Abstract
Recent yeast genetic studies have implicated the ubiquitin-conjugating enzyme and ubiquitin ligase functions of yRad6 and yBre1, respectively, in H2B ubiquitylation. However, there have been no corresponding biochemical analyses demonstrating intrinsic enzyme activities of yRad6 and yBre1 or related mechanistic details. Here, we describe a robust in vitro chromatin ubiquitylation assay that involves purified H2B ubiquitylation factors and natural nucleosomes. Our results indicate that yRad6 has an in vitro ability to nonspecifically ubiquitylate all core histones in the absence of an ubiquitin ligase but that yBre1 functions, through direct interactions with yRad6, to direct the ubiquitin conjugating activity of yRad6 toward the physiological H2B ubiquitylation site. Moreover, a yRad6 domain mapping analysis shows that an intact UBC domain is required for binding to yBre1, whereas the C-terminal acidic tail domain that is not required for a stable yBre1-yRad6 interaction is necessary for full enzyme activity of yRad6. We also find that, analogous to heteromeric complex formation by BRE1 paralogues in other organisms, yBre1 forms a homo-multimeric complex. Of special significance, our detailed biochemical analyses further show that the yBre1 RING finger domain is essential for H2B ubiquitylation but, surprisingly, dispensable for interaction of yBre1 with yRad6. In further support of the genetically identified requirement of the RNA polymerase II-associated yPaf1 complex for H2B ubiquitylation, protein interaction studies reveal that a purified yPaf1 complex directly and selectively interacts with yBre1 and thus serves to link the H2B ubiquitylation and general transcription machineries. These studies provide a more detailed mechanistic basis for H2B ubiquitylation in yeast.
Protein ubiquitylation is mediated by three classes of enzymes: the ubiquitin-activating enzyme (E1),2 ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s). E1 is universal for all protein ubiquitylation, but cognate E2-E3 pairs selectively ubiquitylate specific target proteins (1). Although histone H2B (H2B) ubiquitylation was first reported ∼30 years ago (2), the identification of enzymes for H2B ubiquitylation is rather recent. Thus, deletions or mutations of RAD6 and BRE1 homologues result in the disappearance of H2B ubiquitylation in many organisms (3) but have been studied most extensively in yeast (4). These results have suggested that RAD6 and BRE1 may serve as E2 and E3 enzymes, respectively, for H2B ubiquitylation, although there have been no biochemical analyses that directly demonstrate the enzymatic activities of yeast Rad6 (yRad6) and Bre1 (yBre1) proteins in these functions.
Yeast Rad6 is the first identified E2 shown to possess an in vitro ubiquitin conjugating activity toward free H2B in the absence of E3 (5). In addition to the common E2 signature motif, the UBC (ubiquitin-conjugating enzyme) domain, and unlike RAD6 homologues in other organisms, yRad6 has a highly acidic C-terminal domain that is responsible for histone polyubiquitylation in vitro (6). However, yRad6 was also shown to mediate an in vitro H2A ubiquitylation (6) that is not found in yeast cells, and yCdc34 subsequently was found to have similar histone ubiquitin conjugating activity in vitro (7). A later demonstration of the sole requirement of the yeast RAD6 gene for H2B monoubiquitylation at lysine 123 in vivo (8) strongly suggested the presence of a physiological E3 that restricts yRad6 activity to lysine 123 on H2B. Three years later, two groups independently identified the yeast BRE1 gene as an E3 for H2B ubiquitylation (9, 10). The BRE1 homologues in diverse organisms contain a RING (really interesting new gene) finger domain, one of the E3 signature motifs, that is essential for the ubiquitin ligase activity. Although the exact role of RING fingers in protein ubiquitylation is still unknown, there is a substantial evidence for interactions between E3 RING fingers and cognate E2s (11).
In addition to yRad6 and yBre1, efficient H2B ubiquitylation in yeast requires the yPaf1 (yeast polymerase II-associated factor 1) complex (12, 13) that was first identified as an RNA polymerase II (Pol II)-associated factor (14, 15). From a mechanistic viewpoint, it has been shown that yRad6 is recruited to promoters in a yBre1-dependent manner (13) and to coding regions in an yPaf1 complex-dependent manner (16) and that yPaf1 complex subunits (yPaf1 and yRtf1) are required for intracellular association (in extracts) of yRad6 with Pol II (13, 16). These results suggested that the yPaf1 complex links H2B ubiquitylation and transcription machineries, although direct interactions of Rad6 or Bre1 with Pol II or the Paf1 complex in yeast were not established.
Here we have established robust in vitro chromatin ubiquitylation assay systems with biochemically defined factors to show that yBre1 directs yRad6 to specifically ubiquitylate H2B at the single physiological site. Through extensive domain mapping studies, we further show that the yRad6 acidic C-terminal domain is required for efficient H2B ubiquitylation, that the yBre1 RING finger domain is essential for H2B ubiquitylation but is not required for binding of yBre1 to yRad6, and that yBre1 forms a homomeric complex via multiple domain interactions. Along with the further demonstration of a direct and selective interaction between yBre1 and the yPaf1 complex, these findings provide a detailed biochemical description of the mechanism of H2B ubiquitylation in yeast.
EXPERIMENTAL PROCEDURES
cDNA, Plasmids, Baculoviruses, Recombinant Proteins, and Complexes
The cDNAs for yRad6 (Gene ID 172348), yBre1 (Gene ID 1431087), yCtr9 (Gene ID 1732236), yRtf1 (Gene ID 1841838), yLeo1 (Gene ID 443970), yPaf1 (Gene ID 536721), and yCdc73 (Gene ID 632669) were PCR-amplified from yeast genomic DNA (Invitrogen). For GST- and His-tagged yRad6 proteins, cDNAs were subcloned, respectively, into pGEX4T (Amersham Biosciences) and pET28 (Novagen); expressed in Escherichia coli; and purified, respectively, on glutathione-Sepharose 4B (Amersham Biosciences) and Ni-NTA (Qiagen) beads following the manufacturers' protocols. For baculoviruses, cDNAs were subcloned in pFASTBAC1 vector with or without an epitope tag, and baculoviruses were generated according to the manufacturer's instructions (Invitrogen). For FLAG-tagged yRad6 and yBre1 proteins and for the yPaf1 complex (containing a FLAG-yCdc73 subunit), Sf9 cells were infected with combinations of baculoviruses and proteins/complexes were affinity-purified on M2-agarose as described previously (17). The expression and purification of Xenopus laevis recombinant histones and histone octamers were essentially as described (18). Purification of oligonucleosomes from HeLa nuclear pellet was as described (19). Preparations of FLAG-hE1 and His-pK-HA-Ub proteins were as described (20).
Protein Interaction Assays
For GST pulldown assays, 2 μg of GST or GST-fused proteins immobilized on glutathione-Sepharose 4B beads were incubated with 200 ng of purified factors in binding buffer A (20 mm Tris-Cl, pH 7.9, 150 mm KCl, 0.2 mm EDTA, 20% glycerol, 0.1% Nonidet P-40, 0.5 mg/ml bovine serum albumin, and 0.5 mm PMSF) at 4 °C for 3 h, and then the beads were extensively washed with binding buffer A without bovine serum albumin. For the interaction studies in Fig. 7B, Ni-NTA-agarose preparations previously coupled with 5 μg of purified yPaf1 complex (containing His-yPaf1 subunit) were incubated with 100 ng of purified factors in binding buffer B (20 mm Tris-Cl, pH 7.9, 150 mm KCl, 20% glycerol, 0.05% Nonidet P-40, 0.2 mg/ml bovine serum albumin, and 0.5 mm PMSF) at 4 °C for 3 h, and then the beads were extensively washed with wash buffer (20 mm Tris-Cl, pH 7.9, 150 mm KCl, 20% glycerol, 0.05% Nonidet P-40, 0.5 mm PMSF, and 50 mm immidazole). For the interaction studies in Fig. 6, and after 2 days of Sf9 cell infection with baculoviruses, the total cell extracts were prepared by incubating the cells in binding buffer C (20 mm Tris-Cl, pH 7.9, 300 mm KCl, 0.2 mm EDTA, 20% glycerol, 0.1% Nonidet P-40, and 0.5 mm PMSF). Following clarification by centrifugation, the extracts were incubated with M2-agarose beads at 4 °C for 3 h, and after extensive washing of beads with the binding buffer C, the bound proteins were analyzed by immunoblot.
FIGURE 7.
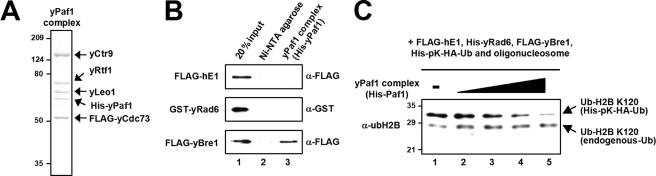
The yPaf1 complex directly interacts with yBre1. A, analysis of the purified yPaf1 complex by SDS-PAGE with Coomassie Blue staining. Components of the reconstituted yPaf1 complex (isolated via FLAG-yCdc73) were expressed from baculovirus vectors. B, binding of H2B ubiquitylation factors to the yPaf1 complex. The purified yPaf1 complex (containing His-yPaf1) bound to Ni-NTA-agarose was incubated with purified FLAG-hE1, GST-yRad6, or FLAG-yBre1. The bound proteins were scored by immunoblots with anti-FLAG or anti-GST antibodies as indicated. Ni-NTA-agarose without any bound His-tagged protein served as a negative control (lane 2). C, effect of the yPaf1 complex on in vitro H2B ubiquitylation. The chromatin ubiquitylation assay was performed as in Fig. 1D. The reactions contained 100 (lane 2), 200 (lane 3), 300 (lane 4), or 600 (lane 5) ng of yPaf1 complex.
FIGURE 6.
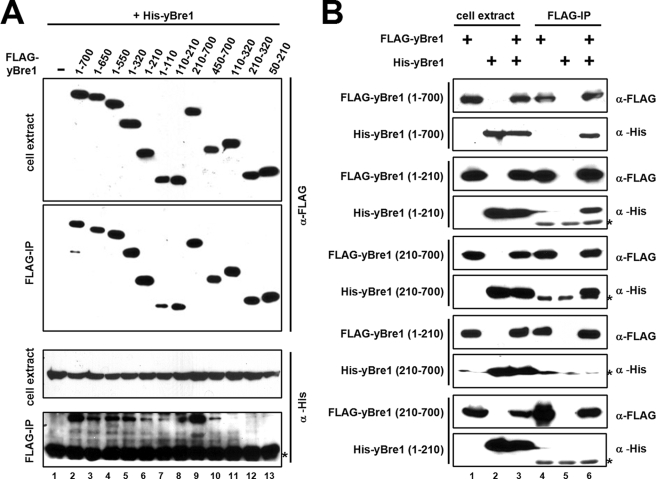
Yeast Bre1 forms a homomeric complex through multiple intermolecular interactions. Insect cells were co-infected with combinations of baculoviruses expressing His-yBre1 and FLAG-yBre1 or FLAG-yBre1 fragments (A) and His-yBre1 and FLAG-yBre1 fragments (B) as indicated. Total cell extracts and complexes purified on M2-agarose (FLAG-IP) were analyzed by immunoblots with indicated antibodies. 30 and 15% of the input (cell extracts) were loaded, respectively, for anti-FLAG and anti-His immunoblots. The asterisks indicate immunoglobulin chains. Note that an extraneous spot in the upper right side of lane 8 in the bottom panel of A partially obscures the absence of a His-yBre1 band. The lack of an interaction of His-yBre1 with the 110–210 fragment was confirmed in another analysis.
In Vitro E3 Ubiquitylation Assays
Reactions containing 100 ng of hE1, 50 ng of His-yRad6 (Figs. 1C and 3E) or 10 ng of FLAG-yRad6 (Fig. 5D), 150 ng of yBre1, 1.3 μg of His-pK-HA-ubiquitin that can be radiolabeled by protein kinase (where pK indicates a protein kinase recognition site; Ref. 21), 50 mm Tris-Cl, pH 7.9, 5 mm MgCl2, 2 mm NaF, 0.4 mm dithiothreitol, and 4 mm ATP in 20 μl were incubated at 37 °C for 1 h, resolved by SDS-PAGE, and subjected to autoradiography.
FIGURE 1.
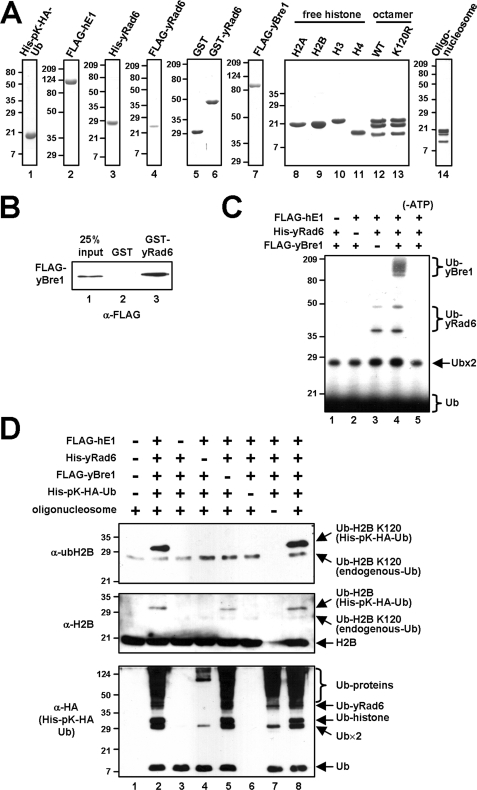
Establishment of an in vitro H2B ubiquitylation assay with purified factors. A, analyses of purified His-pK-HA-ubiquitin, FLAG-hE1, His-yRad6, FLAG-yRad6, GST, GST-yRad6, FLAG-yBre1, recombinant histones, reconstituted histone octamers, and oligonucleosomes by SDS-PAGE with Coomassie Blue staining. B, direct binding of yBre1 to yRad6. Purified FLAG-yBre1 was tested for binding to GST or GST-yRad6 proteins, and bound proteins were scored by immunoblot with anti-FLAG antibody. C, ubiquitylation of yBre1 by yRad6. Purified yBre1 was subjected to an E3 ubiquitylation assay with hE1 and yRad6 in the presence of 32P-labeled ubiquitin. Ubiquitylation of yBre1 was monitored by autoradiography. −ATP indicates the reaction without ATP. Free ubiquitin (Ub), ubiquitin dimer (Ub×2), ubiquitylated yRad6 (Ub-yRad6), and polyubiquitylated yBre1 (Ub-yBre1) are indicated. D, collective requirement of factors for H2B ubiquitylation. The reactions containing the indicated combinations of purified hE1, yRad6, yBre1, ubiquitin, and oligonucleosome were subjected to immunoblot with antibodies indicated on the left of each panel.
FIGURE 3.
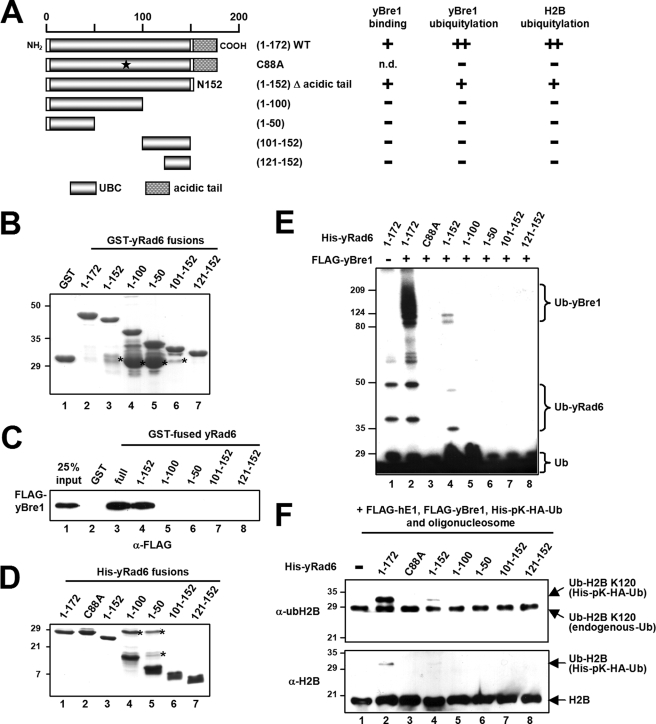
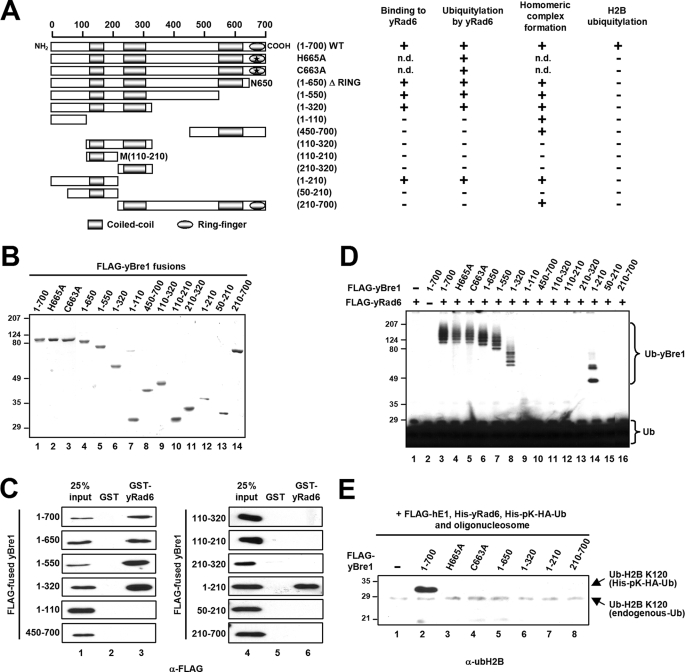
The intact UBC domain of yRad6 is essential for interaction with yBre1 and for H2B ubiquitylation. A, a schematic diagram of full-length yRad6, derived fragments, and a point mutant with predicted UBC and acidic tail domains. Binding of yRad6 proteins to yBre1 and the yBre1 and H2B ubiquitylation activities of the yRad6 proteins are summarized on the right. n.d., not determined. B and D, analyses of purified GST, GST-yRad6, and GST-yRad6 fragments (B) and of purified His-yRad6 and His-yRad6 fragments (D) by SDS-PAGE with Coomassie Blue staining. The point mutation and encoded amino acids within the yRad6 fragments are indicated. The asterisks indicate degradation products (B) or contaminants (D). C, binding of FLAG-yBre1 to GST-yRad6 or GST-yRad6 fragments versus GST. The bound proteins were scored by immunoblots with anti-FLAG antibody. E, yRad6 domain requirement for yBre1 ubiquitylation. Purified yBre1 was analyzed in an E3 ubiquitylation assay as in Fig. 1C with yRad6, the yRad6 point mutant, or the yRad6 fragments as indicated. F, yRad6 domain requirement for H2B ubiquitylation. The chromatin ubiquitylation assay was performed as in Fig. 1D with yRad6, the yRad6 point mutant, or the yRad6 fragments as indicated.
FIGURE 5.
The yBre1 RING finger is dispensable for yBre1 binding to yRad6 but is essential for H2B ubiquitylation. A, a schematic diagram of full-length yBre1, derived fragments, and point mutants with predicted coiled-coil and RING finger domains. Binding of the yBre1 proteins to yRad6, ubiquitylation of the yBre1 proteins by yRad6, homomeric complex formation abilities, and H2B ubiquitylation activities of the yBre1 proteins are summarized on the right. n.d., not determined. B, analyses of purified FLAG-tagged full-length yBre1, fragments, and point mutants by SDS-PAGE with Coomassie Blue staining. Point mutations and encoded amino acids within the yBre1 fragments are indicated. C, binding of FLAG-yBre1 and FLAG-yBre1 fragments to GST-yRad6 versus GST. The bound proteins were scored by immunoblots with anti-FLAG antibody. D, yBre1 domain requirement for ubiquitylation by yRad6. Purified yBre1, yBre1 mutants, and yBre1 fragments were analyzed in an E3 ubiquitylation assay as in Fig. 1C. E, yBre1 domain requirement for H2B ubiquitylation. The chromatin ubiquitylation assay was performed as in Fig. 1D with indicated yBre1, yBre1 mutants, and yBre1 fragments.
In Vitro Histone and Chromatin Ubiquitylation Assays
Reactions containing 5 μg of HeLa cell-derived oligonucleosomes, 100 ng of hE1, 200 ng of yRad6, 600 ng of yBre1, and 2.8 μg of His-pK-HA-ubiquitin, unless otherwise indicated, in 20 μl of reaction buffer (50 mm Tris-Cl, pH 7.9, 5 mm MgCl2, 2 mm NaF, 0.4 mm dithiothreitol, and 4 mm ATP) were incubated at 37 °C for 6 h. The proteins were resolved by SDS-PAGE and subjected to immunoblotting.
Antibodies
The following antibodies were obtained commercially: anti-H2A, anti-H2B, anti-H3, anti-H4 and anti-HA (Abcam); anti-ubH2B (Medlabs); anti-GST (Santa Cruz Biotechnology); anti-FLAG (horseradish peroxidase-conjugated; Sigma); and anti-His (Qiagen).
RESULTS
Yeast Bre1 Is Required for Selective Site-specific H2B Ubiquitylation by yRad6
To establish comprehensive in vitro chromatin ubiquitylation and protein interaction assays, we first prepared basic H2B ubiquitylation factors (recombinant hE1, yRad6, and yBre1) and protein substrates (recombinant histones, histone octamers, and natural oligonucleosomes derived from HeLa cells) (Fig. 1A). The yeast E2-E3 H2B ubiquitylation system has advantages over that of other organisms because the presence of single copy RAD6 and BRE1 genes allows more simplified biochemical analyses (below). Consistent with the phylogenetic sequence conservation of E1 and the universal requirement of E1 for all protein ubiquitylation processes, the hE1 has proved compatible with yeast Rad6 and Bre1 proteins for in vitro ubiquitylation assays (below).
Cognate E2-E3 pairs specifically and directly bind to each other (1). Consistent with co-isolation of yRad6 in an affinity-purified yBre1 complex (10), we confirmed a direct interaction between purified yRad6 and yBre1 proteins (Fig. 1B). Direct interactions between the RING finger E3 and cognate E2 proteins result in both substrate and E3 ubiquitylation (22), as is well documented for the H2A ubiquitylation-specific Ring1B E3 (23). In this regard and further confirming the functional relevance of the physical interaction between yRad6 and yBre1, an E3 ubiquitylation assay revealed ATP-, E1-, and yRad6-dependent polyubiquitylation of yBre1 (Fig. 1C).
To clearly demonstrate direct E2 and E3 functions of yRad6 and yBre1, respectively, we set up an in vitro chromatin ubiquitylation assay. This assay employs natural oligonucleosome arrays derived from HeLa cells because they provide preferred ubiquitylation substrates compared with the chromatin that is assembled with recombinant histones that lack all post-translational histone modifications (see “Discussion”). In this assay, newly synthesized ubiquitylated H2B (modified by His- and HA-tagged ubiquitin) is expected to migrate more slowly than the naturally ubiquitylated H2B that is present in the oligonucleosome substrate. Note also that lysine 120 in vertebrate H2B corresponds to lysine 123 in yeast H2B and is probed here by an anti-ubH2B antibody that specifically recognizes ubH2B (24). Complete reactions containing purified hE1, yRad6, yBre1, ubiquitin, and oligonucleosomes generated a lysine 120-ubiquitylated H2B (Fig. 1D, top panel, lanes 2 and 8), whereas reactions with omission of any of these components did not (top panel, lanes 3–7). Although a significant level of H2B ubiquitylation (probed by an anti-H2B antibody) was detected in the absence of yBre1, this clearly represents H2B ubiquitylation at a site(s) other than lysine 120 (compare top panel, lane 5 versus middle panel, lane 5; see also below). The majority of ubiquitin-conjugated proteins (probed by an anti-HA antibody) are thought to be ubiquitin-conjugated yRad6 because they were mainly observed in reactions containing hE1 and yRad6 (bottom panel, compare lanes 2, 5, 7 and 8 versus lanes 1, 3 and 4).
In a test for efficiency of the in vitro H2B ubiquitylation reactions, we observed a dose-dependent increase in H2B ubiquitylation in response to varying amounts of yBre1 but not of yRad6 (supplemental Fig. S1). This suggests that the E3 is more limiting for H2B ubiquitylation, at least in vitro, and may explain the enhanced H2B ubiquitylation by ectopic E3, but not by ectopic E2, in a cell-based assay (25).
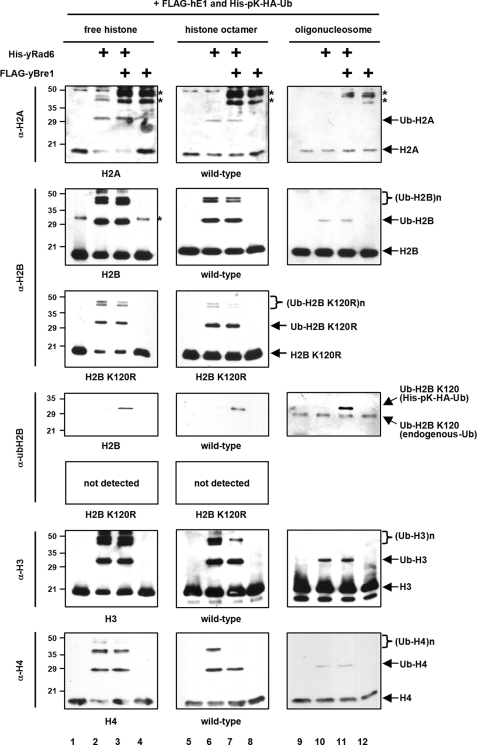
The detection of a significant level of yBre1-independent H2B ubiquitylation by yRad6 in the in vitro chromatin ubiquitylation assay (above) led us to more detailed analyses of substrate and site specificities of yRad6 in the presence and in the absence of yBre1 (Fig. 2). Consistent with the earlier reports of the ability of yRad6 to ubiquitylate H2A and H2B (5, 6), yRad6 was shown to mono- and polyubiquitylate all four core histones both in free and octamer forms in the absence of yBre1 (Fig. 2, lanes 2 and 6, in the anti-H2A, -H2B, -H3, and -H4 immunoblots), and the inclusion of yBre1 did not affect the overall histone ubiquitylation efficiency (Fig. 2, lanes 3 and 7 versus lanes 2 and 6, respectively, in the anti-H2A, -H2B, -H3, and -H4 immunoblots). Importantly, the efficiency of histone ubiquitylation with the nucleosomal substrate decreased significantly compared with the efficiency of histone ubiquitylation with free histones or with histone octamers as substrates (Fig. 2, lanes 9–12 versus lanes 1–4 or lanes 5–8, in the anti-H2A, -H2B, -H3, and -H4 immunoblots). This suggests that nucleosome structure restricts nonspecific access of ubiquitylation factors to the histone core regions. However, and most importantly, H2B ubiquitylation at Lys120 was critically dependent upon yBre1 for all substrates tested (compare lanes 3, 7 and 11 versus lanes 2, 6 and 10, respectively, in the anti-ubH2B immunoblots). This indicates that yBre1 directs the yRad6 ubiquitin conjugating activity to a specific ubiquitylation site (lysine 120) through recognition of the target substrate. In addition, E3-independent histone ubiquitylation by yRad6 reflects a nonspecific yRad6 ubiquitin conjugating activity that is manifested in a purified assay system lacking constraints that normally control accessibility of yRad6 to histones.
FIGURE 2.
Yeast Bre1 is required for H2B ubiquitylation at lysine 120. Reactions containing free histone (300 ng), histone octamer (1.2 μg), or oligonucleosome (5 μg) substrates with hE1, yRad6, yBre1, and ubiquitin, where indicated, were subjected to in vitro ubiquitylation assays. Ubiquitylated histones were scored by immunoblot with the indicated antibodies. Nonspecific bands are indicated by asterisks.
The C-terminal Acidic Tail of yRad6 Is Required for Full H2B Ubiquitin Conjugating Activity
To determine the regions in yRad6 that are required for H2B ubiquitylation and for interaction with yBre1, we generated serial deletion mutants and a C88A mutant (cysteine to alanine substitution at amino acid 88 that abolishes ubiquitin conjugating activity of yRad6 (26)) (Fig. 3A). We first examined the interactions of purified yRad6 fragments (Fig. 3B) with yBre1. Direct binding of the C-terminal acidic tail deletion mutant (N152) (Fig. 3A) to yBre1 was comparable with that of wild type (WT) (Fig. 3C, compare lane 4 versus lane 3), whereas all other deletion mutants within the UBC domain failed to bind to yBre1 (lanes 5–8 versus lane 3). These results indicate that an intact yRad6 UBC domain is essential for interaction with yBre1 but that the yRad6 acidic tail is dispensable for this interaction.
To investigate corresponding H2B ubiquitylation activities with the above-described yRad6 mutant proteins and because GST-fused yRad6 proteins exhibited much weaker ubiquitin conjugating activity compared with that of His-fused yRad6 (data not shown), we newly prepared His-tagged yRad6 fragments (Fig. 3D). Consistent with the absence of detectable interactions with yBre1, none of the partial UBC domain fragments showed any detectable ubiquitin conjugating activity on yBre1 (Fig. 3E, lanes 5–8) or on H2B (Fig. 3F, lanes 5–8). In addition, and as expected, the C88A mutant was catalytically inactive in both yBre1 and H2B ubiquitylation (Fig. 3, E, lane 3, and F, lane 3). Importantly, the N152 mutant lacking the acidic tail showed markedly decreased levels of yBre1 ubiquitylation (Fig. 3E, compare lane 4 versus lane 2) and H2B Lys120 ubiquitylation (Fig. 3F, compare lane 4 versus lane 2) compared with that of intact yRad6. This is consistent with the disappearance of ubH2B in the rad6-149 strain in which WT yRad6 is replaced by a mutant yRad6 that encodes amino acids 1–149 and thus lacks the C-terminal acidic domain (8).
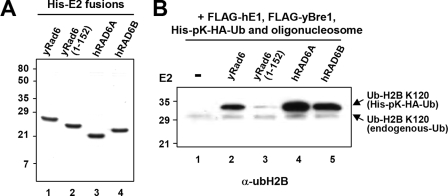
Human RAD6 (hRAD6) homologues show strong amino acid sequence homology to yRad6 and are able to complement the E2 function of yRad6 (20) despite the absence of the C-terminal acidic tail (supplemental Fig. S2). To detail a role of the yRad6 acidic tail in H2B ubiquitylation, we compared the E2 activities of purified yRad6 WT and N152 mutant proteins with those of hRAD6 (Fig. 4A). Consistent with the observed increase of ubH2B in response to ectopic yBre1 expression in human cells (25), both hRAD6A and hRAD6B efficiently ubiquitylated H2B at lysine 120 in the presence of yBre1 (Fig. 4B, lanes 4 and 5). Interestingly, the H2B ubiquitylation level of the yRad6 N152 mutant was significantly lower than the levels observed with yRad6 WT and hRAD6 (compare 3 versus lanes 2, 4 and 5), indicating a specific requirement of the acidic tail for full intrinsic E2 activity of yRad6. Overall these data indicate that the C-terminal acidic domain plays a role in potentiating the enzymatic activity of yRad6.
FIGURE 4.
The acidic tail potentiates the intrinsic E2 activity of yRad6 in H2B ubiquitylation. A, analyses of purified His-E2 proteins by SDS-PAGE with Coomassie Blue staining to ensure comparable levels of protein usage in the in vitro chromatin ubiquitylation assays. B, comparison of the E2 activities of the yeast and human RAD6 proteins. The chromatin ubiquitylation assay was performed as in Fig. 1D with indicated RAD6 proteins.
The yBre1 RING Finger Domain Plays a Role in H2B Ubiquitylation Distinct from the yBre1-yRad6 Interaction
To determine the yBre1 domains that are responsible for H2B ubiquitylation and interaction with yRad6, we generated serial deletion mutants as well as mutants (H665A and C663A) with point mutations in residues critical for RING finger formation (10, 27) (Fig. 5A). There are many instances in which RING finger domains are required for E3 ubiquitin ligase activities through direct interactions with cognate E2s (11) and result in RING finger-dependent E3 ubiquitylation (22). However, and strikingly, protein interaction studies with yRad6 and purified yBre1 fragments (Fig. 5B) revealed, first, that the yBre1 RING finger deletion mutant (N650) (Fig. 5A) has a binding capacity comparable with that of WT yBre1 and, second, that an N-terminal yBre1 domain (residues 1–210) is minimally responsible for the yRad6 interaction (Fig. 5C). Consistent with these results, all of the yBre1 fragments containing residues 1–210 were ubiquitylated by yRad6 in an E3 ubiquitylation assay (Fig. 5D, lanes 3–8 and 14), whereas other fragments lacking all or part of residues 1–210 were unable to be ubiquitylated (lanes 9–13, 15, and 16). As expected from the results with the N650 mutant, abrogation of the RING finger structure (H665A and C663A) did not affect yBre1 ubiquitylation by yRad6 (lanes 4 and 5). These results indicate that, in contrast to other E2s, yRad6 binds to yBre1 through a yBre1 N-terminal region rather than through the yBre1 RING finger domain.
Next, in a chromatin ubiquitylation assay with yBre1 and the yBre1 mutants, we found that those yBre1 fragments that interact with yRad6 in the absence of the RING finger (Fig. 5E, lanes 5–7), as well as the yBre1 fragment that is unable interact with yRad6 (lane 8), all fail to mediate ubiquitylation of H2B. This suggests that a direct interaction of yBre1 with yRad6 is insufficient for H2B ubiquitylation. In a further analysis, H2B ubiquitylation was abrogated by point (Fig. 5E, lanes 3 and 4) or deletion (lane 5) mutations of the RING finger, thus demonstrating that the RING finger is critical for the ubiquitin ligase activity of yBre1. Overall, these results indicate an essential function for the yBre1 RING finger in H2B ubiquitylation, but not through a mechanism that is primarily involved in the yBre1-yRad6 interaction (see “Discussion”).
Yeast Bre1 Forms a Homomeric Complex through Association of Multiple Regions
In contrast to yeast, several organisms contain two BRE1 paralogues that form a multimeric complex (20, 28–31), and complex formation is essential for interaction with RAD6 and for H2B ubiquitylation (20). In this regard, we tested whether yBre1 is also able to form a multimeric complex, and, in such a case, which region(s) are responsible for complex formation. To this end and following FLAG- and His-tagged co-expression of yBre1 proteins in insect cells, we examined the presence of His-yBre1 in the affinity-purified (M2-agarose) FLAG-yBre1 complex. The protein expression levels (Fig. 6A, top panel) and binding efficiencies on M2-agarose (second panel) for all FLAG-yBre1 fragments were comparable with those of WT yBre1. Significantly, His-yBre1 was bound to M2-agarose only in the presence of FLAG-yBre1 (Fig. 6, A, bottom panel, compare lane 2 versus lane 1, and B, top two panels), thus demonstrating the ability of yBre1 to form a homomeric complex. Further mapping revealed that homomeric association of yBre1 is minimally dependent upon two separate regions (residues 1–110 and 450–700) (Fig. 6A, bottom panel, lanes 7 and 10) and further stabilized by adjacent regions (lane 7 versus lanes 5–6 and lane 10 versus lane 9) but independent of an internal region (residues 110–320) (lanes 8 and 11–13).
Next, in a test for how yBre1 proteins are aligned in the yBre1 complex (Fig. 6B), the N-terminal yBre1 fragment (residues 1–210) was found to associate with another copy of this fragment (third and fourth panels) but not with the C-terminal yBre1 fragment (residues 210–700) (seventh and eighth panels). Overall these data indicate that yBre1 forms a homo-multimeric complex through intermolecular associations of both an N-terminal region and a C-terminal region.
Yeast Bre1 Directly Interacts with the yPaf1 Complex
Although the yPaf1 complex is also essential for H2B ubiquitylation in yeast, the molecular basis for this function is not well understood (Introduction). To address this question, the yPaf1 complex was reconstituted in insect cells by co-infection with baculoviruses that individually express yCtr9, yRtf1, yLeo1, His-yPaf1, and FLAG-yCdc73 components. FLAG-yCdc73-associated proteins were purified by affinity chromatography on M2-agarose, and the resulting complex showed stoichiometric association of all five subunits (Fig. 7A). In a test for interactions between highly purified H2B ubiquitylation factors (hE1, yRad6, and yBre1) and the yPaf1 complex, yBre1 was shown to be solely responsible for association with the yPaf1 complex (Fig. 7B).
In view of the direct association of yBre1 with the yPaf1 complex, we tested whether the yPaf1 complex directly stimulates H2B ubiquitylation (Fig. 7C). However, the yPaf1 complex was found to decrease yRad6-yBre1-mediated H2B ubiquitylation (lanes 2–5 versus lane 1), presumably because competitive binding of the yPaf1 complex prevents yBre1 from recognizing histones. Our results suggest that the yPaf1 complex plays a role in H2B ubiquitylation other than stimulating catalysis by yRad6-yBre1 (see “Discussion”).
DISCUSSION
Genetic analyses have established functions for the Rad6 E2 and the Bre1 E3, as well as the Paf1 elongation complex, in H2B ubiquitylation in yeast (Introduction). In an extension of our recent biochemical analyses of the human RAD6 and BRE1 homologues (20), and through systematic analyses with purified wild type and mutant proteins, the present study establishes the intrinsic enzymatic activities of yeast Rad6 and Bre1 and domains important for their interactions and functions in H2B ubiquitylation. This study further underscores the key role of yBre1 in H2B ubiquitylation site selection, the noncanonical (RING finger-independent) association of Rad6 with Bre1, and a direct interaction between H2B ubiquitylation and transcription elongation factors in yeast.
Establishment of a Robust in Vitro H2B Ubiquitylation Assay with a Chromatin Substrate
To establish a reliable in vitro H2B ubiquitylation assay, it was essential to overcome two major difficulties. First, as highlighted in Figs. 1 and 2, the in vitro chromatin ubiquitylation reaction generates a large number of protein-ubiquitin conjugates that include a ubiquitin dimer, ubiquitylated yBre1, ubiquitylated yRad6, and ubiquitylated histones. Especially significant is our observation that all core histones are nonspecifically ubiquitylated by yRad6 in the absence of the yBre1 E3, probably because of uncontrolled accessibility of yRad6 to histones in the defined system. This made it almost impossible to detect H2B that is specifically ubiquitylated at the single physiological site (ubH2B) with anti-ubiquitin or anti-H2B antibodies. Remarkably, the recent development of an anti-ubH2B monoclonal antibody (24) enabled us to solve this problem and to clearly demonstrate the requirement of yBre1 for H2B ubiquitylation at the physiological site. Second, our initial attempt using recombinant chromatin as a substrate failed to generate ubH2B, but it was later found that natural oligonucleosomes serve as preferential substrates, relative to recombinant chromatin with unmodified histones, for H2B ubiquitylation in vitro (supplemental Fig. S3). This raises the possibility that transcriptionally active chromatins (contained in natural oligonucleosome arrays with associated histone modifications) may serve as preferential substrates for H2B ubiquitylation in vitro. This also fits our recent demonstration that on-going transcription is required for efficient H2B ubiquitylation (20). Apart from describing catalytically active yBre1 and yRad6 enzymes, this report also describes the preparation of a catalytically active recombinant hE1 protein (supplemental Fig. S4) that may serve as a convenient source for all in vitro protein ubiquitylation assays.
Architecture of Homomeric yBre1 Complex
Consistent with the formation of heteromeric complexes containing BRE1 paralogues in other organisms (20, 28–31), we found that yBre1 forms a homomeric complex (Fig. 6). Size exclusion chromatography has further suggested that the yBre1 complex is a homo-tetramer.3 Consistent with this observation, the hBRE1 complex (composed of hBRE1A/RNF20 and hBRE1B/RNF40) was shown to elute at ∼600 kDa on a gel filtration column (28), indicating that the hBRE1 complex consists of two copies of hBRE1A and hBRE1B. These observations suggest that, regardless of the BRE1 gene copy number, the overall architecture and mode of action of the BRE1 complex in H2B ubiquitylation are conserved from yeast to human. In addition, the current study demonstrates yBre1 complex formation through two interacting regions (Fig. 6), whereas our previous study suggested an hBRE1A-hBRE1B association through N-terminal regions (20). These results raise the possibility that the heterotetrameric human BRE1 complex may be composed of hBRE1A·hBRE1A and hBRE1B·hBRE1B pairs, each resulting from multiple interactions, that interact through heterologous N-terminal domains to form the hBRE1 complex.
Related to homo-multimeric complex formation, yBre1 has a putative leucine zipper domain (leucine residue heptad repeats at amino acid positions 139, 146, 153, 160, and 167) that is also found in human and plant BRE1 homologues (but not in fission yeast). However, in a test for the possible requirement of these residues for yBre1 complex formation, we found that, consistent with the dispensability of the M(110–210) (Fig. 5A) fragment containing this region (Fig. 5A) for association with another copy of yBre1 (Fig. 6A, lane 8), yBre1 with point mutations at these sites still forms a homomeric complex (20). These results suggest that this putative leucine zipper domain may play another role in yBre1 function.
Role of yBre1 RING Fingers in H2B Ubiquitylation
The RING finger domain is generally believed to serve as a binding platform for a cognate E2 (11, 22). However, the yUbr1 N-end rule E3 (32) and the DNA repair-related yRad18 E3 (33) interact with their cognate E2, yRad6, in a RING finger-independent manner. In addition, we found that BRE1 interacts with RAD6 in a RING finger-independent manner both in yeast (Fig. 5) and in human (20). It is noteworthy that RAD6 serves as the E2 in all of the above instances. These results indicate that RAD6 is a noncanonical E2 that binds to its cognate E3 via regions other than the RING finger and, importantly, further suggest that the role of the RING finger in E3 is not merely to recruit E2 to vicinity of the target proteins.
Related to a RING finger role other than E2 binding in ubiquitylation processes, a structural analysis of the Bmi1-Ring1B H2A ubiquitylation ligase led to the proposal that an association of the Bmi1 RING finger stimulates the E3 activity of Ring1B through an enhanced binding to the nucleosomal substrate (34). Interestingly, we also observed that a RING finger deletion results in a significantly decreased interaction of yBre1 with oligonucleosomes.3 Based on the notion that the structure of the RING finger domain is highly similar to that of the PHD finger domain that is frequently found in histone-binding proteins (35), it is plausible that the BRE1 RING finger may play a role in nucleosome recognition.
Transcription Coupled H2B Ubiquitylation
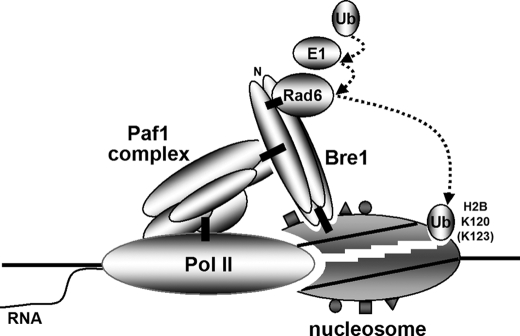
Importantly, our demonstration of direct interactions between yRad6 and yBre1 and between yBre1 and the Paf1 complex significantly extends previous studies and provides a clear explanation of how H2B ubiquitylation factors can be recruited to chromatin (Fig. 8). This relationship (yRad6 → yBre1 → yPaf1 complex) nicely fits earlier yeast genetic studies wherein deletion of either yBre1 or the yPaf1 complex subunit yRtf1 led to a complete loss of yRad6 from promoter and coding regions (13, 16). Because the Paf1 transcription elongation complex associates with Pol II (14), it is plausible that passage of Pol II through nucleosomes allows H2B ubiquitylation factors (recruited by the Paf1 complex) easier access to the ubiquitylation site that is embedded within the H2B C-terminal α-helix (Fig. 8). These results also provide plausible explanations for why natural oligonucleosome arrays, containing a subpopulation of actively transcribed regions, serve as preferential H2B ubiquitylation substrates in vitro and for why H2B ubiquitylation is enriched in gene coding regions (20, 24).
FIGURE 8.
Schematic model of transcription-coupled H2B ubiquitylation. Rad6 interacts directly with the homomeric Bre1 complex, and the resulting Rad6-Bre1 complex is recruited to active chromatin templates through direct interaction of Bre1 with the Paf1 transcription elongation complex that associates with transcribing RNA Pol II. Transcription-coupled alteration of nucleosome structure (depicted as a cleft in the nucleosome) and/or associated post-translational histone modifications (depicted as small rectangles, triangles, and circles on the nucleosome), along with histone chaperones (not shown), may allow ubiquitylation factors easier access to the H2B ubiquitylation site. Verified direct interactions are depicted by thick bars. The dashed arrows indicate ubiquitin transfer pathway from ubiquitylation factors to H2B Lys120 (Lys123). Although not required for Bre1 binding to Rad6, the C-terminal RING finger of Bre1 is required for efficient H2B ubiquitylation by Rad6.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA129325 and DK071900 (to R. G. R.). This work was also supported by a Leukemia and Lymphoma Society Specialized Centers of Research grant and by the Starr Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- Pol II
- RNA polymerase II
- WT
- wild type
- H2A
- histone H2A
- H2B
- histone H2B
- H3
- histone H3
- H4
- histone H4
- ubH2B
- lysine 120-ubiquitylated H2B
- GST
- glutathione S-transferase
- Ni-NTA
- nickel-nitrilotriacetic acid
- PMSF
- phenylmethylsulfonyl fluoride
- HA
- hemagglutinin.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.West M. H., Bonner W. M. (1980) Nucleic Acids Res. 8, 4671–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weake V. M., Workman J. L. (2008) Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 4.Osley M. A. (2004) Biochim. Biophys. Acta 1677, 74–78 [DOI] [PubMed] [Google Scholar]
- 5.Jentsch S., McGrath J. P., Varshavsky A. (1987) Nature 329, 131–134 [DOI] [PubMed] [Google Scholar]
- 6.Sung P., Prakash S., Prakash L. (1988) Genes Dev. 2, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 7.Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. (1988) Science 241, 1331–1335 [DOI] [PubMed] [Google Scholar]
- 8.Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 9.Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. (2003) Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 10.Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 11.Joazeiro C. A., Weissman A. M. (2000) Cell 102, 549–552 [DOI] [PubMed] [Google Scholar]
- 12.Ng H. H., Dole S., Struhl K. (2003) J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 13.Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 14.Shi X., Finkelstein A., Wolf A. J., Wade P. A., Burton Z. F., Jaehning J. A. (1996) Mol. Cell. Biol. 16, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade P. A., Werel W., Fentzke R. C., Thompson N. E., Leykam J. F., Burgess R. R., Jaehning J. A., Burton Z. F. (1996) Protein Expr. Purif. 8, 85–90 [DOI] [PubMed] [Google Scholar]
- 16.Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Mol. Cell. Biol. 25, 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T., Levenstein M. E., Fyodorov D. V., Kutach A. K., Kobayashi R., Kadonaga J. T. (1999) Genes Dev. 13, 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 19.Utley R. T., Owen-Hughes T. A., Juan L. J., Côté J., Adams C. C., Workman J. L. (1996) Methods Enzymol. 274, 276–291 [DOI] [PubMed] [Google Scholar]
- 20.Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J., Kim J. H., Lee S. H., Kim D. H., Kang H. Y., Bae S. H., Pan Z. Q., Seo Y. S. (2002) J. Biol. Chem. 277, 24530–24537 [DOI] [PubMed] [Google Scholar]
- 22.Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 24.Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. (2008) Nat. Cell Biol. 10, 483–488 [DOI] [PubMed] [Google Scholar]
- 25.Kim J., Hake S. B., Roeder R. G. (2005) Mol. Cell 20, 759–770 [DOI] [PubMed] [Google Scholar]
- 26.Sung P., Prakash S., Prakash L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2695–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson P. K., Eldridge A. G., Freed E., Furstenthal L., Hsu J. Y., Kaiser B. K., Reimann J. D. (2000) Trends Cell Biol. 10, 429–439 [DOI] [PubMed] [Google Scholar]
- 28.Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 29.Tanny J. C., Erdjument-Bromage H., Tempst P., Allis C. D. (2007) Genes Dev. 21, 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zofall M., Grewal S. I. (2007) J. Biol. Chem. 282, 14065–14072 [DOI] [PubMed] [Google Scholar]
- 31.Cao Y., Dai Y., Cui S., Ma L. (2008) Plant Cell 20, 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Y., Varshavsky A. (1999) EMBO J. 18, 6832–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailly V., Prakash S., Prakash L. (1997) Mol. Cell. Biol. 17, 4536–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Cao R., Wang M., Myers M. P., Zhang Y., Xu R. M. (2006) J. Biol. Chem. 281, 20643–20649 [DOI] [PubMed] [Google Scholar]
- 35.Bienz M. (2006) Trends Biochem. Sci. 31, 35–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.