Abstract
RNase II is the prototype of a ubiquitous family of enzymes that are crucial for RNA metabolism. In Escherichia coli this protein is a single-stranded-specific 3′-exoribonuclease with a modular organization of four functional domains. In eukaryotes, the RNase II homologue Rrp44 (also known as Dis3) is the catalytic subunit of the exosome, an exoribonuclease complex essential for RNA processing and decay. In this work we have performed a functional characterization of several highly conserved residues located in the RNase II catalytic domain to address their precise role in the RNase II activity. We have constructed a number of RNase II mutants and compared their activity and RNA binding to the wild type using different single- or double-stranded substrates. The results presented in this study substantially improve the RNase II model for RNA degradation. We have identified the residues that are responsible for the discrimination of cleavage of RNA versus DNA. We also show that the Arg-500 residue present in the RNase II active site is crucial for activity but not for RNA binding. The most prominent finding presented is the extraordinary catalysis observed in the E542A mutant that turns RNase II into a “super-enzyme.”
Exoribonuclease II (RNase II)5 is one of the major exoribonucleases involved in the degradation of RNA (1). This protein degrades RNA hydrolytically in the 3′ to 5′ direction, releasing 5′-nucleotide monophosphates in a processive manner. Its activity is sequence independent but sensitive to secondary structures, and poly(A) is the preferential substrate for this enzyme (2–6). In Escherichia coli RNase II is responsible for 90% of the hydrolytic activity in crude extracts (7). Moreover, its expression is differentially regulated at the transcriptional and post-transcriptional levels (8–10), and the protein can be regulated by environmental conditions (10). The E. coli enzyme is the prototype of a widespread family of exoribonucleases, and RNase II homologues can be found in prokaryotes and eukaryotes (11). In the nucleus and in the cytoplasm of eukaryotic cells, RNase II homologues (Rrp44/Dis3) are part of the exosome, an essential multiprotein complex of exoribonucleases involved in processing, turnover, and quality control of different types of RNAs (12). Rrp44 is the only catalytically active nuclease in the human and yeast core exosome (13, 14). Moreover, it was recently demonstrated that apart from the RNB catalytic domain, Rrp44p has a second nuclease domain, the PINc domain, that confers endonucleolytic activity to the protein (15, 16). In prokaryotic cells, apart from RNase II, there is an additional RNase II-like enzyme, RNase R, that is involved in RNA degradation and RNA and protein quality control (17–20). RNase R has also been shown to be required for virulence (21).
The determination of E. coli RNase II structure (22–24) showed that, contrary to the sequence predictions, RNase II consisted of four domains; they are two N-terminal cold shock domains (CSD1 and CSD2) involved in RNA binding, one central RNB catalytic domain, and a third RNA binding domain at the C terminus, the S1 domain (Fig. 1A). Inside RNase II the RNA contacts the enzyme at two different and non-contiguous regions, the anchoring region (formed by the three RNA binding domains) and the catalytic region (buried inside the catalytic domain) (22, 25). The shortest RNA molecule able to retain contacts with these two regions simultaneously is a 10-nucleotide fragment, and this is also the minimum length necessary to maintain the processivity of the enzyme (22, 25, 26). Therefore, simultaneous binding of substrate to both sites is necessary for a processive degradation. Moreover, the structure of the RNA-bound complex revealed a tight packing of the five 3′-terminal nucleotides in the catalytic cavity, mediated by the conserved aromatic residues Tyr-253 and Phe-358 (22). When the RNA substrate is shorter than 5 nucleotides the required packing of the bases can no longer occur, impeding the translocation of the RNA and generating the typical 4-nt fragment as the end-product for RNase II. A mutational analysis of E. coli RNase II has been reported (26), confirming the crucial role of Tyr-253 in setting the end-product of RNase II degradation. In addition, this analysis showed that conserved acidic residues in the active site can have a different role during the degradation mechanism (26).
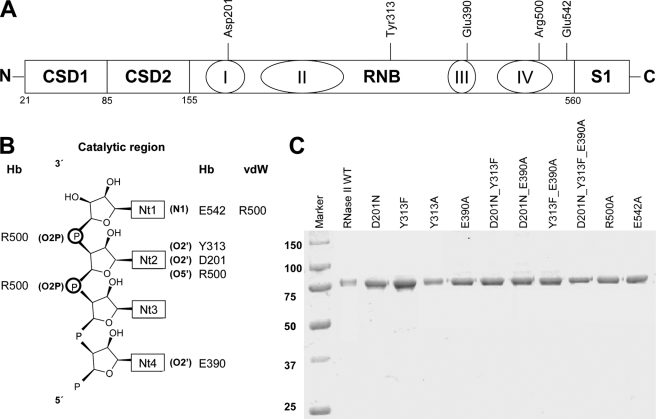
FIGURE 1.
RNase II and mutant proteins. A, linear representation of RNase II domains with the specification of the position of the conserved residues within RNB domain. B, atomic interactions scheme between RNA and protein residues. Hb indicates hydrogen bonds up to 3.2 Å, and vdW indicates van der Waals interactions up to 3.6 Å. The labeling of the nucleotides starts from the 3′-end of the RNA, inversely to previous reports where the nucleotides at the 3′-end was labeled as nt 13 (22). C, purity of the enzymes. 0.5 μg of purified His6-RNase II, His6-D201N, His6-Y313F, His6-Y313A, His6-E390A, His6-D201N/Y313F, His6-D201N/E390A, His6-Y313F/E390A, His6-D201N/Y313F/E390A, His6-R500A, and His6-E542A were applied and visualized by Coomassie Blue staining. Molecular weights of standard proteins are indicated on the left.
RNase II is specific for RNA. However, although it does not degrade DNA, it is still able to bind it. DNA oligonucleotides have been shown to act as reaction inhibitors of the E. coli RNase II, suggesting that both DNA and RNA compete for the same binding sites of the enzyme (25). Previous reports have demonstrated that the ribose specificity of RNase II is not for the scissile bond but for the nearby nucleotides (22, 25). The presence of a ribose between positions 2 through 5 from the 3′-end of the RNA substrate is required for cleavage to occur (25). The crystal structure of the RNase II-RNA-bound complex (22) revealed direct contacts between the residues Asp-201, Tyr-313, and Glu-390 and the O2′ ribose oxygens of nucleotides in positions 2, 4, and 6 (Fig. 1B). Such interactions seem to be responsible for the proper orientation of the RNA at the catalytic cavity and may account for the specificity of RNA versus DNA in RNase II.
Structural data also revealed that there are two other conserved residues at the active site that seem to be crucial for RNase II activity, Arg-500 and Glu-542 (22). In the RNA-complexed structure, the Arg-500 is positioned between the two nucleotides at the 3′-end interacting with their phosphate backbones (Fig. 1B). It was postulated that Arg-500 could assist in catalysis by fixing the phosphodiester bond at the cleavage position and enhancing the susceptibility of the leaving phosphorous atom to a nucleophilic attack (22). Glu-542 is in close proximity to the nitrogen atoms of the leaving nucleotide (Fig. 1B) and was suggested to facilitate the elimination of this nucleotide upon phosphor-ester cleavage by pulling it out of the base-stacked position (22).
In this report we analyze the role of some of these highly conserved amino acids in RNase II activity. We studied the function of Asp-201, Tyr-313, and Glu-390 in the cleavage discrimination of RNA versus DNA. Our results showed that only Tyr-313 and Glu-390 are crucial for RNA specificity of RNase II. We also demonstrate that Arg-500 is essential for RNase II activity, but it is not involved in RNA binding. Moreover, the most prominent findings presented are the extraordinary catalysis and binding abilities observed in the E542A mutant that turns RNase II into a super-enzyme.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes, T4 DNA ligase, Pfu DNA polymerase, and T4 polynucleotide kinase were purchased from Fermentas. Unlabeled oligonucleotide primers were synthesized by STAB Vida, Portugal.
Strains
The E. coli strains used were DH5α (F′ fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17a) (27) for cloning experiments and BL21(DE3) (F− rB− mB− gal ompT (int∷PlacUV5 T7 gen1 imm21 nin5) (28) for expression and purification of enzymes.
Construction of RNase II Mutants by PCR Overlapping
The point mutations Y313F, Y313A, E390A, R500K, R500A, and E542A, the double mutations D201N/Y313F, D201N/E390A, and Y313F/E390A, and the triple mutation D201N/Y313F/E390A were introduced into pFCT6.9 (10) or in the pFCT6.9/D201N (26) by PCR overlapping (29).
The primers used in this study were the following (base changes are indicated in small letters): forward primer Y313Fa, 5′-GCAAAGCTtGTGTtTGACCAGGTTTCTGAC-3′, and reverse primer Y313Fb; 5′-GTCAGAAACCTGGTCAaACACaAGCTTTGC-3′; forward primer, Y313Aa, 5′-GCAAAGCTtGTGgcTGACCAGGTTTCTGAC-3′, and reverse primer, Y313Ab; 5′-GTCAGAAACCTGGTCAgcCACaAGCTTTGC-3′, forward primer, E390Aa, 5′-CCAACCGTATCGTCGctGAAGCGATGATTGCC-3′, and reverse primer, E390Ab, 5′-GGCAATCATCGCTTCagCGACGATACGGTTGG-3′; forward primer, R500Ka, 5′-CCACCTGGACgTCGCCGATCaagAAATATG-3′, and reverse primer, R500Kb, 5′-CATATTTcttGATCGGCGAcGTCCAGGTGG-3′; forward primer, R500Aa, 5′-CGCCACCTGGACTTCGCCGATCgcTAAATA-3′, and reverse primer, R500Ab, 5′-CCATATTTAgcGATCGGCGAAGTCCAGGTG-3′; forward primer, E542Aa, 5′-CGCCGgCTCAACCGGATGGCAGcACGTGATGTT-3′, and reverse primer, E542Ab, 5′-AACATCACGTgCTGCCATCCGGTTGAGcCGGCG-3′. All mutant constructs were confirmed by DNA sequencing at STAB Vida, Portugal.
Overexpression and Purification of Wild Type RNase II and Mutants
The plasmid used for expression of wild-type E. coli histidine-tagged RNase II protein was pFCT6.9 plasmid (10). This plasmid contains the rnb gene cloned into pET-15b vector (Novagen) under the control of ϕ10 promoter, allowing the expression of the His6-tagged RNase II fusion protein.
All other plasmids bearing mutations were transformed into BL21(DE3) E. coli strain (Novagen) to allow the expression of the recombinant proteins. Cells were grown at 37 °C in 100 ml of LB medium supplemented with 150 μg/ml ampicillin to an A600 of 0.45 and then induced for 2 h by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. Cell cultures were pelleted by centrifugation and stored at −80 °C.
Purification of all proteins was performed by histidine affinity chromatography using HiTrap Chelating HP columns (GE Healthcare) and the AKTA fast protein liquid chromatography system (GE Healthcare) following the protocol previously described (30, 31). Briefly, cell suspensions were lysed using a French press at 9000 p.s.i. in the presence of 0.1 mm phenylmethylsulfonyl fluoride. The crude extracts were treated with Benzonase (Sigma) to degrade the nucleic acids and clarified by a 30-min centrifugation at 10,000 × g. The clarified extracts were then added to a HiTrap chelating-Sepharose 1-ml column equilibrated in buffer A (20 mm Tris-HCl, 0.5 m NaCl, pH 8, 20 mm imidazole and 2 mm β-mercaptoethanol). Protein elution was achieved by a continuous imidazole gradient (from 20–500 mm) in buffer A. The fractions containing the purified protein were pooled together, and buffer was exchanged to buffer B (20 mm Tris-HCl, pH 8, 100 mm KCl, and 2 mm β-mercaptoethanol) using a desalting 5-ml column (GE Healthcare). Eluted proteins were concentrated by centrifugation at 15 °C with Amicon Ultra Centrifugal Filter Devices of 30,000 molecular weight cutoff (Millipore). Protein concentration was determined by spectrophotometry, and 50% (v/v) glycerol was added to the final fractions before storage at −20 °C. 0.5 μg of each purified protein was applied in a 8% SDS-PAGE and visualized by Coomassie Blue staining (Fig. 1C).
Activity Assays
Exoribonucleolytic activity was assayed using different RNA oligoribonucleotides as substrates (30): a poly(A) chain of 35 nt as a single-stranded substrate, a 30-mer oligoribonucleotide (5′-CCCGACACCAACCACUAAAAAAAAAAAAAA-3′) annealed to the complementary 16-mer oligodeoxyribonucleotide (5′-AGTGGTTGGTGTCGGG-3′) as a double-stranded substrate with a 3′-single-stranded extension, and the DNA-RNA chimeric substrates Chi1 (5′-dTdTdTdTdTdTdTdTdTdTdTrCrCdTdT-3′) and Chi2 (5′-dTdTdT dTdTdTdTdTdTdTdTrCdTrCdT-3′). All the oligoribonucleotides were labeled at the 5′-end with [γ-32ATP] and T4 polynucleotide kinase and further purified with Microcon YM-3 Centrifugal Filter Devices (Millipore) to remove the non-incorporated nucleotides. The hybridization between 30-mer and 16-mer oligomers was performed in a 1:1 (mol:mol) ratio in the Tris component of the activity by 5 min of incubation at 68 °C followed by 45 min as room temperature. The exoribonucleolytic reactions were carried out in a final volume of 10 μl containing 30 nm concentrations of substrate, 20 mm Tris-HCl, pH 8, 100 mm KCl, 1 mm MgCl2, and 1 mm dithiothreitol. The amount of each enzyme added to the reaction was adjusted to obtain linear conditions and is indicated in the respective figures (Figs. 2–4). Reactions were started by the addition of the enzyme and incubated at 37 °C. Samples were withdrawn at the time points indicated in the figures (Figs. 2–4), and the reaction was stopped by adding formamide-containing dye supplemented with 10 mm EDTA. Reaction products were resolved in a 20% polyacrylamide, 7 m urea and analyzed by autoradiography. Autoradiograms were scanned, and the densities of the bands were quantified using ImageQuant 5.0 software. The exoribonucleolytic activity of the enzymes was determined by measuring and quantifying the disappearance of the substrate from gels in several distinct experiments, and each value obtained represents the mean of at least three independent assays. The specific activity of each enzyme is given as the nmol of substrate consumed/min/nmol of protein at 37 °C. Results obtained are shown in Table 1.
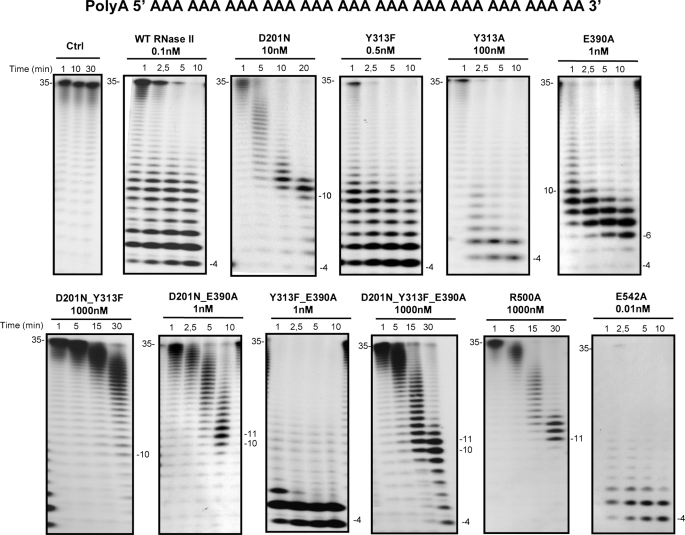
FIGURE 2.
Exoribonuclease activity with single-stranded RNA substrate; comparison of wild-type and mutant proteins. Activity assays were performed as described under “Experimental Procedures” using a poly(A) chain of 35 nt as substrate. The mutants used and their respective protein concentrations are shown. The wild-type enzyme was used as control. Samples were taken during the reaction at the time points indicated, and reaction products were analyzed in a 20% polyacrylamide, 7 m urea gel. Control reactions with no enzyme added (Ctrl) were incubated at the maximum reaction time for each protein. Length of substrates and degradation products are indicated in the figure.
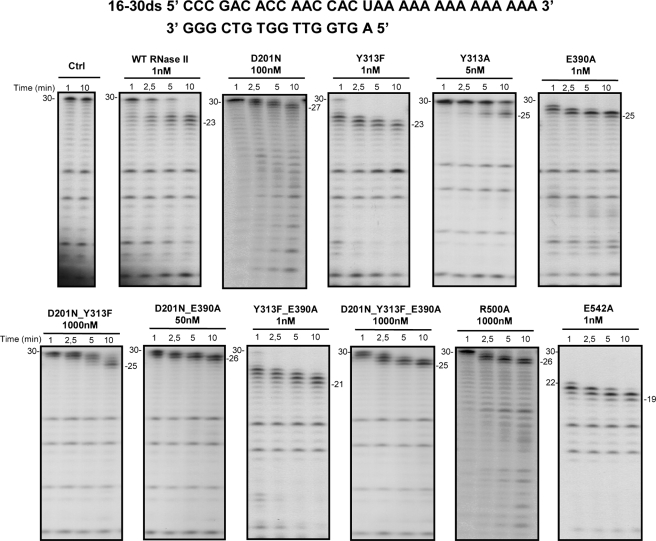
FIGURE 3.
Exoribonuclease activity with ds16–30 substrate; comparison of wild-type and mutant proteins. Activity assays were performed as described under “Experimental Procedures” using a 30-mer oligoribonucleotide hybridized to the complementary 16mer oligodeoxyribonucleotide, thus obtaining the corresponding double-stranded substrate 16–30ds. The mutants used and their respective protein concentrations are shown. The wild-type enzyme was used as control. Samples were taken during the reaction at the time points indicated, and reaction products were analyzed in a 20% polyacrylamide, 7 m urea gel. Control reactions with no enzyme added (Ctrl) were incubated at the maximum reaction time for each protein. Length of substrates and degradation products are indicated in the figure.
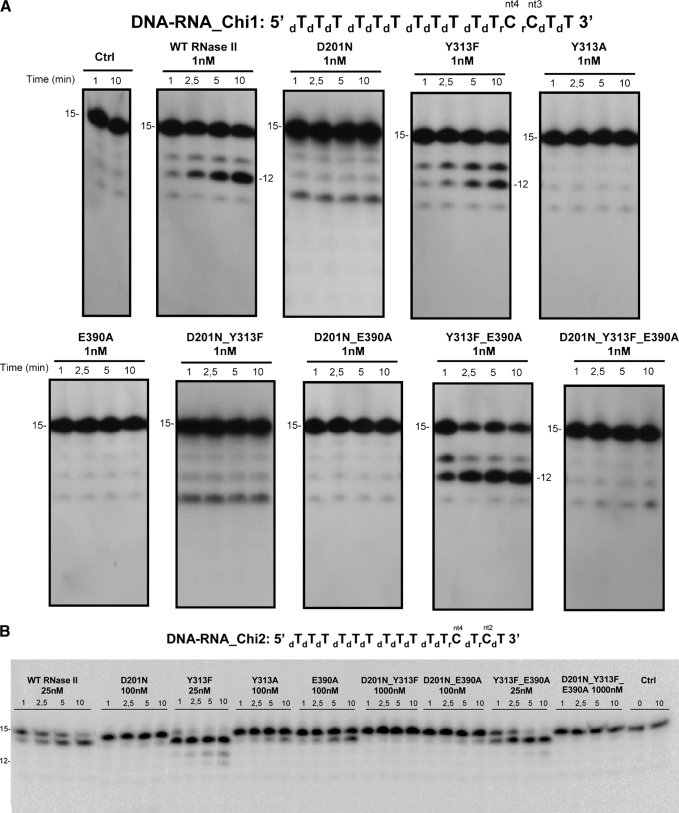
FIGURE 4.
Exoribonuclease activity with DNA-RNA chimeric substrates; comparison of wild-type and mutant proteins. Activity assays were performed using two 15-mer chimeric substrates as described under “Experimental Procedures.” A, activity assays with Chi 1 (5′-dTdTdTdTdTdTdTdTdTdTdTrCrCdTdT-3′) were the 3rd and 4th positions from the 3′-end are occupied by ribonucleotides (rC), and the other 13 are deoxyribonucleotides (dT). B, activity assays with Chi2 (5′-dTdTdTdTdTdTdTdTdTdTdTrCdTrCdT-3′) were the 2nd and 4th positions from the 3′-end are occupied by ribonucleotides, and the other 13 are deoxyribonucleotides. The mutants used and their respective protein concentrations are shown. The wild-type enzyme was used as control. Samples were taken during the reaction at the time points indicated, and reaction products were analyzed in a 20% polyacrylamide, 7 m urea gel. Control reactions with no enzyme added (Ctrl) were incubated at the maximum reaction time for each protein. Length of substrates and degradation products are indicated in the figure.
TABLE 1.
Specific exoribonucleolytic activity of wild-type and mutant enzymes
Exoribonucleolytic activity was assayed using a 35-nt poly(A) chain as substrate. Activity assays were performed in triplicate as described under “Experimental Procedures.” Each value represents nmol of substrate oligoribonucleotide consumed per min and per nmol of protein, and the exoribonucleolytic activity of the wild-type enzyme was taken as 100%.
| Protein | Protein activity | Relative activity |
|---|---|---|
| nmol min−1nmol−1 | % | |
| WT RNase II | 0.30 ± 0.04 | 100 |
| Y313F | 0.35 ± 0.04 | 117 |
| Y313A | <0.01 | 1 |
| E390A | 0.36 ± 0.03 | 120 |
| D201N | <0.01 | 0.2 |
| D201N/Y313F | <0.01 | <0.1 |
| D201N/E390A | 0.31 ± 0.06 | 103 |
| Y313F/E390A | 0.33 ± 0.01 | 110 |
| D210N/Y313F/E390A | ≪0.01 | ≪0.1 |
| E542A | 33.75 ± 3.90 | 11,250 |
| R500A | ≪0.01 | ≪0.1 |
Formal Kinetic Analysis
To determine the 3′-5′ exoribonuclease rates of the WT and the E542A mutant, the reactions were carried out in 20 mm Tris-HCl, pH 8, 100 mm KCl, 1 mm MgCl2, and 1 mm dithiothreitol using the 35-nt poly(A) RNA oligonucleotide as a substrate. Preliminary assays (not shown) showed that product formation proceeded linearly at the protein concentrations and for the incubation times used in the experiments reported here. As such, the protein activities were measured at the following substrate concentrations: 10, 25, 50, 100, 150, 200, 250 and 300 nm. The protein concentrations used were 5 pm and 0.1 fm for the WT and the E542A mutant, respectively. The reactions were carried out at 37 °C for 2 min with these protein concentrations as under these conditions less than 20% of substrate was degraded. Reactions were stopped by adding formamide-containing dye supplemented with 10 mm EDTA. Reaction products were resolved in a 20% polyacrylamide, 7 m urea and analyzed by autoradiography. Autoradiograms were scanned, and the densities of the bands were quantified using ImageQuant 5.0 software. The amount of product was calculated by using the original substrate concentration and the final ratio of remaining substrate and product formed. Lineweaver-Burk plots (32) were used to estimate kinetic parameters: Km, Vmax, and kcat. Each value obtained represents the mean of at least three independent assays, and the S.D. was calculated from these data sets.
Surface Plasmon Resonance Analysis-BIACORE
Biacore SA chips were obtained from Biacore Inc. (GE Healthcare). The flow cells of the SA streptavidin sensor chip were coated with a low concentration of the following substrates. On flow cell 1 no substrate was added so this cell could be used as the control blank cell. On flow cell 2 a 5′-biotinylated 25-nucleotide RNA oligomer (5′-CCCGACACCAACCACUAAAAAAAAA-3′) was added to allow the study of the protein interaction with a single-stranded RNA molecule. On flow cell 3 a 5′-biotinylated 16-nucleotide DNA oligomer (5′-AGTGGTTGGTGTCGGG-3′) was added to allow the study of the protein interaction with a single-stranded DNA molecule. On flow cell 4 a 5′-biotinylated 35-nucleotide poly(A) oligomer was added to allow the study of the protein interaction with a single-stranded RNA molecule composed only of adenosines. The target RNA and DNA substrates were immobilized on flow cells 2 and 3 by injecting 20 μl of a 500 nm solution of the target RNA or DNA in 1 m NaCl at a 10 μl/min flow rate as described in previous reports (31, 33). The biosensor assay was run at 4 °C in 20 mm Tris-HCl, pH 8, 100 mm KCl, 1 mm dithiothreitol, and 25 mm EDTA. The proteins were injected over the flow cells for 2 min at concentrations of 10, 20, 30, 40, and 50 nm using a flow rate of 20 μl/min. All experiments included triple injections of each protein concentration to determine the reproducibility of the signal and control injections to assess the stability of the RNA or DNA surface during the experiment. Bound protein was removed with a 60-s wash with 2 m NaCl, which did not damage the substrate surface. Data from flow cell 1 were used to correct for refractive index changes and nonspecific binding. Rate constants and equilibrium constants were calculated using the BIA EVALUATION 3.0 software package according to the fitting model 1:1 Languimir binding.
Modeling of Wild-type RNase II and E542A Mutant
Using standard comparative modeling methods and the software DeepView (34), three-dimensional models of wild-type E. coli RNase II (RNB/ECOLI) and E542A mutant proteins were built up from the x-ray structure (PDB code 2IX1 (22)) of RNase II D209N mutant complexed with a 13-nt poly(A) RNA.
Molecular Dynamics (MD) Simulation of the E542A Mutant and RNase II Models Based on the 2ix1 Structure
To obtain a fine-grained theoretical model of wild-type RNase II and E542A mutant enzymes bound to a 13-nt poly(A) RNA, a 4-ns MD simulation was performed. To readjust side chains and domain conformations, the recreation was calculated using the PMEMD module of the AMBER9 package (35, 36) and the parm99 parameter set from this distribution. Dynamically stabilized 2IX1 crystal structure model of RNase II in complex with RNA described elsewhere (26) was taken into consideration for subsequent molecular dynamics trajectory analyses. The details on MD methodology applied to the models were described (26).
MD Models of the Wild-type RNase II and E542A Mutant Proteins Bound to RNA
Theoretical models of RNase II wild-type and E542A mutant enzymes bound to a 13-nt poly(A) RNA were, respectively, built up from an energy-minimized average structure of the stabilized molecular dynamics simulation of the x-ray structure of RNase II D209N mutant complexed with a 13-nt poly(A) RNA (PDB code 2IX1 (22)), previously published (26). To obtain biophysically consistent models, 4-ns MD simulations were performed with each model. Calculations were done by using the PMEMD module and the parm99 parameter set in the AMBER9 package (35, 37, 38). MD simulations of both RNase II systems included the four RNase II domains (CSD1, CSD2, RNB, and S1), the RNA molecule, and the magnesium atom of the active center reported in the real structure of D209N mutant. To neutralize the electrostatic charge of the system, Na+ counterions were placed in a shell around the system using a grid of coulombic potentials. Electrostatically neutralized complexes where then embedded in a truncated octahedron solvation box, keeping a distance of 12 Å between the limits of the box and the closest atom of the solute. Both counterions and solvent were added using the LEAP module in AMBER9. Initial relaxation of each complex was completed by performing 10,000 steps of energy minimization with a cut-off of 10.0 Å. Initial heating and equilibration were performed simultaneously by raising the temperature in two stages; a first one from 0 to 5 K to induce a slow adaptation of the system to the force field represented in parm99 and a second up to 298 K in 200-ps continuous heating phases. During this procedure stage velocities were reassigned at each new temperature according to the Maxwell-Boltzmann distribution, and positions of the Cα and P atoms of the solute were constrained with a force constant of 20 Kcal·mol−1 to impede spurious disorganizations of protein and RNA backbone structures. During the last 100 ps of the equilibration phase of the MD, the force constant was reduced stepwise down to 0 for all constrained atoms to progressively allow the stabilization of the system. Consecutively, a MD simulation of 4 ns over the complete systems was completed.
RESULTS
Comparing the Exoribonucleolytic Activity of the Different Mutant Proteins
E. coli RNase II is the prototype of the RNase II superfamily of exoribonucleases, and this protein has shown to be a good study model for all family members, as they share a similar mode of action (15, 26, 39–41). To better understand the reaction mechanism of E. coli RNase II and the processes underlying the specificity of RNA cleavage, we constructed a set of single, double, and triple mutants of RNase II and studied their effect in RNA binding and in catalytic activity. We introduced the following point mutations in RNase II: Y313F, Y313A, E390A, R500A, R500K, and E542A, the double mutations D201N/Y313F, D201N/E390A, and Y313F/E390A, and the triple mutation D201N/Y313F/E390A. All these mutant proteins were introduced in E. coli BL21(DE3) and were overproduced by isopropyl 1-thio-β-d-galactopyranoside induction. The optimal induction conditions were standardized for each mutant by analysis of protein production and solubility at different times of isopropyl 1-thio-β-d-galactopyranoside treatment. All the mutants were shown to be more than 80% soluble after induction, except for R500K, which was mainly in the insoluble fraction (data not shown). All the mutant proteins, with the exception of R500K were, purified (Fig. 1C) as described under “Experimental Procedures.” The exoribonucleolytic activity of the purified mutant proteins was analyzed and compared with the wild type by performing activity assays using two different types of RNA substrates as described under “Experimental Procedures.”
As previously demonstrated, the wild-type enzyme was able to degrade the single-stranded RNA substrate, and the end-product was a 4-nt fragment (26, 42) (Fig. 2). With the R500A mutant the shortest product observed was a 11 nt, even with 1000 nm concentration after a 30-min reaction. The double mutant D201N/Y313F was only able to degrade a few nucleotides in the conditions tested (1000 nm concentration of protein in a 30-min reaction). Previous reports had shown that D201N mutant accumulated a 10–11-nt fragment as a major degradation product, although longer reaction times resulted in the usual 4-nt fragment as a secondary product (26). This is probably because of a higher dissociation rate of RNA fragments shorter than 11 nt. A similar behavior was also observed for the double D201/E390A and triple D201N/Y313F/E390A mutants, which confirm the important role played by Asp-201 in the RNase II activity. Y313F, Y313A, Y313F/E390A, and E542A generated a 4-nt fragment like wild-type RNase II (Fig. 2). In the conditions tested (1 nm concentrations of protein in a 10-min reaction), the E390A mutant was able to degrade the substrate until it reached a 6-nt fragment (Fig. 2). However, in more extreme conditions (i.e. longer reaction times and/or higher enzyme concentrations) the mutant was also able to reach the typical final end-product of 4 nt (data no shown).
When the substrate tested was a double-stranded molecule with a 3′-single-stranded extension (16–30ds), the wild-type enzyme degraded the single-stranded portion, generating 23–25-nt oligomers (30). This shows that the enzyme stalls 7–9 nt before reaching the double-stranded region (30, 42) (Fig. 3). Similarly, most of the mutant enzymes rendered products ranging between 21 and 25 nt in length behaving like the wild-type does against secondary structures. The D201N, D201N/E390A, and R500A mutants, however, generated longer products than the wild type, with 26–27 nt in length. The E542A mutant was able to go closer to the double-stranded portion of the RNA, rendering a 19-nt product.
The exoribonucleolytic activity of wild-type and mutant enzymes was determined by measuring the substrate disappearance from the activity gels (Table 1). The results obtained suggested that although fully conserved in all domains of life, Glu-390 is not essential for RNase II catalysis because the activity of E390A was very similar to that of the wild-type enzyme (0.30 and 0.36 nmol·min−1·nmol−1 for the wild-type and E390A mutant, respectively) (Table 1). In the case of Tyr-313, its substitution by Phe did not alter the enzymatic activity, whereas its replacement by an Ala resulted in a 100-fold reduction (Table 1). These observations imply that the aromatic moiety of Tyr-313, but not the hydroxyl group, is important for the maintenance of the enzymatic activity. Accordingly, the exoribonucleolytic activity of the double mutant Y313F/E390A was not affected by the presence of the two mutations simultaneously (Table 1). As previously reported, the activity of D201N was highly impaired (only 0.2% of the activity of wild-type enzyme), confirming the importance of this residue in RNase II activity (26). This effect was also reflected in the D201N/Y313F and the D201N/Y313F/E390A mutants, which showed less than 0.1% of the activity present in the wild type (Table 1). However, the D201N mutation did not have such a pronounced effect on the activity of the double mutant D201N/E390A, which had a very similar specific activity to the wild type (0.30 and 0.31 nmol·min−1·nmol−1 for the wild type and D201N/E390A double mutant, respectively) (Table 1). This mutant was able to degrade the poly(A) substrate in a very processive manner until generating the 10-nt oligomer, as does the wild-type enzyme. However, further degradation of this 10-nt oligomer into shorter products seemed to occur with much more difficulty, and higher concentrations of enzyme were required to generate the typical 4-nt product. One explanation for the results obtained is the possible reconstitution of the catalytic site around the Mg2+ atom and the D201N, induced by the E390A mutation. The electrostatic environment in the catalytic center and in the cleft has to be preserved for the enzyme to continue to function; as such, an uncertain number of casual rearrangements might have taken place.
The Arg-500 residue seems to be very important for the RNase II activity, as the R500A mutant showed more than a 40,000-fold reduction in activity when compared with the wild type (7.0 × 10−6 versus 0.30 nmol·min−1·nmol−1) (Table 1). In fact, the side chain of this residue interacts with the phosphate backbone of the two nucleotides at the 3′-end of the RNA molecule, and it was postulated to assist in catalysis by fixing the phosphodiester bond at the cleavage position and enhancing the susceptibility of the leaving phosphorous atom to a nucleophilic attack (22). Its substitution by Ala would, therefore, prevent such an interaction, resulting in the inactivation of the enzyme.
Finally, the substitution of the Glu-542 by Ala rendered a mutant version of the RNase II that was much more active than the wild-type enzyme, with more than a 100-fold increase in the exoribonucleolytic activity. This residue is in close proximity to the leaving nucleotide and was suggested to facilitate the elimination of this nucleotide upon phosphor-ester cleavage by pulling it out of the base-stacked position (22). The elimination of the cleaved nucleotide from the catalytic cavity is essential for the enzymatic process to continue, and an alanine residue in this position seems to facilitate this process even more.
Determination of RNA Dissociation Constants (KD) by Surface Plasmon Resonance Analysis
To determine the contribution of each residue in RNA binding, we calculated the dissociation constants (KD) of the wild-type and the mutant proteins by surface plasmon resonance analysis with Biacore 2000 using two different single-stranded RNA substrates as described under “Experimental Procedures.” The results obtained are presented in Table 2. The dissociation constants for the 25-mer single-stranded RNA substrate of the wild-type RNase II and D201N enzymes were previously determined (6.5 ± 0.4 and 11.4 ± 0.7 nm, respectively) (26). When assaying the 25-mer single-stranded RNA substrate, almost all the mutant proteins tested in this work showed KD values similar to that of the wild-type enzyme, except for the E542A mutant. This mutant enzyme presented a significant increase in RNA binding affinity with a much lower KD value when compared with the wild type (0.5 ± 0.08 versus 6.5 ± 0.4 nm) (Table 2). When a poly(A) substrate was used almost all enzymes showed a moderate reduction on KD values, indicating a higher affinity for this substrate. When compared with the wild type, most of the mutant enzymes showed no significant differences in KD values except for the triple mutant, which showed a 9-fold reduction in poly(A) binding affinity. Once more, the E542A mutant presented a significant increase in RNA affinity for poly(A), with a ∼20-fold higher KD value when compared with that of the wild type (Table 2). The same result was obtained by electrophoretic mobility shift assay, with this mutant showing RNA-protein complex formation at much lower protein concentration than wild-type enzyme (supplemental Fig. S1). This means that the high level of activity of this mutant is, at least in part, because of a higher RNA binding affinity than the wild type.
TABLE 2.
RNA binding affinity of wild-type and mutant enzymes
The dissociation constants (KD) were determined by surface plasmon resonance using BIACORE 2000 with a 25-nt RNA oligomer (5′-Biotin-CCC GAC ACC AAC CAC UAA AAA AAA A-3′) and 35-nt poly(A) RNA oligomer.
| Proteins | 25-mer ssRNA |
Poly(A) ssRNA |
||
|---|---|---|---|---|
| KD | Relative KD | KD | Relative KD | |
| nm | nm | |||
| WT RNase II | 6.5 ± 0.4 | 1.0 | 1.3 ± 0.4 | 1.0 |
| D201N | 11.4 ± 0.7 | 1.8 | 1.1 ± 0.1 | 0.8 |
| Y313F | 12.9 ± 2.4 | 2.0 | 4.4 ± 0.1 | 3.4 |
| Y313A | 17.1 ± 0.8 | 2.6 | 4.2 ± 0.7 | 3.2 |
| E390A | 8.7 ± 1.4 | 1.3 | 2.3 ± 0.4 | 1.8 |
| D201N/Y313F | 3.4 ± 0.3 | 0.5 | 4.4 ± 0.6 | 3.4 |
| D201N/E390A | 17.8 ± 1.7 | 2.7 | 1.6 ± 0.1 | 1.2 |
| Y313F/E390A | 7.3 ± 0.3 | 1.1 | 6.0 ± 0.7 | 4.6 |
| D201N/Y313F/E390A | 22.1 ± 7.2 | 3.4 | 12.0 ± 2.9 | 9.2 |
| R500A | 10.9 ± 1.2 | 1.7 | 3.3 ± 0.6 | 2.5 |
| E542A | 0.5 ± 0.08 | 0.1 | 0.06 ± 0.005 | 0.05 |
Kinetic Analyses of Wild-type and E542A Mutant
Given the extraordinary properties of the E542A mutant, we wanted to better characterize its catalytic properties. With this aim, we performed a kinetic analysis of this E542A mutant and the wild-type enzyme and determined their kinetic parameters. The experiments were carried out using the poly(A) RNA substrate as described under “Experimental Procedures.” The results obtained were plotted, and the reaction proceeded according to a Michaelis-Menten equation, as indicated by linearity in the Lineweaver-Burk plots (32) (data not shown). From this plot we determined the kinetic parameters, Vmax, Km, and kcat for the wild type and the E542A mutant, which are presented in Table 3. The Km values obtained were 1.25 ± 0.17 μm for the wild type and 0.30 ± 0.05 μm for E542A (Table 3), suggesting that the E542A mutation significantly increased the affinity of RNase II enzyme for the poly(A) substrate and confirming the data obtained from the KD values. In addition, the kcat value of the E542A mutant was also much higher than that of the wild type (∼200,000-fold), thus giving a final kcat/Km ratio of ∼1,000,000-fold higher in the E542A mutant than in the wild type (Table 3). All these data explain why the E542A mutant is much more efficient in catalysis than the wild-type enzyme, confirming that, in fact, substitution of Glu-542 by Ala resulted in a super-enzyme.
TABLE 3.
3′-5′-Exoribonuclease kinetic constants of wild-type and E542A mutant enzymes
The exoribonuclease rates were measured at different substrate concentrations using a 35-nt poly(A) chain as substrate, as indicated under “Experimental Procedures.”
| Proteins | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm.s−1 | μm | s−1 | μm−1.s−1 | |
| WT RNase II | 0.25 ± 0.02 | 1.25 ± 0.17 | 0.41 ± 0.01 | 0.23 ± 0.06 |
| E542A | (1.14 ± 0.48) × 104 | 0.30 ± 0.05 | (8.08 ± 0.84) × 104 | (2.36 ± 0.38) × 105 |
Can the Discrimination of RNA Versus DNA by RNase II Be Dictated by Three Conserved Residues?
It was previously described that residues Asp-201, Tyr-313, and Glu-390 are involved in ribose binding (22); therefore, they are probably important for the specificity of RNA cleavage. Particularly and starting from the 3′-end of the RNA molecule, Tyr-313 and Asp-201 interact with the O2′-ribose oxygen of the second nucleotide, and Glu-390 interacts with the O2′-ribose oxygen of the fourth nucleotide (Fig. 1B). To determine the role of these residues in the discrimination of RNA versus DNA, the activity of these mutants was assayed using two different 15-nt chimeric DNA-RNA oligomers. In the first chimeric substrate (Chi1), the 3rd and 4th positions from the 3′-end are occupied by ribonucleotides (rC) and the other 13 are deoxyribonucleotides (dT) (5′-dTdTdTdTdTdTdTdTdTdTdTrCrCdTdT-3′), whereas in the second substrate (Chi2) the ribonucleotides are located in the 2nd and 4th positions from the 3′-end (5′-dTdTdTdTdTdTdTdTdTdTdTrCdTrCdT-3′). Our data showed that RNase II is only able to cleave DNA bases when having a ribose in the 2nd or the 4th positions. The results demonstrate that, with the Chi1 substrate, wild-type RNase II was only able to cleave three nucleotides, rendering a final degradation product of 12 nt (Fig. 4A). After the first cleavage event the RNA translocates, and the riboses now occupy the 2nd and 3rd positions. Because one of the riboses is still in the 2nd position, cleavage can pursue. The riboses then move to positions 1 and 2, and cleavage is still allowed. Finally, once there is only one ribose present, in the 1st position, cleavage halts. This explains why after three cleavage events the 12-nt fragment is released. With the Chi2 substrate only one cleavage event occurred with wild-type RNase II, converting the 15-mer into a 14-mer oligonucleotide (Fig. 4B and supplemental Fig. S2). After this first and unique cleavage event, the RNA translocates, the two riboses then occupy the 1st and the 3rd positions, and the enzyme is not able to continue degradation. Therefore, RNase II has a strict requirement for a ribose in the second and/or the fourth nucleotides from the 3′-end of the molecule and not in other positions, as previously described (43). This fact confirms that the specific contacts observed in the RNase II structure with the O2′ oxygen at these positions are essential for RNase II activity.
The substitution of the Asp-201 by Asn inhibited the degradation activity with both DNA-RNA substrates used (Fig. 4). However, this mutant has also been shown to be highly inactive in degradation of a poly(A) RNA substrate (with 0.2% of the wild-type activity) because of the role of Asp-201 in Mg2+ coordination (22, 26) (Fig. 2 and Table 1). Its role as an Mg2+ ligand at the active site of RNase II is so critical for the activity of the enzyme that we are unable to see the actual contribution of this residue in the interaction with the second ribose. The E390A mutant presented a similar degradation efficiency over an RNA substrate, like that of the wild type. However, this mutant was not able to degrade the Chi1 DNA-RNA substrate (Fig. 4A). The substitution of Glu-390 by Ala, which probably prevents the interaction with the 4th ribose, abolished the activity, confirming that the presence of a ribonucleotide in the 3rd position cannot support activity. With the Chi2 substrate, the E390A behaved like the wild type, although with less efficiency (Fig. 4B). Therefore, it seems that the single establishment of contacts with the ribose of 2nd nucleotide is enough to support catalysis to a certain extent. The importance of contacts between protein residues and the ribose of the 2nd nucleotide of the substrate for RNA recognition and cleavage was confirmed by mutations introduced in Tyr-313. Based on the crystal structure, the side chain of this residue is hydrogen-bonded to the O2′ oxygen of ribose of the 2nd nucleotide. Its substitution by Ala prevented the degradation of the Chi1 substrate, and the ability of the enzyme to degrade the Chi2 substrate was highly reduced, even at high protein concentrations (Fig. 4). This reduction seems to be similar to that observed with the poly(A) RNA substrate (Table 1). However, Y313F was shown to be highly active in degradation of both chimeric substrates (Fig. 4), even more active than the wild type over the Chi2. Furthermore, with this second substrate, Y313F was able to perform more than one cleavage event, behavior that was also observed in the Y313F/E390A double mutant (Fig. 4B and supplemental Fig. S2). Therefore, it seems that the absence of the side-chain hydroxyl group in Phe-313 could favor the degradation of certain substrates. The presence of a Phe instead of a Tyr may induce a local rearrangement of the nucleotides and/or the protein residues at the catalytic cavity, thus allowing the establishment of new contacts with a ribose in the 3rd or the 1st positions of the substrate. Such contacts, which are not present in the wild type, allow the enzyme to proceed in degradation of the Chi2 substrate even in the absence of the canonical interactions with the 2nd or the 4th riboses.
Determination of DNA Dissociation Constants (KD) by Surface Plasmon Resonance Analysis
We wanted to explore the effect of these residues, Asp-201, Tyr-313, and Glu-390, in general DNA binding affinity. For this purpose we determined the KD values by surface plasmon resonance using a DNA substrate. The results obtained (Table 4) showed that all mutants tested have a similar DNA affinity as the wild type. with the exceptions of E390A and Y313A derivatives, which showed a ∼9-fold and a >100-fold reduction, respectively (Table 4). These results confirm that the contact of Tyr-313 and Glu-390 with the 2nd and 4th riboses are important not only for catalysis but also for a proper substrate binding at the catalytic cavity in the absence of the canonical interactions.
TABLE 4.
DNA binding affinity of wild-type and mutant enzymes
The dissociation constants (KD) were determined by surface plasmon resonance using BIACORE 2000 with a 16-nt DNA oligomer (5′-Biotin-AGT GGT TGG TGT CGG G-3′).
| Proteins | 16-mer ssDNA |
|
|---|---|---|
| KD | Relative KD | |
| nm | ||
| WT RNase II | 8.7 ± 0.7 | 1.0 |
| D201N | 15.9 ± 2.4 | 1.8 |
| Y313F | 6.9 ± 0.3 | 0.8 |
| Y313A | ≫100 | ≫100 |
| E390A | 75.7 ± 8.8 | 8.7 |
| D201N/Y313F | 6.4 ± 0.5 | 0.7 |
| D201N/E390A | 12.8 ± 2 | 1.5 |
| Y313F/E390A | 12.0 ± 0.4 | 1.4 |
| D201N/Y313F/E390A | 10.7 ± 0.3 | 1.2 |
| R500A | 14.2 ± 0.2 | 1.6 |
| E542A | 6.1 ± 0.3 | 0.7 |
Our data indicate that inside the cavity the unique specific contacts for ribose established by RNase II are those with the 2nd and 4th nucleotides from the 3′-end of the RNA molecule. Moreover, these contacts are necessary and sufficient for cleavage to occur, and therefore, they seem to be responsible for the RNA specificity versus DNA in RNase II.
DISCUSSION
E. coli RNase II is the model of the RNase II family of enzymes, whose homologues are present in all three domains of life (1, 11, 12, 44, 45). The resolution of the structure of E. coli RNase II in the RNA-free and bound complex constituted a significant breakthrough (22, 24). The structural study together with biochemical analysis helped to explain certain aspects of the enzyme activity and led to the proposal of a model for RNA degradation by RNase II that can be extrapolated to other family members (13, 15, 22, 26, 41). However, some essential features remain unknown.
The structure of the D209N mutant complexed with a 13-nt poly(A) oligomer revealed specific contacts of several residues at the active site with the RNA oligomer, with most of these residues highly conserved in RNase II-like enzymes of all domains of life (22). To identify the specific role of these amino acids and verify their precise function in RNase II activity, we introduced several single, double, and triple mutations in residues Asp-201, Tyr-313, Glu-390, Arg-500, and Glu-542 and studied the exoribonucleolytic activity and substrate binding ability of the corresponding mutant proteins.
The results obtained in this report revealed that, except for the Glu-542, none of the residues analyzed is crucial for RNA binding, as only slight differences in RNA binding affinity (KD values) were observed upon mutation (Table 2). However, the Glu-542 residue is very important in the prevention of the binding to the substrate, as its substitution by an Ala causes the protein to bind RNA more tightly than the wild-type enzyme. Moreover, our data demonstrate that, among the residues mutated in this study, Arg-500 plays a central role in catalysis. The R500A mutation practically inactivated the enzyme, although the protein was still able to bind RNA efficiently. A similar result had previously been obtained when mutating the conserved Asp-209 into an Asn (6, 26). Comparable results were obtained with the double mutant D201N/Y313F and the triple mutant D201N/Y313F/E390A, which also have their activities highly impaired.
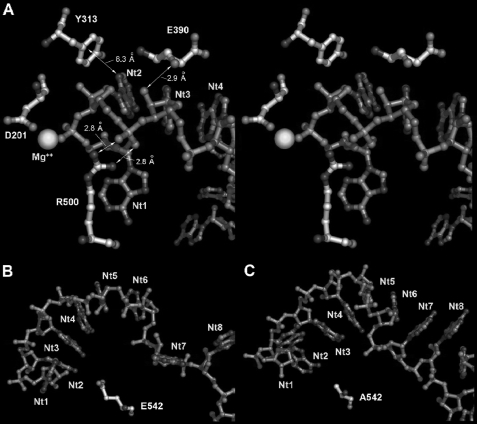
The results obtained with R500A support the essential role of this residue in assisting catalysis (22). Arg-500 has been described as interacting with the phosphate backbone of the two nucleotides at the 3′-end of the substrate (Fig. 1B). It was suggested that the role of Arg-500 could be to fix the phosphodiester bond at the cleavage position, enhancing the susceptibility of the phosphorous atom of the leaving nucleotide to a nucleophilic attack. To confirm our experimental results, we performed a computational model of the putative binding mode of a 13-nt poly(A) RNA fragment to RNase II wild-type enzyme based on the x-ray structure of RNase II D209N-RNA-bound complex (22). The model obtained gave us precious information about the interactions established between the RNA substrate and protein residues and helped to clarify certain aspects (Fig. 5A). For instance, the computational model predicts that in the wild-type enzyme, the guanidinium group in the Arg-500 residue would contact directly with the phosphate of nt 2 from the 3′-end, stabilizing its position. This conformation keeps the enzyme catalytically competent and allows the outgoing nucleotide to remain coordinated with the magnesium atom and the rest of the active center elements. Thus, the model shows that changes in this position could induce an extensive reorganization of the region that would turn the enzyme catalytically incompetent, which correlates with the experimental results obtained for mutant R500A and confirms the hypothesis previously postulated (22).
FIGURE 5.
Modeling of RNase II and E542A mutant proteins in RNA-bound conformations. To obtain biophysically consistent models, minimal energy average structures of both models were obtained from stabilized trajectories of 4-ns molecular dynamics. Figures in sticks were depicted with Pymol© (DeLano Scientific LLC, San Carlos, CA). A, stereo diagram of wild-type protein active center showing Glu-390, Tyr-313, Arg-500, and Asp-201 residues from the RNB domain. Significant polar contacts distances and relative positions of Mg2+ atom and outgoing RNA nucleotides (nt 1) are also represented. Hydrogen bonding between the Glu-390 backbone trace carbonyl oxygen and nt 3 ribose 2′-OH group could be responsible for RNA versus DNA recognition. Mutation to alanine would have introduced soft changes in local RNB conformation that could affect RNA discrimination in chimeric substrates. The guanidinium phosphate-hydrogen-bonding network between Arg-500 and nt 2 would essentially result in stabilizing the catalytic complex while keeping the outgoing nucleotide conformation correct. Catalytic competence of mutant R500A would result in being completely abolished. Long range hydrogen-π facial interactions between nucleotide nt 12 and Tyr-313 or Phe-313 mutant aromatic side chains could contribute to avoiding harmful interactions in the active center during procession and/or catalysis. B and C, modeled conformations of the poly(A) RNA strand in the RNB domain procession cleft in the wild-type and Ala-542 mutant RNase II proteins, respectively. As depicted in B, after 4 ns of MD simulation, the RNB domain of RNase II Ala-542 mutant could be able to deploy 6 stacked nucleotides (from nt 8 to 2) in progression, ready for cleavage. As shown in C, the same region in the wild-type model could only allocate 4 nucleotides (from nt 5 to 2) simultaneously in the cleft during a catalytic event. Substitution in position 542 of the negatively charged glutamic side chain for the smaller neutral methyl group of alanine could significantly reduce both electrostatic and steric surfaces in the RNA binding interface.
The RNase II structure showed that Tyr-313, Asp-201, and Glu-390 hydrogen-bonded with the O2′-ribose oxygen of two nucleotides in the RNA molecule (Fig. 1B). Because this oxygen is absent in a deoxyribose, the three residues were postulated to be involved in RNA discrimination versus DNA.
Previous studies showed that Asp-201 residue was very important for RNase II activity but not for substrate binding (26). In this report we performed new experiments with the previously constructed D201N mutant, and we demonstrated that this mutant enzyme was unable to degrade DNA-RNA chimeras. The computational model performed in this report helped to explain all experimental data obtained with the D201N mutant, showing that different groups of the aspartate participate in different contacts. The carboxylic of its side chain coordinates the Mg2+ ion that is essential for catalysis, and therefore, its substitution by asparagine led to the loss of activity. However, interaction with the O2′-ribose is mediated by polar contacts with the carbonyl oxygen in the backbone trace, indicating that any other residue could account for such interaction as long as the substrate and protein residue conformation is conserved. Therefore, although the actual contribution of Asp-201 to the RNA cleavage specificity cannot be deciphered with the D201N mutant because of its inactivation, computational model confirms the postulated contact between this residue and the 2nd ribose of the RNA molecule. According to our results Tyr-313 and Glu-390 seemed to be two essential contacts that RNase II establishes with the ribose of the substrate, demonstrating that these residues are responsible for the RNA cleavage specificity.
Substitution of Glu-390 by Ala caused a significant loss of activity over the chimeric substrates but did not affect degradation of a RNA molecule. The model predicts polar contacts between the ribose 2′-OH and the carbonyl oxygens of the glutamic. This means that its replacement by Ala should not destabilize the interaction. However, Glu-390 is also involved in polar contacts with neighboring arginine residues through its side chain. Therefore, E390A mutation will probably induce local conformational changes that may prevent the hydrogen bond formation between Ala (smaller residue than Glu) and the second ribose oxygen. The loss of this interaction would have a drastic effect in degradation of the chimeric substrate, where other canonical interactions may not be occurring. In the case of Tyr-313, our experimental results suggested that its aromatic moiety but not its hydroxyl group is crucial for activity and for RNA specificity of RNase II, as the Y313A but not Y313F mutation produced a drastic reduction of RNA and DNA-RNA substrate degradation efficiency. In fact, the computational modeling predicts contacts between the Tyr-313 aromatic ring and the OH of the ribose, but the hydroxyl group of the Tyr-313 does not participate in such interactions, confirming this hypothesis. In addition, our results demonstrate that the contact with Tyr-313 (or Phe) is critical for the degradation of DNA-RNA chimeric substrate and that this single contact is able to support catalysis of DNA bases. Therefore, the presence of a ribose in positions 2 or 4 from the 3′-end of the nucleic acid is the unique requirement for RNase II to perform degradation. RNA specificity shown by this ribonuclease resides in these two positions and interaction with Tyr-313 and Glu-390.
Finally, Glu-542 was shown to play a very important function in RNase II. The carboxylic group of this glutamate is in close proximity to the nitrogen atoms of the leaving nucleotide, and the establishment of one or more hydrogen-bonds between them could facilitate the elimination of this nucleotide after cleavage, thus allowing the degradation process to continue. Thus, Glu-542 seemed to be very important for the degradation to occur. Our experimental results demonstrate that its substitution by Ala resulted in a 110-fold increase in RNA degradation efficiency compared to the wild-type enzyme (Table 1) and a 10–20-fold increase in RNA affinity (Table 2). Moreover, the kinetic data suggest that this mutant not only has a much higher affinity for the poly(A) RNA substrate but also presents a much higher catalytic rate, confirming its role in the catalytic event. Such intriguing results led us to perform computational modeling with the E542A mutant enzyme with the RNA bound and compared it with the model of the wild type. The model predicts that substitution of the negatively charged glutamic acid side chain for a short quasi-apolar alanine residue seems to induce a subtle conformational change in the Cα backbone of the RNB domain itself that leads to a reorganization of its RNA binding interface (Fig. 5B). Comparing the E542A mutant and wild-type models in this region (Fig. 5B), the RNA bound to E542A mutant shows that nucleotides in positions ranging from 2 to 8 are set out in a stable stacked conformation. In comparison, the wild-type model presents only nucleotides 2–6 in a similar conformation. The efficiency of E542A mutant to deploy 50% more residues within the catalytic domain in the model could determine an increased capability of binding the RNA substrate by the RNB domain during catalysis than the RNase II wild type. In addition, the higher degree of organization and density of packing of the nucleotide chain in the E542A mutant could favor the RNA translocation upon cleavage, leading to higher degradation efficiency. These two aspects together may, therefore, be responsible for the high enzymatic activity experimentally determined of this mutant.
The results we present in this study substantially improve the RNase II model for RNA degradation. We have identified the residues that are responsible for the discrimination of cleavage of RNA versus DNA, which are fully conserved in all domains of life. We also show that the Arg-500 residue, present in RNase II active site, is crucial for activity but not for RNA binding. Finally, we report a very interesting mutant that acts as a super-enzyme, in which the substitution of Glu-542 by Ala leads to an outstanding catalytic efficiency, because of the high increase of both the exoribonucleolytic activity and substrate binding.
Because Rrp44/Dis3 protein (an RNase II homologue) is the only catalytically active nuclease in the exosome, the understanding of the degradation mechanism will have a large impact in future studies of the exosome. Moreover, the recent determination of the structure of yeast Rrp44 showed that E. coli RNase II is a good study model (39, 40). Also, yeast Rrp44/Dis3 has a similar linear arrangement of domains in the sequence when compared with E. coli RNase II. Both proteins share a high degree of identity as their conservation is the highest at the active site. This suggests that these two exoribonucleases share a similar hydrolytic mechanism (40). As such, the results from this report on RNase II mutants can be extrapolated for the comprehension of the mode of action of other members of the RNase II family.
Supplementary Material
Acknowledgments
We thank Ambro Van-Hoof for critical reading. We also thank to Biomol-Informatics SL for bioinformatics consulting.
The work was supported by Ministerio de Educación y Ciencia, Spain, Grant SAF2007-61926, an institutional grant from the “Fundación Ramón Arecesm,” and by Fundação para a Ciência e a Tecnologia, Portugal.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- RNase
- ribonuclease
- CSD
- cold shock domain
- RNB
- RNase II catalytic domain
- MD
- molecular dynamics
- WT
- wild type
- nt
- nucleotide(s).
REFERENCES
- 1.Andrade J. M., Pobre V., Silva I. J., Domingues S., Arraiano C. M. (2009) Prog. Mol. Biol. Transl. Sci. 85, 187–229 [DOI] [PubMed] [Google Scholar]
- 2.Gupta R. S., Kasai T., Schlessinger D. (1977) J. Biol. Chem. 252, 8945–8949 [PubMed] [Google Scholar]
- 3.McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. (1991) J. Mol. Biol. 221, 81–95 [PubMed] [Google Scholar]
- 4.Coburn G. A., Mackie G. A. (1996) J. Biol. Chem. 271, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 5.Marujo P. E., Hajnsdorf E., Le Derout J., Andrade R., Arraiano C. M., Régnier P. (2000) RNA 6, 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amblar M., Arraiano C. M. (2005) FEBS J. 272, 363–374 [DOI] [PubMed] [Google Scholar]
- 7.Deutscher M. P., Reuven N. B. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3277–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zilhão R., Cairrão F., Régnier P., Arraiano C. M. (1996) Mol. Microbiol. 20, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 9.Zilhão R., Camelo L., Arraiano C. M. (1993) Mol. Microbiol. 8, 43–51 [DOI] [PubMed] [Google Scholar]
- 10.Cairrão F., Chora A., Zilhão R., Carpousis J., Arraiano C. M. (2001) Mol. Microbiol. 276, 19172–19181 [DOI] [PubMed] [Google Scholar]
- 11.Mian I. S. (1997) Nucleic Acids Res. 25, 3187– 3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. (1997) Cell 91, 457–466 [DOI] [PubMed] [Google Scholar]
- 13.Dziembowski A., Lorentzen E., Conti E., Séraphin B. (2007) Nat. Struct. Mol. Biol. 14, 15–22 [DOI] [PubMed] [Google Scholar]
- 14.Liu Q., Greimann J. C., Lima C. D. (2006) Cell 127, 1223–1237 [DOI] [PubMed] [Google Scholar]
- 15.Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., Sanchez-Rotunno M., Arraiano C. M., van Hoof A. (2009) Nat. Struct. Mol. Biol. 16, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebreton A., Tomecki R., Dziembowski A., Séraphin B. (2008) Nature 456, 993–996 [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z. F., Deutscher M. P. (2005) Mol. Cell 17, 313–318 [DOI] [PubMed] [Google Scholar]
- 18.Cairrão F., Cruz A., Mori H., Arraiano C. M. (2003) Mol. Microbiol. 50, 1349–1360 [DOI] [PubMed] [Google Scholar]
- 19.Andrade J. M., Cairrão F., Arraiano C. M. (2006) Mol. Microbiol. 60, 219–228 [DOI] [PubMed] [Google Scholar]
- 20.Cairrão F., Arraiano C. M. (2006) Biochem. Biophys. Res. Commun. 343, 731–737 [DOI] [PubMed] [Google Scholar]
- 21.Cheng Z. F., Zuo Y., Li Z., Rudd K. E., Deutscher M. P. (1998) J. Biol. Chem. 273, 14077–14080 [DOI] [PubMed] [Google Scholar]
- 22.Frazão C., McVey C. E., Amblar M., Barbas A., Vonrhein C., Arraiano C. M., Carrondo M. A. (2006) Nature 443, 110–114 [DOI] [PubMed] [Google Scholar]
- 23.McVey C. E., Amblar M., Barbas A., Cairrão F., Coelho R., Romão C., Arraiano C. M., Carrondo M. A., Frazão C. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 684–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo Y., Vincent H. A., Zhang J., Wang Y., Deutscher M. P., Malhotra A. (2006) Mol. Cell 24, 149–156 [DOI] [PubMed] [Google Scholar]
- 25.Cannistraro V. J., Kennell D. (1994) J. Mol. Biol. 243, 930–943 [DOI] [PubMed] [Google Scholar]
- 26.Barbas A., Matos R. G., Amblar M., López-Viñas E., Gomez-Puertas P., Arraiano C. M. (2008) J. Biol. Chem. 283, 13070–13076 [DOI] [PubMed] [Google Scholar]
- 27.Taylor R. G., Walker D. C., McInnes R. R. (1993) Nucleic Acids Res. 21, 1677–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 29.Higuchi R. (1990) in PCR Protocols. A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. eds) Academic Press, Inc., Harcourt Brace Jovanovich, Publishers, San Diego, CA [Google Scholar]
- 30.Amblar M., Barbas A., Fialho A. M., Arraiano C. M. ( 2006) J. Mol. Biol. 360, 921– 933 [DOI] [PubMed] [Google Scholar]
- 31.Arraiano C. M., Barbas A., Amblar M. (2008) Methods Enzymol. 447, 131–160 [DOI] [PubMed] [Google Scholar]
- 32.Lineweaver H., Burk D. (1934) J. Am. Chem. Soc. 56, 658–666 [Google Scholar]
- 33.Park S., Myszka D. G., Yu M., Littler S. J., Laird-Offringa I. A. (2000) Mol. Cell. Biol. 20, 4765–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 35.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Case D. A., Darden T., Cheatham T. E., 3rd, Simmerling C., Wang J., Duke R. E., Luo R., Merz K. M., Jr., Wang B., Pearlman D. A., Crowley M., Brozell S., Tsui V., Gohlke H., Mongan J., Hornak V., Cui G., Beroza P., Schafmeister C., Caldwell J. W., Ross W. S., Kollman P. A. (2004) AMBER 8, University of California, San Francisco [Google Scholar]
- 37.Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., DeBolt S., Ferguson D., Seibel G., Kollman P. ( 1995) Comput. Phys. Commun. 91, 1– 41 [Google Scholar]
- 38.Case D. A., Darden T. E., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. (2006) AMBER 9, University of California, San Franscisco [Google Scholar]
- 39.Wang H. W., Wang J., Ding F., Callahan K., Bratkowski M. A., Butler J. S., Nogales E., Ke A. ( 2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16844– 16849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorentzen E., Basquin J., Tomecki R., Dziembowski A., Conti E. (2008) Mol. Cell 29, 717–728 [DOI] [PubMed] [Google Scholar]
- 41.Schneider C., Anderson J. T., Tollervey D. (2007) Mol. Cell 27, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amblar M., Barbas A., Gomez-Puertas P., Arraiano C. M. (2007) RNA 13, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cannistraro V. J., Kennell D. (2001) Methods Enzymol. 342, 309–330 [DOI] [PubMed] [Google Scholar]
- 44.Grossman D., van Hoof A. (2006) Nat. Struct. Mol. Biol. 13, 760–761 [DOI] [PubMed] [Google Scholar]
- 45.Zuo Y., Deutscher M. P. (2001) Nucleic Acids Res. 29, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.