Abstract
Chromatin insulators are DNA–protein complexes with broad functions in nuclear biology. Drosophila has at least five different types of insulators; recent results suggest that these different insulators share some components that may allow them to function through common mechanisms. Data from genome-wide localization studies of insulator proteins indicate a possible functional specialization, with different insulators playing distinct roles in nuclear biology. Cells have developed mechanisms to control insulator activity by recruiting specialized proteins or by covalent modification of core components. Current results suggest that insulators set up cell-specific blueprints of nuclear organization that may contribute to the establishment of different patterns of gene expression during cell differentiation and development.
Keywords: Drosophila, insulator, transcription, nucleus
CHROMATIN INSULATORS AND NUCLEAR FUNCTION
Patterns of transcription required for cell differentiation are initially established by specific transcription factors. The maintenance of these patterns of gene expression is then carried out by alterations in chromatin structure that are epigenetically inherited between cell generations. These changes in chromatin organization take place at the level of the 10 nm fibre and include covalent histone modifications, DNA methylation and alterations induced by ATP-dependent remodelling complexes. In addition, more recent evidence suggests that the three-dimensional organization of the genome within the nucleus of eukaryotic cells may also be critical for achieving proper spatio-temporal patterns of gene expression during development. The factors and processes involved in the establishment, maintenance and regulation of specific states of nuclear organization are largely unknown but insulators are emerging as likely candidates to play this crucial role [1].
Insulators are DNA–protein complexes that are experimentally defined by their ability to block enhancer–promoter interactions and/or serve as barriers against the spreading of the silencing effects of heterochromatin. Drosophila has been a particularly good model system in which to analyze insulator function; several different insulators have been identified in this organism whereas vertebrates appear to mostly rely on one type of insulator [2]. The apparent paradox created by the lack of a correlation between insulator complexity and genome size is an important issue for future study. At the same time, the complexity of Drosophila insulators may offer the opportunity of easily dissecting different aspects of insulator function.
Here we summarize the current status of the insulator field in Drosophila with special emphasis on recent genome-wide localization studies. We conclude by proposing that the primary role of insulators may not be to regulate enhancer–promoter interactions or heterochromatin spreading. Rather, insulators may mediate intra- and inter-chromosomal interactions with the primary goal of organizing the eukaryotic genome into epigenetically-inheritable states. This insulator-mediated organization may be important to regulate DNA function at multiple levels, including transcription initiation, elongation and DNA recombination. These different functions, which in vertebrates may be carried out by the CTCF and/or SINE B2 insulators [3], may have been allocated to different insulator subclasses in Drosophila.
DROSOPHILA INSULATORS: SEQUENCES AND PROTEINS
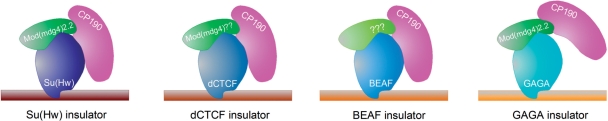
There are at least five types of insulators in Drosophila that have been studied in detail. They include the scs and scs′ sequences flanking the heat shock hsp70 locus at chromosome subdivision 87A [4, 5], the gypsy insulator first found in the gypsy retrotransposon [6], the Fab 8 insulator located in the bithorax complex and the SF1 insulator described in the Antennapedia complex [7]. Each of these insulators consists of a DNA sequence and a specific DNA-binding protein that interacts with this sequence (Figure 1).
Figure 1:
Diagram showing the different subclasses of Drosophila insulators for which components other than the DNA-binding protein are known. Each subclass contains a different DNA binding protein that may define the specific function of the corresponding subclass. All insulators share the common protein CP190, although the role of this protein in the function of the GAGA insulator has not been demonstrated experimentally. In addition, all subclasses may also have one Mod(mdg4) isoform. The gypsy/Su(Hw) insulator contains Mod(mdg4)2.2. The dCTCF insulator lacks this isoform but contains a different variant of Mod(mdg4) (T. Gerasimova, E. Lei and V. Corces, unpublished observations). It is not known whether the BEAF insulator contains a Mod(mdg4) variant but GAGA has been shown to interact with Mod(mdg4)2.2.
In the case of the scs insulator, the DNA-binding protein component is Zeste-White 5 (ZW5), which is a Zn finger protein required for cell viability. Null mutations in the zw5 gene are recessive lethal, but hypomorphic alleles display a variety of pleiotropic effects on wing, bristle and eye development [8]. The nature of other proteins that interact with ZW5 to elicit insulator function is unknown at this time. The scs′ sequences interact with a protein called Boundary Element Associated Factor 32 (BEAF 32). The BEAF-32 gene encodes two different proteins named BEAF-32A and BEAF-32B, which are present at hundreds of sites on Drosophila polytene chromosomes [5]. The two isoforms differ at the N terminal DNA-binding domain (BED Finger domain). The common C-terminal region is involved in protein–protein interactions between the two isoforms. Analysis of mutations in the BEAF 32 gene shows that BEAF 32B is required for viability whereas BEAF 32A mutations do not show significant phenotypic defects. Expression of a dominant negative form of BEAF 32 results in changes in chromosome structure and cell viability [9–11].
The gypsy insulator contains binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein, which is a 12 zinc finger DNA-binding protein. Mutations in the Su(Hw) gene cause female sterility but do not result in lethality [12]. Su(Hw) interacts with two other components of the gypsy insulator, Mod(mdg4)2.2 and CP190 [13, 14]. Mod(mdg4)2.2 does not bind to DNA directly but interacts with Su (Hw) through its carboxy-terminal domain. In addition, Mod(mdg4)2.2 also contains a BTB domain in the N-terminal region that mediates homo- and hetero-multimerization with other insulator components. The mod(mdg4) gene encodes ∼29 different isoforms that arise by alternative cis- and trans-splicing [15, 16]; null mutations in the gene result in lethality, but mutations affecting the Mod(mdg4)2.2 isoform are viable and show defects in gypsy insulator function [17]. CP190 also contains a BTB domain as well as three zinc fingers and it interacts with both Su(Hw) and Mod(mdg4)2.2. CP190 binds DNA with low affinity and specificity but it does not interact directly with insulator sequences present in the gypsy retrotransposon, where it is recruited through interactions with Su(Hw) and Mod(mdg4)2.2 instead. Mutations in the CP190 gene are lethal [18].
The bithorax complex of Drosophila contains an intricate collection of transcriptional regulatory sequences that orchestrate the complex spatio-temporal expression of the three genes present in the complex. The proper interplay between these regulatory sequences requires the function of several insulators of which Fab 8 has been studied in most detail. Fab 8 sequences interact with the Drosophila homolog of the vertebrate CTCF insulator protein. Drosophila CTCF (dCTCF) has 12 zing fingers. Mutations in dCTCF are lethal and show abdominal hometic phenotypes [19, 20]. dCTCF is also found in the Mcp and Fab 6 insulators present in the bithorax complex but not Fab 7 [21]. Fab 7 may represent a fourth class of insulators that use the GAGA factor (GAF) as a DNA-binding protein that also contains a BTB domain. Mutations in the trl gene, which encodes GAF, affect Fab 7 insulator activity [22]. In addition, GAF is present and required for the function of the SF1 insulator found in the Antennapedia complex [7].
SHARED PROTEIN COMPONENTS AMONG DROSOPHILA INSULATORS
Vertebrates have at least two types of insulators characterized thus far, the CTCF insulator and the SINE B2 element [2] and therefore it seems at first puzzling that Drosophila, with a less complex genome, has at least five different types of insulators. One possibility is that vertebrates have additional insulators that have not yet been discovered. Alternatively, the vertebrate CTCF and SINE B2 insulators may have captured the functions of the five present in Drosophila. If this is the case, and the functions of all Drosophila insulators have converged into one or two, one would expect to find some shared components among Drosophila insulators. This is indeed the case. The CP190 protein, first found in the gypsy/Su(Hw) insulator, also interacts with dCTCF [19, 20]. Genome-wide mapping of dCTCF and CP190 sites also support this conclusion [23, 24]. These studies have also shown that BEAF and CP190 co-localize at hundreds of sites throughout the genome [24]. These results suggest that the insulators defined by these three different DNA-binding proteins, Su(Hw), dCTCF and BEAF share the BTB domain-containing protein CP190 and may therefore use similar mechanisms to effect their insulator function. On the other hand, GAF does not appear to interact directly with CP190 but has been shown to interact with Su(Hw) and Mod(mdg4)2.2 [25]; since these two proteins can in turn interact with CP190, GAF insulators may act mechanistically like the other three types (Figure 1).
INSULATORS MEDIATE INTRA- AND INTER-CHROMOSOMAL INTERACTIONS
All Drosophila insulators, with perhaps the exception of scs/ZW5 for which data are unavailable, share the BTB domain proteins CP190 and likely one isoform of Mod(mdg4). The BTB domains of these proteins as well as GAF can interact in various in vitro or in vivo assays, suggesting that insulator proteins may mediate intra- and inter-chromosomal interactions among insulator sites throughout the genome. Various types of observations support this conclusion. For example, Su(Hw), Mod(mdg4)2.2, dCTCF and CP190 show a punctuate distribution in the nuclei of diploid cells. These sites, called insulator bodies, appear to contain multiple individual insulator sequences and their morphology is disrupted by mutations in insulator components [18, 20, 26]. Furthermore, FISH experiments have shown that DNA sequences contained between two insulators form a loop, which becomes two smaller loops when a new insulator is inserted in the middle of the DNA [27]. Although insulator bodies are present throughout the nucleus, they seem to localize preferentially in the nuclear periphery. This localization may be mediated by the protein dTopors, which has been shown to interact with Su(Hw) and Mod(mdg4)2.2 as well as with Lamin Dm0 [28, 29]. Therefore, dTopors may serve as an anchor to attach insulator sites to the nuclear lamina or nuclear matrix [28]. In addition, results from 3C experiments have shown that scs and scs′ sequences are present in close proximity in the nucleus forming a loop of the intervening sequence. These contacts may be mediated by CP190 or may be direct, since ZW5 and BEAF-32 have been shown to interact both in vitro and in vivo in Drosophila embryos [30].
MECHANISMS OF REGULATION OF INSULATOR FUNCTION
If insulators mediate inter- or intra-chromosomal interactions that result in the formation of chromatin loops, which in turn are attached to the nuclear matrix, it is possible that the resulting structures determine a particular pattern of nuclear organization that may be important for gene expression. For example, it is possible that as cells differentiate, insulator-mediated changes in nuclear organization precede or accompany cell differentiation and may be crucial in the establishment and/or maintenance of specific patterns of gene expression. If this is the case, cells must possess mechanisms to regulate insulator activity in order to establish distinct nuclear architectures that are cell fate-specific.
Evidence for the existence of mechanisms to control insulator function comes in part from genome-wide studies of insulator protein localization in Drosophila cell lines of different tissue origin. Studies in Kc cells, which have a neural origin, indicate that there are 3747 Su(Hw), 2266 dCTCF, 2995 BEAF and 5272 CP190 sites where these proteins are present in the genome. Of these, 47% of Su(Hw), 62% of dCTCF and 71% of BEAF sites colocalize with CP190 sites [24]. Since CP190 is required for insulator function, this observation suggests that cells may control the activity of these various insulators by regulating the recruitment of CP190. In addition, comparison of the genomic location of different insulator proteins in Kc and Mbn2 cells (a hematopoietic cell line), revealed that while many sites are constant, a fraction of the localization sites for each of the four insulator proteins is different between the two lines. For example, 18% of Su(Hw) sites in Kc cells and 5% of Su(Hw) sites in Mbn2 cells are cell type-specific. This is also the case for dCTCF, for which 18% of sites in Kc cells and 37% in Mbn2 cells are cell-unique, whereas the number of cell type-specific BEAF sites is 11% in Kc cells and 11% in Mbn2 cells. In the case of CP190, which is found at all three insulator subclasses, 17% of sites present in Kc cells and 14% in Mbn2 cells were found to be cell type specific [24]. These results suggest that cells may regulate insulator activity by controlling the recruitment of the DNA-binding proteins to their target sites in the genome in addition to controlling the recruitment of CP190.
Several proteins have been characterized in Drosophila that may play a role in regulating insulator function. dTopors, in addition to serving as an attachment point for insulators to the nuclear lamina, has E3 ubiquitin ligase activity. This activity is required for proper insulator function. Its substrate has not been clearly identified but Su(Hw) is a likely candidate, since over-expression of dTopors enzymatic activity reverses the effect of mod(mdg4) mutations on the ability of Su(Hw) to interact with chromatin [28]. In addition, modification of Mod(mdg4)2.2 and CP190 by sumoylation inhibits insulator function. Disruption of the SUMO conjugation pathway improves the enhancer-blocking function of a partially active insulator, indicating that SUMO modification acts to negatively regulate the activity of the gypsy insulator. Sumoylation does not affect the ability of CP190 or Mod(mdg4)2.2 to bind chromatin [29]. Interestingly, dTopors inhibits sumoylation of Mod(mdg4)2.2 and CP190. Therefore, this protein may have a dual effect on insulator function by ubiquitinating some insulator components and inhibiting the sumoylation of others.
A second candidate protein with a possible role in regulating insulator function is the Rm62 RNA helicase. Insulator activity decreases in the presence of mutations in components of the RNAi machinery; insulator function is restored by mutations in Rm62. These observations have led to a model suggesting that insulator bodies contain RNA whose synthesis requires RNAi proteins. Rm62 may interact with this RNA to decrease insulator function [31].
dTopors and Rm62 have been only shown to affect the function of the gypsy/Su(Hw) insulator but their potential role in regulating the activity of other insulator subclasses has not been tested. There are currently no characterized mechanisms that control the activity of the dCTCF insulator. O-glycosylation of BEAF can be detected in Drosophila embryonic cells in a domain of the protein that is required for association with the nuclear matrix; however, it is not clear whether glycosylation is required for scs′ insulator function [32]. A second possible candidate to regulate the BEAF insulator is the DREF protein. DREF has been characterized as a transcription factor that shares binding sites with BEAF-32. It is possible that DREF regulates BEAF binding through competition for the same DNA sequences [33].
DIFFERENT DROSOPHILA INSULATOR SUBCLASSES MAY HAVE SPECIALIZED FUNCTIONS
The existence of three and perhaps four insulator subclasses with different DNA-binding proteins but sharing other functional components raises the question of whether they all have the same role in the regulation of gene expression or whether there is a functional specialization in their tasks. The possibility of such specialization is highlighted by results showing differential localization of insulator subclasses with respect to genomic landmarks. For example, Su(Hw) and dCTCF are preferentially excluded from exonic regions (mostly 5′ and 3′ UTRs), with only 8, 16 and 17% of sites found within exons, respectively, whereas BEAF sites are enriched in UTRs [24]. When the location of these proteins is compared with the location of genes, few Su(Hw)-binding sites are found in the 1 kb regions flanking genes. However, dCTCF and BEAF show a distribution that is highly skewed toward the 5′ end of genes and is enriched in the first 200 bp just upstream of the transcription start site. Insulator proteins also show a compartmentalized distribution in relation to the level of gene expression. For example, 83% of dCTCF sites and 89% of BEAF sites at the 5′ end of genes localize to genes that are highly expressed. However, Su(Hw)-binding sites are most often found near genes with low expression levels. Finally, different insulator proteins appear to associate with genes involved in different cellular processes. Genes containing dCTCF in the 200-bp upstream of their transcription start site are mostly involved in developmental processes, whereas genes containing BEAF in this region are mostly involved in metabolic processes. Both dCTCF- and BEAF-containing genes also show an enrichment for cell cycle genes, whereas Su(Hw)-containing genes show little significant clustering based on biological process [24, 34, 35]. These observations suggest a division of labour among Drosophila insulators, both with respect to gene function as well as specific aspects of cell function. Given the fact that all insulators share CP190 and perhaps Mod(mdg4), it is likely that all of them use the same mechanism to perform their function, namely to bring together different regions of the genome. Nevertheless, Drosophila cells appear to use a variety of DNA-binding proteins to recruit insulator components to mediate these interactions. Therefore, the specific outcome of these interactions may be determined by where in the genome the binding sites for each of these proteins are localized. In the case of Su(Hw) and a subset of dCTCF sites, their localization in intergenic regions suggests that their role may be to form loops that may represent independent functional domains. The rest of dCTCF sites and BEAF sites are located around promoter regions and their function may rely on the same type of interactions to bring these regions of genes to specific nuclear compartments such as transcription factories.
PERSPECTIVE
Chromatin insulators are important regulatory sequences present in the genome of most (probably all) eukaryotes. Although they are defined experimentally based on their ability to affect enhancer–promoter interactions and interfere with the spreading of repressive signals from heterochromatin, their role appears to be more general. Intra- and inter-chromosomal interactions mediated by insulator proteins may establish a web of contacts between individual insulator sites that give rise to specific patterns of nuclear organization. Insulator-mediated nuclear structures may be regulatable by controlling the interaction between insulator DNA-binding proteins and their cognate sequences. In addition, recruitment of insulator components involved in mediating inter-insulator interactions may represent a second level of controlling the function of these elements. These two levels of regulation may be the result of specific covalent modifications of insulator proteins. The specific outcome of inter-insulator interactions is a consequence of the location of the particular insulator sequences with respect to specific genome features, and interference with enhancer–promoter interactions may be just one of these outcomes. Understanding the nature of these different insulator roles as well as the more general function in nuclear organization remain the main issues in the field for future investigation.
Key Points.
Chromatin insulators are characterized experimentally by their effect on enhancer–promoter interactions and their ability to buffer the effects of heterochromatin. Nevertheless, their primary role in the cell may be broader, and they may be involved in mediating intra- and inter-chromosomal interactions to create specific patterns of nuclear organization.
Drosophila has at least five different types of insulators whereas vertebrate cells appear to have only two. These different insulators vary in the DNA-binding protein component but they share other proteins involved in mediating inter-insulator contacts.
Different insulator subclasses are located in different regions of the genome with respect to gene features and therefore they may have distinct roles in gene regulation.
Analyses of genome-wide localization patterns of insulator proteins suggest that genomic site occupancy varies from one cell type to another. Cells appear to regulate insulator activity by either controlling the recruitment of the insulator DNA-binding proteins or controlling the other insulator components involved in inter-insulator contacts.
Cells appear to employ different auxiliary regulatory proteins or different covalent modifications of the core structural proteins to regulate insulator activity.
FUNDING
National Institutes of Health [grant number GM35463].
Acknowledgements
The authors would like to thank La Madre Maravillas for help with difficult experiments and the Corces lab for inspirational discussions.
Biographies
B.V. Gurudatta is a postdoctoral fellow in the Biology Department of Emory University. His current research interest is the analysis of the BEAF insulator and its role in gene expression.
Victor G. Corces is an HHMI Professor in the Biology Department of Emory University. His research interests include the role of chromatin structure and nuclear organization in the control of cell fate and gene expression.
References
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–31. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–89. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- Spana C, Harrison DA, Corces VG. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988;2:1414–23. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. Embo J. 2003;22:3113–21. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Tan YY, Hart CM. A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol Genet Genom. 2007;277:273–86. doi: 10.1007/s00438-006-0187-8. [DOI] [PubMed] [Google Scholar]
- Gilbert MK, Tan YY, Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–75. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Gilbert MK, Hart CM. Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics. 2007;176:801–13. doi: 10.1534/genetics.106.068056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, Gdula DA, Coyne RS, Corces VG. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993;7:1966–78. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. Embo J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–17. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K, Roth P, Schotta G, et al. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–57. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG. Protein determinants of insertional specificity for the Drosophila gypsy retrovirus. Genetics. 2001;158:1101–10. doi: 10.1093/genetics/158.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, et al. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–97. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. Embo J. 2007;26:4203–14. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–72. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, et al. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, et al. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–84. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, et al. Active promoters and insulators are marked by the centrosomal protein 190. Embo J. 2009;28:877–88. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–50. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Juge F, Gruzdeva N, et al. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci USA. 2004;101:14806–11. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–35. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–74. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–16. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Capelson M, Corces VG. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. Embo J. 2006;25:1906–14. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Pathak RU, Rangaraj N, Kallappagoudar S, et al. Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol Cell Biol. 2007;27:4796–806. doi: 10.1128/MCB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–83. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of BEAF binding sites in Drosophila links BEAF to transcription. Mol Cell Biol. 2009;29:3556–68. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Schuettengruber B, et al. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]