Summary
Retroviral reverse transcriptases possess both a DNA polymerase and an RNase H activity. The linkage with the DNA polymerase activity endows the retroviral RNases H with unique properties not found in the cellular counterparts. In addition to the typical endonuclease activity on a DNA/RNA hybrid, cleavage by the retroviral enzymes is also directed by both DNA 3' recessed and RNA 5' recessed ends, and by certain nucleotide sequence preferences in the vicinity of the cleavage site. This spectrum of specificities enables the retroviral RNases H to carry out a series of cleavage reactions during reverse transcription to degrade the viral RNA genome after minus strand synthesis, precisely generate the primer for the initiation of plus strands, facilitate the initiation of plus strand synthesis, and remove both plus- and minus-strand primers after they have been extended.
Keywords: RNase H, reverse transcriptase, reverse transcription, human immunodeficiency virus, type 1 (HIV-1), Moloney murine leukemia virus (M-MLV), DNA/RNA hybrids, polypurine tract (PPT), catalytic mechanism
Introduction
At the time of its discovery in 1970, the presence of an RNA-dependent DNA polymerase activity in retrovirus particles provided strong and exciting support for the hypothesis that the single-stranded RNA genome of a retrovirus is replicated through a DNA intermediate [1,2]. Not only did this discovery of reverse transcriptase (as it was dubbed) challenge the existing dogma concerning the flow of genetic information in biology, it raised the critical question as to how the DNA/RNA hybrid created when the viral genome RNA is used as a template by reverse transcriptase is further processed. In retrospect, it is not surprising that an RNase H activity that degrades the RNA strand of a DNA/RNA hybrid is required to free the newly-made DNA strand (called the minus strand since it is complementary to the plus genome RNA) for use as a template in the synthesis of the second or plus strand of DNA. However, it was a surprise when the retroviral-specific RNase H activity turned out to be present in the same protein molecule as the polymerase activity [3]. This intimate association of the DNA polymerase and RNase H activities in reverse transcriptase has profound effects on the activities and capabilities of both enzymes.
This review article will provide a summary of the salient features of retroviral RNases H with a focus on how the shared substrate binding sites for the two activities of reverse transcriptase endow the retroviral RNases H with features not found in the cellular counterparts, and how these unusual properties are crucial for the multiple roles played by RNase H in reverse transcription. Although occasional reference will be made to other retroviral enzymes, the primary focus will be on the well-studied RNase H activities associated with human immunodeficiency virus, type 1 (HIV-1) and Moloney murine leukemia virus (M-MLV) reverse transcriptases. The reader is directed to other excellent reviews that describe the older literature and cover other recent aspects of retroviral RNases H [4–8].
Structure-function considerations
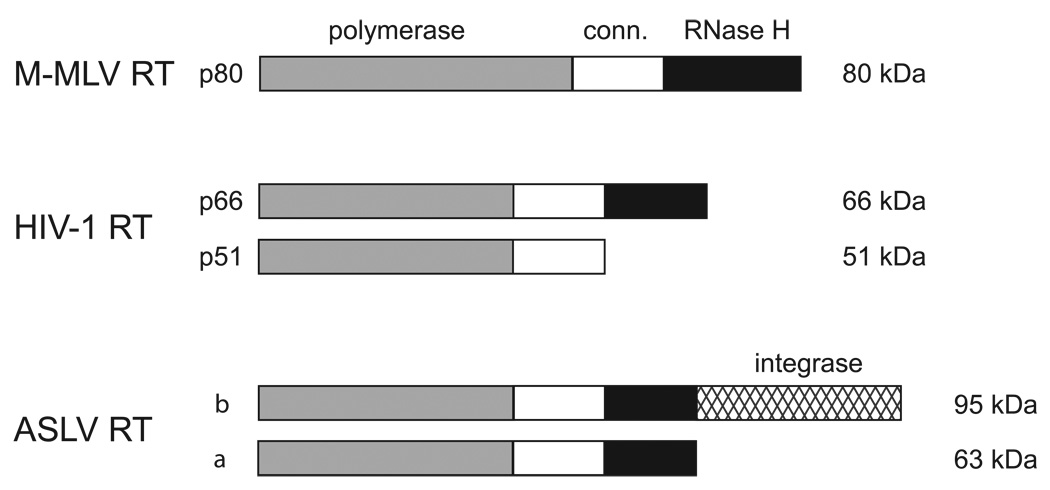
Although the reverse transcriptases from murine, human, and avian retroviruses display different subunit structures, the relative orientations and sizes of the DNA polymerase, connection, and RNase H domains within a given polypeptide chain are similar for the different proteins (Fig. 1). M-MLV reverse transcriptase is a 80 kDa monomer in which the DNA polymerase activity occupies the N-terminal ~55% and the RNase H domain occupies the C-terminal ~25% of the protein, with the connection domain accounting for the remainder. HIV-1 reverse transcriptase is a heterodimer made up of a p66 subunit containing the active forms of both the polymerase and the RNase H arranged similarly to that of the M-MLV monomer, and a p51 subunit that is derived by proteolysis of p66 and is missing the C-terminal RNase H domain (see Fig. 2). The p51 subunit is enzymatically inactive and simply plays a structural role in the protein. The avian sarcoma-leukosis virus (ASLV) reverse transcriptase is also a heterodimer, but the larger β subunit, in addition to possessing both the polymerase and RNase H domains found in the α subunit, also contains a C-terminal region corresponding to the viral integrase.
Fig. 1.
Subunit and domain structure of retroviral reverse transcriptases. Reverse transcriptase from M-MLV is a monomer, whereas the HIV-1 and ASLV reverse transcriptases are both heterodimeric. The subunit designations and their sizes (kDa) are indicated along the left and right sides of the figure, respectively. The approximate sizes of the polymerase, connection (conn.) and RNase H domains are shown in gray, white, and black, respectively. The larger β subunit of the ASLV reverse transcriptase also contains the integrase domain depicted by crosshatching.
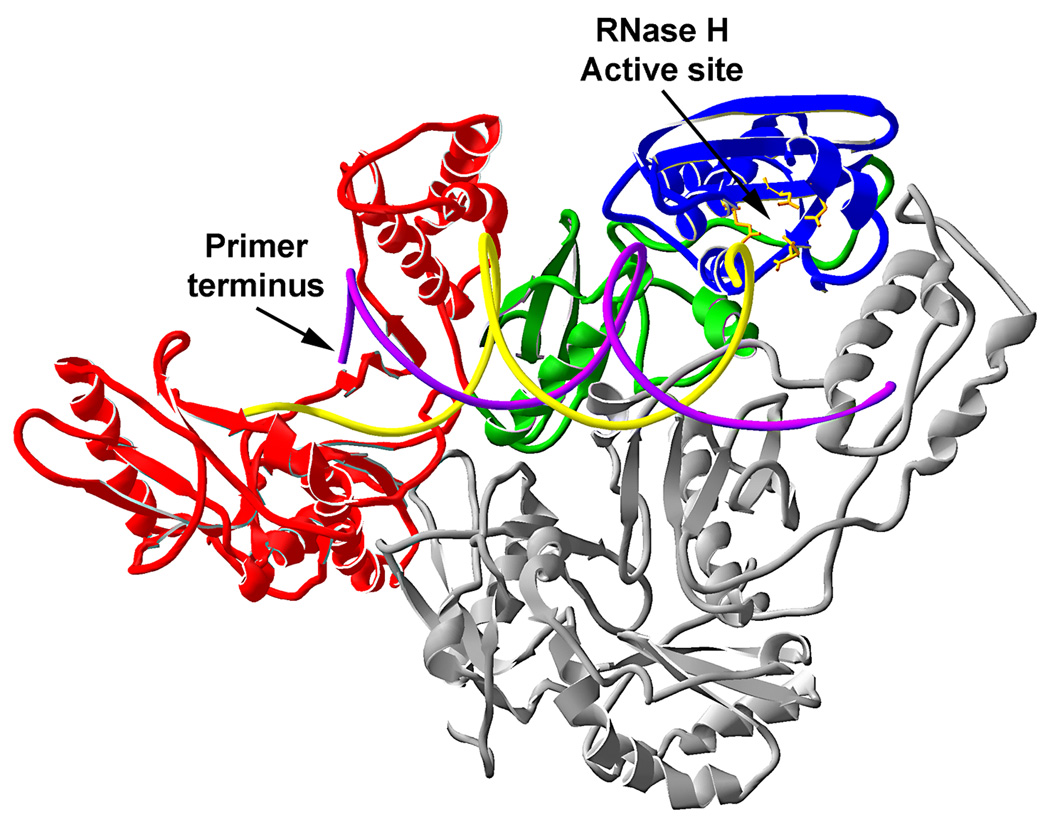
Fig. 2.
Ribbon diagram of the co-crystal structure of HIV-1 reverse transcriptase with a bound RNA template and DNA primer (pdb entry 1HYS) [19]. The polymerase (p66 residues 1–318), connection (p66 residues 319–437) and RNase H (p66 residues 438–553) domains are drawn in red, green, and blue, respectively with the p51 subunit shown in gray. The RNase H active site is indicated with the four key acidic residues drawn in yellow ball and stick. The primer terminus of the DNA primer strand (purple) is indicated with the RNA template strand shown in yellow. The drawing was created using Swiss-Pdb Viewer software (v3.7) (GlaxoSmithKline).
The isolated RNase H domain of M-MLV reverse transcriptase is enzymatically active, but the activity is low and exhibits a greatly relaxed substrate specificity [9–11]. The isolated HIV-1 RNase H domain is inactive, but the addition of various N-terminal extensions restores some RNase H activity [12–18]. The reduced specificity of the isolated RNase H domains underscores the importance of the polymerase and connection domains for substrate binding and selectivity. Structural models support this conclusion by showing that a DNA/RNA hybrid substrate gains access to the RNase H active site by associating with the same binding cleft utilized by the polymerase for binding a primer-template [19] (Fig. 2). Some of the structural features within and outside the RNase H domain that are important for substrate selectivity are highlighted in the remainder of this section.
The polymerase domain has been directly implicated in RNase H specificity through the mutagenesis of individual amino acids. Notable examples include changes at Trp266 and Phe61 in HIV-1 reverse transcriptase, both of which render the RNase H incapable of generating the polypurine tract (PPT) primer or removing the PPT primer once it has been extended [20–22].
The RNase H domains of M-MLV and HIV-1 reverse transcriptases are structurally very similar to the Escherichia coli and Bacillus halodurans RNases H, and to human RNase H1, and these similarities provide key insights concerning substrate recognition and catalysis by the retroviral enzymes. One conspicuous difference among these enzymes is a positively charged helix called the C-helix that is present in the M-MLV, human, and E. coli RNases H, but absent in the RNases H from HIV-1 and B. halodurans [23–27,19,28–30]. Structure-function studies with the E. coli and M-MLV RNases H implicate the C-helix in substrate recognition and catalytic activity, and a mutant form of the M-MLV reverse transcriptase in which the C-helix has been deleted is replication defective [31–33]. Despite the apparent absence of a C-helix in the RNase H domain of HIV-1 reverse transcriptase, modeling studies comparing the C-helix of M-MLV RNase H with the p66 subunit of the HIV-1 enzyme suggest that a series of positively charged residues in the p66 connection domain may functionally substitute for the C-helix in the HIV-1 reverse transcriptase [34]. Mutagenesis studies with HIV-1 reverse transcriptase identify additional residues within the connection domain that contribute to the activity of the RNase H [35] and linker scanning mutagenesis of the M-MLV connection domain indicate that this region is essential for viability of the virus [36].
The RNase H primer grip is a region near the RNase H active site that contacts the nucleotides in the DNA strand of the hybrid substrate that are base paired with RNA nucleotides at positions −4 to −9 relative to the site of cleavage, which is defined as occurring between the −1 and +1 RNA nucleotides [19,34]. For HIV-1 reverse transcriptase, this region includes residues found in the polymerase, RNase H, and connection domains of p66 and also two residues present in the p51 subunit. The RNase H primer grip is important for binding the DNA/RNA hybrid substrate since point mutations in this region not only reduce RNase H activity, but also affect the specificity of the enzyme [37–39,35,40]. Primer grip residue Tyr501 in HIV-1 reverse transcriptase (Tyr586 in M-MLV) appears to be a particularly important substrate contact residue as changes at this site profoundly affect both the RNase H activity and proper substrate recognition [37,39,41,42,40]. Gln475 in HIV-1 reverse transcriptase is also a critical primer grip residue that not only interacts with the DNA strand but also contacts the RNA strand at positions −2 and +1. Mutagenesis studies indicate that Gln475 is particularly important for the cleavage specificity of the enzyme [39].
Based on co-crystal structures of HIV-1 reverse transcriptase with DNA duplexes or DNA/RNA hybrids [25,27,19], the physical distance between the 3' end of a primer located in the polymerase active site and the region of the substrate in close contact with the RNase H active site corresponds to 17–18 base pairs (Fig. 2). This relationship helps explain some of the observations concerning the effects of recessed DNA 3' and RNA 5' ends on RNase H specificity as described in the sections to follow.
Enzyme activity and catalysis
Retroviral RNases H are partially processive endonucleases that cleave the RNA strand of a DNA/RNA hybrid in a Mg++-dependent reaction to produce 5' phosphate and 3' hydroxyl termini [43,44]. It has been shown that the RNases H associated with both HIV-1 and M-MLV reverse transcriptases are capable of cleaving RNA/RNA duplexes, an activity that has been termed RNase H* [45–47]. However, since the RNase H* activity is only manifest in the presence of the less biologically-relevant divalent cation, Mn++, it is doubtful that this activity plays a role during reverse transcription in vivo. Given a substrate in which one strand is entirely DNA and the other strand is RNA at the 5' end followed by a stretch of DNA, the HIV-1 and M-MLV retroviral RNases H strongly prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction rather than precisely at the junction itself [48]. As discussed below, the most dramatic example of this preference is the finding that a single ribo A is left on the 5' end of the DNA during tRNA primer removal by HIV-1 RNase H [49,50,14]. However, this preference to cleave one nucleotide away from the RNA-DNA junction is not absolute since in the presence of other specificity determinants, the retroviral RNases H will cleave precisely at an RNA-DNA junction [51,52,49].
Two recent co-crystal structures of the B. halodurans and human RNases H with bound substrate [29,53,54,30] provide key insights regarding the role of divalent cations in the catalytic mechanism of the structurally similar RNase H domains of HIV-1 and M-MLV reverse transcriptases. Thus, the current model for hydrolytic cleavage by the retroviral RNases H invokes a two-Mg++-ion catalytic mechanism [8]. In HIV-1 RNase H, four highly conserved acidic amino acids (Asp443, Glu478, Asp498, and Asp549) coordinate the binding of two Mg++ ions. The corresponding active site amino acids in the M-MLV enzyme are Asp524, Glu562, Asp583, and Asp653. Catalysis involves activation of the nucleophilic water by one of the Mg++ ions with transition-state stabilization apparently being achieved by both Mg++ ions.
Substrate specificity
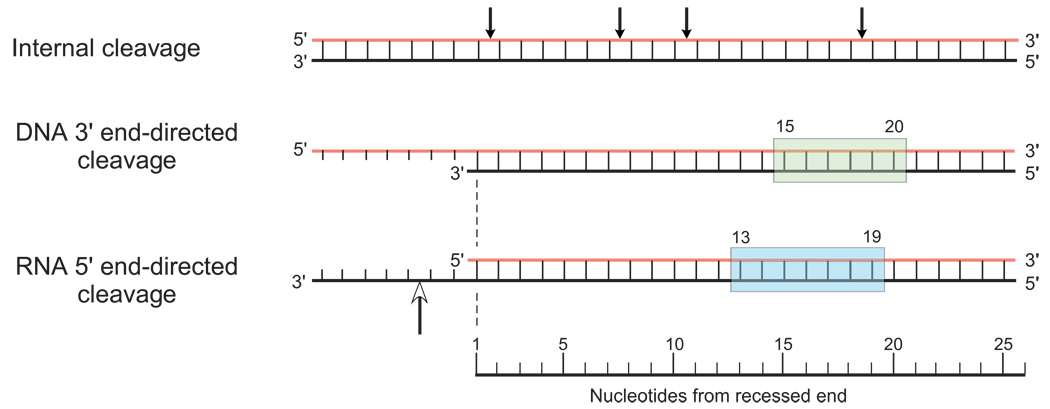
Three distinct cleavage modes have been described for retroviral RNases H that are referred to as internal, DNA 3' end-directed, and RNA 5' end-directed cleavages. The two end-directed modes are unique to the retroviral RNases H and derive from the presence of the associated polymerase domain. In the internal cleavage mode, the RNases H behave as typical endonucleases and cleave the RNA along the length of a DNA/RNA hybrid substrate in the absence of any "end" effects. In the two end-directed modes of cleavage, the interaction of the enzyme with the substrate involves recognition of a recessed RNA 5' or a recessed DNA 3' end.
Internal Cleavage
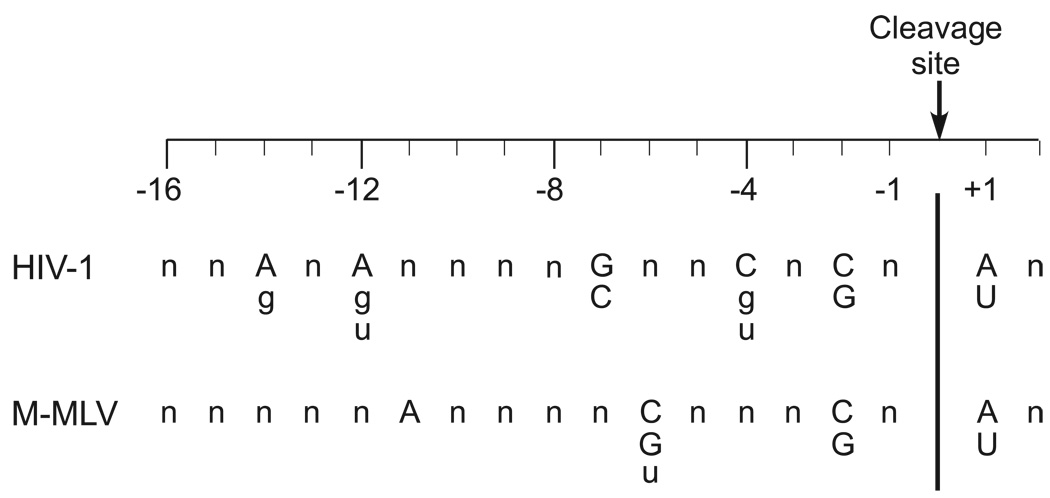
Although cleavage at internal sites on an extended DNA/RNA hybrid has been inferred from a variety of studies over the years, only recently has it been recognized that nucleotide sequence preferences play an important role in this mode of cleavage. HIV-1 and M-MLV RNase H cleavage sites that were too far from an end to be either DNA 3' or RNA 5' end-directed were mapped on a long DNA/RNA hybrid and the nucleotide sequences surrounding the scissile phosphate (designated as between the −1 and +1 positions) were aligned. A statistical analysis of the frequency of nucleotides on both sides of the cleavage site revealed that HIV-1 RNase H prefers certain nucleotides at positions +1, −2, −4, −7, −12, and −14. For M-MLV, the preferred positions are located at +1, −2, −6, and −11 (summarized in Fig. 3) [55,8]. Notably, the preferred nucleotides at the +1 (A or U) and −2 (C or G) positions are the same for the two enzymes. The preferred positions all fall within a region of the substrate contacted by the enzymes as defined by the co-crystal structure containing a DNA/RNA hybrid [19] and by DNase I footprinting studies [56–58]. The structural basis for these sequence preferences remains for the most part obscure, but the contact between Gln475 in the HIV-1 enzyme and the −2 guanine base in the RNA strand likely contributes to the preference at this position [19].
Fig. 3.
Sequence preferences for internal cleavage by retroviral RNases H. For the purposes of site alignment, RNase H cleavage is designated as occurring between nucleotides −1 and +1. The preferred nucleotides at positions −14, −12, −7, −4, −2 and +1 are shown for HIV-1 RNase H and at positions −11, −6, −2 and +1 for M-MLV RNase H. The strongest preferences are indicated in upper case letters with the weaker preferences in lower case letters.
DNA 3' end-directed cleavage
A recessed DNA 3' end in a DNA/RNA hybrid is recognized by the polymerase activity of reverse transcriptase as a primer terminus and is utilized for the synthesis of a DNA strand complementary to the RNA. In the absence of dNTPs or at a pause site during polymerization, the active site of the RNase H activity would be predicted, based on structural models, to be positioned 17–18 nucleotides away from the DNA primer terminus (Fig. 4) [25,27,19]. Results from a number of laboratories indicate that RNase H cleavage of a hybrid with a recessed DNA 3' end, or at pause sites during polymerization, actually occurs within a window ~15–20 nucleotides away from the primer terminus (Fig. 4) [59–64]. Notably, the cleavage window centers on the distance predicted from the crystal structures, but extends in both directions by 2–3 base pairs, presumably owing to some degree of structural variation in the substrate and flexibility within the protein.
Fig. 4.
Three cleavage modes for retroviral RNases H. DNA/RNA hybrids are drawn with RNA strands in red and DNA strands in black. In the internal cleavage mode, the arrows mark the sites of cleavage along the length of the hybrid where nucleotide sequence alone determines the cleavage site. The cleavage window for the DNA 3' end-directed cleavage mode (15–20 nucleotides from the recessed DNA end) is highlighted in green. The corresponding cleavage window for RNA 5' end-directed cleavage (13–19 nucleotides from end) is highlighted in blue. The open-headed arrow in the RNA 5' end-directed cleavage mode indicates the position of the DNA phosphate that appears to be bound near the active site pocket in the polymerase domain normally occupied by the 3' DNA primer terminus during DNA polymerization.
RNA 5' end-directed cleavage
Unexpectedly, reverse transcriptase will bind to a hybrid duplex containing a recessed RNA 5' end and cleave the RNA ~13–19 nucleotides from the RNA end (Fig. 4) [65,66,60,67–72]. RNase H cleavage only occurs at sites within the window that conform to the nucleotide sequence preferences for internal cleavage that are proximal to the active site of the enzyme [72]. It is not known why the window for RNA 5' end-directed cleavage is two nucleotides closer to the recessed end than the window for DNA 3' end-directed cleavage. However, based on this difference, a phosphate residue in the single-stranded DNA that extends beyond the recessed RNA 5' end (Fig. 4, open-headed arrow) would be predicted to occupy the position in the polymerase active site normally occupied by the primer terminus during DNA synthesis. Presumably some feature of the polymerase active site region interacts with the recessed 5' RNA end to facilitate this unique binding configuration to the primer-template binding cleft of reverse transcriptase.
In some studies, cleavage in the RNA 5' end-directed mode has been observed as close as 7 base pairs and as many as 21 base pairs from the recessed end [73,74,67,75–78], possibly resulting from sliding of the enzyme after the initial binding event. Importantly, an RNA 5' end at a nick is not recognized for this mode of cleavage by the HIV-1 and M-MLV RNases H. However, cleavage will occur by this mode if a gap of 2–3 nucleotides is present upstream of the RNA 5' end.
Roles of RNase H in reverse transcription
Starting with the retroviral plus-strand genome, the process of reverse transcription produces a double-stranded DNA product that is integrated into the host cell genome and ultimately serves as a template for the production of more genome RNAs [79,80]. The RNase H activity of reverse transcriptase is required for several stages of the reverse transcription process (reviewed in [4,6,7]), making it an essential enzyme activity for viral replication [32,81]. While all retroviruses have diploid genomes and template switching between genomes has been observed during reverse transcription, only a single genome strand will be considered in the following discussion. The key steps in M-MLV and HIV-1 reverse transcription are summarized below with an emphasis on the multiple roles played by RNase H in the process (Fig. 5).
Fig. 5.
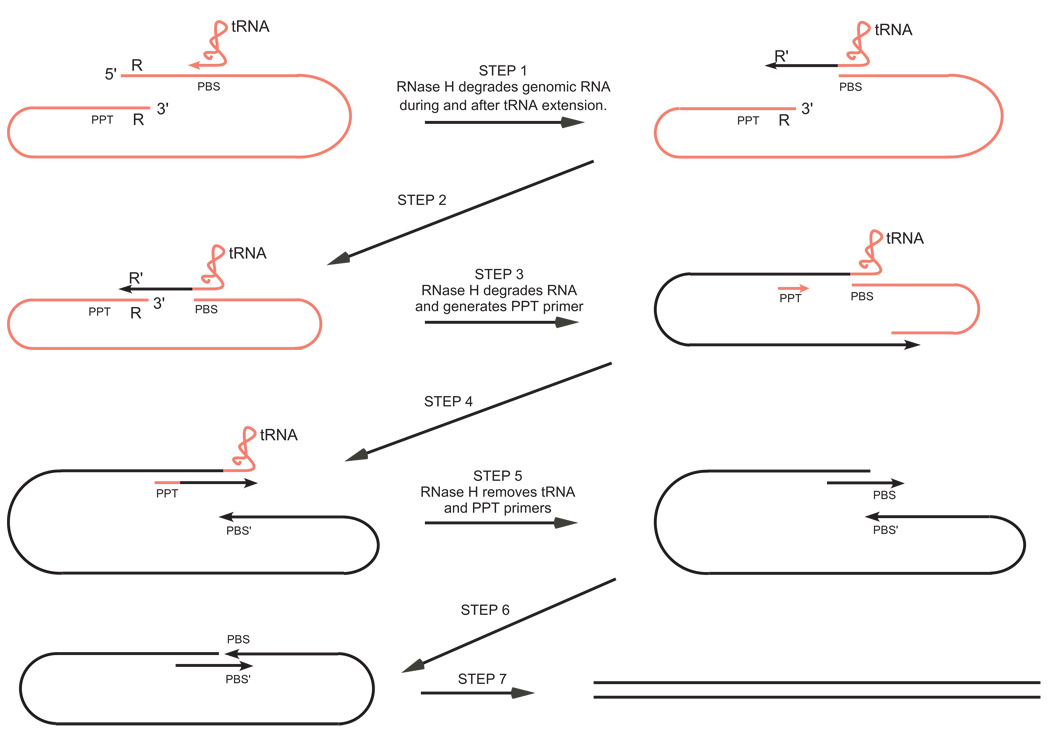
Roles of RNase H in reverse transcription. The retroviral genome and the associated cell-derived tRNA bound to the primer binding site (PBS) are shown in red with the DNA strands produced during reverse transcription shown in black. A repeated sequence denoted R is located at both ends of the retroviral genome. The sequences complementary to PBS and R are denoted PBS' and R', respectively. The polypurine tract (PPT) serves as the primer for plus-strand synthesis. The steps at which RNase H plays a role are highlighted. See the text for a detailed explanation.
STEP 1
Early after infection a subviral particle enters the cytoplasm containing, in addition to the viral RNA associated with the nucleocapsid protein (NC), a host-derived tRNA bound to the genome at the 18 nucleotide-long primer binding site (PBS), 50–100 molecules of reverse transcriptase and the integrase. As shown in Fig. 5, the polymerase activity of reverse transcriptase initiates reverse transcription by extending the tRNA primer to copy the 5' repeat sequence (R) at the end of the genome and produce what is called the minus strong-strop DNA. Concomitant with polymerization and presumably at pause sites [63,64], the RNase H activity utilizes the DNA 3' end-directed cleavage mode to cleave the RNA strand of the resulting hybrid. However, for HIV-1 and M-MLV reverse transcriptases, such cleavages occur on average only once for every 100–120 nucleotides polymerized, a frequency that is insufficient to degrade the RNA to small enough fragments to render the newly synthesized DNA free of RNA [62,82]. Therefore, complete degradation of the template RNA likely requires multiple internal cleavages to generate gaps that subsequently enable degradation by the RNA 5' end-directed mode of cleavage.
STEP 2
When the polymerase reaches the end of the RNA template, the RNase H cleavages nearest to the 5' end of the RNA would be expected to be determined by the cleavage window for whichever end-directed cleavage mode applies to a blunt-ended substrate. In either case, a short RNA oligonucleotide would likely remain base paired with the 3' end of the nascent DNA chain. In fact, for HIV-1, it has been observed that in the presence of the NC protein, a 14 nucleotide-long RNA remains associated with the DNA (not shown in Fig. 5) and, importantly this association prevents self-priming caused by the DNA hairpin (the complement of the RNA TAR structure) that otherwise could form at the 3' end of the nascent DNA [83,84]. Since this residual RNA fragment is short relative to the R sequence (R is 98 nucleotides for HIV-1 and 68 nucleotides for M-MLV), it does not interfere with the first template switch mediated by base pairing between the R' sequence found at the 3' end of the minus strong-stop DNA and the R sequence found at the 3' end of the genome RNA. Once these complementary sequences pair, branch migration displaces the short RNA oligonucleotide, positioning the 3' end of the nascent DNA to act as a primer for the completion of minus strand synthesis.
STEP 3
The first template switch enables continued synthesis of the minus-strand DNA (Fig. 5). RNase H degradation of the genome RNA follows the same pattern as described above, beginning with the occasional DNA 3' end-directed cleavage during polymerization, followed by sequential internal and RNA 5' end-directed cleavages. It is likely that some longer RNA fragments remain base-paired to the minus DNA and must be removed by displacement synthesis during polymerization of the plus-strand DNA [85,82].
Once the PPT region of the genome has been copied, a specific RNase H cleavage near the 3' end of the polypurine sequence generates the primer for plus-strand initiation (for review, see [8]). Underscoring the importance of this specific cleavage event is the fact that the initiation site of the plus-strand DNA determines the left end of the linear product of reverse transcription (Fig. 5) which is a substrate for the viral integrase. Although cleavage at the PPT site is very efficient in the internal cleavage mode for M-MLV, HIV-1 reverse transcriptase is less efficient in this mode and cleavage may instead occur through the DNA 3' end-directed mode at a pause site during HIV-1 minus-strand synthesis [86,52,87,66,88–92,21,93,48]. A possible explanation for the reduced efficiency of cleavage by the HIV-1 enzyme is that while the M-MLV PPT sequence conforms to the preferred nucleotide pattern for internal cleavage described above (Fig. 3), there is an A instead of the preferred G or C at the −7 position of the HIV-1 PPT sequence. A variety of studies have identified the nucleotide positions within the PPT that are critical for proper cleavage and although some of these overlap with the more general preferences for internal cleavage, other positions do not. Thus, for proper PPT primer generation by M-MLV RNase H, positions −1, −2, −4, −5, −6, −7, −10, and −11 are important [94,51,95], while positions +1, −2, −4, −5, −7 have been found to be critical for HIV-1 PPT primer formation [89,38,96,40,97,98].
Step 4
The PPT primer is utilized to initiate plus-strand synthesis which then continues until it reaches the 18th nucleotide in the tRNA where further synthesis is blocked by a methylated base (Fig. 5). This product has been referred to as plus strong-stop DNA. At least for M-MLV, a nick within the PPT that generates the correct primer terminus for plus-strand initiation is poorly utilized in the displacement synthesis mode by the polymerase activity of reverse transcriptase [99]. Efficient utilization of the PPT primer requires at least a small gap and indeed there exists a series of internal RNase H cleavage sites just downstream of the PPT that would appear to fulfill this role.
Step 5
Continued synthesis of the minus and plus strands requires removal of the extended tRNA primer from the end of the minus DNA (Fig. 5). With further extension temporarily blocked by a methylated base at position 19 in the tRNA, the tRNA-DNA junction is within the 15–20 nucleotide window required for DNA 3' end-directed cleavage. As mentioned previously, the RNase H activity of reverse transcriptases strongly prefers to cleave one nucleotide away from an RNA-DNA junction and indeed for HIV-1, tRNA primer removal is observed to cleave the RNA between the 17th and 18th nucleotides from the nascent DNA 3' end to leave a single ribo A on the 5' end of the minus-DNA strand [49,50,14]. Furthermore, cleavage precisely at the RNA-DNA junction by the HIV-1 enzyme, although still within the cleavage window, would appear to be disfavored by the presence of a dC residue at the +1 position rather than the preferred A or U. For M-MLV, cleavage to leave a single ribo A as well as junctional cleavage are both observed, presumably owing to the presence of favored nucleotides at the critical positions flanking both cleavage sites [100,11].
Removal of the PPT primer appears to occur by an internal cleavage event precisely at the RNA-DNA junction [51,52,101,90,91,48]. Apparently the same sequence features responsible for PPT primer generation determine the site of primer removal and override the natural tendency of the RNase H to cleave one ribonucleotide away from an RNA-DNA junction.
Steps 6 and 7
Once the tRNA primer has been removed, the second template switch is effected by the pairing of the complementary PBS and PBS' sequences. A combination of nondisplacement and displacement synthesis [102] converts the circular intermediate into the final linear product of reverse transcription (see Fig. 5).
Perspectives
The specificity determinants for the RNase H activities associated with retroviral reverse transcriptases derive not just from the RNase H domain itself, but also from the polymerase and connection domains. These determinants endow the enzymes with the ability to cleave DNA/RNA hybrids in the three cleavage modes described above. During reverse transcription, these specificities enable the RNase H to carry out a remarkable series of diverse cleavage reactions that lead to the degradation of the genome RNA after minus-strand synthesis, the precise generation of the PPT primer, the facilitation of plus-strand initiation, and the removal of both primers after they have been extended.
Acknowledgements
This work was supported by NIH grant CA51605.
Abbreviations used
- HIV-1
human immunodeficiency virus, type 1
- M-MLV
Moloney murine leukemia virus
- PPT
polypurine tract
- ASLV
avian sarcoma-leukosis virus
- NC
nucleocapsid protein
- PBS
primer binding site.
References
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 3.Molling K, Bolognesi DP, Bauer H, Busen W, Plassmann HW, Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971;234:240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- 4.Champoux JJ. In: Reverse Transcriptase. Skalka AM, Goff SP, editors. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 103–118. [Google Scholar]
- 5.Telesnitsky A, Goff SP. In: Reverse Transcriptase. Skalka AM, Goff SP, editors. Plainview, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 49–83. [Google Scholar]
- 6.Arts EJ, LeGrice SFJ. Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Prog Nucleic Acid Res Mol Biol. 1998;58:339–393. doi: 10.1016/s0079-6603(08)60041-0. [DOI] [PubMed] [Google Scholar]
- 7.Rausch JW, Le Grice SF. 'Binding, bending and bonding': polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int J Biochem Cell Biol. 2004;36:1752–1766. doi: 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Schultz SJ, Champoux JJ. RNase H activity: Structure, specificity, and function in reverse transcription. Virus Res. 2008 doi: 10.1016/j.virusres.2007.12.007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanese N, Goff SP. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz SJ, Champoux JJ. RNase H domain of Moloney murine leukemia virus reverse transcriptase retains activity but requires the polymerase domain for specificity. J Virol. 1996;70:8630–8638. doi: 10.1128/jvi.70.12.8630-8638.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhan X, Crouch RJ. The isolated RNase H domain of murine leukemia virus reverse transcriptase. Retention of activity with concomitant loss of specificity. J Biol Chem. 1997;272:22023–22029. doi: 10.1074/jbc.272.35.22023. [DOI] [PubMed] [Google Scholar]
- 12.Evans DB, Brawn K, Deibel MR, Jr., Tarpley WG, Sharma SK. A recombinant ribonuclease H domain of HIV-1 reverse transcriptase that is enzymatically active. J Biol Chem. 1991;266:20583–20585. [PubMed] [Google Scholar]
- 13.Hostomsky Z, Hostomska Z, Hudson GO, Moomaw EW, Nodes BR. Reconstitution in vitro of RNase H activity by using purified N-terminal and C-terminal domains of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1991;88:1148–1152. doi: 10.1073/pnas.88.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JS, Roth MJ. Specificity of human immunodeficiency virus-1 reverse transcriptase- associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3) J Biol Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 15.Smith JS, Roth MJ. Purification and characterization of an active human immunodeficiency virus type 1 RNase H domain. J Virol. 1993;67:4037–4049. doi: 10.1128/jvi.67.7.4037-4049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JS, Gritsman K, Roth MJ. Contributions of DNA polymerase subdomains to the RNase H activity of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1994;68:5721–5729. doi: 10.1128/jvi.68.9.5721-5729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith CM, Potts WB, 3rd, Smith JS, Roth MJ. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology. 1997;229:437–446. doi: 10.1006/viro.1997.8454. [DOI] [PubMed] [Google Scholar]
- 18.Smith CM, Leon O, Smith JS, Roth MJ. Sequence requirements for removal of tRNA by an isolated human immunodeficiency virus type 1 RNase H domain. J Virol. 1998;72:6805–6812. doi: 10.1128/jvi.72.8.6805-6812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr., Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao HQ, Boyer PL, Arnold E, Hughes SH. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 21.Powell MD, Beard WA, Bebenek K, Howard KJ, Le Grice SF, Darden TA, Kunkel TA, Wilson SH, Levin JG. Residues in the alphaH and alphaI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J Biol Chem. 1999;274:19885–19893. doi: 10.1074/jbc.274.28.19885. [DOI] [PubMed] [Google Scholar]
- 22.Mandal D, Dash C, Le Grice SF, Prasad VR. Analysis of HIV-1 replication block due to substitutions at F61 residue of reverse transcriptase reveals additional defects involving the RNase H function. Nucleic Acids Res. 2006;34:2853–2863. doi: 10.1093/nar/gkl360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Hendrickson WA, Crouch RJ, Satow Y. Structure of ribonuclease H phased at 2 Å resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 24.Davies JF, 2nd, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 25.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr., Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayanagi K, Okumura M, Morikawa K. Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 Å resolution: proof for a single Mg(2+)-binding site. Proteins. 1993;17:337–346. doi: 10.1002/prot.340170402. [DOI] [PubMed] [Google Scholar]
- 27.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 28.Das D, Georgiadis MM. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure (Camb) 2004;12:819–829. doi: 10.1016/j.str.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 29.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Kanaya S, Katsuda-Nakai C, Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J Biol Chem. 1991;266:11621–11627. [PubMed] [Google Scholar]
- 32.Telesnitsky A, Blain SW, Goff SP. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli RNase H. J Virol. 1992;66:615–622. doi: 10.1128/jvi.66.2.615-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim D, Orlova M, Goff SP. Mutations of the RNase H C helix of the Moloney murine leukemia virus reverse transcriptase reveal defects in polypurine tract recognition. J Virol. 2002;76:8360–8373. doi: 10.1128/JVI.76.16.8360-8373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim D, Gregorio GG, Bingman C, Martinez-Hackert E, Hendrickson WA, Goff SP. Crystal structure of the moloney murine leukemia virus RNase H domain. J Virol. 2006;80:8379–8389. doi: 10.1128/JVI.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J Virol. 2003;77:8548–8554. doi: 10.1128/JVI.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puglia J, Wang T, Smith-Snyder C, Cote M, Scher M, Pelletier JN, John S, Jonsson CB, Roth MJ. Revealing domain structure through linker-scanning analysis of the murine leukemia virus (MuLV) RNase H and MuLV and human immunodeficiency virus type 1 integrase proteins. J Virol. 2006;80:9497–9510. doi: 10.1128/JVI.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arion D, Sluis-Cremer N, Min KL, Abram ME, Fletcher RS, Parniak MA. Mutational analysis of Tyr-501 of HIV-1 reverse transcriptase. Effects on ribonuclease H activity and inhibition of this activity by N-acylhydrazones. J Biol Chem. 2002;277:1370–1374. doi: 10.1074/jbc.M110254200. [DOI] [PubMed] [Google Scholar]
- 38.Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci U S A. 2002;99:9515–9520. doi: 10.1073/pnas.142123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SF. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 40.McWilliams MJ, Julias JG, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Combining mutations in HIV-1 reverse transcriptase with mutations in the HIV-1 polypurine tract affects RNase H cleavages involved in PPT utilization. Virology. 2006;348:378–388. doi: 10.1016/j.virol.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 41.Zhang WH, Svarovskaia ES, Barr R, Pathak VK. Y586F mutation in murine leukemia virus reverse transcriptase decreases fidelity of DNA synthesis in regions associated with adenine-thymine tracts. Proc Natl Acad Sci U S A. 2002;99:10090–10095. doi: 10.1073/pnas.152186199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mbisa JL, Nikolenko GN, Pathak VK. Mutations in the RNase H primer grip domain of murine leukemia virus reverse transcriptase decrease efficiency and accuracy of plus-strand DNA transfer. J Virol. 2005;79:419–427. doi: 10.1128/JVI.79.1.419-427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krug MS, Berger SL. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc Natl Acad Sci U S A. 1989;86:3539–3543. doi: 10.1073/pnas.86.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeStefano JJ, Buiser RG, Mallaber LM, Bambara RA, Fay PJ. Human immunodeficiency virus reverse transcriptase displays a partially processive 3' to 5' endonuclease activity. J Biol Chem. 1991;266:24295–24301. [PubMed] [Google Scholar]
- 45.Ben-Artzi H, Zeelon E, Le-Grice SF, Gorecki M, Panet A. Characterization of the double stranded RNA dependent RNase activity associated with recombinant reverse transcriptases. Nucleic Acids Res. 1992;20:5115–5118. doi: 10.1093/nar/20.19.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blain SW, Goff SP. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 47.Hostomsky Z, Hughes SH, Goff SP, Le Grice SF. Redesignation of the RNase D activity associated with retroviral reverse transcriptase as RNase H. J Virol. 1994;68:1970–1971. doi: 10.1128/jvi.68.3.1970-1971.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz SJ, Zhang M, Kelleher CD, Champoux JJ. Analysis of plus-strand primer selection, removal, and reutilization by retroviral reverse transcriptases. J Biol Chem. 2000;275:32299–32309. doi: 10.1074/jbc.M000021200. [DOI] [PubMed] [Google Scholar]
- 49.Furfine ES, Reardon JE. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA(Lys3)-primer excision. Biochemistry. 1991;30:7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- 50.Pullen KA, Ishimoto LK, Champoux JJ. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rattray AJ, Champoux JJ. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J Mol Biol. 1989;208:445–456. doi: 10.1016/0022-2836(89)90508-1. [DOI] [PubMed] [Google Scholar]
- 52.Huber HE, Richardson CC. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1990;265:10565–10573. [PubMed] [Google Scholar]
- 53.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 55.Schultz SJ, Zhang M, Champoux JJ. Recognition of internal cleavage sites by retroviral RNases H. J Mol Biol. 2004;344:635–652. doi: 10.1016/j.jmb.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 56.Wohrl BM, Georgiadis MM, Telesnitsky A, Hendrickson WA, Le Grice SF. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 57.Wohrl BM, Tantillo C, Arnold E, Le Grice SF. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:5343–5356. doi: 10.1021/bi00016a005. [DOI] [PubMed] [Google Scholar]
- 58.Winshell J, Champoux JJ. Structural alterations in the DNA ahead of the primer terminus during displacement synthesis by reverse transcriptases. J Mol Biol. 2001;306:931–943. doi: 10.1006/jmbi.2001.4439. [DOI] [PubMed] [Google Scholar]
- 59.Furfine ES, Reardon JE. Reverse transcriptase.RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 60.Gopalakrishnan V, Peliska JA, Benkovic SJ. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 62.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Quantitative analysis of RNA cleavage during RNA-directed DNA synthesis by human immunodeficiency and avian myeloblastosis virus reverse transcriptases. Nucleic Acids Res. 1994;22:3793–3800. doi: 10.1093/nar/22.18.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suo Z, Johnson KA. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12468–12476. doi: 10.1021/bi971218+. [DOI] [PubMed] [Google Scholar]
- 64.Suo Z, Johnson KA. Effect of RNA secondary structure on the kinetics of DNA synthesis catalyzed by HIV-1 reverse transcriptase. Biochemistry. 1997;36:12459–12467. doi: 10.1021/bi971217h. [DOI] [PubMed] [Google Scholar]
- 65.Schatz O, Mous J, Le Grice SF. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'→ 5' exonuclease activity. EMBO J. 1990;9:1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wohrl BM, Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- 67.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 1993;21:4330–4338. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gotte M, Fackler S, Hermann T, Perola E, Cellai L, Gross HJ, Le Grice SF, Heumann H. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 1995;14:833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palaniappan C, Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 70.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci U S A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao G, Goff SP. Replication defect of moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schultz SJ, Zhang M, Champoux JJ. Sequence, distance, and accessibility are determinants of 5'-end-directed cleavages by retroviral RNases H. J Biol Chem. 2006;281:1943–1955. doi: 10.1074/jbc.M510504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu TB, Taylor J. When retroviral reverse transcriptases reach the end of their RNA templates. J Virol. 1992;66:4271–4278. doi: 10.1128/jvi.66.7.4271-4278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Artzi H, Zeelon E, Amit B, Wortzel A, Gorecki M, Panet A. RNase H activity of reverse transcriptases on substrates derived from the 5' end of retroviral genome. J Biol Chem. 1993;268:16465–16471. [PubMed] [Google Scholar]
- 75.DeStefano JJ. The orientation of binding of human immunodeficiency virus reverse transcriptase on nucleic acid hybrids. Nucleic Acids Res. 1995;23:3901–3908. doi: 10.1093/nar/23.19.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc Natl Acad Sci U S A. 2000;97:11978–11983. doi: 10.1073/pnas.210392297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. The sequential mechanism of HIV reverse transcriptase RNase H. J Biol Chem. 2000;275:37664–37671. doi: 10.1074/jbc.M007381200. [DOI] [PubMed] [Google Scholar]
- 78.Wisniewski M, Chen Y, Balakrishnan M, Palaniappan C, Roques BP, Fay PJ, Bambara RA. Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J Biol Chem. 2002;277:28400–28410. doi: 10.1074/jbc.M201645200. [DOI] [PubMed] [Google Scholar]
- 79.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 80.Telesnitsky A, Goff SP. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 81.Telesnitsky A, Goff SP. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelleher CD, Champoux JJ. RNA degradation and primer selection by Moloney murine leukemia virus reverse transcriptase contribute to the accuracy of plus strand initiation. J Biol Chem. 2000;275:13061–13070. doi: 10.1074/jbc.275.17.13061. [DOI] [PubMed] [Google Scholar]
- 83.Driscoll MD, Golinelli MP, Hughes SH. In vitro analysis of human immunodeficiency virus type 1 minus-strand strong-stop DNA synthesis and genomic RNA processing. J Virol. 2001;75:672–686. doi: 10.1128/JVI.75.2.672-686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao HQ, Sarafianos SG, Arnold E, Hughes SH. RNase H cleavage of the 5' end of the human immunodeficiency virus type 1 genome. J Virol. 2001;75:11874–11880. doi: 10.1128/JVI.75.23.11874-11880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelleher CD, Champoux JJ. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- 86.Finston WI, Champoux JJ. RNA-primed initiation of Moloney murine leukemia virus plus strands by reverse transcriptase in vitro. J Virol. 1984;51:26–33. doi: 10.1128/jvi.51.1.26-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo GX, Sharmeen L, Taylor J. Specificities involved in the initiation of retroviral plus-strand DNA. J Virol. 1990;64:592–597. doi: 10.1128/jvi.64.2.592-597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wohrl BM, Moelling K. Coupling of reverse transcriptase and RNase H during HIV-1 replication. Behring Inst Mitt. 1991:100–107. [PubMed] [Google Scholar]
- 89.Pullen KA, Rattray AJ, Champoux JJ. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1993;268:6221–6227. [PubMed] [Google Scholar]
- 90.Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase. J Biol Chem. 1995;270:28169–28176. doi: 10.1074/jbc.270.47.28169. [DOI] [PubMed] [Google Scholar]
- 91.Powell MD, Levin JG. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Klarmann GJ, Yu H, Chen X, Dougherty JP, Preston BD. Discontinuous plus-strand DNA synthesis in human immunodeficiency virus type 1-infected cells and in a partially reconstituted cell-free system. J Virol. 1997;71:9259–9269. doi: 10.1128/jvi.71.12.9259-9269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schultz SJ, Zhang M, Kelleher CD, Champoux JJ. Polypurine tract primer generation and utilization by Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1999;274:34547–34555. doi: 10.1074/jbc.274.49.34547. [DOI] [PubMed] [Google Scholar]
- 94.Rattray AJ, Champoux JJ. The role of Moloney murine leukemia virus RNase H activity in the formation of plus-strand primers. J Virol. 1987;61:2843–2851. doi: 10.1128/jvi.61.9.2843-2851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robson ND, Telesnitsky A. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J Virol. 2000;74:10293–10303. doi: 10.1128/jvi.74.22.10293-10303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J Virol. 2004;78:13315–13324. doi: 10.1128/JVI.78.23.13315-13324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones FD, Hughes SH. In vitro analysis of the effects of mutations in the G-tract of the human immunodeficiency virus type 1 polypurine tract on RNase H cleavage specificity. Virology. 2007;360:341–349. doi: 10.1016/j.virol.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Rausch JW, Le Grice SF. Purine analog substitution of the HIV-1 polypurine tract primer defines regions controlling initiation of plus-strand DNA synthesis. Nucleic Acids Res. 2007;35:256–268. doi: 10.1093/nar/gkl909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schultz SJ, Zhang M, Champoux JJ. Specific cleavages by RNase H facilitate initiation of plus-strand RNA synthesis by Moloney murine leukemia virus. J Virol. 2003;77:5275–5285. doi: 10.1128/JVI.77.9.5275-5285.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schultz SJ, Whiting SH, Champoux JJ. Cleavage specificities of Moloney murine leukemia virus RNase H implicated in the second strand transfer during reverse transcription. J Biol Chem. 1995;270:24135–24145. doi: 10.1074/jbc.270.41.24135. [DOI] [PubMed] [Google Scholar]
- 101.Randolph CA, Champoux JJ. The use of DNA and RNA oligonucleotides in hybrid structures with longer polynucleotide chains to probe the structural requirements for moloney murine leukemia virus plus strand priming. J Biol Chem. 1994;269:19207–19215. [PubMed] [Google Scholar]
- 102.Whiting SH, Champoux JJ. Strand displacement synthesis capability of Moloney murine leukemia virus reverse transcriptase. J Virol. 1994;68:4747–4758. doi: 10.1128/jvi.68.8.4747-4758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]