n=6) and shRNA-Runx2-MDA-MB-231group (n=6)) were injected with 150mg_kg D-Luciferin (Xenogen, Alameda, CA) in PBS. Bioluminescence images were acquired by using the IVIS Imaging System (Xenogen) 2–10 min after 12 days of cell injections. Images were set at the indicated pseudocolor scale to show relative bioluminescent. (D) Quantitation of luciferase activity shows inhibition of tumor growth under mammary fat pad in shRNA-Runx2 animals compared to control group. Analysis was performed with LIVINGIMAGE software (Xenogen) by measuring photon f lux (measured in photons/sec/cm2/steradian) using a region of interest drawn around the bioluminescence signal to be measured (n=6). Data were normalized to the signal obtained right after xenografting (day 0). SD. *, P<0.005; **, P<0.001 compared with empty vector controls; P values calculated using Student’s t test.
We also performed mammary fat pad injections of firefly luciferase labeled MDA-MB-231 cells stably expressing either shRNA-Runx2 or control shRNA in the SCID mouse model. Tumor growth was visualized by luciferase bioluminescence imaging 12 days after cell injections (Fig. 4C). Animals injected (n=6) with MDA-MB-231 cells expressing shRNA-Runx2 showed a significant reduction in tumor growth compared to a control group (expressing empty vector) as shown by quantitation of luciferase activity (Fig. 4D). Examining tumor growth up to 4 weeks revealed a 41% decrease in tumor volume in shRNA-Runx2 animals (1.8 cm3, n=6) compared to controls (3.1 cm3, n=6). Thus, our results from in vitro and in vivo studies show that depleting Runx2 levels in metastatic breast cancer cells restores mammary gland organization by regulating cell proliferation and polarization and reduces tumor growth in mammary fat pads.
Discussion
Our results provide evidence that elevated Runx2 expression in the 3D culture model of normal mammary epithelial cells, activates several key cancer-related genes and disorganizes acinar architecture to resemble a tumorigenic phenotype. A striking feature of the loss of glandular architecture and formation of proliferating cell aggregates by Runx2 overexpression is the absence of β4 integrin, an essential component for organization of glandular epithelium (28). We demonstrate that depletion of endogenous Runx2 in highly aggressive metastatic breast cancer cells limits tumor formation in the mammary gland in vivo and reverts cancer cell aggregates into more normal acini-like structures in vitro. Combined with our previous reports (17, 18, 21) and based on present findings, we propose that Runx2 not only promotes metastatic properties of cancer cells, but also can initiate tumorigenic properties in normal mammary epithelial cells.
We identified mechanisms by which ectopic Runx2 expression contributes to the altered phenotype of MCF-10A cells, including increased proliferation and decreased apoptosis. Runx factors have been linked to apoptosis (29, 30). Our data show that Runx2 induces Bcl-2 expression in MCF-10A cells, which likely contributes to luminal filling via anti-apoptotic events and loss of glandular structure. Also, co-expression of Bcl-2 with CyclinD1 in MCF-10A cells is known to result in luminal filling (31). Runx2 stimulates expression of the cell cycle regulator cyclinD1, an essential factor for transduction to proliferation and oncogene identified in metastatic breast cancer cells (21). Apoptosis may also be decreased due to the synergy between Runx2 and c-Myc which causes a low rate of apoptosis and proliferative advantage of lymphoma cells (32). The ineffectiveness of the subnuclear targeting deficient Runx2 mutant in causing acini disruption may likely be due to the loss of interacting co-regulatory factors in organized nuclear foci with Runx2. Runx2 facilitates TGFβ mediated-growth effects on breast cancer cells by interaction with Smads (20). Our results are consistent with other findings that Runx2 functions as an oncogene when abnormally expressed in tumor cells (20, 32). This property of Runx2 is in contrast to its tumor suppressor function in normal cells (33).
Our results demonstrate that expression of Runx2 causes loss of basement membrane and cell polarity leading to disorganized acini. The IGF1 signaling pathway has been implicated in disorganization of acinar morphogenesis in 3D cultures (1), and this pathway stimulates Runx2 activity (34). Activation of the type 1 IGF receptor increases proliferation, survival signaling, loss of apico-basal polarity in mammary epithelial cells and promotes early breast cancer lesions (35). In breast cancer cells inhibition of PI3K or MAPK results in nearly complete reversion to acini structure (36). We show that Runx2 depletion in metastatic breast cancer cells restores more normal appearing acini-like structures in 3D cultures. Studies have shown loss of the Runx2 C-terminus leads to elimination of domains that contain PI3K/MAPK phosphorylation sites (34, 37). Interestingly, Runx2 synergizes with PI3K–Akt signaling to enhance cell migration (38). Thus Runx2 may be responding in vivo to IGF and/or PI3K signaling pathways that lead to mammary gland disorganization during early tumor progression.
Profiling cDNAs in breast cancer cells has revealed a set of genes highly expressed in response to mitogenic signals that enhance tumorigenic properties (39). Runx2 activates MMP-2, MMP-9, and osteopontin involved in migration and invasion of MCF-10A cells through p38 MAPK signaling in response to TGFβ (40, 41). Recent studies identified Runx2 as an essential component of TGFβ mediated activation of the vicious cycle operative in the bone microenvironment during breast cancer metastasis (18, 21). Therefore, we propose increased expression of Runx2 and its target genes related to tumor growth and metastasis in the MCF-10A cells are promoting the tumor phenotype in the 3D culture model. This conclusion is supported by the suppression of the tumor cell phenotype of the MDA-MB-231 cell in matrigel by Runx2 shRNA and Runx2 subnuclear targeting mutants. In this study we further identified Bcl-2 and interleukin-8 as novel Runx2 target genes. Bcl-2 was shown to be induced in hematopoietic cells by the leukemic factor AML1(Runx1)-ETO (42). IL-8 enhances cell proliferation, survival, and MMP expression in CXCR1- and CXCR2-expressing endothelial cells and regulates angiogenesis (43). Our chromatin immunoprecipitation studies demonstrate that Runx2 associates with promoters of these genes in regions with consensus sequences for Runx regulatory element. Therefore, these studies indicate that depletion of Runx2 and the resulting down-regulation of Runx2 target genes can promote differentiation of breast cancer cells into more normal epithelial cells as we have observed in the 3D culture model.
Taken together, our studies have established that the induction of Runx2 levels in normal mammary epithelial cells enhances tumorigenic properties and promotes disorganization of acini-like structures formed by these cells. We propose that Runx2 activates cancer related genes in response to deregulated cell signaling pathways during early stages of breast cancer. As such, Runx2 is a novel therapeutic target that takes advantage of unique subnuclear targeting properties for a broad spectrum of biological activities in tumor cells.
Acknowledgments
We thank Judy Rask for editorial assistance, Microscopy Core for structural analysis of acini and members of the participating laboratories for valuable suggestions throughout the study. Studies reported were in part supported by National Institutes of Health grants P01CA082834, R03CA123599, and P30DK32520. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- 1.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ. Glandular structure and gene expression. Lessons from the mammary gland. Ann N Y Acad Sci. 1998;842:1–6. doi: 10.1111/j.1749-6632.1998.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 3.Harris RA, Eichholtz TJ, Hiles ID, Page MJ, O'Hare MJ. New model of ErbB-2 over-expression in human mammary luminal epithelial cells. Int J Cancer. 1999;80:477–484. doi: 10.1002/(sici)1097-0215(19990129)80:3<477::aid-ijc23>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reginato MJ, Mills KR, Becker EB, et al. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyth K, Cameron ER, Neil JC. The runx genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 7.Zaidi SK, Pande S, Pratap J, et al. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 9.Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 10.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 11.Choi J-Y, Pratap J, Javed A, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci, USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas DM, Johnson SA, Sims NA, et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–934. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan SS, Pereira BP, Zhou YF, et al. Elevated expression of Runx2 as a key parameter in the etiology of osteosarcoma. Mol Biol Rep. 2009;36:153–158. doi: 10.1007/s11033-008-9378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvamurugan N, Partridge NC. Constitutive expression and regulation of collagenase-3 in human breast cancer cells. Mol Cell Biol Res Commun. 2000;3:218–223. doi: 10.1006/mcbr.2000.0215. [DOI] [PubMed] [Google Scholar]
- 15.Barnes GL, Hebert KE, Kamal M, et al. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases associated osteolytic disease. Cancer Res. 2004;64:4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- 16.Inman CK, Shore P. The osteoblast transcription factor Runx2 is expressed in mammary epithelial cells and mediates osteopontin expression. J Biol Chem. 2003;278:48684–48689. doi: 10.1074/jbc.M308001200. [DOI] [PubMed] [Google Scholar]
- 17.Javed A, Barnes GL, Pratap J, et al. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci , USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pratap J, Javed A, Languino LR, et al. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brubaker KD, Vessella RL, Brown LG, Corey E. Prostate cancer expression of runt-domain transcription factor Runx2, a key regulator of osteoblast differentiation and function. Prostate. 2003;56:13–22. doi: 10.1002/pros.10233. [DOI] [PubMed] [Google Scholar]
- 20.Pratap J, Lian JB, Javed A, et al. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 21.Pratap J, Wixted JJ, Gaur T, et al. Runx2 transcriptional activation of Indian hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Underwood JM, Imbalzano KM, Weaver VM, Fischer AH, Imbalzano AN, Nickerson JA. The ultrastructure of MCF-10A acini. J Cell Physiol. 2006;208:141–148. doi: 10.1002/jcp.20639. [DOI] [PubMed] [Google Scholar]
- 23.Afzal F, Pratap J, Ito K, et al. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- 24.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 25.Zaidi SK, Javed A, Pratap J, et al. Alterations in intranuclear localization of Runx2 affect biological activity. J Cell Physiol. 2006;209:935–942. doi: 10.1002/jcp.20791. [DOI] [PubMed] [Google Scholar]
- 26.Javed A, Afzal F, Bae JS, et al. Specific residues of RUNX2 are obligatory for formation of BMP2-Induced RUNX2-SMAD complex to promote osteoblast differentiation. Cells Tissues Organs. 2009;189:133–137. doi: 10.1159/000151719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Weaver VM, Petersen OW, Wang F, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano T, Ito K, Fukamachi H, et al. The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol Cell Biol. 2006;26:4474–4488. doi: 10.1128/MCB.01926-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eliseev RA, Dong YF, Sampson E, et al. Runx2-mediated activation of the Bax gene increases osteosarcoma cell sensitivity to apoptosis. Oncogene. 2008;27:3605–3614. doi: 10.1038/sj.onc.1211020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 32.Blyth K, Vaillant F, Hanlon L, et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- 33.Galindo M, Pratap J, Young DW, et al. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao M, Shapiro P, Kumar R, Passaniti A. Insulin-like growth factor-1 regulates endogenous RUNX2 activity in endothelial cells through a phosphatidylinositol 3-kinase/ERK-dependent and Akt-independent signaling pathway. J Biol Chem. 2004;279:42709–42718. doi: 10.1074/jbc.M404480200. [DOI] [PubMed] [Google Scholar]
- 35.Yanochko GM, Eckhart W. Type I insulin-like growth factor receptor over-expression induces proliferation and anti-apoptotic signaling in a three-dimensional culture model of breast epithelial cells. Breast Cancer Res. 2006;8:R18. doi: 10.1186/bcr1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Hansen RK, Radisky D, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277:36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- 38.Fujita T, Azuma Y, Fukuyama R, et al. Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol. 2004;166:85–95. doi: 10.1083/jcb.200401138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 40.Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375–1382. [PubMed] [Google Scholar]
- 41.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 42.Klampfer L, Zhang J, Zelenetz AO, Uchida H, Nimer SD. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci U S A. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nor JE, Christensen J, Liu J, et al. Up-Regulation of Bcl-2 in microvascular endothelial cells enhances intratumoral angiogenesis and accelerates tumor growth. Cancer Res. 2001;61:2183–2188. [PubMed] [Google Scholar]