Abstract
We developed a digital RNA allelotyping method for quantitatively interrogating allele-specific gene expression. This method involves ultra-deep sequencing of padlock captured SNPs from the transcriptome. We characterized four cell lines established from two human subjects in the Personal Genome Project. Approximately 11–22% of the heterozygous mRNA-associated SNPs show allele-specific expression in each cell line; and 4.3–8.5% are tissue-specific, suggesting the presence of tissue-specific cis-regulation. When applied to two pairs of sibling human embryonic stem cell lines, the sibling lines were more similar in allele-specific expression than were the genetically unrelated lines. We found that the variation of allelic ratios in gene expression among different cell lines is primarily explained by genetic variations, much more so than by specific tissue types or culturing conditions. Comparison of expressed SNPs on the sense and anti-sense transcripts suggested that allelic ratios are primarily determined by cis-regulatory mechanisms on the sense transcripts.
INTRODUCTION
Recent advances in the search of genetic determinants of common human diseases can be attributed to the advances in high-throughput genotyping technologies, which enabled the comprehensive mapping of linkage disequilibrium (LD) in the human genome. The block-like distribution of LD allows researchers to quickly home in on the genomic regions associated with a given phenotype using a set of common SNPs. While this approach allows a general association between genotype and phenotype, determining the causal genetic variants remains difficult due to the strong LD structure within the human population. Although a limited success has been reported on screening coding variants, candidate SNPs often do not fall within a protein coding region. Regulatory polymorphisms have been shown to play a role in common diseases, but such variants are more difficult to identify.
Cis-regulatory polymorphisms can modulate gene expression by a variety of means including alteration of DNA binding sites for cis-regulators (transcription factors, enhancers, repressors, and miRNA binding sites), copy number variations, or DNA methylation. In individuals heterozygous for a cis-regulatory polymorphism, an unequal expression of the two alleles would be expected, resulting in allele-specific gene expression (ASE)1. Since its readout directly reflects the effect of functional cis-regulatory variants, systematic analysis of allele-specific gene expression in human tissues may facilitate the identification of many causal non-coding variants2.
Existing methods for genome-scale quantification of ASE rely mostly on microarray hybridization, which produces analog read-outs3–8. Full transcriptome resequencing (RNA-seq) has recently been used in the digital characterization of transcriptome and alternatively spliced transcripts9,10. However, due to the size and complexity of the transcriptome, the wide dynamic range of gene expression, and the low density of transcribed heterozygous SNPs (approximately one per 3.3 kb), most informative SNPs were not covered at the sequencing depth sufficient to make accurate allelic quantification. Here we report digital RNA allelotyping based on the integration of large-scale synthesis of padlock probes11 on programmable microarrays, multiplexed capture of transcribed SNPs in a single reaction, and deep sequencing. This strategy allows us to focus sequencing efforts only on a specific fraction of the transcriptome carrying SNPs. It combines the sensitivity and the quantitative accuracy of digital expression measurements (i.e. RNA-seq) with the efficiency of targeted sequencing. We demonstrated the utility of this assay by characterizing the spectrum of allele-specific gene expression in three different adult cell types from two Personal Genome Project donors (PGP1 and PGP9), as well as two pairs of sibling human embryonic stem cell lines.
RESULTS
Digital allelotyping
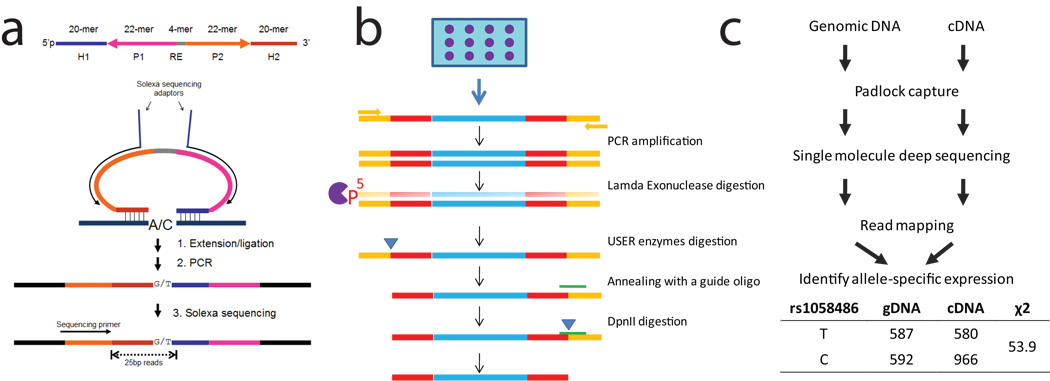
We designed single-stranded DNA probes to capture SNPs from the human genome and transcriptome for sequencing (Fig. 1a). Each probe contains two terminal capturing arms (H1 and H2), that can anneal to the flanking region of the targeted SNPs with a gap of one or more nucleotide bases. In the capturing reaction similar to Molecular Inversion Probes12, the gap is filled by a DNA polymerase and closed by a DNA thermal ligase. The capturing arms are connected by a common linker DNA sequence. This linker contains priming sites for multiplex PCR amplification of the circularized single-stranded DNA probes. After the circularization reaction and PCR amplification, the resulting libraries are sequenced with Illumina Genome Analyzer. As demonstrated previously, circularization of padlock probes is extremely specific; >10,000 targets can be captured simultaneously in a single tube12–15.
Figure 1.
Digital allelotyping with padlock probes. (a) The design of padlock probe (top) and a schematic diagram of padlock capturing experiments (bottom). (b) (c) The experimental and analytic workflow of digital allelotyping assay.
We developed a restriction-free method for making large libraries of padlock probes from Agilent’s programmable microarray (Fig. 1b, Online Methods). We designed and synthesized a library of probes targeting 27,000 SNPs (minor allele frequency > 0.07) located within 10,345 genes in the human genome. The probe design and synthesis, as well as padlock capture has been optimized, such that the representation bias, capturing efficiency and quantification accuracy was dramatically improved compared with the previous protocols14 (Supplementary Note).
We performed SNP capture and single-molecule sequencing on both genomic DNA and cDNA of the same individuals (Fig. 1c). We made genotyping calls on the SNPs that were covered by at least 20 reads using a “best-p” method (see online Methods). With approximately 6–9 millions mappable reads obtained from one lane of the Illumina sequencing flow cell, we made genotyping calls on 68–82% of the SNPs. We compared the genotypes between the digital allelotyping assay and Affymetrix 500k SNP chip, and found 98.4% of calls consistent between the two assays. We made RNA allelotyping calls on heterozygous SNPs that were sequenced at least 50 times. For the convenience of cross-sample comparison, RNA allelic ratios (Fref) were calculated for the common alleles based on the NCBI dbSNP annotation. We normalized the RNA allelic ratios based on the allelic counts from genomic DNAs, such that the quantification is robust in the presence of copy number variations or systematic capturing bias. To validate the allele ratios determined by digital allelotyping, we obtained 76 measurements with quantitative Sanger sequencing16. The results were consistent between the two assays (Supplementary Fig. 1).
Spectrum of allele-specific gene expression in Personal Genome Project cell lines
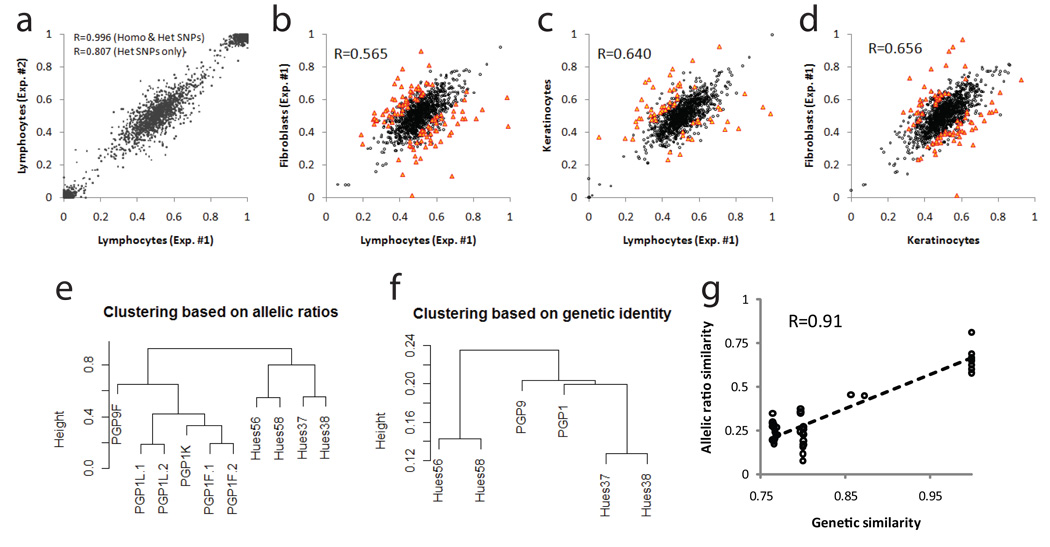
Tissue-specific regulation of gene expression is a well-known phenomenon. Analysis of cis-regulation in the tissue type (adipose tissues) directly relevant to the phenotype (obesity) was shown to be more informative than on unrelated tissue type (blood)17. However, disease related human tissues are often difficult to obtain for research purposes. To characterize the extent of ASE in different cell types from the same individual, we performed RNA allelotyping on three cell lines derived from a male donor PGP1 from the Personal Genome Project (PGP): EBV transformed B-lymphocytes (PGP1L), primary fibroblasts (PGP1F) and primary keratinocytes (PGP1K). To compare the allele-specific gene expression of the same cell type with different genetic background, we also included another primary fibroblast line (PGP9F) from a female donor (PGP9). In estimating the measurement variability of the assay, we performed two technical replicates on PGP1L, and two biological replicates on PGP1F. The allelic ratios are highly correlated between technical replicates (Pearson R=0.811) and biological replicates (Pearson R=0.809), indicating the robustness of the allelotyping assay (Supplementary Fig. 2). Distribution of RNA allelic ratios follows a bell-shape continuous distribution in all nine experiments on seven cell lines (Supplementary Fig. 3 and 4). No biologically meaningful threshold seems to exist to separate SNPs that are in “allelic balance” or “allelic imbalance”. ASE is more appropriate to be treated as a quantitative trait instead of a binary trait. We performed χ2 tests on the raw allelic counts from genomic DNA and cDNA with a cutoff of 6.64 (p-value =0.01). A fraction of SNPs were sequenced very deep (>1000x) in our assay such that a very small allelic drift in expression could be detected as highly significant even though it might not have any biological relevance. Therefore, we also required that the magnitude of allelic drift has to be no less than 0.1 (allelic ratio < 0.4 or >0.6) to be considered allele-specific. Using these criteria we found that 11–22% of SNPs show allele-specific expression (Table 1), among which 4.3–7.7% are likely false positive calls. Allelic ratios between the two technical replicates (PGP1L.1 and PGP1L.2) are highly correlated (Pearson R=0.996 for all SNPs; R=0.811 for heterozygous SNPs, Fig. 2a). A similar correlation was observed between the two biological replicates of PGP1F (R=0.809 for heterozygous SNPs, Supplementary Fig. 2b).
Table 1.
Summary of digital allelotyping experiments.
| Samples | # mapped reads | # SNP mapped | # SNPs above thresholds | # SNP called | # hetSNPs | # genes | # snp with ASE | FDR | % SNP with ASE | # autosomal SNPs with mono-allelic expression |
|---|---|---|---|---|---|---|---|---|---|---|
| PGP1G | 6,390,846 | 25,265 | 19,582 | 19,561 | 4,761 | |||||
| PGP1L1 | 2,884,606 | 15,328 | 5,657 | 1,387 | 1,075 | 180 | 7.7% | 12.0% | 11 | |
| PGP1L2 | 4,092,513 | 15,875 | 6,513 | 1,586 | 1,198 | 217 | 7.3% | 12.7% | 14 | |
| PGP1K | 4,096,083 | 16,239 | 6,332 | 1,541 | 1,201 | 204 | 7.6% | 12.2% | 10 | |
| PGP1F1 | 7,392,218 | 17,355 | 7,686 | 1,785 | 1,333 | 317 | 5.6% | 16.8% | 10 | |
| PGP1F2 | 8,104,770 | 18,476 | 8,045 | 1,871 | 1,406 | 363 | 5.2% | 18.4% | 15 | |
| PGP9G | 8,613,616 | 25,748 | 22,106 | 21,993 | 5,539 | |||||
| PGP9F | 7,301,815 | 15,883 | 7,763 | 1,939 | 1,443 | 452 | 4.3% | 22.3% | 12 | |
| Hues37G | 7,919,357 | 24,578 | 18,505 | 18,450 | 4,493 | |||||
| Hues37C | 7,941,706 | 22,315 | 10,232 | 2,315 | 1,705 | 280 | 8.3% | 11.1% | 10 | |
| Hues38G | 8,059,825 | 24,553 | 18,565 | 18,546 | 4,467 | |||||
| Hues38C | 6,060,550 | 20,774 | 8,802 | 1,961 | 1,505 | 288 | 6.8% | 13.7% | 3 | |
| Hues56G | 8,921,985 | 25,450 | 20,695 | 20,627 | 5,389 | |||||
| Hues56C | 6,491,835 | 18,864 | 7,785 | 2,013 | 1,602 | 392 | 5.1% | 18.5% | 10 | |
| Hues58G | 7,145,720 | 24,621 | 18,651 | 18,617 | 4,931 | |||||
| Hues58C | 3,752,161 | 16,888 | 5,871 | 1,493 | 1,243 | 301 | 5.0% | 19.2% | 8 |
Note: PGP1G, PGP9G, Hues37G, Hues38G, Hues56G and Hues58G are genomic DNA, the others are cDNAs.
Figure 2.
Allele-specific expression in human cell lines of various degrees of genetic and phenotypic similarities. (a) Consistency of allelic ratios between two technical replicates. (b–d) Tissue-specific ASE among three PGP1 cell lines of the same genetic background. (e, f) Hierarchical clustering of samples based on allelic ratios or genetic identity. (g) Correlation between the allelic ratio similarity and genetic similarity.
We next sought to identify SNPs that show different allele-specific expression pattern in different cell types. With the same threshold for discovery as above, we found that the allelic ratios of 20 out of 1379 heterozygous SNPs were different in the two PGP1L technical replicates (Supplementary Fig. 2a). These SNPs are false positives due to the measurement variability of the assay. When adjusted based on the estimated false positive rate, we found that 4.3–8.5% of SNPs show tissue-specific allelic ratios among the three cells lines from the same individuals (Fig. 2 b–d, Table 2), which means approximately 1/3-1/2 of allele-specific SNPs are also tissue specific. From the two biological replicates of PGP1F, we estimated that approximately 2% of the allele-specific SNPs are due to the biological variability (Supplementary Fig 2b). However, the differences of allelic ratios for the same SNPs between two cell types were larger than those between biological replicates. Therefore, the tissue-specific allelic biases are likely due to presence of tissue-specific cis-regulators, but not the biological variability or variable culturing artifacts.
Table 2.
Summary of allelic-specific, tissue-specific and individual-specific gene expression.
| Line 1 | Line 2 | # shared het. SNPs | # tsASE calls | % SNPs with tsASE |
|---|---|---|---|---|
| PGP1L.1 | PGP1L.2 | 1379 | 20 | 1.5% |
| PGP1F.1 | PGP1F.2 | 1586 | 55 | 2.0% |
| PGP1L.1 | PGP1F.1 | 1106 | 110 | 8.5% |
| PGP1L.2 | PGP1F.1 | 1218 | 103 | 7.0% |
| PGP1L.1 | PGP1F.2 | 1158 | 115 | 8.5% |
| PGP1L.2 | PGP1F.2 | 1280 | 110 | 7.1% |
| PGP1F.1 | PGP1K | 1252 | 88 | 5.6% |
| PGP1L.1 | PGP1K | 1087 | 62 | 4.3% |
| Hues37 | Hues38 | 1153 | 141 | 10.8% |
| Hues37 | Hues58 | 370 | 76 | 19.1% |
| Hues56 | Hues38 | 475 | 97 | 19.0% |
| Hues56 | Hues58 | 793 | 125 | 14.3% |
Genetically related hESC lines share common allele-specific gene expression
A large panel of hESC lines that included 18 sibling lines has recently been derived18. As a first step towards dissecting cis-regulatory effects in hES cells, we characterized the patterns of ASE on two pairs of sibling cell lines HUES37/38 and HUES56/58 (each pair contains a female line and a male line). We hypothesized that, if allele-specific gene expression is caused by cis-regulatory SNPs, rather than by epigenetic mechanisms or by stochastic processes, then genetically related cell lines should be more similar in ASE. Similar to the adult tissue-derived PGP lines, 11–18% of heterozygous SNPs show allele-specific expression in these hES lines (Table 1). Sibling cell lines share less line-specific ASE than genetically unrelated lines (Supplementary Fig. 5), which is consistent with our hypothesis.
Based on the similarity of allelic ratios, we performed hierarchical clustering on all the ten samples characterized in this study (Fig. 2e). The five PGP1 samples were grouped in a clade, within which the two pairs of replicates are closer to each other than with different cell types. The two pairs of hES sibling lines were also grouped as expected. To further illustrate this finding, we calculated the genetic similarity between these cell lines based on the genotypes of approximately 18,000–22,000 coding SNPs determined by the allelotyping assay on genomic DNA, and generated a cluster dendrogram from the similarity matrix (Fig. 2f). The similarities of allelic ratios and genetic similarities are highly correlated (Fig. 2g). ANOVA analysis showed that 82.5% of the variation in the similarity of allelic ratios between two lines can be explained by the genetic similarities between the lines (p<2.2e-16), and that the effect of cell type is not statistically significant (p=0.176). This observation suggests that variation of ASE among different cell lines is primarily determined by genetic instead of epigenetic or environmental factors, which are predominantly trans-acting.
Strand-specific ASE
Recently antisense transcription was found to be more common than was previously thought19. Antisense transcription initiates from different starting sites than the sense transcripts. The allelic ratios measured by conventional array-based assays could be the averages of the sense and antisense transcripts, which are likely to be regulated by different cis-regulatory mechanisms. Unlike array-based assays, padlock capture can be strand-specific. The 27k probe set contains roughly half of the probes targeting the sense strand, and another half for the anti-sense strand. However, since the capturing experiments were performed on double-stranded cDNA, all the probes are functional even in the absence of antisense expression. To distinguish the sense and antisense expression, we did two additional capturing and sequencing experiments on the first-strand cDNA of PGP1L and PGP1F. We calculated the ratios of read counts for each captured SNP between the single-stranded cDNA and double-stranded cDNA (SS/DS ratios) from the same cell line. For the probes targeting the antisense transcripts, the expected SS/DS ratios are zero if there is no antisense transcription or the antisense transcripts do not contain poly-A tails. We saw different distribution of the SS/DS ratios between the probes targeting the sense strand, and the probes for the antisense strand. Roughly half of the antisense SNPs were not detected; the majority of detectable antisense SNPs expressed at a much lower level than sense SNPs (Supplementary Fig. 6). Therefore, the allelic ratios measured from double-stranded DNA should represent cis-regulatory effects mostly on the sense transcripts. This is consistent with the clustering analysis, in which strand-specific allelic ratios from PGP1L and PGP1F were closely grouped together with the allelic ratios from corresponding double-stranded cDNAs (Fig. 2e).
X inactivation in hESC lines
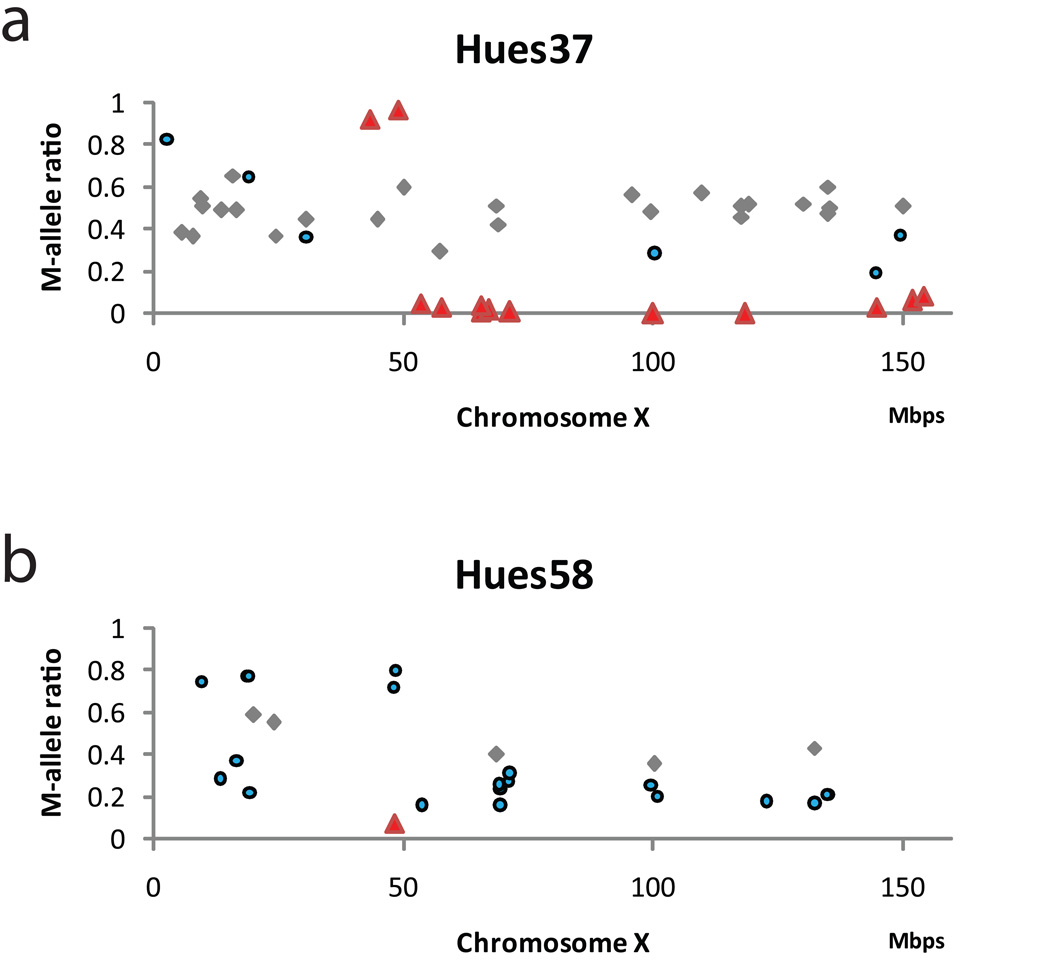
One X chromosome in adult female cells is randomly inactivated, but the developmental stage at which X inactivation happens in human is still not known. It has been reported that hESCs vary in X-inactivation status from one line to another20–22. The inclusion of two female hESC lines allowed us to characterize X-inactivation status across the chromosome based on mono-allelic gene expression. We identified 49 and 27 heterozygous SNPs on the X chromosome in HUES37 and HUES58. Using an arbitrary thresholds for allelic ratios of <0.1 or >0.9, thirteen SNPs (12 genes) have mono-allelic expression in HUES37, but only one SNP was found in HUES58 (Fig. 3).
Figure 3.
X-inactivation in female human embryonic stem cell lines (HUES37 and HUES58). HUES37 (a). and HUES58 (b). The x-axes are chromosomal positions for the SNPS and the y-axes are the allelic ratios for the M-alleles. Red triangles represent the SNPs that were called monoallelic; blue circles are the SNPs called as allele-specific; and the gray diamonds are the SNPs with bi-allelic expression.
To investigate whether a particular X chromosome from one of the parents was inactivated, we took advantage of the inclusion of the male sibling lines. Recombination occurs on average once per chromosome per meiosis, therefore male and female sibling lines tend to share very long segment of haplotypes on the X chromosome inherited from the mothers. For all heterozygous SNPs on chromosome X of the female line, we plotted along the chromosome the ratios for the “M-alleles”, which are the alleles presented in the sibling male line and hence very likely came from the mother. In HUES37, we found that the M-alleles of the 10 SNP spanning a 100Mb region including the entire q-arm are silent (Fig. 3a), indicating that one particular X chromosome was inactivated in HUES37. The other two SNPs on the p-arm showed dominant expression (Fig. 3a), presumably because there was a meiotic crossover at ~50Mb from the p-terminal and the M-alleles of these two SNPs actually came from the other parent. Such a pattern would be observed only when the cell line is clonal and one X chromosome is inactivated. The expression of many other chromosome X SNPs are not monoallelic, suggesting that X-inactivation was probably not complete in HUES37.
Although only one SNP with mono-allelic expression was found in HUES58 (Fig. 3b), an unusually high fraction of SNPs was detected as allele-specific (21/27, compared with 281/1261 in autosomes), suggesting that this line could be in the very early stage of X-inactivation. Similar to HUES37, the ratios for the M-alleles biased towards zero across most of the p-arm. Therefore, one particular X chromosome was less active transcriptionally in HUES58 even though not silenced. In summary, the difference in the distributions of allelic ratios between HUES37 and HUES58 is consistent with the previous observation that different hESC lines vary in the degree of X-inactivation. In addition, the observation of multiple SNPs with mono-allelic expression in HUES37 suggests that X-inactivation could be initiated from multiple locations in the X chromosome.
DISCUSSION
In this report we demonstrated that accurate digital allelotyping can be achieved with the combination of padlock capture and single molecule sequencing. The high capturing specificity associated with padlock probes makes it possible to focus sequencing efforts on a subset of most informative regions. The HuRef genome contains 10,842 heterozygous SNPs in the 31,185 Ensembl genes (35.6 Mb)23, which means roughly 1.1% of sequencing reads (36 bp) in a typical RNA-seq experiment would contain heterozygous SNPs. In comparison, every 36 bp read in the padlock captured libraries covers a SNP, and ~25% of the SNPs are heterozygous, which translates to the reduction of sequencing by ~20 fold. In addition, the relative abundance of different transcripts in RNA-seq libraries varies across a range of 105, while most padlock captured fragments are in a range of 103–104. We were able to make allelotyping calls on an average of 1,789 SNPs (in 1,371 genes) with ≥50x coverage using ~5.8 million reads per sample. Extrapolated from the distribution of “reads per kilobase of exon model per million mapped reads” (RPKM) in the mouse transcriptome10, approximately 203 genes were sequenced at ≥50x coverage across the full length with the same amount of 36bp reads, and only 1/3 of such genes contain heterozygous SNPs. Therefore, padlock capture provides an advantage over RNA-seq in the efficiency of the assay for ASE quantitation. Padlock capture of expressed SNP can be further improved in several aspects. First, our current probe set was designed for capturing exonic SNPs. The inclusion of intronic SNPs will extend the assay to genes that do not contain common exonic SNPs. However mRNA is preferable since it has higher abundance and reflects more steps that can be regulated post-transcriptionally. Second, with the set of 27,000 probes we typically obtained allelotyping measurements from less than 2000 SNPs. In addition to the fact that only ~25% of the SNPs are heterozygous and informative, we also missed 50%–70% heterozygous SNPs because of the low expression of the corresponding genes. A subsetting strategy we recently developed for normalizing bisulfite sequencing library can be used to adjust the relative concentration of different targets in the sequencing libraries15. This would enable us to detect the ASE in less abundant transcripts.
Although allele-specific gene expression has been reported by many studies in both human and mouse1,2,8, and several related cis-regulatory polymorphisms have been identified, some other reports also argued that epigenetic mechanisms, especially DNA methylation, also play a critical role24,25. Using a unique panel of eight cell lines with various degree of genetic similarity, we found that roughly 82.5% of the global variation in ASE is determined by genetic factors. Gimbelbrant et al recently reported that roughly 300 of 4000 human genes are subject to random monoallelic expression in clonal lymphocyte and fibroblast lines26. We did not observe the similar patterns in this study, which could be due to the fact that all the four PGP lines we used are not clonal, and the four HUES lines are pluripotent although they are mostly clonal. At the single cell level, randomly allelic drift could be dominating for a fraction of genes in the genome. However for a population of non-clonal cells, stochastic epigenetic effects could be averaged out, and genetic factors become dominating. In addition, genetic effects and epigenetic effects might not be mutually exclusive. A cis-regulatory variant could result in the change of binding affinity of either a protein or RNA regulator, which could directly or indirectly recruit the protein complexes related to DNA methylation or histone modifications and lead to the change of local epigenetic status. With the integration of global allele-specific assays for gene expression and DNA methylation (or histone modifications), such hypotheses are testable on a genome scale.
Treating gene expression as quantitative traits (eQTLs), the genetic determinants associated with the variation of gene expression in human population have been identified in several recent studies27,28. The success of these studies raised the hope that the genetic variants that were mapped to eQTLs could potentially be considered as candidates for complex human diseases. However, limited by the availability of human samples, many studies focused on EBV-transformed B-lymphocytes. One exception is the recent study by Emilsson et al., which demonstrated the dramatically improved power of detecting cis-regulatory variants when analyzing adipose tissues instead of lymphocytes from obesity patients17. The prevalence of tissue-specific ASE revealed in our study suggests that many cis-regulatory variants function in a tissue-specific manner. Therefore, eQTL analysis performed on B-lymphocytes could be of limited utility to many human diseases that affect other human cell types. Recent revolutionary advances in the reprogramming of adult cells29 and the efficient differentiation into cell-types affected by disease30 have created new opportunities to perform population genomic studies on disease relevant tissues. Finally, mapping cis-regulatory polymorphisms to ASE genes would require a much smaller sample size than typical GWAS studies, because candidate SNPs are restricted to only ~300 sites in vicinity of the gene of interest. Once the candidate is narrowed down to a short list of SNPs that show genotype-phenotype correlation, proof of causality can be achieved by replacing a single allele using homologous recombination. Therefore, treating ASE as an intermediate phenotype could be a very efficient way to identify causal polymorphisms responsible for complex human diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Colleen Ludka and Narimene Lekmine for assistance for Illumina sequencing.
K.Z. and J.B.L. developed and optimized the digital allelotyping method; J.D., Z.L. and J.L. participated in the experiments; D.E and K.E provided DNA/RNA of hES cells; E.L. provided oligonucleotide libraries; Y.G. and B.X. performed Illumina sequencing. K.Z., J.B.L. and J.A. performed data analysis; K.Z and G.M.C. oversaw the project.
This study was supported by NHGRI (CEGS), NHLBI, the Broad Institute (SPARC) and PGP donations (to G.M.C.), and UCSD new faculty startup fund (to K.Z.). J.D. was sponsored by a CIRM post-doctoral fellowship.
REFERENCES
- 1.Yan H, Yuan W, Velculescu VE, et al. Science. 2002;297(5584):1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 2.Pastinen T, Hudson TJ. Science. 2004;306(5696):647. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 3.Lo HS, Wang Z, Hu Y, et al. Genome Res. 2003;13(8):1855. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight JC, Keating BJ, Rockett KA, et al. Nat Genet. 2003;33(4):469. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 5.Serre D, Gurd S, Ge B, et al. PLoS Genet. 2008;4(2):e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maynard ND, Chen J, Stuart RK, et al. Nat Methods. 2008;5(4):307. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 7.Pant PV, Tao H, Beilharz EJ, et al. Genome Res. 2006;16(3):331. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milani L, Gupta M, Andersen M, et al. Nucleic Acids Res. 2007;35(5):e34. doi: 10.1093/nar/gkl1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cloonan N, Forrest AR, Kolle G, et al. Nat Methods. 2008;5(7):613. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 10.Mortazavi A, Williams BA, McCue K, et al. Nat Methods. 2008;5(7):621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson M, Malmgren H, Samiotaki M, et al. Science. 1994;265(5181):2085. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 12.Hardenbol P, Baner J, Jain M, et al. Nat Biotechnol. 2003;21(6):673. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 13.Hardenbol P, Yu F, Belmont J, et al. Genome Res. 2005;15(2):269. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porreca GJ, Zhang K, Li JB, et al. Nat Methods. 2007;4(11):931. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 15.Deng J, Shoemaker R, Xie B, et al. Nat Biotechnol. 2009;27(4):353. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge B, Gurd S, Gaudin T, et al. Genome Res. 2005;15(11):1584. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emilsson V, Thorleifsson G, Zhang B, et al. Nature. 2008;452(7186):423. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 18.Chen AE, Egli D, Niakan K, et al. Cell Stem Cell. 2009;4(2):103. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Vogelstein B, Velculescu VE, et al. Science. 2008;322(5909):1855. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman LM, Hall L, Batten JL, et al. Stem Cells. 2005;23(10):1468. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Matsuno Y, Fouse SD, et al. Proc Natl Acad Sci U S A. 2008;105(12):4709. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva SS, Rowntree RK, Mekhoubad S, et al. Proc Natl Acad Sci U S A. 2008;105(12):4820. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng PC, Levy S, Huang J, et al. PLoS Genet. 2008;4(8):e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milani L, Lundmark A, Nordlund J, et al. Genome Res. 2008 [Google Scholar]
- 25.Bjornsson HT, Albert TJ, Ladd-Acosta CM, et al. Genome Res. 2008;18(5):771. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimelbrant A, Hutchinson JN, Thompson BR, et al. Science. 2007;318(5853):1136. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 27.Dixon AL, Liang L, Moffatt MF, et al. Nat Genet. 2007 doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 28.Stranger BE, Nica AC, Forrest MS, et al. Nat Genet. 2007 doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, Tanabe K, Ohnuki M, et al. Cell. 2007;131(5):861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Dimos JT, Rodolfa KT, Niakan KK, et al. Science. 2008;321(5893):1218. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.