Abstract
Hypoxia-inducible factor 1 (HIF-1) is a central regulator of the hypoxic response in many cell types. In endothelial cells, HIF-1 induces the expression of key proangiogenic factors to promote angiogenesis. Recent studies have identified Kruppel-like factor 2 (KLF2) as a potent inhibitor of angiogenesis. However, the role of KLF2 in regulating HIF-1 expression and function has not been evaluated. KLF2 expression was induced acutely by hypoxia in endothelial cells. Adenoviral overexpression of KLF2 inhibited hypoxia-induced expression of HIF-1α and its target genes such as interleukin 8, angiopoietin-2, and vascular endothelial growth factor in endothelial cells. Conversely, knockdown of KLF2 increased expression of HIF-1α and its targets. Furthermore, KLF2 inhibited hypoxia-induced endothelial tube formation, whereas endothelial cells from mice with haploinsufficiency of KLF2 showed increased tube formation in response to hypoxia. Consistent with this ex vivo observation, KLF2 heterozygous mice showed increased microvessel density in the brain. Mechanistically, KLF2 promoted HIF-1α degradation in a von Hippel-Lindau protein-independent but proteasome-dependent manner. Finally, KLF2 disrupted the interaction between HIF-1α and its chaperone Hsp90, suggesting that KLF2 promotes degradation of HIF-1α by affecting its folding and maturation. These observations identify KLF2 as a novel inhibitor of HIF-1α expression and function. Therefore, KLF2 may be a target for modulating the angiogenic response in disease states.
Hypoxia is an imbalance between oxygen supply and demand that occurs in a wide variety of diseases including ischemic cardiovascular disease and cancer (1). Hypoxia triggers the induction of an adaptive gene program that regulates critical processes such as angiogenesis, metabolism, and cell proliferation/survival in a variety of cell types and tissues. Hypoxia-inducible factor 1 (HIF-1)2 is a central regulator of hypoxia-mediated gene expression (2) that was originally identified as the transcription factor that mediates hypoxia-induced erythropoietin expression (3, 4). HIF-1 is a heterodimer that consists of a constitutively expressed HIF-1β subunit (also known as ARNT) and a HIF-1α subunit (5, 6). Under normoxic conditions, newly synthesized HIF-1α protein is rapidly degraded by the ubiquitin-proteasome system. The degradation of HIF-1α is mediated by its interactions with the von Hippel-Lindau (VHL) protein, a tumor suppressor that functions as an E3 ubiquitin ligase for HIF-1α (7, 8). The interaction between HIF-1α and VHL depends on the enzymatic hydroxylation of two prolyl residues (Pro402 and Pro564) in the oxygen degradation domain of HIF-1α (9, 10). During hypoxia, HIF-1α protein is stabilized, translocates into the nucleus, and dimerizes with HIF-1β and binds the hypoxic responsive elements on target promoters to initiate gene transcription.
Accumulating evidence has revealed that HIF-1 plays a critical role in hypoxia-mediated angiogenesis. Angiogenesis is a complex process that involves the coordinated interaction of multiple hypoxia-inducible genes (e.g. VEGF (11, 12), VEGF receptor (13), IL-8 (14, 15), and angiopoietin-2 (16, 17)) across numerous cell types.
The Sp/Kruppel-like factor (KLF) family of transcription factors is a subclass of the zinc finger family of transcriptional regulators implicated in the regulation of cellular growth and differentiation (18, 19). To date 21 members have been identified that include four Sp factors (Sp1–4) and 17 KLF factors (KLF1–17) (20, 21). Members of this family can bind with varying affinities to the same DNA sequences (termed GC box or CACCC element) and exert diverse transcriptional functions. Furthermore, members of this family can modulate each other's function through a number of distinct mechanisms, such as regulating each other's expression or through direct interaction (20, 22, 23). One member of this family, KLF2 (24), is strongly expressed in endothelial cells and is required for normal vessel formation (25). More recently, our group has provided evidence that KLF2 serves as a “molecular switch” in regulating endothelial function (26–28). These studies demonstrate that overexpression of KLF2 can potently attenuate the cytokine-mediated induction of pro-inflammatory targets such as vascular cellular adhesion molecule 1, E-selectin, and tissue factor while increasing anti-inflammatory targets such as endothelial nitric-oxide synthase and thrombomodullin (26–28). In addition, our group has demonstrated that KLF2 potently inhibits VEGF-A-mediated angiogenesis via a reduction in VEGF receptor 2 expression (29). However, the role of KLF2 in hypoxia-mediated signaling has been less well understood. In this study, we demonstrate that KLF2 is a novel inhibitor of HIF-1α expression/function and hypoxia-mediated angiogenesis.
EXPERIMENTAL PROCEDURES
Reagents
Common chemicals, solvents, and general reagents were obtained from Sigma. Human Hsp90 overexpression plasmid was purchased from OriGene. Human HIF-1α overexpression plasmid was kindly provided by Dr. Eric Huang (30). MG132, lactacystin, N-acetyl-leucyl-leucyl-norleucinal, and Z-Leu-Leu-CHO were purchased from Calbiochem. The adenoviral constructs were generated by the Harvard Gene Therapy Initiative.
Cell Culture and Hypoxia Treatment
Human vascular endothelial cells (HUVECs) and human microvascular endothelial cells (HMVECs) were purchased from Lonza. HUVECs and HMVECs were maintained with EBM-2 and supplemental growth factors (Lonza). For experiments, the cells were cultured in growth factor-starved medium and then were exposed to hypoxia for the indicated time. HUVECs of passages 2–4 and HMVECs of passages 4 and 5 were used in all experiments. RCC4 VHL+/+ and VHL−/− cells were kind gifts from Dr. Celeste Simon (University of Pennsylvania) and were maintained as previously described (31). Ts20 cells were kindly provided by Dr. Harvey Ozer (New Jersey Medical School) and were maintained as previously described (32). HCT116 p53+/+ and p53−/− cells were kindly provided by Dr. Bert Vogelstein (The Johns Hopkins University) and maintained as previously described (33). Primary mouse embryonic fibroblasts (MEFs) were kindly provided by Dr. Jerry Lingrel (University of Cincinnati) and were isolated from KLF2 wild-type and KLF2 null embryos at embryonic day 11.5 as described previously (34). Primary MEFs were immortalized by utilizing SV40 Large T antigen (kindly provided by Dr. Diana Ramirez, Case Western Reserve University). For hypoxic treatment, the cells were transferred to a MIC-101 modular incubator chamber (Billups-Rosenberg) that was flushed with 1% O2, 5% CO2, 94% N2, sealed, and placed at 37 °C for the indicated time.
siRNA-mediated Knockdown of KLF2
siRNA-mediated knockdown of KLF2 was performed as previously described (35). HUVECs were plated 1 day before transfection in antibiotic-free EBM-2 medium. On the day of transfection, 100 nmol/liter of specific siRNA targeting human KLF2 or nonspecific siRNA was incubated with Lipofectamine 2000 (Invitrogen) at room temperature for 30 min before it was added to the HUVECs in Opti-MEM (Invitrogen). 3 h later, the medium was replaced by growth factor-starved EBM-2 and cultured for an additional 48 h. The cells were exposed to hypoxia for the indicated time and harvested for experiments. Pooled siRNA (four individual duplex siRNA) against human KLF2 (catalog number M-006928-01), and the negative control-pool (catalog number D-001206-13-20) were products of Dharmacon. The sequences of siRNA are: 5′-GCACCGACGACCUCAA-3′, 5′-ACAUGAAACGGCACAUGUA-3′, 5′-UGCUGGAGGCCAAGCCAAA-3′, and 5′-ACCAAGAGUUCGCAUCUGA-3′. Pooled siRNA against human VHL and control siRNA were purchased from Santa Cruz.
Matrigel Angiogenesis Assay
All of the experiments were performed using growth factor-reduced Matrigel at a concentration of 1 mg/ml (BD Biosciences). 60 μl of Matrigel was added to each well of a 96-well plate on ice and then placed in a humidified incubator at 37 °C. HUVECs (2 × 104 cells/well) infected with adenovirus were added to the Matrigel-coated plates in a final volume of 200 μl. The cells were then exposed to hypoxia or normoxia for 24 h and photographed with a Leica EC3 photomicroscopic camera. Total tube length was determined by image analysis software (Image J; National Institutes of Health).
Primary Endothelial Cell Isolation
Mouse primary lung microvascular endothelial cells were isolated as previously described (36). For Matrigel tube formation assays, 1 × 105 of cells were plated on growth factor-reduced Matrigel (BD Biosciences) and were exposed to normoxia or hypoxia for 16 h. Quantification analysis was performed as described before.
Immunoprecipitation and Immunoblot Assays
For immunoprecipitation assays, 293T cells were transfected with the indicated expression plasmids (HIF-1α, Hsp90, and KLF2) using FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol. 48 h later, the cells were exposed to hypoxia for 4 h with treatment of MG132. The cells were lysed in 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.1% Nonidet P-40, 1 mm dithiothreitol, protease inhibitor mixture, sodium orthovanadate, and sodium fluoride. To stabilize Hsp90 protein interactions, 20 mm sodium molybdate was added. A total of 5 μg of antibody (anti-HIF-1α antibody, NB100–105; Novus Biological) was incubated with 1 mg whole cell lysates overnight at 4 °C followed by incubation with protein A/G-agarose beads (Santa Cruz) for 2 h at 4 °C. The beads were washed with phosphate-buffered saline four times, eluted in SDS sample buffer, and subjected to SDS-PAGE. For immunoblot assays, the cells were lysed in radioimmune precipitation assay buffer (Sigma) and centrifuged at 14,000 rpm for 10 min at 4 °C. The cellular lysates were subjected to SDS-PAGE followed by immunoblot assays. Nuclear protein was extracted by use of NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer's protocol (Pierce). For HIF-1α detection, either primary rabbit polyclonal anti-HIF-1α antibodies raised against the COOH-terminal peptide (kindly provided by Dr. Faton Agani, Case Western Reserve University) or BD Pharmingen's mouse monoclonal antibody was used, followed by secondary antibody detection with enhanced chemiluminescence (Amersham Biosciences). Anti-HIF-2α polyclonal antibody was purchased from Novus Biologicals. Anti-KLF2 polyclonal antibody was provided by Dr. Ng (Genome Institute of Singapore). Anti-VHL polyclonal antibody was purchased from Cell Signaling. Anti-Hsp90 monoclonal antibody was purchased from Stressgen. Anti-β-actin and anti-tubulin antibodies were purchased from Santa Cruz and Sigma, respectively.
Animals
KLF2 heterozygous mice (generously provided by Dr. Jeffry Leiden) were generated as previously described (25, 37). All of the animal studies were performed with littermate controls. Animal care and procedures were performed according to National Institutes of Health guidelines.
Brain Capillary Density
Immunocytochemical staining for glucose transporter 1 (GLUT1) was performed to assess brain microvascular density as previously described (38). Five-μm-thick sections were cut, deparaffinized, hydrated, and subjected to antigen retrieval at 90 °C for 10 min using 0.01 m sodium citrate buffer. Subsequently, sections were incubated with a solution of 5% normal rabbit serum and 0.3% Triton X-100 in phosphate-buffered saline, stained using goat polyclonal anti-GLUT1 antibody (1:200, Santa Cruz) and a biotinylated secondary antibody (Vector Laboratories). Color detection was carried out with the use of avidin-biotin horseradish peroxidase solution, ABC kit, and the diaminobenzidine peroxidase substrate kit (Vector Laboratories). Images from the cortex were taken with the Leica AS LMD V4 laser microdissection microscope with a 20× objective. Image J software was used to determine the number of GLUT1-positive capillaries/field. The obtained values from several animals were averaged, and the results were expressed, for each region, as the mean number of capillaries/field ± S.D.
ELISA
Human VEGF, IL-8, and angiopoietin-2 ELISA kits were purchased from R & D Systems. For human IL-8, angiopoietin-2, and VEGF, HUVECs were cultured in EBM-2 with 5% fetal bovine serum (no other growth factors) in 6-well plates and exposed to hypoxia for indicated times. The supernatants were harvested and assayed for VEGF, IL-8/CXCL8, and angiopoietin-2 by ELISA according to the manufacturer's instructions.
RNA Isolation, Northern Blot, and Quantitative Real Time Transcription PCR
Total RNA was extracted with TRIzol (Invitrogen) following the manufacturer's instructions. Equal amounts (10 μg) of total RNA were electrophoresed in 1.2% agarose-formaldehyde gels and transferred to nylon membranes. The membrane was then UV cross-linked, prehybridized with Quickhyb solution (Stratagene) containing salmon sperm DNA, and then hybridized for 1 h at 68 °C with random prime-labeled (Stratagene) DNA probes. Quantitative real time PCR was performed in a Mx3005P real time PCR system (Stratagene), using Brilliant SYBR Green QPCR Master Mix (Stratagene), according to the manufacturer's protocol. The specific primers used in these reactions are as follows: human prepro-adrenomedullin (forward 5′-AAGAAGTGGAATAAGTGGGCTCT-3′; reverse 5′-GGCCGAATAAGGGTCTGGG-3′), human GLUT1 (forward 5′-CTTTTCTGTTGGGGGCATGAT-3′; reverse 5′-CCGCAGTACACACCGATGAT-3′), human SDHA (forward 5′-TGGGAACAAGAGGGCATCTG-3′; reverse 5′-CCACCACTGCATCAAATTCATG-3′), human KLF2 (forward, 5′-TGCGGCAAGACCTACACCAAGAGT-3′; reverse, 5′-AGCCGCAGCCGTCCCAGTT-3′), human KLF4 (forward, 5′-ACCAGGCACTACCGTAAACACA-3′; reverse 5′-GGTCCGACCTGGAAAATGCT-3′), human KLF6 (forward 5′-GAGCCCTGCTATGTTTCACG-3′; reverse 5′-CGGATTCCTCCTTTTTCTCC-3′), mouse KLF2 (forward, 5′-ACCAAGAGCTCGCACCTAAA-3′; reverse, 5′-GTGGCACTGAAAGGGTCTGT-3′), mouse VEGF (forward, 5′-CCACGTCAGAGAGCAACATCA-3′; reverse 5′-TCATTCTCTCTATGTGCTGGCTTT-3′), and mouse 36B4 (forward, 5′-GCTCCAAGCAGATGCAGCA-3′; reverse 5′-CCGGATGTGAGGCAGCAG-3′). Relative levels of gene expression were normalized to the SDHA or 36B4 gene, which are not regulated by O2 tension (39, 40), using the comparative Ct method as previously described (41).
Statistics
All of the results are shown as the means ± S.D. Two-tailed, unpaired Student's t test was used for comparison of parameters. A p value of <0.05 was considered statistically significant.
RESULTS
Regulation of KLF2 by Hypoxia
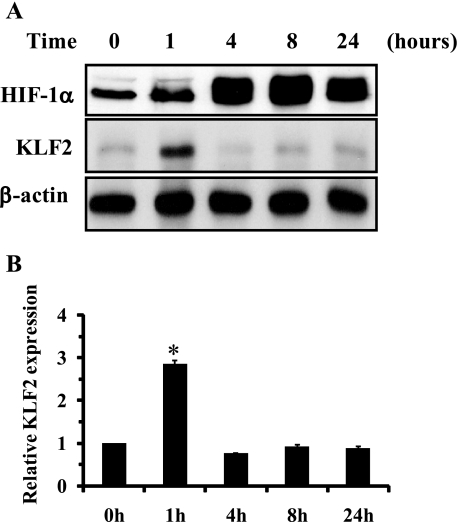
To understand the role of KLF2 in hypoxia-mediated angiogenesis, we first performed a time course study and assessed KLF2 regulation in endothelial cells. HUVECs were exposed to hypoxia (1% O2) for 1, 4, 8, and 24 h, and cell lysates were assessed for HIF-1α protein. As shown in Fig. 1, HIF-1α protein expression was induced at 1 h and peaked 8 h after hypoxic stimulation. KLF2 protein expression was transiently up-regulated by hypoxia at 1 h and subsequently returned to basal levels.
FIGURE 1.
A, regulation of KLF2 during hypoxia. HUVECs were exposed to hypoxia (1% O2) for the indicated time. The cell were harvested and followed by Western blot analysis as described under “Experimental Procedures.” A representative blot is shown of three independent experiments. B, densitometric analysis of KLF2 expression levels was performed. *, p < 0.01.
KLF2 Regulates Hypoxia-mediated Gene Induction
We next explored whether KLF2 can regulate the expression of the hypoxia-responsive gene using both overexpression and knockdown studies (supplemental Fig. S1, A and B) and assessed the effect of KLF2 on hypoxia-mediated gene induction. IL-8 is a potent proangiogenic factor that is regulated by hypoxia. To study the effect of KLF2 on IL-8 mRNA expression and secretion, HUVECs were infected with control (Ad-GFP) or KLF2 (Ad-K2) virus and subsequently exposed to normoxia (21% O2) or hypoxia (1% O2) for 24 h. The cells and culture media were harvested and subjected to Northern blot analysis and ELISA, respectively. As shown in Fig. 2A (top left panel), KLF2 overexpression inhibits both basal and hypoxia-induced IL-8 mRNA expression. Consistent with this observation, ELISA revealed that KLF2 inhibited IL-8 protein secretion from HUVECs (Fig. 2A, top right panel). Next, HUVECs were transfected with nonspecific or specific siRNA targeting human KLF2 (siK2) for 48 h followed by treatment with hypoxia for an additional 24 h. Northern blot analysis revealed that knockdown of KLF2 enhanced hypoxia-induced IL-8 mRNA expression (Fig. 2A, bottom left panel) and secretion (Fig. 2A, bottom right panel). These observations suggest that KLF2 levels modulate expression of angiogenic factors during hypoxia. We then examined the effect of KLF2 on other hypoxia-regulated factors such as angiopoietin-2, VEGF, adrenomedullin, and GLUT1. As shown in Fig. 2 (B and C, left panels), KLF2 overexpression inhibited the hypoxia-mediated secretion of both angiopoietin-2 and VEGF. Conversely, knockdown of KLF2 in HUVECs resulted in enhanced secretion of angiopoietin-2 and VEGF in response to hypoxia (Fig. 2, B and C, right panels). Real time PCR revealed that KLF2 inhibited hypoxic induction of prepro-adrenomedullin and GLUT1 mRNA expression in endothelial cells (Fig. 2, D and E, left panels). Conversely, knockdown of KLF2 resulted in enhanced mRNA levels of adrenomedullin and GLUT1 in response to hypoxia (Fig. 2, D and E, right panels). Taken together, these findings indicate that KLF2 negatively regulates expression of genes involved in both hypoxia-mediated angiogenesis and hypoxia adaptive metabolism.
FIGURE 2.
KLF2 regulates hypoxia-mediated gene induction. A, effect of KLF2 on IL-8 induction by hypoxia. HUVECs infected with control (Ad-GFP) or KLF2 (Ad-K2) virus were exposed to normoxia (21% O2) or hypoxia (1% O2) for 24 h. Cells and culture media were harvested and followed by Northern blot analysis and ELISA (n = 3), respectively (top left and top right panels). HUVECs were transfected with siRNA targeting human KLF2 (siK2) or nonspecific siRNA (NS). 48 h later, the cells were exposed to normoxia or hypoxia for an additional 24 h (bottom left and bottom right panels). The cells and supernatants were collected and followed by Northern blot and ELISA (n = 3), respectively. Representative Northern blots are shown of three independent experiments. B, effect of KLF2 on angiopoietin-2 secretion by hypoxia. HUVECs were treated as described above, and culture media were subjected to ELISA (n = 6). C, effect of KLF2 on VEGF secretion in response to hypoxia. HUVECs infected with indicated virus or transfected with indicated siRNA were exposed to hypoxia for 6 h. The supernatants were collected, and VEGF secretion levels were determined by ELISA (n = 6). D, effect of KLF2 on prepro-adrenomedullin mRNA expression. HUVECs infected with indicated virus or transfected with indicated siRNA were exposed to hypoxia for 24 h. The cells were harvested, and RNA was isolated followed by real time PCR (n = 3). E, effect of KLF2 on GLUT1 mRNA expression during hypoxia. HUVECs infected with indicated virus or transfected with indicated siRNA were exposed to hypoxia for 24 h. GLUT1 mRNA levels were assessed by real time PCR (n = 3). *, p < 0.01.
KLF2 Regulates Angiogenesis under Hypoxic Conditions
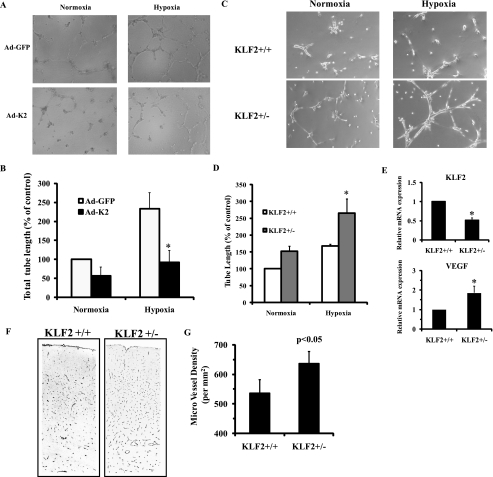
Because KLF2 inhibits the expression of hypoxia-inducible angiogenic genes, we next sought to study the effect of KLF2 levels on endothelial tube formation on Matrigel. HUVECs infected with control (Ad-GFP) or KLF2 (Ad-K2) virus were plated on Matrigel and subject to hypoxia (1% O2) for 24 h. As shown in Fig. 3(A and B), KLF2 overexpression significantly inhibited hypoxia-induced endothelial tube formation. Subsequently, we attempted to study the effect of siRNA-mediated knockdown of KLF2 on endothelial tube formation. However, we found knockdown of KLF2 (more than 80% knockdown) following exposure to hypoxia inhibited tube formation caused by loss of cellular viability, whereas knockdown of KLF2 did not increase cell death in regular tissue culture conditions. A significant body of evidence suggests that KLF2 is necessary for cell viability under pro-angiogenic conditions (25, 42, 43) and that significant reductions in KLF2 levels may render the endothelial cell incapable of tube formation under hypoxic conditions. Given this limitation with Matrigel tube formation assays using siRNA-mediated KLF2 knockdown, we next performed tube formation assays using primary lung microvascular endothelial cells from mice with haploinsufficiency of KLF2 (37). As shown in Fig. 3 (C–E), hemizygous deficiency of KLF2 resulted in increased VEGF expression and enhanced tube formation on Matrigel in response to hypoxia. To extend these observations in vivo, we next studied angiogenesis in KLF2 heterozygous mice. Because brain parenchyma is known to be a low oxygen microenvironment (PtO2 15–30 Torr) (44), we assessed numbers of microvessels in cerebral tissue sections from KLF2 heterozygous mice versus wild-type littermates. Consistent with our in vitro findings, KLF2 heterozygous mice showed increased numbers of microvessels in the cerebral cortex (Fig. 3, F and G), suggesting that KLF2 deficiency promotes hypoxia-mediated angiogenesis in vivo.
FIGURE 3.
Effect of KLF2 on hypoxia-mediated angiogenesis in vitro and in vivo. A, HUVECs were infected with indicated virus for 24 h. Then the cells were plated on Matrigel and were exposed to normoxia or hypoxia (1% O2) for 24 h as described under “Experimental Procedures.” Representative pictures of three independent experiments are shown. B, every well was divided into three different parts, and the length of tubes was measured in each part using Image J software (n = 6). *, p < 0.01. C, primary microvascular endothelial cells isolated from KLF2 wild-type (KLF2+/+) or KLF2 heterozygous (KLF2+/−) mice were plated on Matrigel and exposed to normoxia or hypoxia for 24 h. D, quantification analysis was performed as described under “Experimental Procedures” (n = 4). *, p < 0.01. E, KLF2 mRNA expression is down-regulated in KLF2+/− endothelial cells (top). VEGF mRNA expression is up-regulated in KLF2+/− endothelial cells (bottom). *, p < 0.01. KLF2 and VEGF mRNA levels in KLF2+/+ or KLF2+/− endothelial cells were determined by real time PCR (n = 4). F, KLF2 wild-type and KLF2 heterozygous (8 weeks old, littermates) mice were sacrificed, and the brains were harvested. Immunostaining for GLUT1 was performed in brain cortex. Representative pictures are shown in the left panel. G, quantification analysis was performed as described under “Experimental Procedures” (n = 3).
KLF2 Inhibits HIF-1α Accumulation in Response to Hypoxia
Because KLF2 regulates the hypoxia-mediated angiogenic response in endothelial cells, we next explored whether KLF2 has a role in regulating HIF-1α. Because we found a modest effect of KLF2 on HIF-1α mRNA expression in endothelial cells (data not shown), we hypothesized that KLF2 regulates HIF-1α levels mainly through affecting its protein stability. HUVECs infected with control (Ad-GFP) or KLF2 (Ad-K2) virus were exposed to hypoxia (1% O2) for 4 h, and lysates were subjected to Western blot analysis. As shown in Fig. 4A, KLF2 significantly blunted the hypoxia-induced accumulation of HIF-1α protein but did not alter HIF-2α (Fig. 4A) and HIF-1β expression (data not shown). Similarly, KLF2 inhibited HIF-1α but not HIF-2α protein expression in human microvascular endothelial cells (supplemental Fig. S2). We then tested the effects of KLF2 deficiency using both siRNA knockdown in HUVEC (Fig. 4B) and KLF2 null MEF cells (Fig. 4C and supplemental Fig. S1C). In these two settings, we observed enhanced HIF-1α and levels under both normoxia and hypoxia. Taken together, these data demonstrate that KLF2 directly regulates HIF-1α protein levels in cultured endothelial cells.
FIGURE 4.
Effect of KLF2 on HIF-1α protein expression. A, overexpression of KLF2 inhibits HIF-1α expression in response to hypoxia. HUVECs were infected with indicated virus and were exposed to normoxia or hypoxia (1% O2) for 4 h. The cell lysates were harvested and subjected to Western blot analysis. Representative blots of three independent experiments are shown. B, siRNA-mediated KLF2 knockdown enhances HIF-1α induction by hypoxia. HUVECs were transfected with indicated siRNA. 48 h later, the cells were stimulated with hypoxia (1% O2) for 2 h. Representative blots of three independent experiments are shown. C, deficiency of KLF2 accelerated HIF-1α accumulation in response to hypoxia. Immortalized MEFs isolated from KLF2 wild-type (WT) or KLF2 null (KO) embryos were exposed to hypoxia (1% O2) for 2 h. Nuclear extracts were isolated and followed by Western blot. Representative blots of three independent experiments are shown.
KLF2-mediated HIF-1α Degradation Is Independent of VHL and p53 but Mediated by the Proteasome System
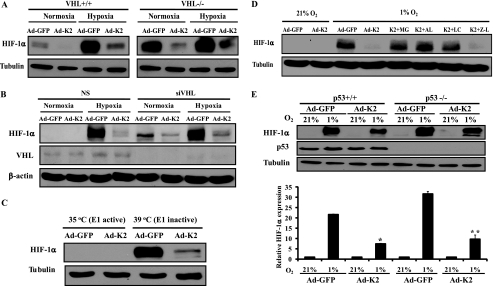
To further understand the mechanisms by which KLF2 decreases HIF-1α protein levels, we examined whether this effect is dependent on either VHL (an E3 ligase for HIF-1α) or the proteasome system. For the former, we used RCC4 VHL−/− cells, which express HIF-1α constitutively (31). As expected, KLF2 overexpression inhibited hypoxia-induced accumulation of HIF-1α in RCC4 VHL+/+ cells (Fig. 5A, left panel). Interestingly, KLF2 overexpression also inhibited HIF-1α protein expression in VHL−/− cells both under normoxic and hypoxic conditions (Fig. 5A, right panel). We next investigated whether KLF2 can promote HIF-1α protein degradation in the absence of VHL in endothelial cells. As expected, siRNA-mediated knockdown of VHL resulted in an induction of HIF-1α protein under normoxic conditions. Intriguingly, KLF2 inhibited HIF-1α protein expression in the absence of VHL under normoxic and hypoxic conditions (Fig. 5B). These data indicate that KLF2 promotes HIF-1α degradation in a VHL-independent fashion.
FIGURE 5.
HIF-1α degradation by KLF2 is VHL/p53-independent but mediated by the proteasome system. A, RCC4 VHL wild-type (+/+) or deficient (−/−) cells were infected with control (Ad-GFP) or KLF2 (Ad-K2) virus for 48 h. Then the cells were exposed to normoxia or hypoxia (1% O2) for 4 h. The cell lysates were extracted and subjected to Western blot. Representative blots of three independent experiments are shown. B, KLF2 inhibits HIF-1α protein expression in a VHL-independent fashion in endothelial cells. HUVECs transfected with siRNA against VHL were infected with indicated virus for 24 h. Then the cells were exposed to normoxia and hypoxia (1% O2) for 4 h. Representative blots of three independent experiments are shown. C, ubiquitin is not absolutely required for KLF2-mediated HIF-1α degradation. Thermo-sensitive E1-deficient cells (Ts 20 cells) were infected with indicated virus for 48 h and were cultured at indicated temperature for an additional 24 h. The cell lysates were subjected to Western blot analysis. Representative blots of three independent experiments are shown. D, proteasome inhibitors rescue KLF2-mediated HIF-1α degradation. HUVECs infected with indicated virus were exposed to normoxia or hypoxia (1% O2) for 4 h in the absence or presence of proteasome inhibitors (MG, MG132 5 μm; AL, N-acetyl-leucyl-leucyl-norleucinal 30 μm; LCT, lactacystin 2 μm). Z-Leu-Leu-CHO (Z-L, 50 μm) was used as a negative control. Representative blots of three independent experiments are shown. E, HCT116 p53 wild-type (+/+) or deficient (−/−) cells were infected with indicated virus for 48 h. The cells were exposed to normoxia or hypoxia (1% O2) for 4 h. The cell lysates were extracted and subjected to Western blot analysis. Representative blots of three independent experiments are shown (upper panel). Densitometric analysis of HIF-1α expression levels was performed (lower panel). *, p < 0.01 (compared with lane 2); **, p < 0.01 (compared with lane 6).
The rapid degradation of HIF-1α in normoxia requires VHL-mediated ubiquitination to target HIF-1α for proteasomal degradation. Given the ability of KLF2 to promote HIF-1α degradation independent of VHL, we next assessed whether this effect was dependent on ubiquitin conjugation itself (possibly by an alternative E3 ligase). We utilized Ts20 cells, which lose the ability to ubiquitinate proteins in the setting of hyperthermia because of the presence of a temperature-sensitive ubiquitin-activating enzyme, E1(32). In these cells, elevation of the temperature to 39 °C deactivates E1, leading to inhibition of ubiquitination and accumulation of HIF-1α, even under normoxic conditions. We adenovirally overexpressed KLF2 in Ts20 cells and cultured the cells at 35 °C (E1 active) and 39 °C (E1 inactive) under normoxia. Fig. 5C shows that in Ts20 cells cultured at 39 °C, HIF-1α was induced as expected. Interestingly, KLF2 inhibited HIF-1α induction in the absence of E1 enzyme activity. This indicates that the ubiquitin system is not an absolute requirement for the proteasomal degradation of HIF-1α by KLF2. We then explored the role of the proteasome system in KLF2-induced HIF-1α degradation by use of pharmacologic proteasome inhibitors. Fig. 5D shows that MG132 rescued HIF-1α degradation by KLF2. Furthermore, similar rescue was observed using highly specific proteasome inhibitors lactacystin and N-acetyl-leucyl-leucyl-norleucinal but not by Z-Leu-Leu-CHO (a calpain inhibitor).
Because p53 has also been implicated in the degradation of HIF-1α (45), we overexpressed KLF2 in HCT 116 p53+/+ and p53−/− cells. As shown in Fig. 5E, KLF2 inhibited hypoxic induction of HIF-1α protein in both p53+/+ and p53−/− cells, suggesting that KLF2 does not require p53 to promote HIF-1α degradation. Taken together, these findings indicate that KLF2-mediated HIF-1α degradation is independent of VHL, ubiquitin ligation, and p53 pathways but remains proteasome-dependent.
KLF2 Disrupts the Interaction Between HIF-1α and Hsp90
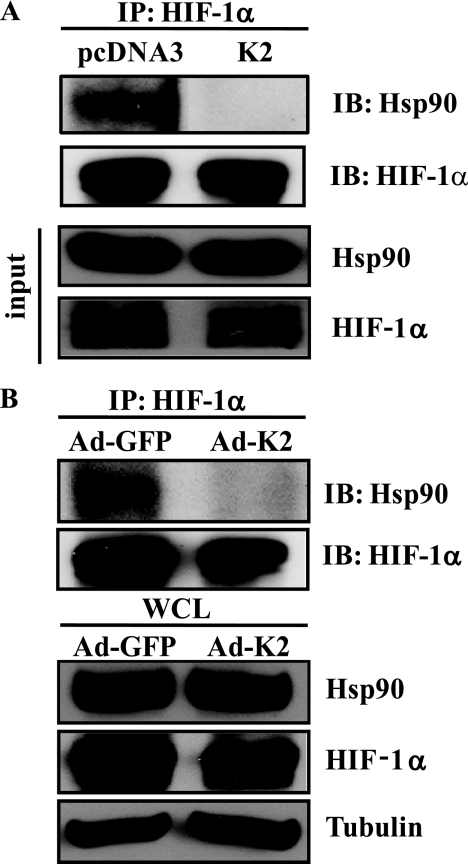
Newly synthesized HIF-1α protein is known to bind Hsp90 (46), which serves as HIF-1α chaperone. Because HIF-1α is a known Hsp90 client protein, we investigated the effect of KLF2 on the interaction between HIF-1α and Hsp90. We transfected 293T cells with expression plasmids for Hsp90, HIF-1α, and KLF2. The cells were exposed to hypoxia for 4 h with treatment of MG132 to reverse HIF-1α protein degradation by KLF2. Immunoprecipitation with anti-HIF-1α antibody and Western blot with an antibody against Hsp90 showed that KLF2 overexpression inhibited the ability of HIF-1α to form a complex with Hsp90 (Fig. 6A). Furthermore, we verified this effect on endogenous protein in RCC4 VHL-deficient cells. These cells were infected with control (Ad-GFP) or KLF2 (Ad-K2) virus and were exposed to hypoxia for 4 h in the absence of MG132. Endogenous HIF-1α was immunoprecipitated, and Western blotting with anti-Hsp90 antibody revealed that KLF2 overexpression also disrupted the endogenous HIF-1α/Hsp90 complex (Fig. 6B). These findings suggest that KLF2 promotes degradation of HIF-1α by disrupting its ability to interact with the Hsp90 chaperone complex, thereby affecting its folding and maturation.
FIGURE 6.
KLF2 disrupts the interaction between HIF-1α and Hsp90, a chaperone of HIF-1α. A, 293T cells were transfected with pcDNA3 or pcDNA3-KLF2 (K2) as well as Hsp90 and HIF-1 overexpression plasmid. 48 h later, the cells were stimulated by hypoxia (1% O2) for 6 h in the presence of MG132 (5 μm). The cell lysates were immunoprecipitated (IP) by anti-HIF-1α antibody. The precipitate was immunoblotted (IB) with anti-HIF-1α and anti-Hsp90 antibodies. B, RCC4 VHL deficient cells were infected with indicated virus for 48 h. Then the cells were exposed to hypoxia (1% O2) for 6 h in the absence of MG132 (5 μm). The cell lysates were immunoprecipitated by anti-HIF-1α antibody, and the precipitates were immunoblotted with anti-HIF-1α and anti-Hsp90 antibodies.
DISCUSSION
HIF-1α is a transcriptional regulator that is a central regulator of the cellular response to hypoxia. By inducing expression of angiogenic factors, it triggers the neovascularization of tissues under physiologic and pathologic conditions. In this study, we provide evidence that KLF2 is a novel regulator of HIF-1α expression and function.
KLF2 has been implicated as an anti-angiogenic factor by its ability to inhibit VEGF-mediated angiogenesis. However, the current study is the first to report the role of KLF2 in hypoxia-mediated angiogenesis and HIF-1α signaling. We found KLF2 is transiently up-regulated at 1 h after hypoxic treatment. Given the ability of KLF2 to inhibit HIF-1α expression and function, early induction of KLF2 by hypoxia may reflect an acutely protective effect of KLF2 against hypoxia and a potentially unchecked angiogenic response.
Tube formation assay demonstrated that KLF2 potently inhibits hypoxia-mediated angiogenesis. To determine the effect of systemic deficiency of KLF2 on angiogenesis during hypoxia, we studied brain capillary density in KLF2 wild-type and heterozygous mice because brain tissue represents a relatively low oxygen microenvironment (15–30 Torr) (44). Our observation of increased brain capillary density in KLF2 heterozygous mice indicates that KLF2 negatively regulates angiogenesis during hypoxia. These observations indicate that KLF2 is a negative regulator of hypoxia-mediated angiogenesis in vitro and in vivo.
HIF-1 activates gene expression through binding hypoxic responsive elements on target promoters. In addition to the HIF-1-dependent mechanisms, there are also HIF-1-independent mechanisms involved in hypoxia-mediated gene induction. For example, IL-8 is known to be activated through the NF-κB pathway as well as the HIF-1-dependent pathway under hypoxic conditions (15). We have previously demonstrated that KLF2 exerts NF-κB function through inhibiting co-activator recruitment (26, 47). As such, we speculate that KLF2 may exert its anti-angiogenic effects in both HIF-1α-dependent and HIF-1α-independent manners (e.g. by effects on NF-κB). Understanding how KLF2 modulates these parallel hypoxic signaling pathways and their relative roles in angiogenesis will be of significant interest.
VHL functions as an E3 ligase and plays an important role in HIF-1 degradation under normoxic conditions. Interestingly, KLF2 promoted HIF-1α degradation in a VHL-independent manner. In addition, HIF-1α degradation under normoxic conditions is regulated by Hsp90, a molecular chaperone that protects client proteins from misfolding and degradation. Hsp90 binds to the HIF-1α PAS domain, and the Hsp90 inhibitors geldanamycin and 17-allylaminogeldanamycin induce proteasomal degradation of HIF-1α even in VHL-deficient cells (48–51). In addition to Hsp90 inhibitors, several other factors promote VHL-independent HIF-1α degradation. Trichostatin A, FK506, and cyclosporin A have been shown to promote HIF-1α degradation in ubiquitin/VHL-independent pathway (52, 53). Furthermore, trichostatin A has been shown to disrupt the interaction between Hsp90 and HIF-1α (52). A recent report demonstrated that RACK1 (receptor of activated protein kinase C), a HIF-1α-interacting protein, promotes VHL-independent proteasomal degradation of HIF-1α by competing with Hsp90 for binding to HIF-1α (54). We tested whether KLF2 interacts with Hsp90 or HIF-1α but were unable to show clear interaction with KLF2 (data not shown), suggesting that KLF2 does not disrupt the interaction between HIF-1α and Hsp90 through competing with Hsp90 for binding HIF-1α. KLF2 may affect effectors of Hsp90 function because KLF2 does not alter Hsp90 expression and does not interact with Hsp90 itself (data not shown). Further studies will be interesting to elucidate the mechanism underlying this observation. p53 promotes Mdm2-mediated ubiquitination and proteasomal degradation (45). Our results in p53−/− cells and E1-deficient cells both suggest that KLF2 functions in a p53-independent manner and likely does not involve Mdm2 or other ubiquitin ligases. Given that a number of tumors are p53- or VHL-deficient, the observation that KLF2 can inhibit HIF-1α independent of these factors may provide novel insights into anti-angiogenic cancer therapy.
In summary, we demonstrate here for the first time that KLF2 is a novel inhibitor of HIF-1α and the angiogenic response. Given the central role of KLF2 in endothelial cell biology, further elucidation of its precise role in the molecular regulation of angiogenesis will be a critical step in the novel therapies for cardiovascular disease, cancer, and diabetes.
Supplementary Material
Acknowledgments
We thank Dr. Faton Agani (Case Western Reserve University) for providing anit-HIF-1α antibody. We also thank Drs. Celeste Simon (University of Pennsylvania), Harvey Ozer (New Jersey Medical School), Bert Vogelstein (The Johns Hopkins University), and Jerry Lingrel (University of Cincinnati) for providing RCC4 VHL-deficient cells, Ts20 cells, HCT116 p53-deficient cells, and KLF2 null MEF cells, respectively. We thank Constantinos Tsipis (the Laboratory of Experimental Stroke and Cerebrovascular Disease, Case Western Reserve University) for the brain capillary density studies.
This work was supported, in whole or in part, by National Institutes of Health Grants HL72952, HL75427, HL76754, HL086548, HL084154, and P01 HL048743 (to M. K. J.); HL087595 (to Z. L.); HL088740 (to G. B. A.); HL083090 (to A. H.); HL086614 (to S. H.); and NS38632 (to J. C. L.). This work was also supported by an Alliance for Cancer Gene Therapy grant (to M. K. J.), a Robert Wood Johnson/Harold Amos Medical Faculty Development grant (to G. B. A.), a Dominic Visconsi Scholar Award (to G. B. A., A. H., and S. H.), American Heart Association Grants 0635579T (to Z. L.) and 0725297B (to D. K.), and a Kanae Foundation for the Promotion of Medical Science grant (to D. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HIF
- hypoxia-inducible factor
- KLF
- Kruppel-like factor
- IL
- interleukin
- VEGF
- vascular endothelial growth factor
- VHL
- von Hippel-Lindau
- E1
- ubiquitin-activating enzyme
- E3
- ubiquitin-protein isopeptide ligase
- Z
- benzyloxycarbonyl
- CHO
- Chinese hamster ovary
- HUVEC
- human vascular endothelial cell
- HMVEC
- human microvascular endothelial cell
- MEF
- mouse embryonic fibroblast
- siRNA
- small interfering RNA
- GLUT
- glucose transporter
- ELISA
- enzyme-linked immunosorbent assay
- GFP
- green fluorescent protein.
REFERENCES
- 1.Hirota K., Semenza G. L. (2006) Crit. Rev. Oncol. Hematol. 59, 15–26 [DOI] [PubMed] [Google Scholar]
- 2.Ke Q., Costa M. (2006) Mol. Pharmacol. 70, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 3.Semenza G. L., Nejfelt M. K., Chi S. M., Antonarakis S. E. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5680–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg M. A., Dunning S. P., Bunn H. F. (1988) Science 242, 1412–1415 [DOI] [PubMed] [Google Scholar]
- 5.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza G. L. (2003) Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 7.Srinivas V., Zhang L. P., Zhu X. H., Caro J. (1999) Biochem. Biophys. Res. Commun. 260, 557–561 [DOI] [PubMed] [Google Scholar]
- 8.Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) EMBO J. 20, 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 10.Min J. H., Yang H., Ivan M., Gertler F., Kaelin W. G., Jr., Pavletich N. P. (2002) Science 296, 1886–1889 [DOI] [PubMed] [Google Scholar]
- 11.Shweiki D., Itin A., Soffer D., Keshet E. (1992) Nature 359, 843–845 [DOI] [PubMed] [Google Scholar]
- 12.Namiki A., Brogi E., Kearney M., Kim E. A., Wu T., Couffinhal T., Varticovski L., Isner J. M. (1995) J. Biol. Chem. 270, 31189–31195 [DOI] [PubMed] [Google Scholar]
- 13.Gerber H. P., Condorelli F., Park J., Ferrara N. (1997) J. Biol. Chem. 272, 23659–23667 [DOI] [PubMed] [Google Scholar]
- 14.Mizukami Y., Jo W. S., Duerr E. M., Gala M., Li J., Zhang X., Zimmer M. A., Iliopoulos O., Zukerberg L. R., Kohgo Y., Lynch M. P., Rueda B. R., Chung D. C. (2005) Nat. Med. 11, 992–997 [DOI] [PubMed] [Google Scholar]
- 15.Kim K. S., Rajagopal V., Gonsalves C., Johnson C., Kalra V. K. (2006) J. Immunol. 177, 7211–7224 [DOI] [PubMed] [Google Scholar]
- 16.Pichiule P., Chavez J. C., LaManna J. C. (2004) J. Biol. Chem. 279, 12171–12180 [DOI] [PubMed] [Google Scholar]
- 17.Simon M. P., Tournaire R., Pouyssegur J. (2008) J. Cell Physiol. 217, 809–818 [DOI] [PubMed] [Google Scholar]
- 18.Feinberg M. W., Lin Z., Fisch S., Jain M. K. (2004) Trends Cardiovasc. Med. 14, 241–246 [DOI] [PubMed] [Google Scholar]
- 19.Atkins G. B., Jain M. K. (2007) Circ. Res. 100, 1686–1695 [DOI] [PubMed] [Google Scholar]
- 20.Black A. R., Black J. D., Azizkhan-Clifford J. (2001) J. Cell Physiol. 188, 143–160 [DOI] [PubMed] [Google Scholar]
- 21.van Vliet J., Crofts L. A., Quinlan K. G., Czolij R., Perkins A. C., Crossley M. (2006) Genomics 87, 474–482 [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Shields J. M., Sogawa K., Fujii-Kuriyama Y., Yang V. W. (1998) J. Biol. Chem. 273, 17917–17925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang D. T., Zhao W., Mahatan C. S., Geiman D. E., Yang V. W. (2002) Nucleic Acids Res. 30, 2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson K. P., Kern C. B., Crable S. C., Lingrel J. B. (1995) Mol. Cell Biol. 15, 5957–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo C. T., Veselits M. L., Barton K. P., Lu M. M., Clendenin C., Leiden J. M. (1997) Genes Dev. 11, 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SenBanerjee S., Lin Z., Atkins G. B., Greif D. M., Rao R. M., Kumar A., Feinberg M. W., Chen Z., Simon D. I., Luscinskas F. W., Michel T. M., Gimbrone M. A., Jr., García-Cardeña G., Jain M. K. (2004) J. Exp. Med. 199, 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Z., Kumar A., SenBanerjee S., Staniszewski K., Parmar K., Vaughan D. E., Gimbrone M. A., Jr., Balasubramanian V., García-Cardeña G., Jain M. K. (2005) Circ. Res. 96, e48–57 [DOI] [PubMed] [Google Scholar]
- 28.Lin Z., Hamik A., Jain R., Kumar A., Jain M. K. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1185–1189 [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya R., Senbanerjee S., Lin Z., Mir S., Hamik A., Wang P., Mukherjee P., Mukhopadhyay D., Jain M. K. (2005) J. Biol. Chem. 280, 28848–28851 [DOI] [PubMed] [Google Scholar]
- 30.Huang L. E., Arany Z., Livingston D. M., Bunn H. F. (1996) J. Biol. Chem. 271, 32253–32259 [DOI] [PubMed] [Google Scholar]
- 31.Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Mol. Cell Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chowdary D. R., Dermody J. J., Jha K. K., Ozer H. L. (1994) Mol. Cell Biol. 14, 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 34.Wu J., Srinivasan S. V., Neumann J. C., Lingrel J. B. (2005) Biochemistry 44, 11098–11105 [DOI] [PubMed] [Google Scholar]
- 35.Sen-Banerjee S., Mir S., Lin Z., Hamik A., Atkins G. B., Das H., Banerjee P., Kumar A., Jain M. K. (2005) Circulation 112, 720–726 [DOI] [PubMed] [Google Scholar]
- 36.Mahabeleshwar G. H., Somanath P. R., Byzova T. V. (2006) Methods Mol. Med. 129, 197–208 [DOI] [PubMed] [Google Scholar]
- 37.Atkins G. B., Wang Y., Mahabeleshwar G. H., Shi H., Gao H., Kawanami D., Natesan V., Lin Z., Simon D. I., Jain M. K. (2008) Circ. Res. 103, 690–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanaan A., Farahani R., Douglas R. M., Lamanna J. C., Haddad G. G. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1105–1114 [DOI] [PubMed] [Google Scholar]
- 39.Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., Påhlman S. (2006) Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]
- 40.Hosogai N., Fukuhara A., Oshima K., Miyata Y., Tanaka S., Segawa K., Furukawa S., Tochino Y., Komuro R., Matsuda M., Shimomura I. (2007) Diabetes 56, 901–911 [DOI] [PubMed] [Google Scholar]
- 41.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 42.Wani M. A., Means R. T., Jr., Lingrel J. B. (1998) Transgenic Res. 7, 229–238 [DOI] [PubMed] [Google Scholar]
- 43.Wu J., Bohanan C. S., Neumann J. C., Lingrel J. B. (2008) J. Biol. Chem. 283, 3942–3950 [DOI] [PubMed] [Google Scholar]
- 44.Ndubuizu O., LaManna J. C. (2007) Antioxid. Redox. Signal 9, 1207–1219 [DOI] [PubMed] [Google Scholar]
- 45.Ravi R., Mookerjee B., Bhujwalla Z. M., Sutter C. H., Artemov D., Zeng Q., Dillehay L. E., Madan A., Semenza G. L., Bedi A. (2000) Genes Dev. 14, 34–44 [PMC free article] [PubMed] [Google Scholar]
- 46.Whitesell L., Lindquist S. L. (2005) Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 47.Das H., Kumar A., Lin Z., Patino W. D., Hwang P. M., Feinberg M. W., Majumder P. K., Jain M. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6653–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gradin K., McGuire J., Wenger R. H., Kvietikova I., fhitelaw M. L., Toftgård R., Tora L., Gassmann M., Poellinger L. (1996) Mol. Cell Biol. 16, 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isaacs J. S., Jung Y. J., Mimnaugh E. G., Martinez A., Cuttitta F., Neckers L. M. (2002) J. Biol. Chem. 277, 29936–29944 [DOI] [PubMed] [Google Scholar]
- 50.Isaacs J. S., Jung Y. J., Neckers L. (2004) J. Biol. Chem. 279, 16128–16135 [DOI] [PubMed] [Google Scholar]
- 51.Mimnaugh E. G., Xu W., Vos M., Yuan X., Isaacs J. S., Bisht K. S., Gius D., Neckers L. (2004) Mol. Cancer Ther. 3, 551–566 [PubMed] [Google Scholar]
- 52.Kong X., Lin Z., Liang D., Fath D., Sang N., Caro J. (2006) Mol. Cell Biol. 26, 2019–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong X., Lin Z., Caro J. (2006) FEBS Lett. 580, 6182–6186 [DOI] [PubMed] [Google Scholar]
- 54.Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.