Abstract
Epithelial Na+ channels facilitate the transport of Na+ across high resistance epithelia. Proteolytic cleavage has an important role in regulating the activity of these channels by increasing their open probability. Specific proteases have been shown to activate epithelial Na+ channels by cleaving channel subunits at defined sites within their extracellular domains. This minireview addresses the mechanisms by which proteases activate this channel and the question of why proteolysis has evolved as a mechanism of channel activation.
Many ion channels are silent at rest and are activated in response to a variety of factors, including membrane potential, external ligands, and intracellular signaling processes. The ENaC2 has evolved as a channel that is thought to reside primarily in an active state, facilitating the bulk movement of Na+ out of renal tubular or airway lumens. The regulated insertion and retrieval of channels at the plasma membrane have important roles in modulating ENaC-dependent Na+ transport (1). A number of factors also have a role in regulating ENaC activity via changes in channel Po or gating. In this regard, it has become increasingly apparent that proteolysis of ENaC subunits has a key role in this process (2). This minireview addresses several questions regarding the role of ENaC subunit proteolysis in regulating channel gating. (i) Where are ENaC subunits cleaved? (ii) Which proteases mediate ENaC cleavage? (iii) Why are channels activated by proteolysis? (iv) Is proteolysis responsible, in part, for the highly variable channel Po that has been noted for ENaC? (v) Why have ENaCs evolved as channels that require proteolysis for activation?
Where Are ENaC Subunits Cleaved?
Reports in the early 1980s that serine protease inhibitors reduced transepithelial Na+ transport across toad urinary bladder suggested that proteases have a role in activating ENaC (3). A series of studies over the past decade have confirmed that proteases activate ENaC and have begun to elucidate the mechanism by which this occurs. Following the observation that ENaC activity was significantly reduced in epithelial cells treated with aprotinin and that low concentrations of external trypsin rapidly activated ENaC in aprotinin-pretreated cells, a series of CAPs were identified that activated ENaC when coexpressed in heterologous expression systems (4–6). Furthermore, channels with a very low Po responded to external trypsin with a dramatic increase in Po (7).
What is the target of these proteases? ENaC is composed of three structurally related subunits (α, β, and γ) that have two membrane-spanning domains connected by a large extracellular loop composed of ∼450 residues. Early reports suggested that ENaC subunits or closely associated proteins were the protease target (5). Subsequent studies demonstrated that the α and γ subunits of ENaC were processed by proteases (8–11). The presence of full-length forms as well as faster migrating forms of the α and γ subunits on SDS-polyacrylamide gels, both in cell lysates and at the plasma membrane, provided the first clue that channel subunits were processed by proteases. Furthermore, the size of the cleaved fragments helped to define the sites of proteolysis (12).
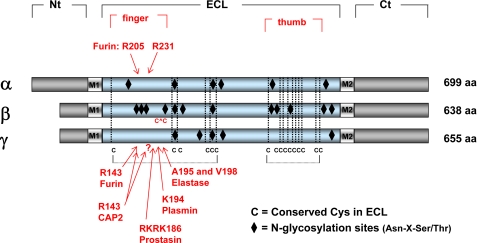
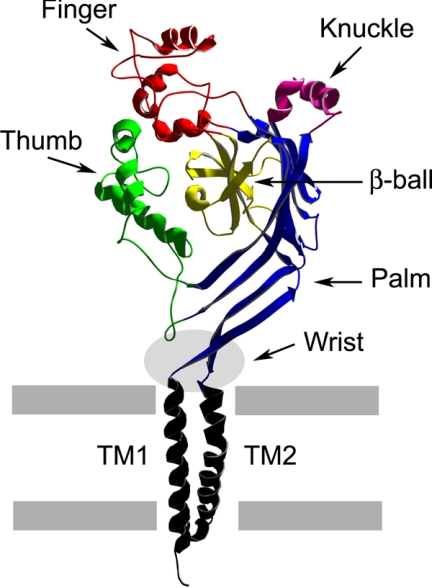
Functionally relevant cleavage sites were identified within the proximal regions of the extracellular domains of the α and γ subunits, as mutations of putative protease consensus cleavage sites prevented both subunit cleavage and channel activation (Fig. 1) (12–14). Subsequent studies also showed that proteolytic processing of subunits within a channel complex was an all-or-none event (9). Jasti et al. (15) recently resolved the crystal structure of the acid-sensing ion channel ASIC1, a member of the ENaC/degenerin ion channel family. This structure has provided important insights into the structural organization of ASIC and related family members, such as ENaC. The extracellular domain of ASIC1 has a highly ordered structure that resembles an outstretched hand containing a ball and has defined subdomains termed wrist, finger, thumb, palm, β-ball, and knuckle (Fig. 2) (15). ASIC1 proton-dependent gating has been proposed to occur in conjunction with conformational changes within the thumb and finger domains, which are transmitted to the wrist region and eventually to the transmembrane domains, where the channel's gate likely resides (15). Sites of ENaC subunit proteolysis that have been shown to be functionally relevant are within the “finger” domains and are likely located at peripheral sites that would be expected to be accessible to proteases (Fig. 2). As the finger domains are not conserved among members of the ENaC/degenerin family (15), the structures of the finger domains of ENaC subunits are likely to differ significantly from the resolved structure of the ASIC1 finger domain. Even within the three ENaC subunits, there are notable differences within the finger domains. For example, the finger domain of the β subunit lacks protease cleavage sites and instead exhibits three consensus sites for N-linked glycosylation and a unique pair of Cys residues (Fig. 1).
FIGURE 1.
Linear models of the ENaC subunits. The cytoplasmic N-terminal (Nt) and C-terminal (Ct) tails, the first (M1) and second (M2) transmembrane domains, the large extracellular loop (ECL), and the predicted finger and thumb domains are denoted for the three subunits. The sites for α subunit cleavage by furin and γ subunit cleavage by furin, prostasin (CAP1), CAP2, elastase (neutrophil and pancreatic), and plasmin are within the finger domain in the large extracellular loop. This alignment also reveals that the finger domain of the β subunit (i) lacks the consensus motifs for protease recognition, (ii) has additional sites for N-linked glycan addition, and (iii) has an additional Cys pair. aa, amino acids.
FIGURE 2.
Structure of an ASIC1 subunit. The extracellular domain of ASIC1 is a highly ordered structure that resembles an outstretched hand containing a ball (15). Defined subdomains are highlighted. Sites of proteolysis are primarily within the corresponding finger domain of ENaC. TM1 and TM2, first and second transmembrane domains.
Additional cleavage sites within the distal regions of the extracellular domains of ENaC subunits have been described (11, 16, 17). Recent studies examining the regulation and processing of ENaC subunits by the protease CAP2 (or TMPRSS4) have located cleavage sites within the “palm” domains of all three subunits (18). However, mutations that prevent cleavage in the palm domains do not affect ENaC function, suggesting that cleavage at these sites is not involved in regulating channel activity. Given the compact structure of the palm domain in ASIC1 that is stabilized by multiple hydrogen bonds, it is possible that cleavage at selected sites within these domains in ENaC subunits does not perturb its structure.
Which Proteases Mediate ENaC Cleavage?
The sizes of the cleaved α and γ subunit fragments suggested that proteolysis occurred within a region of the extracellular loop that is rich in basic amino acid residues and that contains furin consensus cleavage sites (12). Furin is a member of the proprotein convertase family of serine proteases that resides primarily within the trans-Golgi network and cleaves substrates immediately after the minimal consensus sequence of Arg-X-X-Arg, where X is any residue (19). Analyses of ENaC subunits with mutations of key Arg residues within the furin consensus sites revealed that α subunits were cleaved at two furin sites and that γ subunits were cleaved at a single furin site (12). Na+ currents in oocytes were reduced by up to ∼90% when these sites were mutated to prevent cleavage, suggesting that proteolysis was required for channels to be active (12). Furin site mutations in the α subunit alone inhibited ENaC activity by up to 85%, whereas a modest reduction in current was found when the γ subunit furin site was mutated (12). However, channel activity was rescued when oocytes expressing these mutant channels were exposed to the protease trypsin, suggesting that the mutant channels were at the plasma membrane in a functionally inactive state (12). Whole cell Na+ currents were also reduced by ∼90% when ENaC was expressed in furin-deficient Chinese hamster ovary cells compared with control cells expressing ENaC, and channel activity was rescued by coexpression of ENaC and furin (12). Furthermore, furin inhibitors reduced Na+ currents in cells expressing endogenous ENaC (12). Although furin appears to have a role in the processing of ENaC subunits, it is likely that other members of the proprotein convertase family cleave and activate ENaC.
Additional proteases have been shown to process the γ subunit and further activate the channel. Prostasin (or CAP1) is a glycosylphosphatidylinositol-anchored serine protease likely localized on the surface of renal and airway epithelia (4, 20). Prostasin-induced cleavage of the γ subunit at a defined site, distal to the previously identified furin cleavage site, further activates mouse ENaC (13) and is required to fully activate rat ENaC (18). Other proteases have been shown to both activate ENaC and cleave the γ subunit at sites distal to the furin site, including elastase, CAP2 (TMPRSS4), and plasmin (14, 18, 21, 22). CAP2 was also shown to enhance cleavage at the “furin” consensus site of the rat γ subunit (18). Moderate channel activation was still observed when this site was mutated, consistent with CAP2 cleavage of the rat γ subunit at other sites (18). Mutation of a Lys residue in the prostasin cleavage site of the rat γ subunit results in channel activation as well as cleavage at this site by an endogenous protease, perhaps furin (23). Matriptase (or CAP3) activates the channel, although it has not been shown that this protease directly cleaves ENaC (6). Trypsin activates ENaC and enhances subunit cleavage at sites that are in the vicinity of the furin cleavage sites (24). Studies with mice that lack kallikrein expression suggest that this protease may have a role in processing the γ subunit under basal conditions (25), whereas other proteases also have a role in processing the γ subunit and activating ENaC in the setting of volume depletion (10, 11, 26).
Clearly, a number of proteases can cleave and activate ENaC. It is likely that furin, prostasin, CAP2, matriptase, kallikrein, and elastase do not encompass the complete repertoire of proteases that cleave and activate ENaC. In addition, proteases are likely to activate ENaC indirectly by cleaving and activating other proteases that subsequently cleave the channel. For example, prostasin is translated as a proenzyme that must be cleaved to be active but does not undergo autocatalytic cleavage (27). Matriptase (CAP3) is one of the proteases that cleaves and activates prostasin (28). Bengrine et al. (29) have suggested that cleavage of other proteins, perhaps protease-activated receptors, may also have a role in regulating ENaC activity. Although proteases certainly have an important role in cleaving and activating ENaC, a number of important questions remain to be resolved. (i) Which are the key proteases that cleave and activate ENaC in vivo? (ii) Are there tissue-specific proteases that cleave and activate ENaC? (iii) Is ENaC proteolysis a regulated process? (iv) Is there differential expression of proteases that cleave and activate ENaC under physiologic and pathologic conditions?
Several studies have begun to address the question of whether ENaC proteolysis is a regulated process. The expression of prostasin as well as protease nexin-1, an inhibitor of prostasin and other serine proteases, may be regulated by aldosterone (30, 31). ENaC residence time at the plasma membrane affects the extent of α and γ subunit cleavage (24, 32). Longer residency times were associated with a greater degree of subunit proteolysis. Several factors, including aldosterone-dependent signaling processes, increase ENaC residency time at the plasma membrane (1, 33). Furthermore, rates of Na+ entry or changes in intracellular [Na+] influence proteolytic processing of the α and γ subunits (34). A reduced rate of Na+ entry led to enhanced cleavage of channel subunits. Mechanisms by which rates of Na+ entry alter channel cleavage remain to be defined.
Investigators have also begun to address the question of whether the activities of specific proteases that cleave and activate ENaC are altered in particular diseases. Two recent studies suggest that ENaC activation by plasmin may contribute to the renal Na+ retention and volume expansion that are observed in nephrotic syndrome (14, 35). Plasmin activates ENaC by cleaving the γ subunit at a site distal to the furin site. Furthermore, plasminogen, as well as plasmin, is excreted in the urine of both rats and humans with nephrotic syndrome (14, 35). These studies suggest that when the glomerular filtration barrier is damaged, plasminogen is filtered and converted to activate plasmin by urokinase that is expressed in the lumen of renal tubules.
Several studies have suggested that there is enhanced cleavage of ENaC subunits in airway epithelia in the setting of cystic fibrosis (17, 36) as well as in the distal nephron in the setting of volume depletion and aldosterone administration (10, 26). ENaC activation in cystic fibrosis airway contributes to the reduction in airway surface liquid volume and the impairment in mucociliary clearance (37). The proteases responsible for the enhanced cleavage of ENaC subunits in these settings remain to be defined.
Why Are Channels Activated by Proteolysis?
Once it became clear that proteases activate ENaC by cleaving its subunits, the next question to address was how channel activation occurs. Was there simply a physical constraint, where an increase in flexibility following subunit cleavage would facilitate transitions from a closed to an open state? If so, cleavage at a single site should be sufficient to activate the channel. Surprisingly, cleavage at a single furin site in the α subunit was not sufficient to activate ENaC (24, 38). α subunits had to be cleaved at both furin sites for channels to exhibit “normal” activity, suggesting that the tract between the furin cleavage sites in the α subunit functions as an inhibitor that stabilizes the channel in the closed conformation (38). In support of this hypothesis, channels lacking both α subunit furin cleavage sites and the tract between these sites were found to be active, although the α subunit was not cleaved (38). Furthermore, a synthetic 26-residue peptide corresponding to the tract that is presumably released from the α subunit by furin cleavage reversibly inhibited ENaC by reducing channel Po (38). The key inhibitory region within these 26 residues was subsequently identified as an 8-residue tract that is highly conserved among species (39).
Does the γ subunit also need to be cleaved twice to activate the channel? Studies published to date support this hypothesis. Channels with a γ subunit lacking the furin and prostasin cleavage sites and the intervening 43-residue tract exhibited markedly increased activity due to a very high Po, although the γ subunit was not cleaved (13). A synthetic peptide corresponding to the fragment presumably released from the γ subunit by furin and prostasin cleavage was also a reversible ENaC inhibitor (13). As discussed above, multiple proteases appear to activate ENaC by cleaving the γ subunit at sites distal to the furin cleavage site (13, 21, 22, 25). Although one group has argued that the primary mechanism by which CAP2 activates the channel is by enhancing γ subunit cleavage at the furin site, their data also support a primary role for cleavage of the γ subunit at sites distal to the furin site in activating ENaC (18).
If proteolysis activates channels by excising inhibitory tracts from the α and γ subunits, how do these tracts inhibit the channel? As mentioned above, these inhibitory tracts are located within the finger regions of the extracellular domains (Fig. 2). Jasti et al. (15) have proposed that conformational changes within the thumb and finger domains are required for ASIC1 gating. If this is also true for ENaC, perhaps the inhibitory tracts constrain movement of the thumb and finger domains.
Although prostasin activates ENaC by inducing γ subunit cleavage at a defined site, prostasin mutants that should have little or no proteolytic activity also activate the channel (13, 40). If proteolysis is required to activate ENaC, why do these prostasin mutants activate the channel? Does very limited proteolytic activity allow for sufficient cleavage to activate the channel (13)? Alternatively, perhaps mutant prostasin binds to channels where γ subunits have been processed by furin, displacing the inhibitory tract from its normal site within the ENaC complex and facilitating a transition to an open state.
Is Proteolysis Responsible, in Part, for the Highly Variable Open Probability That Has Been Noted for ENaC?
When studied at a single channel level in epithelia, ENaCs display a wide range of Po values, from <0.1 to 0.9 (41). Why is ENaC Po so variable? What are the cellular mechanisms that control this widely variable Po? A number of cellular factors, such as specific kinases and anionic lipids, influence channel Po (42, 43). Channel subunits lacking cleavage have been observed at the plasma membrane in vivo as well as in heterologous expression systems (9, 22, 24, 26). Channels with non-cleaved subunits have a very low Po and provide a reserve pool of poorly functional channels that could be readily activated by proteases in post-Golgi compartments (7, 44). Channel cleavage by furin removes the α subunit inhibitory tract and moves the channel to an intermediate Po state (2, 12, 38). Furin also primes the channel for further activation by cleaving the γ subunit once. Subsequent cleavage of the γ subunit at sites distal to the furin site moves the channel to a high Po state (2, 13).
If excising inhibitory tracts from the α and γ subunits activates the channel, does the processing of one subunit have a dominant role in activating ENaC? Recent work suggests that simply removing the γ subunit inhibitory tract may be sufficient to transition the channel to a high Po state, even in the absence of α subunit cleavage (45).
Why Have ENaCs Evolved as Channels That Require Proteolysis for Activation?
ENaCs evolved from a family of channels that are activated in response to factors within their external environment. For example, Mec4/Mec10 channels in Caenorhabditis elegans are activated by mechanical forces; a family of ENaC-related channels in marine snails are activated by peptides; and ASICs are activated by external acidification (46, 47). These channels reside primarily in the closed state and transition transiently to an open state in response to external cues. On the other hand, ENaC facilitates the bulk movement of Na+ across an epithelial layer. For this process to occur, it is necessary for ENaC to be constitutively active. We propose that proteolytic processing of ENaC subunits provided a mechanism that has allowed ENaCs to evolve from channels that reside primarily in the closed state to constitutively active channels that facilitate transepithelial Na+ transport.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R56 DK065161. This work was also supported by a Carl W. Gottschalk award from the American Society of Nephrology (to M. D. C.). This is part of the Thematic Minireview Series on Proteolytic Enzymes. The first article was published in the November 7, 2008 issue; the second and third articles were published in the May 22, 2009 issue; and the fourth and fifth articles are published in this issue. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
- ENaC
- epithelial Na+ channel
- Po
- open probability
- CAP
- channel-activating protease
- ASIC
- acid-sensing ion channel.
REFERENCES
- 1.Snyder P. M. (2005) Endocrinology 146, 5079–5085 [DOI] [PubMed] [Google Scholar]
- 2.Hughey R. P., Carattino M. D., Kleyman T. R. (2007) Curr. Opin. Nephrol. Hypertens. 16, 444–450 [DOI] [PubMed] [Google Scholar]
- 3.Orce G. G., Castillo G. A., Margolius H. S. (1980) Am. J. Physiol. Renal Physiol. 239, F459–F465 [DOI] [PubMed] [Google Scholar]
- 4.Vallet V., Chraibi A., Gaeggeler H. P., Horisberger J. D., Rossier B. C. (1997) Nature 389, 607–610 [DOI] [PubMed] [Google Scholar]
- 5.Chraïbi A., Vallet V., Firsov D., Hess S. K., Horisberger J. D. (1998) J. Gen. Physiol. 111, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., Rossier B. C. (2002) J. Gen. Physiol. 120, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell R. A., Boucher R. C., Stutts M. J. (2004) Am. J. Physiol. Cell Physiol. 286, C190–C194 [DOI] [PubMed] [Google Scholar]
- 8.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 9.Hughey R. P., Bruns J. B., Kinlough C. L., Kleyman T. R. (2004) J. Biol. Chem. 279, 48491–48494 [DOI] [PubMed] [Google Scholar]
- 10.Masilamani S., Kim G. H., Mitchell C., Wade J. B., Knepper M. A. (1999) J. Clin. Invest. 104, R19–R23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ergonul Z., Frindt G., Palmer L. G. (2006) Am. J. Physiol. Renal Physiol. 291, F683–F693 [DOI] [PubMed] [Google Scholar]
- 12.Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 13.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 14.Passero C. J., Mueller G. M., Rondon-Berrios H., Tofovic S. P., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 36586–36591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 16.Michlig S., Harris M., Loffing J., Rossier B. C., Firsov D. (2005) J. Biol. Chem. 280, 38264–38270 [DOI] [PubMed] [Google Scholar]
- 17.Myerburg M. M., Butterworth M. B., McKenna E. E., Peters K. W., Frizzell R. A., Kleyman T. R., Pilewski J. M. (2006) J. Biol. Chem. 281, 27942–27949 [DOI] [PubMed] [Google Scholar]
- 18.García-Caballero A., Dang Y., He H., Stutts M. J. (2008) J. Gen. Physiol. 132, 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donaldson S. H., Hirsh A., Li D. C., Holloway G., Chao J., Boucher R. C., Gabriel S. E. (2002) J. Biol. Chem. 277, 8338–8345 [DOI] [PubMed] [Google Scholar]
- 21.Adebamiro A., Cheng Y., Rao U. S., Danahay H., Bridges R. J. (2007) J. Gen. Physiol. 130, 611–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris M., Firsov D., Vuagniaux G., Stutts M. J., Rossier B. C. (2007) J. Biol. Chem. 282, 58–64 [DOI] [PubMed] [Google Scholar]
- 23.Diakov A., Bera K., Mokrushina M., Krueger B., Korbmacher C. (2008) J. Physiol. 586, 4587–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabra R., Knight K. K., Zhou R., Snyder P. M. (2008) J. Biol. Chem. 283, 6033–6039 [DOI] [PubMed] [Google Scholar]
- 25.Picard N., Eladari D., El Moghrabi S., Planès C., Bourgeois S., Houillier P., Wang Q., Burnier M., Deschenes G., Knepper M. A., Meneton P., Chambrey R. (2008) J. Biol. Chem. 283, 4602–4611 [DOI] [PubMed] [Google Scholar]
- 26.Frindt G., Ergonul Z., Palmer L. G. (2008) J. Gen. Physiol. 131, 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shipway A., Danahay H., Williams J. A., Tully D. C., Backes B. J., Harris J. L. (2004) Biochem. Biophys. Res. Commun. 324, 953–963 [DOI] [PubMed] [Google Scholar]
- 28.Netzel-Arnett S., Currie B. M., Szabo R., Lin C. Y., Chen L. M., Chai K. X., Antalis T. M., Bugge T. H., List K. (2006) J. Biol. Chem. 281, 32941–32945 [DOI] [PubMed] [Google Scholar]
- 29.Bengrine A., Li J., Hamm L. L., Awayda M. S. (2007) J. Biol. Chem. 282, 26884–26896 [DOI] [PubMed] [Google Scholar]
- 30.Narikiyo T., Kitamura K., Adachi M., Miyoshi T., Iwashita K., Shiraishi N., Nonoguchi H., Chen L. M., Chai K. X., Chao J., Tomita K. (2002) J. Clin. Invest. 109, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakida N., Kitamura K., Tuyen D. G., Maekawa A., Miyoshi T., Adachi M., Shiraishi N., Ko T., Ha V., Nonoguchi H., Tomita K. (2006) Kidney Int. 70, 1432–1438 [DOI] [PubMed] [Google Scholar]
- 32.Knight K. K., Olson D. R., Zhou R., Snyder P. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2805–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 34.Knight K. K., Wentzlaff D. M., Snyder P. M. (2008) J. Biol. Chem. 283, 27477–27482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svenningsen P., Bistrup C., Friis U. G., Bertog M., Haerteis S., Krueger B., Stubbe J., Jensen O. N., Thiesson H. C., Uhrenholt T. R., Jespersen B., Jensen B. L., Korbmacher C., Skøtt O. (2009) J. Am. Soc. Nephrol. 20, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarran R., Trout L., Donaldson S. H., Boucher R. C. (2006) J. Gen. Physiol. 127, 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mall M., Grubb B. R., Harkema J. R., O'Neal W. K., Boucher R. C. (2004) Nat. Med. 10, 487–493 [DOI] [PubMed] [Google Scholar]
- 38.Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 39.Carattino M. D., Passero C. J., Steren C. A., Maarouf A. B., Pilewski J. M., Myerburg M. M., Hughey R. P., Kleyman T. R. (2008) Am. J. Physiol. Renal Physiol. 294, F47–F52 [DOI] [PubMed] [Google Scholar]
- 40.Andreasen D., Vuagniaux G., Fowler-Jaeger N., Hummler E., Rossier B. C. (2006) J. Am. Soc. Nephrol. 17, 968–976 [DOI] [PubMed] [Google Scholar]
- 41.Pácha J., Frindt G., Antonian L., Silver R. B., Palmer L. G. (1993) J. Gen. Physiol. 102, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pochynyuk O., Bugaj V., Stockand J. D. (2008) Curr. Opin. Nephrol. Hypertens. 17, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blazer-Yost B. L., Nofziger C. (2004) Adv. Exp. Med. Biol. 559, 359–368 [DOI] [PubMed] [Google Scholar]
- 44.Sheng S., Carattino M. D., Bruns J. B., Hughey R. P., Kleyman T. R. (2006) Am. J. Physiol. Renal Physiol. 290, F1488–F1496 [DOI] [PubMed] [Google Scholar]
- 45.Carattino M. D., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mano I., Driscoll M. (1999) BioEssays 21, 568–578 [DOI] [PubMed] [Google Scholar]
- 47.Lingueglia E., Deval E., Lazdunski M. (2006) Peptides 27, 1138–1152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.