Abstract
Serpins form an enormous superfamily of 40–60-kDa proteins found in almost all types of organisms, including humans. Most are one-use suicide substrate serine and cysteine proteinase inhibitors that have evolved to finely regulate complex proteolytic pathways, such as blood coagulation, fibrinolysis, and inflammation. Despite distinct functions for each serpin, there is much redundancy in the primary specificity-determining residues. However, many serpins exploit additional exosites to generate the exquisite specificity that makes a given serpin effective only when certain other criteria, such as the presence of specific cofactors, are met. With a focus on human serpins, this minireview examines use of exosites by nine serpins in the initial complex-forming phase to modulate primary specificity in either binary serpin-proteinase complexes or ternary complexes that additionally employ a protein or other cofactor. A frequent theme is down-regulation of inhibitory activity unless the exosite(s) are engaged. In addition, the use of exosites by maspin and plasminogen activator inhibitor-1 to indirectly affect proteolytic processes is considered.
Serpins are ubiquitously found in all multicellular organisms and even in some viruses and bacteria (1, 2). They are 40–60-kDa proteins present both extra- and intracellularly that function mostly as serine and cysteine proteinase inhibitors (1). Well known examples are antithrombin, the principal inhibitor of blood coagulation proteinases; PAI-1,3 an inhibitor of the plasminogen activators tPA and uPA; and α1PI, the principal inhibitor of neutrophil elastase. A characteristic of the processes regulated by these serpins is that they involve mostly proteinase cascades that need to be regulated with respect to both the site where they occur and their duration of action.
Serpin Branched Pathway Mechanism
All serpin structures determined so far have the same basic fold, composed of three major β-sheets, eight to nine α-helices, and an exposed reactive center loop (RCL) that contains the primary recognition site for attacking proteinases (1). Based on secondary structure predictions and the presence of ∼51 conserved, mostly interior residues, all serpins probably adopt this fold (3). Extensive biochemical studies over the past 30 years have established that serpins inhibit proteinases by a branched pathway suicide substrate inhibition mechanism (1). RCL residues are recognized as a suitable substrate for an attacking proteinase, which binds to form an initial Michaelis-like complex (1). For serine and cysteine proteinases, substrate hydrolysis involves initial cleavage of the scissile bond and formation of an acyl ester (1). Most unusually, the serpin fold represents a metastable conformation. Consequently, upon cleavage of the scissile bond, the N-terminal portion of the cleaved RCL spontaneously and irreversibly inserts into β-sheet A as a middle strand through expansion of the sheet and, in so doing, drags the covalently linked proteinase to the bottom of the serpin (1), where compression results in distortion of the active site and kinetic trapping of the covalent intermediate (1, 4). The two competing branches of the pathway are thus (i) hydrolysis of the acyl intermediate to yield a cleaved serpin and free proteinase and (ii) kinetic trapping of the proteinase through translocation and concomitant distortion of the enzyme active site. Because such distortion requires only a covalent linkage between serpin and proteinase, the mechanism works for serine proteinases of both chymotrypsin and subtilisin folds and for cysteine proteinases of the caspase and cathepsin families. Because once started, loop insertion is essentially irreversible and rapid, the rate-determining step(s) for inhibition involve formation of the initial noncovalent Michaelis complex and/or the subsequent acylation step (1). This minireview focuses on this initial complex-forming phase.
Primary Determinants of Specificity
Given this serpin mechanism, it is not surprising that the primary specificity determinants are those at P1 and the immediately adjacent residues. The resulting interactions are thus very like those in noncovalent complexes of proteinases with Kunitz, Kazal, and Bowman-Birk inhibitors and involve backbone–backbone H-bonds between the RCL in extended β-conformation and the proteinase, and between the side chains of P1 and immediately adjacent residues with S1, S2, etc., pockets of the proteinase (1). In keeping with this, antithrombin, with Arg at P1, is an inhibitor of the Arg-specific proteinases IIa and fXa, and CrmA, with Asp at P1, is an inhibitor of caspases. Likewise, mutagenesis of the P1 Arg of antithrombin to Trp turns it into an effective chymotrypsin inhibitor (5).
Although there may be several potential cleavage sites within the RCL, only those that are 16–17 residues C-terminal from the exit point of the RCL from β-sheet A lead to successful proteinase inhibition. This arises directly from the physical restrictions imposed by the mechanism, which requires that the length of the loop that inserts into β-sheet A matches the length of the sheet so that the fully translocated proteinase is compressed against the bottom of the serpin. If the loop length is either too long or too short, the covalent intermediate is short-lived, presumably because active-site distortion is less severe and the proteinase less catalytically compromised (6, 7).
For the 27 inhibitory human serpins, an astonishing 13 have Arg at P1, and two more have Lys (1). A similar pattern is also seen with Drosophila serpins, with 8 of 17 inhibitory members having P1 Arg or Lys (8). Despite human serpins showing a preponderance of basic residues at P1, they appear to have paradoxically high specificity for their cognate proteinases in vivo. Thus, fXa, which has very low substrate specificity beyond P1 (9), is inhibited by antithrombin under physiological conditions 4–5 orders of magnitude faster than by another P1 Arg-containing serpin, PAI-1. Conversely, under physiological conditions, HCII (P1 Leu) inhibits only the Arg-specific proteinase IIa (10), whereas ZPI (P1 Tyr) is a specific inhibitor of membrane-bound fXa (1).
Exosites Allow Refinement of Specificity
The answer to how serpins show high specificity and a high rate of reaction in vivo while having the same P1 residue as many other serpins or else a seemingly inappropriate P1 residue is the use of one or more exosites on the serpin to refine the overall specificity and selectively enhance the reaction rate. This is facilitated by the length and flexibility of the exposed RCL such that a proteinase can simultaneously dock with the P1 residue within the RCL and an exosite elsewhere on the serpin. In addition, unstructured N- and C-terminal extensions on some serpins permit more remote serpin-proteinase interactions, whereas other serpin-proteinase pairs use a cofactor such as heparin or another protein to bring the two reactants together. Because each exosite can modify the overall binding affinity in the Michaelis complex and/or promote catalysis by locking the proteinase into a particular orientation with respect to the scissile bond, rate enhancements can be very large. Moreover, by tightening the Michaelis complex interaction, exosites can ensure that, once a serpin has bound to proteinase, its dissociation is slowed to an extent that commits it to undergoing acylation and conformational change steps that lead to proteinase trapping (11, 12). As a result, most serpin-proteinase encounters proceed to a stable inhibited complex, causing the overall reaction rate to be limited by the rate of diffusional encounter between serpin and proteinase. A frequent overall theme is that, in the absence of the factor that refines the specificity, the serpin-proteinase reaction is deliberately suppressed. Only when full exosite engagement is effected does the rate reach the desired value. In this way, providing the exosite, or access to it, regulates whether or not the serpin will act on its target proteinase. This in turn allows not only specificity but also regulation in time and space by coordinately regulating the provision of the exosite.
Exosite Interactions for Proteinase Inhibition
To date, nine inhibitory human serpins are known to employ one or more exosites. They do so in a variety of ways that involve either binary interaction with the target proteinase or ternary complex formation with another species.
Binary Complexes
Even without an N- or C-terminal extension, some serpins can enhance the rate of proteinase inhibition using an already present exosite on the surface of the protein core. The best example is the inhibition of tPA by PAI-1, in which loop 37 near the active site interacts with residues in the distal portion of the RCL, including Glu at P4′ and P5′ (13). These Glu residues enhance the rate of inhibition through a direct effect on Michaelis complex formation, with reductions of 13-fold in Michaelis complex formation and 5-fold in rate of inhibition upon replacement by Ala (13). The importance of this loop is further illustrated in two related systems. In one, a IIa chimera in which its own loop 37 was replaced by that of tPA was inhibited by PAI-1 1000-fold faster (14). In the second, the snake Trimeresurus stejnegeri has used the opposite strategy of elimination of the loop-exosite interaction through shortening of loop 37 of its own plasminogen activator to slow down inhibition by the prey's PAI-1 and so promote an anticoagulant state (15). Another example is the inhibition of human neutrophil elastase by α1PI. This reaction has a rate constant of 6 × 107 m−1 s−1 even though a peptide with the same P4-P4′ residues reacts 1000-fold slower (1). The involvement of an exosite was demonstrated by replacement of the whole of the RCL of α1-antichymotrypsin from P3 to P3′ with that of α1PI, which resulted in a rate constant for human neutrophil elastase inhibition of only 105 m−1 s−1, i.e. 600-fold lower than the same residues in the context of α1PI (1).
An example of a more complex use of exosites is kallistatin, a specific inhibitor of tissue kallikreins. Kallistatin has a P1 Phe, despite the kallikrein targets being Arg-specific proteinases. However, Phe at P1 confers selectivity for kallikreins over other Arg-specific proteinases without undue loss of reactivity (1). This is due to a basic exosite between helix H and strand 2 of β-sheet C specifically enhancing the binding of kallikrein but not other Arg-specific proteinases (16). Although this parallels the α1PI-human neutrophil elastase case described above, it is more sophisticated in that heparin can also bind to the exosite and block its availability to kallikrein, thereby acting as a negative regulator of kallistatin (17).
Regulation of the ability of plasmin to promote clot lysis should be most effective at the surface of the fibrin clot. α2-Antiplasmin achieves such site-specific inhibition of plasmin by using its N- and C-terminal extensions. The C-terminal extension specifically engages plasmin, using conserved Lys residues to engage the kringle domains of plasmin (18) and to achieve a 30–60-fold rate enhancement to ∼2 × 107 m−1 s−1. Consistent with this, the C-terminal extension lies close to the RCL such that the more distal conserved Lys residues can readily engage the kringle domains (19). To efficiently inhibit plasmin that is close to the fibrin surface, α2-antiplasmin then makes use of its N-terminal tail, which contains a transglutamination site at residue 14 that allows covalent cross-linking to the fibrin surface by factor XIIIa (10).
Ternary Complexes
The quintessential example of a cofactor providing exosites to promote a specific serpin-proteinase interaction is the heparin-dependent reaction of antithrombin with blood coagulation proteinases. Here, heparin not only makes pre-existing exosites on the serpin more accessible to proteinases by allosterically activating the serpin, but itself provides exosites to bind and bring together the serpin and proteinase in a ternary bridging complex (1, 20, 21).
The antithrombin exosites become competent to bind two target proteinases, fIXa and fXa, when a specific heparin pentasaccharide binds to a basic site centered on helix D and conformationally activates the serpin (1). In the low activity state of antithrombin, which is the predominant form in plasma, favorable exosite interactions are offset by unfavorable interactions with the surface surrounding the RCL, which make it repulsive toward an approaching proteinase (22, 23).4 This repulsion is exacerbated by the burial of the RCL hinge in sheet A, which constrains the RCL to lie close to the serpin body. Heparin activation expels the RCL hinge from sheet A and also alters the surface H-bonding network, thereby reducing the repulsive interactions and favoring proteinase interaction with both the RCL and exosite determinants on strand 3 of β-sheet C just below the RCL (21).4 In this heparin-activated state, the exosite residues on antithrombin are positioned to specifically interact with basic residues of the autolysis loop of fXa and fIXa when the proteinases are bound to the serpin RCL (25).
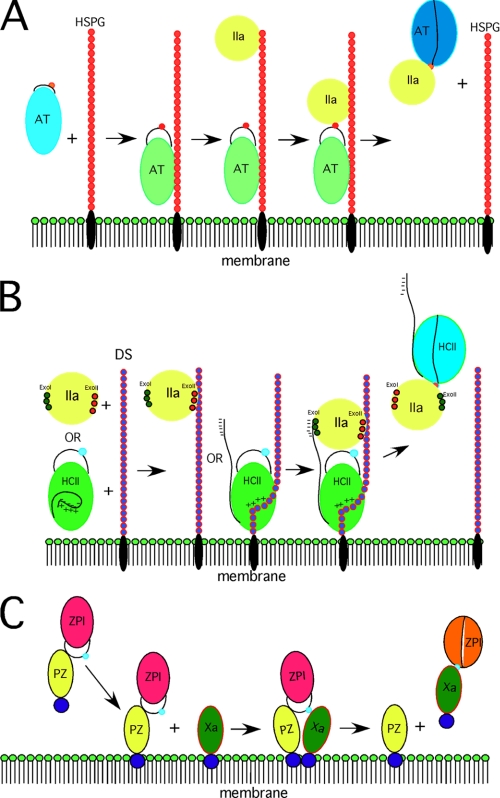
Heparin further promotes binding through bridging exosite interactions involving the basic site on antithrombin and basic exosites on the proteinase that are conserved in fXa, fIXa, and IIa (1). Structures of the heparin-antithrombin-S195A/anhydro-IIa Michaelis complex show that the engagement of the serpin RCL with the proteinase active site aligns the heparin pentasaccharide-binding site on antithrombin with the basic exosite on the proteinase to allow heparin to bridge and stabilize the serpin-proteinase Michaelis complex (Fig. 1) (26, 27). Serpin exosites and heparin-bridging exosites together ensure specific inhibition of the three target proteinases at physiological rates of 107–108 m−1 s−1, representing a 103–105-fold increase (28). It is important, however, that for the Michaelis complex to proceed to the proteinase-translocated covalent complex it be destabilized once the acyl intermediate has formed. Here, the remarkable conformational change represented by RCL insertion into β-sheet A is critical because it reduces the affinity of antithrombin for heparin 1000-fold to cause dissociation from the heparin chain (Fig. 1) (10).
FIGURE 1.
Examples of cofactor involvement. A, antithrombin (AT) inhibition of IIa resulting from bridging heparin as part of a heparan sulfate proteoglycan (HSPG). B, HCII inhibition of IIa at the membrane surface mediated by bridging dermatan sulfate (DS) and the N-terminal tail of HCII, which is displaced by dermatan sulfate binding. ExoI and ExoII, exosites I and II, respectively. C, specific protein Z-bound ZPI inhibition of membrane-associated fXa. Whereas the final complex between ZPI and fXa dissociates from protein Z (PZ), evidence is lacking on whether it also dissociates from the membrane surface (as shown).
The use of exosites rather than the RCL to determine specificity appears to have evolved for a purpose in the case of antithrombin. Thus, the P1 Arg bait is potentially capable of recognizing the anticoagulant proteinase, activated protein C, which shares a trypsin-like specificity for P1 Arg-containing substrates with the procoagulant proteinase targets of antithrombin. However, heparin-activated antithrombin inhibits activated protein C 107-fold slower than IIa, fXa, and fIXa. This poor reactivity results from the antithrombin RCL sequence antagonizing the binding of activated protein C (29), consistent with the idea that the antithrombin RCL sequence evolved to prevent reaction with this anticoagulant proteinase.
Protein C inhibitor (PCI) provides a contrasting example in which a P1 Arg, together with exosite determinants provided by a heparin or protein cofactor, is utilized to specifically inhibit two anticoagulant proteinases. Heparin promotes PCI inhibition of activated protein C by bridging the basic H helix of PCI and loop 70 of the proteinase (30). The endothelial cell receptor, thrombomodulin, transforms IIa into an anticoagulant proteinase and a target of PCI by acting as a bridging cofactor that binds to the basic H helix of PCI and loop 70 of IIa (exosite I) (30–32). In the absence of these cofactors, the positively charged cofactor-binding sites on the serpin and the proteinase antagonize their interaction because of their proximity to the binding interface. Interestingly, in the presence of heparin, PCI efficiently inhibits the procoagulant form of IIa that is not receptor-bound. Heparin again acts as a bridging cofactor to bind the basic helix H site in PCI, but instead of binding to exosite I of thrombin, it binds to exosite II, the same site used in heparin bridging of antithrombin and IIa. The ability of heparin to bridge through either exosite II in IIa or exosite I in activated protein C results from alternative modes of heparin binding to helix H (30). The two binding modes act to align either of the two proteinase exosites with the PCI helix H site with a common basic residue acting as a pivot for these alternative modes. Bridging heparin also enhances the reactivity of PN1 against both IIa and factor XIa, with ∼500-fold enhancement of the former (10) and 20-fold enhancement of the latter (33).
Two other serpins provide examples of an unfavorable P1 residue being used to prevent reaction with P1 Arg-specific proteinases, except for the desired target, through use of favorable exosite interactions. HCII is a specific IIa inhibitor despite having an unfavorable P1 Leu (10). Either of two GAG cofactors, heparin or dermatan sulfate, enables the serpin to specifically inhibit IIa at a physiological rate of ∼107 m−1 s−1 (10). As with antithrombin, binding of the GAG to the serpin presents new exosites through both allosteric activation and bridging mechanisms (Fig. 1). GAG binding to overlapping basic sites in helix D of HCII allosterically activates the serpin by releasing the N-terminal acidic tail from an intramolecular interaction, possibly with the basic GAG-binding site, to promote exosite-exosite interactions between the tail and exosite I of IIa (10, 34). The GAGs may further stabilize the Michaelis complex by bridging the serpin and proteinase through exosite II of IIa. The two types of exosite interactions together position the P1 Leu to interact with the proteinase S1 binding pocket and to compensate for the preference for a P1 Arg (Fig. 1) (34).
A second case of an unfavorable P1 residue is ZPI, which specifically inhibits fXa despite having a P1 Tyr (1). ZPI requires protein Z, phospholipid, and calcium ions to stabilize a protein Z-ZPI-fXa Michaelis complex on a procoagulant membrane surface through exosite-exosite interactions (35). The serpin circulates in plasma as a tight complex with its cofactor, protein Z (1). Interactions between their N-terminal Gla domains promote membrane-specific binding between protein Z and fXa (36). These interactions juxtapose the C-terminal domains of protein Z and fXa, distal from the membrane surface, in a manner that presents the RCL P1 Tyr to the active site so as to overcome the unfavorable P1-S1 interaction (Fig. 1) (36). Mutagenesis studies have suggested that additional serpin-proteinase exosite interactions further stabilize the membrane-associated Michaelis complex (Fig. 1) (37). Presumably, regulation of procoagulant membrane-bound fXa activity by the ZPI-protein Z complex is relevant to fXa bound in procoagulant complexes such as prothrombinase and fXase (38). An important unanswered question concerns the role of the novel N-terminal acidic tail of ZPI in stabilizing the membrane-bound protein Z-ZPI-fXa Michaelis complex and potentially allowing ZPI to compete with prothrombin for fXa bound in the prothrombinase complex. As with the essential weakening of the antithrombin-heparin interaction following progression of the Michaelis complex to the loop-inserted covalent state, the affinity of ZPI for protein Z is greatly reduced upon covalent complex formation (35) to allow the cofactor to act catalytically.
A final example of a surface-localized protein cofactor enhancing the rate of a serpin-proteinase interaction in a site-specific manner is thrombomodulin increasing the rate at which PN1 inhibits IIa. PN1 complexed with thrombomodulin at the surface of endothelial cells is ∼20-fold faster at inhibiting IIa than is free PN1 (39). Although the chondroitin sulfate moiety of thrombomodulin is important for the interaction with PN1, no other details of the interaction are known.
Although not involving a human serpin, the demonstration that the inhibition of human cathepsin V by the intracellular chicken serpin MENT can be accelerated in a template-like manner by double-stranded DNA (40) is useful for showing that inhibition of cysteine proteinase by serpins might be expected to also employ exosites where appropriate to the functional requirements of the interaction. This should not be a surprise given the essentially identical processes involved in forming the initial complexes between serpin and proteinase for cysteine and serine proteinases (7, 41, 42).
Other Relevant Serpin Exosite-Protein Interactions
Whereas the main focus of this minireview has been on how the ability of a serpin to directly inhibit a proteinase can be modulated in rate and location as a function of exosite interactions, there are less direct ways that serpin-protein interactions may influence proteolytic activity. One example is the B-clade serpin maspin, which is a potent tumor suppressor (43). Despite its inability to function as a proteinase inhibitor by the serpin branched pathway, it has been suggested as a possible regulator of enzymes of the plasminogen activator system (44). Although it neither directly interacts with the active site of either tPA or uPA nor affects the ability of these proteinases to act on a variety of macromolecular substrates (45), it does bind to both tPA and uPA at an exosite close to their active sites through an exosite on maspin close to the maspin RCL (46). How this might affect the function of tPA or uPA is not clear. A second example is the PAI-1 promotion of uPA receptor internalization mediated by LRP (low density lipoprotein receptor-related protein), which thereby inhibits urokinase-induced chemotaxis (47) and presumably also uPA activation. In turn, vitronectin can modulate this interaction through binding at or close to the same site on PAI-1 that PAI-1 uses for binding to LRP (48). Here again, the conformational change in the serpin upon RCL insertion modulates the vitronectin effect because vitronectin binds tightly only to the native conformation of PAI-1 (1).
Concluding Remarks
The selection of a serpin rather than a canonical-type proteinase inhibitor is a costly one for the organism. Not only is the serpin much larger, but, being metastable, it is prone to unwanted polymerization (1). Nevertheless, serpins appear to be the inhibitors of choice for regulation of complex proteinase-dependent processes as a result of their capacity to tailor their specificity and rate of reaction in ways that are not open to simple canonical inhibitors. Examples have been presented of the same proteinase being inhibited by two different serpins as circumstances require. Whereas circulating fXa is rapidly inhibited by antithrombin using a heparin cofactor, fXa that is part of the prothrombinase complex is site-specifically inhibited by ZPI by being presented to it at the membrane surface via its interaction with its membrane-associated cofactor, protein Z. Similarly, whereas antithrombin-heparin can inhibit fIXa and fXa as well as IIa, a separate serpin, HCII, has been designed to inhibit only IIa by down-regulating all RCL interactions with these Arg-specific proteinases and allowing only exosite-based up-regulation for the IIa interaction. Here, dermatan sulfate in the subendothelium, where it is abundant, may regulate IIa activity after vascular injury (24). Such deliberate down-regulation except when additional exosite interactions overcompensate is thus critical to achieve serpin specificity and is likely to hold for most inhibitory serpins involved in complex proteolytic cascades. A final advantage of serpins over canonical inhibitors is that they can make use of the conformational change that occurs upon complex formation to terminate the exosite involvement by altering the affinity for the cofactor, as seen with antithrombin-heparin, PAI-1-vitronectin, and ZPI-protein Z.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL49234 and HL79430 (to P. G. W. G.) and HL39888 and HL78827 (to S. T. O.). This is part of the Thematic Minireview Series on Proteolytic Enzymes. The first article was published in the November 7, 2008 issue; the second and third articles were published in the May 22, 2009 issue; and the fourth and fifth articles are published in this issue. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
A. Dementiev, R. Roth, G. Isetti, S. T. Olson, and P. G. W. Gettins, manuscript in preparation.
- PAI-1
- plasminogen activator inhibitor-1
- tPA
- tissue-type plasminogen activator
- uPA
- urokinase-type plasminogen activator
- α1PI
- α1-proteinase inhibitor
- RCL
- reactive center loop
- fXa
- factor Xa
- HCII
- heparin cofactor II
- ZPI
- protein Z-dependent proteinase inhibitor
- fIXa
- factor IXa
- PCI
- protein C inhibitor
- PN1
- proteinase nexin-1
- GAG
- glycosaminoglycan
- IIa
- thrombin.
REFERENCES
- 1.Gettins P. G. (2002) Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 2.Steenbakkers P. J., Irving J. A., Harhangi H. R., Swinkels W. J., Akhmanova A., Dijkerman R., Jetten M. S., van der Drift C., Whisstock J. C., op den Kamp H. J. (2008) Mycol. Res. 112, 999–1006 [DOI] [PubMed] [Google Scholar]
- 3.Irving J. A., Pike R. N., Lesk A. M., Whisstock J. C. (2000) Genome Res. 10, 1845–1864 [DOI] [PubMed] [Google Scholar]
- 4.Dementiev A., Dobó J., Gettins P. G. (2006) J. Biol. Chem. 281, 3452–3457 [DOI] [PubMed] [Google Scholar]
- 5.Chuang Y. J., Swanson R., Raja S. M., Bock S. C., Olson S. T. (2001) Biochemistry 40, 6670–6679 [DOI] [PubMed] [Google Scholar]
- 6.Zhou A., Carrell R. W., Huntington J. A. (2001) J. Biol. Chem. 276, 27541–27547 [DOI] [PubMed] [Google Scholar]
- 7.Tesch L. D., Raghavendra M. P., Bedsted-Faarvang T., Gettins P. G., Olson S. T. (2005) Protein Sci. 14, 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichhart J. M. (2005) Trends Cell Biol. 15, 659–665 [DOI] [PubMed] [Google Scholar]
- 9.Bianchini E. P., Louvain V. B., Marque P. E., Juliano M. A., Juliano L., Le Bonniec B. F. (2002) J. Biol. Chem. 277, 20527–20534 [DOI] [PubMed] [Google Scholar]
- 10.Gettins P. G. W., Patston P. A., Olson S. T. (1996) Serpins: Structure, Function and Biology, R. G. Landes Co., Austin, TX [Google Scholar]
- 11.Izaguirre G., Swanson R., Raja S. M., Rezaie A. R., Olson S. T. (2007) J. Biol. Chem. 282, 33609–33622 [DOI] [PubMed] [Google Scholar]
- 12.Johnson K. A. (2008) J. Biol. Chem. 283, 26297–26301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibarra C. A., Blouse G. E., Christian T. D., Shore J. D. (2004) J. Biol. Chem. 279, 3643–3650 [DOI] [PubMed] [Google Scholar]
- 14.Dekker R. J., Eichinger A., Stoop A. A., Bode W., Pannekoek H., Horrevoets A. J. G. (1999) J. Mol. Biol. 293, 613–627 [DOI] [PubMed] [Google Scholar]
- 15.Braud S., Le Bonniec B. F., Bon C., Wisner A. (2002) Biochemistry 41, 8478–8484 [DOI] [PubMed] [Google Scholar]
- 16.Chen V. C., Chao L., Chao J. (2000) J. Biol. Chem. 275, 40371–49377 [DOI] [PubMed] [Google Scholar]
- 17.Chen V. C., Chao L., Pimenta D. C., Bledsoe G., Juliano L., Chao J. (2001) J. Biol. Chem. 276, 1276–1284 [DOI] [PubMed] [Google Scholar]
- 18.Frank P. S., Douglas J. T., Locher M., Llinás M., Schaller J. (2003) Biochemistry 42, 1078–1085 [DOI] [PubMed] [Google Scholar]
- 19.Law R. H., Sofian T., Kan W. T., Horvath A. J., Hitchen C. R., Langendorf C. G., Buckle A. M., Whisstock J. C., Coughlin P. B. (2008) Blood 111, 2049–2052 [DOI] [PubMed] [Google Scholar]
- 20.Izaguirre G., Zhang W., Swanson R., Bedsted T., Olson S. T. (2003) J. Biol. Chem. 278, 51433–51440 [DOI] [PubMed] [Google Scholar]
- 21.Izaguirre G., Olson S. T. (2006) J. Biol. Chem. 281, 13424–13432 [DOI] [PubMed] [Google Scholar]
- 22.Huntington J. A., McCoy A., Belzar K. J., Pei X. Y., Gettins P. G., Carrell R. W. (2000) J. Biol. Chem. 275, 15377–15383 [DOI] [PubMed] [Google Scholar]
- 23.Whisstock J. C., Pike R. N., Jin L., Skinner R., Pei X. Y., Carrell R. W., Lesk A. M. (2000) J. Mol. Biol. 301, 1287–1305 [DOI] [PubMed] [Google Scholar]
- 24.He L., Giri T. K., Vicente C. P., Tollefsen D. M. (2008) Blood 111, 4118–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manithody C., Yang L., Rezaie A. R. (2002) Biochemistry 41, 6780–6788 [DOI] [PubMed] [Google Scholar]
- 26.Li W., Johnson D. J., Esmon C. T., Huntington J. A. (2004) Nat. Struct. Mol. Biol. 11, 857–862 [DOI] [PubMed] [Google Scholar]
- 27.Dementiev A., Petitou M., Herbert J. M., Gettins P. G. W. (2004) Nat. Struct. Mol. Biol. 11, 863–867 [DOI] [PubMed] [Google Scholar]
- 28.Olson S. T., Swanson R., Raub-Segall E., Bedsted T., Sadri M., Petitou M., Hérault J. M., Björk I. (2004) Thromb. Haemost. 92, 929–939 [DOI] [PubMed] [Google Scholar]
- 29.Hopkins P. C., Pike R. N., Stone S. R. (2000) J. Mol. Evol. 51, 507–515 [DOI] [PubMed] [Google Scholar]
- 30.Li W., Adams T. E., Nangalia J., Esmon C. T., Huntington J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4661–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezaie A. R., Yang L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12051–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L., Manithody C., Walston T. D., Cooper S. T., Rezaie A. R. (2003) J. Biol. Chem. 278, 37465–37470 [DOI] [PubMed] [Google Scholar]
- 33.Knauer D. J., Majumdar D., Fong P. C., Knauer M. F. (2000) J. Biol. Chem. 275, 37340–37346 [DOI] [PubMed] [Google Scholar]
- 34.Baglin T. P., Carrell R. W., Church F. C., Esmon C. T., Huntington J. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11079–11084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X., Swanson R., Broze G. J., Jr., Olson S. T. (2008) J. Biol. Chem. 283, 29770–29783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezaie A. R., Bae J. S., Manithody C., Qureshi S. H., Yang L. (2008) J. Biol. Chem. 283, 19922–19926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezaie A. R., Manithody C., Yang L. (2005) J. Biol. Chem. 280, 32722–32728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han X., Fiehler R., Broze G. J., Jr. (2000) Blood 96, 3049–3055 [PubMed] [Google Scholar]
- 39.Bouton M. C., Venisse L., Richard B., Pouzet C., Arocas V., Jandrot-Perrus M. (2007) Circ. Res. 100, 1174–1181 [DOI] [PubMed] [Google Scholar]
- 40.Ong P. C., McGowan S., Pearce M. C., Irving J. A., Kan W. T., Grigoryev S. A., Turk B., Silverman G. A., Brix K., Bottomley S. P., Whisstock J. C., Pike R. N. (2007) J. Biol. Chem. 282, 36980–36986 [DOI] [PubMed] [Google Scholar]
- 41.Dobó J., Swanson R., Salvesen G. S., Olson S. T., Gettins P. G. (2006) J. Biol. Chem. 281, 38781–38790 [DOI] [PubMed] [Google Scholar]
- 42.Swanson R., Raghavendra M. P., Zhang W., Froelich C., Gettins P. G., Olson S. T. (2007) J. Biol. Chem. 282, 2305–2313 [DOI] [PubMed] [Google Scholar]
- 43.Khalkhali-Ellis Z., Hendrix M. J. (2007) Cancer Res. 67, 3535–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biliran H., Jr., Sheng S. (2001) Cancer Res. 61, 8676–8682 [PubMed] [Google Scholar]
- 45.Bass R., Fernández A. M., Ellis V. (2002) J. Biol. Chem. 277, 46845–46848 [DOI] [PubMed] [Google Scholar]
- 46.Al-Ayyoubi M., Schwartz B. S., Gettins P. G. (2007) J. Biol. Chem. 282, 19502–19509 [DOI] [PubMed] [Google Scholar]
- 47.Degryse B., Sier C. F., Resnati M., Conese M., Blasi F. (2001) FEBS Lett. 505, 249–254 [DOI] [PubMed] [Google Scholar]
- 48.Kamikubo Y., Neels J. G., Degryse B. (2009) Int. J. Biochem. Cell Biol. 41, 578–585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.