Abstract
CK2 phosphorylates a wide variety of substrates, including translation initiation factors. A mass spectrometric approach was used to identify residues phosphorylated by CK2, which may regulate the activity of initiation factors during the translation initiation process in plants. CK2 in vitro phosphorylation sites were identified in wheat and Arabidopsis thaliana eIF2α, eIF2β, eIF5, and wheat eIF3c. Native wheat eIF5 and eIF2α were found to have phosphorylation sites that corresponded to some of the in vitro CK2 phosphorylation sites. A large number of the CK2 sites identified in this study are in conserved binding domains that have been implicated in the yeast multifactor complex (eIF1-eIF3-eIF5-eIF2-GTP-Met-tRNAiMet). This is the first study to demonstrate that plant initiation factors are capable of forming a multifactor complex in vitro. In addition, the interaction of factors within these complexes was enhanced both in vitro and in native extracts by phosphorylation of one or more initiation factors by CK2. The importance of CK2 phosphorylation of eIF5 was evaluated by site-directed mutagenesis of eIF5 to remove CK2 phosphorylation sites. Removal of CK2 phosphorylation sites from eIF5 inhibits the CK2-mediated increase in eIF5 interaction with eIF1 and eIF3c in pulldown assays and reduces the eIF5-mediated stimulation of translation initiation in vitro. These results suggest a functional role for CK2 phosphorylation in the initiation of plant translation.
Translation initiation is a critical, rate-limiting step in eukaryotic gene expression, with a key step early in the pathway being the assembly of the 43 S preinitiation complex (1, 2). This 43 S preinitiation complex is composed of the small 40 S ribosomal subunit, eIF1, eIF1A, the eIF2-GTP-Met-tRNAiMet ternary complex, eIF3, and eIF5. Biochemical data from yeast suggest that a multifactor complex (MFC)2 consisting of eIF3-eIF1-eIF5-eIF2-GTP-Met-tRNAiMet exists free of the ribosome allowing these factors to be simultaneously loaded onto the 40 S ribosomal subunit (3). Mutations that disrupt the interaction of factors within this complex result in substantial reductions in translation initiation (3, 4).
Using a variety of in vitro binding assays, two-hybrid analysis, and in vivo purification of affinity-tagged subunits, an extensive analysis of the subunit interactions within the yeast MFC has resulted in a provisional model of the yeast MFC (5). The yeast eIF3c (NIP1) N-terminal domain plays a particularly important role in binding to other initiation factors in the context of the multifactor complex (6). The NIP1 N-terminal domain was found to bind directly to both eIF1 and eIF5 (7, 8), and it is suspected that this binding coordinates the interaction between eIF1 and eIF5 that is responsible for the inhibition of GTP hydrolysis at non-AUG codons (6). The C-terminal domain (CTD) of yeast eIF5 simultaneously binds to both the N terminus of eIF3c, the N-terminal K-boxes of eIF2β, and eIF1 (3, 9). This creates an indirect link between eIF3c and eIF2β, which is further supported by the direct interaction of eIF3a with eIF2β (10) and the multiple interactions of eIF1 (4). In yeast, eIF1 interacts directly with the N-terminal domains of both eIF2β and eIF3c, in addition to binding to the CTD of eIF5 (4, 6, 9). It is not clear, however, if all of these interactions are conserved in other eukaryotes, as NMR was unable to detect any interaction between human eIF1 and eIF5 (11).
The predicted binding domains from yeast appear to be conserved in plant initiation factors when analyzed by multiple sequence alignment; however, evidence of multifactor complex interactions in plants only comes from the anecdotal observations of co-purification (41). This study presents data to show that plant initiation factors do form multifactor complexes similar to those in yeast.
Because of the number of ionic interactions involved in MFC binding (4, 9, 10, 12), it is suspected that post-translational modifications that alter amino acid charges, such as phosphorylation, may play an influential role in MFC formation. Protein kinase CK2 (formerly known as casein kinase II) is a ubiquitous and highly conserved serine/threonine kinase found in both the nucleus and cytoplasm of all eukaryotic cells (13–16). CK2 is essential for cell viability, and it is involved in processes such as cell proliferation, transcriptional and translational control, cell cycle progression, and apoptosis (14). The mammalian eIF5 CTD is multiply phosphorylated by CK2 in response to cell cycle progression (17). Phosphorylation of the eIF5 C-terminal HEAT domain appears to be conserved in mammals (17), yeast (18), and plants (this study). Expression of mutant mammalian eIF5 with alanine substitutions at CK2 phosphorylation sites reduced the formation of eIF5-eIF2 complexes in vivo, and resulted in significant reductions in growth suggesting regulation by CK2 phosphorylation (19).
Plants have multiple forms of CK2 that are capable of differential phosphorylation of several initiation factors involved in the multifactor complex, including eIF2α, eIF2β, eIF3c, and eIF5 (41). In this study, CK2 phosphorylation sites have been identified in important binding regions of both the wheat eIF3c N terminus and the eIF5 C terminus. Of particular interest are modifications that affect the eIF3c N-terminal domain and the eIF5 C-terminal HEAT domain, both of which have been shown to play a central role in MFC assembly in yeast (5). These critical binding domains appear to be conserved in plant initiation factors, and in this study both domains in wheat initiation factors appear to be phosphorylated by CK2 isoforms on conserved residues. The results presented here provide biochemical evidence that CK2 phosphorylation enhances the interaction of subunits within the plant MFC, and thus suggests that CK2 may have a regulatory role in the initiation of plant protein synthesis.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant Translation Initiation Factors and CK2
To obtain nonphosphorylated plant initiation factors for analysis, Arabidopsis thaliana and wheat initiation factors that were identified as CK2 targets were cloned and expressed in Escherichia coli. All primers used are listed in the supplemental material. Constructs were confirmed by DNA sequencing (DNA Core Facility, Institute for Cell and Molecular Biology, University of Texas). The cloning and expression of wheat eIF1, His-eIF2α, eIF2β, His-eIF3c, and His-eIF5 as well as A. thaliana eIF2α, eIF2β, and eIF5 were described previously (20, 21, 41). Native eIF2 was purified as described previously (22).
eIF3c
To obtain recombinant wild-type wheat eIF3c without a His tag, the full-length cDNA for eIF3c was PCR-amplified from expressed sequence tag BJ257961 and cloned into pET23d(+) using NcoI/SacI restriction sites. This construct was transformed into Arctic Express DE3(RIL) E. coli cells (Stratagene) and expressed as described previously for His-eIF3c (41). Cells were harvested by centrifugation as described previously (21) and resuspended in 30 ml of Buffer B-500 (20 mm HEPES-KOH, pH 7.6, 0.1 mm EDTA, 1 mm DTT, 10% glycerol and KCl as indicated, where Buffer B-500 is Buffer B containing 500 mm KCl) supplemented with 1 Complete Protease Inhibitor tablet (Roche Applied Science), 4.2 mg of soybean trypsin inhibitor (Sigma), and 4.2 mg of phenylmethylsulfonyl fluoride (Sigma) pre-dissolved in 0.5 ml of isopropyl alcohol. The cells were disrupted by sonication, centrifuged, filtered, and diluted with Buffer B to lower the salt concentration to 100 mm KCl as described previously (21). The cell lysate was then loaded onto 2 ml of phosphocellulose (Whatman P11) equilibrated in Buffer B-100. eIF3c was eluted with a 10× gradient (20 ml) from 100 to 500 mm KCl, and 1-ml fractions were collected. Fractions were evaluated by SDS-PAGE, and the peak eIF3c-containing fractions were pooled, concentrated to ∼1 ml using an Amicon Ultra-4 centrifugal filter (Millipore), and stored at −80 °C.
To enrich for the N-terminal domain of eIF3c (eIF3c-NTD-6xHis), the coding region from the wheat-eIF3c-pET23d(+) was cleaved with NcoI/NdeI to produce a 947-bp fragment that coded for the N-terminal 318 amino acids of eIF3c. This fragment was cloned into pET15b(+) using NcoI/NdeI and then re-cut using NcoI/XhoI. The new NcoI/XhoI eIF3c N-terminally truncated fragment was cloned into pET23d(+) vector, such that a C-terminal His6 tag and stop codon were added to the coding sequence. The construct was confirmed by sequencing and transformed into BL21(DE3) E. coli cells for expression and purification using Ni-NTA (Qiagen) as described previously for His-tagged proteins (21).
eIF5
The cloning, expression, and purification of wild-type A. thaliana eIF5 (41) and wheat His-eIF5 (21) have been previously described. To express wild-type wheat eIF5, which did not possess a His6 tag, the eIF5 coding region of His-eIF5-pET15b was cleaved with Nde/BamHI and cloned into pET23b(+). This construct was transformed into BL21(DE3) E. coli, and wild-type wheat eIF5 was expressed and purified as described previously for A. thaliana eIF5 (41).
CK2
The cloning and expression of A. thaliana CK2 subunits have been previously described (41). Because Ni-NTA binding experiments were to be performed in this study, the removal of the His6 tags was desirable. A. thaliana CK2α1 was PCR-amplified from CK2α1-pET23d plasmid and cloned into pET23b using NdeI/BamHI. The new CK2α1-pET23b plasmid was transformed into BL21(DE3) E. coli and was expressed as described above. The cell extract was diluted with Buffer B to 300 mm KCl and loaded onto a 1-ml phosphocellulose column. CK2α1 was then eluted with a 10× gradient (10 ml) of Buffer B containing 0.3–1 m KCl. Fractions were analyzed by SDS-PAGE, and those containing the highest purity and concentration were pooled (∼5 ml). CK2α fractions were dialyzed overnight at 4 °C in Buffer B-100 and frozen at −80 °C. His-CK2β1 was used when holoenzyme was required for the phosphorylation of eIF2β (41).
Site-directed Mutagenesis of Wheat eIF2α, eIF2β, and eIF5
For each CK2 phosphorylation site identified in eIF2α, eIF2β, and eIF5, a forward and reverse primer was designed to alter the phosphorylated serine/threonine to alanine. Double and triple mutants were made for eIF2β (T52A/T85A) and eIF5 (S209A/T240A and S209A/T240A/S451A, the latter of which is abbreviated as eIF5AAA). Primers were also designed to alter eIF5 Ser-451 to glutamate (S451E). Initiation factors containing the desired mutations were expressed in BL21(DE3) E. coli and purified as described previously for the wild-type forms of each protein (20, 21).
NMR Spectroscopy
Recombinant His-eIF5 and His-eIF5AAA were analyzed to evaluate protein folding using one-dimensional NMR spectroscopy. NMR analysis was performed by Dr. David Hoffman, University of Texas Department of Chemistry and Biochemistry. Sample preparation, spectra measurement, and evaluation for one-dimensional NMR spectroscopy were performed as described previously (23).
Purification of Native Wheat eIF2 and eIF5
Native eIF2 and eIF5 were purified from wheat germ using methods similar to those described previously (22). To prevent dephosphorylation of proteins, a phosphatase inhibitor (calyculin A, BioSource) was included during the purification steps. Briefly, wheat germ (200 g) was ground in a cold blender and mixed with 200 ml of ice-cold extraction buffer (50 mm HEPES-KOH, pH 7.6, 120 mm KCl, 2 mm Mg(OAc)2, 2 mm CaCl2, 6 mm β-mercaptoethanol, 0.65 mm soybean trypsin inhibitor, 0.5 mm phenylmethylsulfonyl fluoride, 20 nm calyculin A (BioSource)) and centrifuged as described previously (22). Fractional precipitation was performed to 40% saturation with (NH4)SO4. A second (NH4)SO4 fraction was obtained from 40 to 70% saturation, and the proteins were collected by centrifugation. The proteins that precipitated in the 40–70% fraction were resuspended in ∼25 ml of Buffer B-50 containing 20 nm calyculin A. Dialysis was performed for 8 h with three changes of 500 ml of Buffer B-50 containing 20 nm calyculin A. The supernatant was then applied to a 20-ml Fast Flow DEAE-Sepharose column equilibrated in Buffer B-50 and 20 nm calyculin A. The column was washed with Buffer B-50 and 20 nm calyculin A until the A280 returned to base line and eIF5 was eluted from the column using a 200-ml gradient from 100 to 800 mm KCl in Buffer B containing 20 nm calyculin A. Fractions of 5 ml were collected, and 50-μl aliquots were taken of each fraction for analysis. Each fraction was analyzed by enzyme-linked immunosorbent assay using rabbit antibodies raised to recombinant wheat eIF2β or recombinant wheat eIF5, and fractions containing each protein peak were pooled. The peak eIF2 fractions (∼15 ml) were loaded onto a 1.5-ml phosphocellulose (Whatman P11) column equilibrated in Buffer B-100 plus 20 nm calyculin A. The column was washed with 15 ml of Buffer B-100, followed by 20 ml of Buffer B-300. eIF2 was eluted from the column with a 10× (15 ml) gradient from 300 ml KCl to 500 mm KCl, and 1-ml fractions were collected and analyzed by SDS-PAGE. Similarly, the peak eIF5 fractions (∼20 ml) from the DEAE-Sepharose column were loaded onto a 2-ml phosphocellulose column equilibrated in Buffer B-150 plus 20 nm calyculin A. The column was washed with 20 ml of Buffer B-150 plus 20 nm calyculin A, and eIF5 was eluted with a 10× (20 ml) gradient from 150 to 600 mm KCl. Fractions of 1 ml were collected and analyzed by SDS-PAGE. The peak fractions containing eIF2 and eIF5 were individually pooled and concentrated to ∼1 ml using an Amicon Ultra-4 centrifugal filter (Millipore).
Phosphorylation Site Identification
For in vitro phosphorylation, 2 μg of substrate (rWheIF2α, rWheIF2β, rWheIF3c, rWheIF3c-NTD, rWheIF4B, rWheIF5, rAteIF2α, rAteIF2β, or rAteIF5) was incubated in a 20-μl reaction (50 mm HEPES-KOH, pH 7.6, 5 mm MgCl2, 2.4 mm DTT, 0.2 mm ATP, and 100 mm KCl) with and without 5 pmol of CK2 for 1 h at 30 °C. To optimize phosphorylation, eIF2α, eIF3c, eIF4B, and eIF5 were incubated with recombinant A. thaliana CK2α1. No phosphorylation of eIF2β was detected in the absence of the CK2β subunit; thus a complex of CK2α1β4 was used to phosphorylate eIF2β as described previously (41). The reactions were loaded onto a NuPAGE 12% BisTris gel (Invitrogen), run using MOPS running buffer (Invitrogen), and stained with colloidal Coomassie Blue (Invitrogen). The substrate bands from both the kinase treatment and the no kinase reactions were cut from the gel, and prepared for analysis by mass spectrometry as described previously (41).
For in vivo site identification, the endogenous wheat germ eIF2 and eIF5 preparations were separated on a NuPAGE 12% BisTris gel, and bands containing native wheat eIF2α, eIF2β, and eIF5 were prepared for analysis by mass spectrometry as described previously (41).
Phosphopeptide enrichment and site analysis were performed at the Analytical Instrumentation Facility Core using a MALDI-TOF/TOF instrument (4700 Proteomics Analyzer, Applied Biosystems). The digested protein samples were enriched for phosphopeptides using an immobilized metal affinity chromatography (IMAC) resin (Phos-Select, Sigma). The samples were treated according to the manufacturer's protocol using an elution solution composed of 5% phosphoric acid, 50% acetonitrile. Unseparated digest flow-through (phosphopeptides depleted) and eluant samples (phosphopeptide enriched) were analyzed by MALDI-MS. Prior to MS analysis, each tryptic digest was dried to <5 μl in a SpeedVac (ThermoSavant), desalted with a Ziptip μ-C18 pipette tip (Millipore), and eluted with 1.5 μl of matrix onto three spots on the MALDI target. The matrix was 0.7 mg/ml α-cyano-4-hydroxycinnamic acid (LaserBioLabs Labs) in 67% acetonitrile, 0.2% trifluoroacetic acid. 4700 Proteomics Analyzer Calibration Mixture (Applied Biosystems) was utilized for instrument calibration.
Phosphopeptides were identified using a comparative MALDI-MS strategy described previously (24). The MS spectra were compared between proteins incubated with CK2 or with buffer. MS spectra were obtained over a mass range of 700–4000 and calibrated externally. MS spectra were acquired in linear and reflectron modes. Phosphopeptides were indicated by the presence of new peptides in the CK2-treated samples or the appearance of a neutral loss ion of poor resolution in the reflector MS. Additionally, peptides enriched in the IMAC fraction were tested for the presence of phosphorylation by acquisition of MS/MS, where neutral loss of phosphoric acid is seen as the dominant fragment ion. MS spectra were also analyzed using MASCOT 2.0 (Matrix Science) in the GPS Explorer version 3.5 software (Applied Biosystems), and peptides with masses corresponding to eIF phosphopeptides were subjected to MS/MS analysis. MS/MS fragmentation spectra were acquired for all phosphopeptides to determine the site of modification. Loss of phosphoric acid from the parent and fragment ions was observed with a mass shift of −98 Da. For most peptides, the site(s) of phosphorylation were unambiguously assigned, but in some cases, a stretch of residues contained potential sites and no fragment ions were available to distinguish the exact site. When possible, the MS/MS of the corresponding unphosphorylated peptide was acquired and compared with the phosphopeptide as an additional tool to map the site of phosphorylation.
In Vitro Phosphorylation of eIF2α, eIF2β, eIF3c, and eIF5
Phosphorylation reactions (20 μl) contained 5 μg of substrate, 1 pmol of recombinant CK2α1 or CK2α1β1 (for eIF2β containing reactions) in 50 mm Tris-HCl, pH 7.6, 5 mm MgCl2, 2.4 mm DTT, 0.2 mm [γ-32P]ATP (∼200 cpm/pmol), and 100 mm KCl. Reactions were incubated at 30 °C for 30 min and terminated by the addition of 4× SDS-PAGE loading dye. Proteins were separated by SDS-PAGE, and gels were exposed to a PhosphorImager screen (GE Healthcare) for 12 h. The screen was then analyzed with Molecular Imager FX using Quantity One software (Bio-Rad).
Pulldown Assays
Approximately 400 pmol of C-terminally His6-tagged eIF3c-NTD or N-terminally His6-tagged eIF5 was mixed with the indicated amounts and combinations of wheat reIF1, native eIF2, reIF2α, reIF2β, reIF3c, or reIF5 in 500-μl reactions containing binding buffer (20 mm HEPES-KOH, pH 7.6, 10% glycerol, 150 mm KCl, 5 mm MgCl2, 0.2 mm ATP, 0.1% Nonidet P-40, 20 mm imidazole) and 10 μl of Ni-NTA resin (Qiagen). A similar reaction was conducted in the absence of the His6-tagged protein to control for any interaction between wheat initiation factors and the Ni-NTA matrix. In indicated experiments, the nonhydrolyzable ATP analogue AMP-PCP (Roche Applied Science) was substituted for ATP. The mixture of factors was incubated for 30 min at room temperature with end-over-end mixing followed by up to 12 h at 4 °C. The beads were then extensively washed with binding buffer, and the bound proteins were eluted with 1× SDS gel buffer containing 300 mm imidazole and separated by SDS-PAGE. Assays were either visualized by Coomassie Blue staining or Western analysis.
For pulldowns involving nontagged factors, polyclonal rabbit antibodies were affinity-purified using proteins bound to a nitrocellulose membrane as described previously (25). Native wheat eIF2 (200 pmol) was incubated with 200 pmol of recombinant wheat eIF1 in the presence of 20 mm HEPES-KOH, pH 7.6, 10% glycerol, 150 mm KCl, 5 mm MgCl2, 0.2 mm ATP, 0.1% Nonidet P-40, and CK2 as described above. Protein A-Sepharose beads (10 μl, Sigma) were pre-bound to 5 μg of affinity-purified antibodies to recombinant wheat eIF2β for 1 h at 4 °C. The pre-bound beads were added to each reaction and allowed to incubate for 1 h at 4 °C. The beads were washed extensively with 20 mm HEPES-KOH, pH 7.6, 10% glycerol, 150 mm KCl, 5 mm MgCl2, 0.2 mm ATP, 0.1% Nonidet P-40, and the bound proteins were eluted with 1× SDS loading buffer, separated by SDS-PAGE. eIF1 was visualized using the One-step Western Complete kit (GenScript) using a 1:5000 dilution of affinity-purified rabbit antibodies to recombinant wheat eIF1.
Western Analysis
Antiserum was previously raised to recombinant wheat initiation factors eIF1, eIF2α, eIF2β, His-eIF3c, and His-eIF5 (21). Following separation by SDS-PAGE, proteins were blotted to PVDF and blocked with HNAT (10 mm HEPES-KOH, pH 7.6, 150 mm NaCl, 0.2% bovine serum albumin, 0.2% Tween 20) containing 5% dry milk. Blots were then incubated with a 1:5000 dilution of the primary antibody as indicated for 3 h at room temperature, followed by extensive washes with HNAT/milk solution. The second antibody was a 1:20,000 dilution in HNAT/milk of goat anti-rabbit horseradish peroxidase secondary antibody and incubated for 1 h at room temperature. Following extensive washes with HNAT (>4), antibody-reactive bands were visualized by chemiluminescence (SuperSignal West Pico, Pierce) and exposure to film. To analyze blots with multiple antibodies, the membrane was treated with stripping buffer (5% SDS, 20 mm β-mercaptoethanol, and 125 mm Tris-HCl, pH 6.8) for 30 min at 65 °C, blocked with HNAT containing 5% milk, and incubated with a different primary antibody. PVDF membranes were stripped 3–4 times with no significant loss of signal from bound proteins.
Preparation of eIF5-depleted Wheat Germ Extracts
Wheat germ S30 extracts were prepared as described previously (22) and depleted of eIF5 using protein A beads (Pierce) cross-linked to polyclonal rabbit antibodies raised against recombinant wheat eIF5. A 2-ml protein A anti-wheat eIF5 column was prepared using the protein A IgG Plus orientation kit (Pierce) according to the manufacturer's instructions. The column was equilibrated with 20 ml of Buffer A (20 mm HEPES-KOH, pH 7.6, 5 mm Mg(OAc)2, 120 mm KOAc, 6 mm 2-mercaptoethanol, 10% glycerol). Wheat germ S30 extract (15 ml) was passed over the column, and 1-ml fractions were collected. The S30 flow-through fractions were analyzed by Western blot, and the eIF5-depleted fractions were pooled and stored in 100-μl aliquots at −80 °C.
eIF5-dependent in Vitro Translation Assay
Recombinant eIF5 was assayed for its ability to stimulate translation in the wheat germ S30 assay system (22, 26) using eIF5-depleted wheat germ extracts. Briefly, 50-μl reactions containing 5 pmol of HSP21 mRNA template, recombinant eIF5 (as indicated), 4 pmol of recombinant wheat eIF4F (21), 24 mm HEPES-KOH, pH 7.6, 2.9 mm Mg(OAc)2, 100 mm KOAc, 30 mm KCl, 2.4 mm DTT, 0.1 mm spermine, 1 mm ATP, 0.2 mm GTP, 34 μm [14C]leucine, 50 μm 19 amino acids (minus leucine), 7.8 μm creatine phosphate, 3 μg of creatine kinase, 0.75 A260 units of yeast tRNA, and 20 μl of eIF5-depleted wheat germ extract were used. Prior to performing translation reactions, the eIF5-depleted wheat germ extract was incubated in the presence of 50 nm calyculin A for 15 min on ice. Translation reactions were incubated for 30 min at 27 °C, and the amount of [14C]leucine incorporated into polypeptide was determined as described previously (22, 26).
RESULTS
Identification of CK2 Phosphorylation Sites
Wheat germ eIF2α and eIF3c were known substrates of an endogenous wheat germ kinase (27). This kinase was recently identified as CK2 by this laboratory (41). Further investigation into the phosphorylation of recombinant plant initiation factors revealed that CK2 substrates were not restricted to the eIF2α and eIF3c subunits but also included eIF2β, eIF4B, and eIF5 (41).
Several of these CK2 substrates (eIF2α, eIF2β, eIF3c, and eIF5) are involved in the formation of the MFC that has been extensively studied in yeast (3). To evaluate the potential effect CK2 phosphorylation has on plant translation initiation factors and their possible role in regulation of the MFC, it was necessary to identify the residues that were modified by the kinase. Our analysis of initiation factor phosphopeptides produced by in vitro CK2 phosphorylation has identified numerous phosphorylation sites in eIF2α, eIF2β, eIF3c, and eIF5 (summarized in Table 1). In addition, we have obtained experimental evidence that some of these sites are phosphorylated in preparations of native factors (Table 2).
TABLE 1.
CK2 phosphorylation sites in plant initiation factors
| Substrate | Mass spectrometry CK2 in vitro phosphorylation sites | In silico predicted CK2 phosphorylation sitesa | In vitro CK2 phosphorylation sitesb | Confirmed by mutagenesis | |
|---|---|---|---|---|---|
| eIF2α | Wheat | Ser-318 | 1–4 | 1 | Ser-318 |
| A. thaliana | Ser-317,c Ser-322c | 1–5 | 2 | NDd | |
| eIF2β | Wheat | Thr-52, Thr-85 | 1–2 | 1 | Thr-52, Thr-85 |
| A. thaliana | Ser-42, Ser-80, Ser-112 | 0–4 | 1 | ND | |
| eIF5 | Wheat | Ser-209, Thr-240, Ser-451 | 0–3 | 3 | Ser-209, Thr-240, Ser-451 |
| A. thaliana | Ser-201, Thr-230/Thr-232/ Ser-233, Ser-428, Ser-431, Ser-433 | 0–5 | 3 | ND | |
| eIF3c | Wheat | Ser-11, Ser-13, Ser-22, Ser-26, Ser-28, Ser-31, Ser-53, Ser-56, Ser-198, S Ser-10, Ser-212, Ser-215 | 5–15 | 8 | ND |
a Ranges of the number of potential CK2 sites were predicted using ScanSite 2.0, NetPhosK 1.0, or KinasePhos with low and high stringency cutoff parameters.
b Number of CK2 sites were determined by in vitro phosphorylation (41).
c The site of A. thaliana eIF2α phosphorylation could not be distinguished. The C-terminal peptide could not be visualized due to the lack of any basic amino acids, a high mass, and multiple methionine residues that may be partially oxidized. Based on computational analysis and alignment with the wheat Ser-318 site, it is suspected that AteIF2α is phosphorylated on Ser-322 and Ser-317.
d ND indicates not determined.
TABLE 2.
Observation of wheat CK2 phosphopeptides in vitro and in vivo
|
In vitroa |
In vivob |
|||
|---|---|---|---|---|
| Unmodified | Phosphopeptide | Unmodified | Phosphopeptide | |
| eIF2α | ||||
| Ser-318 | No | Yes | Yes | Yes |
| eIF2β | ||||
| Thr-52 | Yes | Yes | Noc | Noc |
| Thr-85 | Yes | Yes | Noc | Noc |
| eIF5 | ||||
| Ser-209 | Yes | Yes | Yes | Yesd |
| Thr-240 | Yes | Yes | Yes | Yesd |
| Ser-451 | No | Yes | No | Yes |
a Data are from recombinant proteins.
b Proteins were purified from wheat germ extracts in the presence of the phosphatase inhibitor calyculin A.
c Note that wheat germ extracts appear to lack significant amounts of CK2β necessary for phosphorylation of eIF2β.
d Native samples contained peptides 205GAGGSDEEHVSSPR218 + 80 Da and 220DADFAAAADGDDDDDDDVQWATDTSAEAAR248 + 80 Da. A neutral loss of 98 Da was observed in tandem MS for each peptide, with a limited number of fragment ions due to low stoichiometry of phosphorylation (see supplemental material for details).
eIF2α
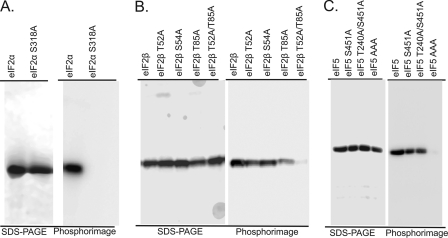
Wheat eIF2α has 1–4 predicted CK2 consensus sites (Ser-97, Ser-166, Ser-318, and Thr-324); however, only Ser-318 is of high stringency. Mass spectrometry of the tryptic peptide 297LLNAQLDTLVEQNAEVAGDDDSEDEEDTGMGDIDLTNSGVHAD339 of in vitro phosphorylated recombinant eIF2α identified Ser-318 as the only CK2 phosphorylation site. To further substantiate Ser-318 as the only wheat eIF2α CK2 phosphorylation site, a mutant that cannot be phosphorylated (eIF2α S318A) was prepared. As shown in Fig. 1A, only the wild-type wheat eIF2α and not the mutant eIF2α S318A is phosphorylated by CK2. This result established that there is only one CK2 phosphorylation site in wheat eIF2α, and this site is conserved across plants (Fig. 2).
FIGURE 1.
Phosphorylation of wild-type and mutant eIF2α, eIF2β, and eIF5 by CK2. Recombinant wheat eIF2α, eIF2α S318A, eIF2β, eIF2β T52A, eIF2β S54A, eIF2β T85A, eIF2β T52A/T85A, eIF5, eIF5 S451A, and eIF5AAA (S209A, T240A, and S451A) were all incubated in the presence of 1 pmol of CK2α1 or CK2α1β1 as described under “Experimental Procedures.” Reactions were separated by 12.5% SDS-PAGE, stained with Coomassie Blue, and analyzed by phosphorimaging.
FIGURE 2.
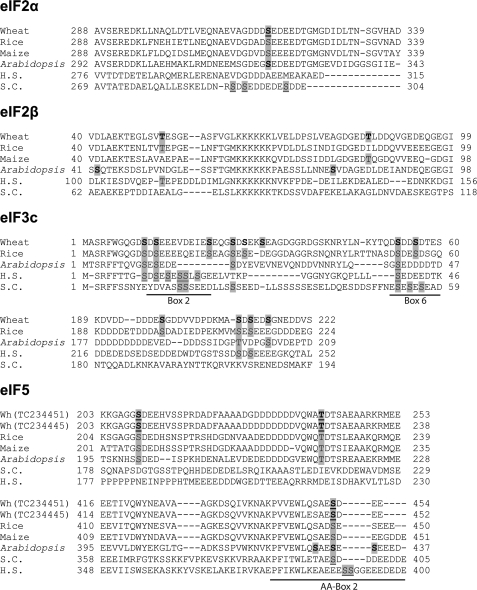
Multiple sequence alignment of eIF2α, eIF2β, eIF3c, and eIF5 amino acid sequences. Sequences for eIF2α from rice (EAY89622), maize (TC318839), A. thaliana (AAK29673), human (H.S.) (P05198), and Saccharomyces cerevisiae (S.C.) (EDN63334) that align with residues 288–339 of wheat eIF2α (TC263298) are shown. Sequences for eIF2β are shown for rice (Os07g06841000), maize (TC318899), A. thaliana (AAK29672), human (NP_003899), and S. cerevisiae (SUI3) and aligned with residues 42–99 of wheat eIF2β (0Z4473). N-terminal residues 1–60 of wheat eIF3c (TC235391) as well as well as residues 189–222 are aligned with residues from rice (EAZ38556), A. thaliana (AAC83464), S. cerevisiae (END64250), and human eIF3c (AAD03462). Segments in the yeast eIF3c (NIP1) implicated in binding eIF1 and eIF5 (Box 2 and 6) are indicated (6). No complete maize eIF3c sequence was found. Shown are eIF5 sequences of wheat (TC234451 and TC234445), maize (CAA10616), rice (EAZ02236), A. thaliana (NP177907), S. cerevisiae (P38431), and human (CAG32993). AA-Box 2 from the eIF5 C-terminal HEAT domain is indicated. In vitro CK2 phosphorylation sites in wheat and A. thaliana factors that were identified in this study are indicated in bold. Conservation of similar CK2 sites are highlighted. In vivo wheat factor phosphorylation sites observed in this study are underlined, as well as the in vivo phosphorylation of the eIF3c N terminus at a CK2 consensus site reported by large scale phosphoproteomic analysis for A. thaliana (28) or mammals (37). CK2 phosphorylation sites in the C terminus of both S. cerevisiae and human eIF5 (18, 40) have also been observed both in vitro and in vivo and are also underlined in their respective sequences.
Mass spectrometric analysis of native wheat eIF2 purified in the presence of the phosphatase inhibitor calyculin A suggests that eIF2α from wheat germ is at least partially phosphorylated on this residue in vivo, as both phosphorylated and unphosphorylated isoforms of eIF2α Ser-318 were observed in our analysis (Table 2 and supplemental Fig. A).
A. thaliana eIF2α contains up to five predicted CK2 phosphorylation sites (Ser-185, Ser-295, Thr-276, Ser-317, and Ser-322). Of these sites only Ser-322, which aligns with wheat Ser-318, is of high stringency. Quantitative phosphorylation of AteIF2α suggests two phosphorylation sites are present (41). Although clearly phosphorylated in the presence of CK2 and [32P]ATP, no phosphopeptides were detected during the mass spectrometric analysis of AteIF2α tryptic fragments. The tryptic fragment containing Ser-295 was not observed even in unphosphorylated samples. This is likely because it is less than 700 Da. The peptide containing Ser-317 and Ser-322 was also not observed either phosphorylated or triphosphorylated. This C-terminal peptide lacks any basic amino acids, has a high mass, and contains multiple methionines that may be partially oxidized. These factors all likely contribute to the difficulty in detecting it by mass spectrometry. Although no sites could be unambiguously identified, based on computational analysis, alignment with wheat eIF2α Ser-318, and the elimination of phosphorylation by a C-terminal truncation of AteIF2α (data not shown), it is likely that AteIF2α is phosphorylated on Ser-317 and Ser-322.
eIF2β
Wheat eIF2β contains 1–2 predicted CK2 consensus sites (Thr-52 and Thr-112). The peptide containing Thr-112 was only detected in the nonphosphorylated form, and thus this residue does not appear to be a bona fide CK2 site. Analysis of the tryptic peptides found that the mass of peptide 46TEGLSVTESGEASFVGLK63 is shifted by a single phosphate; unfortunately, the MS/MS spectra for this region could not differentiate between Thr-52 and Ser-54. Although only Thr-52 is a consensus CK2 site, based on the spectra alone it cannot be ruled out that Ser-54 is phosphorylated by CK2 in vitro. Using site-directed mutagenesis, Thr-52 was unambiguously identified as the site of phosphorylation, as only the wheat eIF2β T52A mutant exhibited a reduction in phosphorylation by CK2, whereas eIF2β S54A is phosphorylated by CK2 to a similar extent as wild-type eIF2β (Fig. 1B). In addition to Thr-52, Thr-85 was also observed partially phosphorylated. The double eIF2β mutant (eIF2β T52A/T85A) is not phosphorylated by CK2 in vitro (Fig. 1B). Quantitative analysis of eIF2β phosphorylation suggests that ∼1 mol of phosphate is incorporated into each mole of recombinant wheat eIF2β in vitro (41). It was thus surprising that two unique phosphorylation sites were identified by mass spectrometry and mutagenesis (Thr-52 and Thr-85). Taken together, this suggests that the recombinant eIF2β is not homogeneously phosphorylated by CK2, and each site may only be partially phosphorylated. Some portion of the recombinant enzyme, although soluble, may not be properly folded in the absence of the other two eIF2 subunits (α and γ). Improper folding could result in the observed reduction in CK2 phosphorylation. An analysis of native eIF2β from wheat germ did not reveal any phosphopeptides (Table 2). Because wheat germ extracts do not appear to contain CK2β subunit, which is required for eIF2β phosphorylation (41), this result is not surprising.
AteIF2β is predicted to have up to four CK2 phosphorylation sites. Sites at Ser-42, Ser-80, and Ser-112 were identified by mass spectrometry. In addition, a weak di-phosphopeptide containing Ser-41, Ser-42, and Thr-44 was also detected, suggesting that either Ser-41 or Thr-44 is weakly phosphorylated by CK2 in addition to Ser-42. Both the phosphorylated and unphosphorylated versions of each peptide were observed during mass spectrometric analysis. Quantitative analysis of AteIF2β phosphorylation suggests that ∼1 mol of phosphate is incorporated into each mole of substrate (Table 1), suggesting that like wheat eIF2β, the phosphorylation of AteIF2β is not homogeneous. Interestingly, there is poor alignment of the wheat and Arabidopsis CK2 sites in eIF2β compared with other initiation factor CK2 substrates.
eIF3c
Wheat eIF3c has 5–15 predicted CK2 consensus sites. Initial attempts to identify the phosphorylation sites using full-length recombinant wheat eIF3c failed because of poor spectral coverage in the N terminus. The N-terminal region of eIF3c is not only highly acidic, but it also contains a large number of CK2 phosphorylation sites and has few tryptic cleavage sites. The large negative charge imparted by this combination makes the peptides from this region difficult to analyze using mass spectrometry. Two critical peptides, amino acids 5–38 and 45–62, could be clearly observed in the nonphosphorylated sample, but they were undetectable in phosphorylated samples of full-length eIF3c. Even in negative ion mode, no phosphorylated version of the peptide was seen. This is likely due to the formation of adducts with Na+ or Fe2+ in the presence of multiple phosphorylation events. To overcome this problem and enrich for peptides specifically in this region, a 315-amino acid N-terminal truncation mutant of eIF3c was created (eIF3c-NTD) containing a His6 tag. Using this construct 12 CK2 phosphorylation sites were identified in eIF3c (Table 1).
The MS spectra for eIF3c-NTD phosphorylation was also complicated by the presence of multiple phosphorylation states in combination with a variable number of Fe2+ adducts, which resulted in the appearance of peptide-adduct peak clusters with varying amounts of phosphorylation. The strongest peak formed by the phosphorylated peptide 5FWGQGDSDSEEEVDEIESEQGSDSEKSEAGDGGR38 occurs at 3889 Da, suggesting three phosphorylation sites; however, there are peaks for this cluster that correspond with the addition of up to six phosphates and Fe2+, suggesting that all six serines within this region (Ser-11, Ser-13, Ser-22, Ser-26, Ser-28, and Ser-31) can be phosphorylated by CK2 in vitro. The phosphorylation of Ser-31 appears to inhibit trypsin cleavage at Lys-30. Two phosphates also appeared to be incorporated into the peptide 49YTQDSDDSDTESHR62, and the MS/MS for this peptide identified Ser-53 and Ser-56 as in vitro CK2 phosphorylation sites. The mass of the eIF3c-NTD peptide from 174CRENPESFEDDVADDKDVDDDDDESGDDVVDPDK207 was found to shift in the phosphorylated sample because of the presence of a single phosphorylation site, which was identified at Ser-198. Similarly, peptide 208MASDSEDSGNEDDVSQDGGAWEK230 was found to contain three phosphorylation sites, which were identified in the MS/MS at Ser-210, Ser-212, and Ser-215. Because of the large number of phosphorylation sites and the difficulties that presents to mass spectrometry, confirmation by mutagenesis and the analysis of native eIF3c from wheat germ was not undertaken in this study. A. thaliana eIF3c has not been expressed and purified; however, 7 of the 12 sites identified in wheat eIF3c are conserved in AteIF3c (Fig. 2). AteIF3c does appear to be phosphorylated in vivo, as a large scale phosphoproteomic analysis recently observed AteIF3c to be phosphorylated on the consensus CK2 site Ser-40 (28).
eIF5
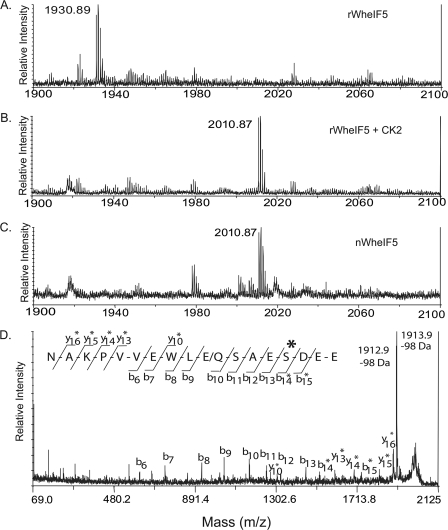
Wheat eIF5 has up to three predicted CK2 consensus sites, and quantitative phosphorylation suggests that there are three sites per molecule (Table 1). Using mass spectrometry, we have unambiguously identified three distinct CK2 in vitro phosphorylation sites in recombinant wheat eIF5 at Ser-209, Thr-240, and Ser-451. All three sites are conserved across plants (Fig. 2). The linear MS spectra produced from recombinant wheat eIF5 peptide 438NAKPVVEWLQSASDEE454 exhibits the predicted mass of 1930.89 (Fig. 3A). Following in vitro phosphorylation by CK2, the mass of this peptide shifts by +80 Da to 2010.87 (Fig. 3B).
FIGURE 3.
Mass spectrometric analysis of wheat eIF5. Shown are spectra from MS analysis of wheat eIF5 peptides following IMAC enrichment. MALDI-TOF spectra of peptides from recombinant wheat eIF5 (A), recombinant wheat eIF5 phosphorylated by CK2 in vitro (B), and native wheat eIF5 (C) are shown. A shift in peptide mass from 1930.89 to 2010.87 by +80 Da is shown. This peptide sequence corresponds to the wheat eIF5 C-terminal tryptic peptide 438NAKPVVEWLESAESDEE454. This peptide exhibits the expected mass of 1930.89 Da in recombinant wheat eIF5 (A). Following in vitro phosphorylation by CK2 (B), this peptide shifts by +80 Da to 2010.87, representing the addition of one phosphate. Native wheat eIF5 purified in the presence of phosphatase inhibitor calyculin A exhibits a similar peak at 2010.87 and the absence of any peak at 1930.89, suggesting it is fully phosphorylated. D, native wheat eIF5 was analyzed by MALDI-MS/MS following IMAC sample enrichment. This sample contained two distinct eIF5 isoforms, represented by the MS/MS of 2010.9 and 2011.9, that correspond to peptide sequences NAKPVVEWLESAES(Pi)DEE (m/z 2011.9) and NAKPVVEWLQSAES(Pi)DEE (m/z 2010.9). Fragment ion peaks from the two isoforms show monoisotopic doublets representative of peptide doublet shifted by 1 Da. Fragment ions shifted by the addition of a phosphate (+80 Da) or loss of phosphoric acid (−98 Da) are indicated with an asterisk. Based on the shift of b- and y-ions, peptides from both isoforms of native wheat eIF5 are phosphorylated on the residue corresponding to Ser-451.
Native wheat eIF5 was purified from wheat germ in the presence of the protein phosphatase inhibitor calyculin A. Following IMAC enrichment, the native sample exhibits a peak representing the phosphorylated C-terminal tryptic fragment plus one phosphate for the peptide containing Ser-451 (Fig. 3C). Mass spectrometric analysis of wheat germ eIF5 revealed two distinct isoforms of eIF5 (TC234451 and TC234445), and both were nearly 100% phosphorylated at the C-terminal residue corresponding to Ser-451 (Fig. 3). There is also evidence for the partial phosphorylation of both Ser-209 and Thr-240 in this preparation (see supplemental Figs. B and C). There are strong signals in the native sample representing peptides 205GAGGSDEEHVSSPR218 + 80 Da and 220DADFAAAADGDDDDDDDVQWATDTSAEAAR248 + 80 Da, but unfortunately there is only a neutral loss of 98 Da with no other fragment ions in the tandem mass spectra (see supplemental Figs. B and C). It cannot be ruled out that Ser-215, Ser-216, Thr-242, or Ser-243 could also be phosphorylated; however, on both peptides only one residue is phosphorylated, and the corresponding in vitro studies point to phosphorylation of Ser-209 and Thr-240. Based on the low stoichiometry of phosphorylation at these sites, it is difficult to resolve the identity of the phosphorylated residues in these two fragments. It is possible that the use of other phosphatase inhibitors would improve the stoichiometry observed for phosphorylation in vivo at Ser-209 and Thr-240. No phosphorylation sites could be detected in native wheat eIF5 when purified in the absence of the phosphatase inhibitor calyculin A.
To confirm Ser-209, Thr-240, and Ser-451 as CK2 phosphorylation sites, various mutants containing alanine substitution at one or more of the phosphorylation sites were prepared and expressed. Wild-type wheat eIF5, which contains three phosphorylation sites, is more heavily phosphorylated than eIF5 S451A, suggesting loss of a CK2 site (Fig. 1C). The triple alanine mutant eIF5 S209A/T240A/S451A is no longer phosphorylated by CK2 in vitro (Fig. 1C), confirming that there are only three CK2 phosphorylation sites in wheat eIF5.
Several phosphopeptides were also observed in CK2-treated A. thaliana eIF5. The peptide 198HSSDEDISPK208 was observed with one phosphorylation event, which is either on Ser-200 or Ser-201, or partial phosphorylation of both. There were only weak fragment ions in the MS/MS, so these adjacent sites could not be distinguished, but the Ser-201 site aligns with the unambiguous Ser-209 CK2 site identified in wheat eIF5 (Fig. 3). The peptide from 209HDENALEVDEDEDDDDGVEWQTDTSR234 was observed partially phosphorylated on Thr-230, Thr-232, or Ser-233. Thr-230 aligns with the unambiguous Thr-240 CK2 site from wheat eIF5. The peptide from 418NVKPFVEWLQSAESESEEED437 is observed, most intensely as a diphosphopeptide but also very weakly as a triphosphopeptide. The MS/MS of the disphosphopeptide shows that Ser-431 and Ser-433 are phosphorylated, whereas Ser-428 is phosphorylated in the triphosphopeptide. The peptide appears to be completely phosphorylated at the Ser-431 and Ser-433 sites, as the unphosphorylated peptide is not observed.
Additional CK2 Substrates
Two additional protein synthesis machinery CK2 substrates, Co-eIF2α (29) and Co-eIF2β (30), were identified by in vitro phosphorylation and mass spectrometry. Tryptic peptides identified Co-eIF2α as dehydrin and Co-eIF2β as nucleolin (data not shown). Nucleolin has been identified as a CK2 substrate in mammals (31), and dehydrin was identified as a CK2 substrate in maize (32). Co-eIF2α and Co-eIF2β stimulate the formation of a ternary complex in vitro by different means, and their role in protein synthesis initiation is not clear (29, 30). Further analysis of phosphorylation sites was not pursued.
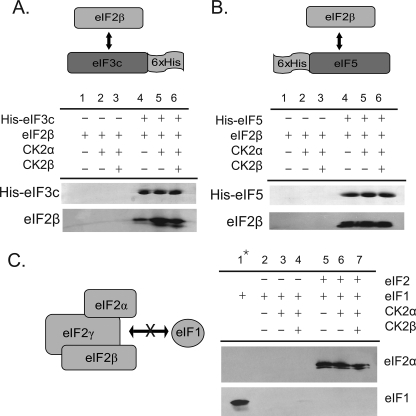
Interaction of Wheat Initiation Factors with eIF3c-NTD in Vitro
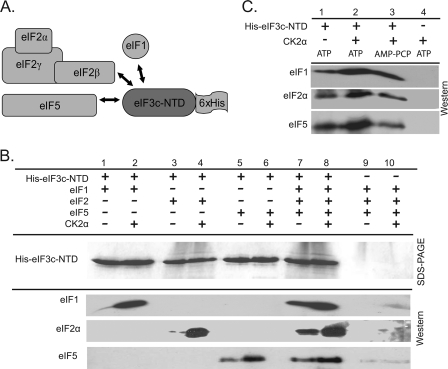
The interaction of eIF1, eIF2, and eIF5 with the N-terminal domain of wheat eIF3c was evaluated using a series of Ni-NTA pulldowns (Fig. 4A). His-eIF3c-NTD directly interacts in vitro with eIF1, eIF2, and eIF5 (Fig. 4B, lanes 1, 3, and 5, respectively) in the apparent formation of a multifactor complex. In addition, the ability of His-eIF3c-NTD to bind to individual factors is enhanced in the presence of all three factors (Fig. 4B, compare lanes 1 and 3 and 5–7). This cooperative binding may be due to additional bridging interactions between the various subunits or the result of conformational changes that might occur in the eIF3c N terminus upon interaction with factors. The interaction of recombinant wheat eIF1 with the eIF3c-NTD occurs modestly under the in vitro binding conditions used in Fig. 4. This assay was conducted at 150 mm KCl, and at lower salt concentrations (∼50 mm KCl) the eIF3c/eIF1 interaction is more robust, suggesting an ionic interaction is responsible (results not shown).
FIGURE 4.
CK2 enhances interaction of MFC components with His-eIF3c-NTD in vitro. A, potential interactions of wheat initiation factors with His-eIF3c-NTD were evaluated by a series of Ni-NTA pulldowns. B, His-eIF3c-NTD (400 pmol) was mixed with ∼400 pmol of the following initiation factors as indicated: recombinant wheat eIF1, native wheat eIF2 (22), and/or recombinant wheat eIF5. 10 pmol of recombinant A. thaliana CK2α1 was used to phosphorylate initiation factors as indicated. Control reactions (lanes 9 and 10) were conducted in the absence of the His-tagged protein to evaluate the interaction between wheat initiation factors/CK2α and the Ni-NTA beads. The eluted proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining or transferred to PVDF and analyzed with rabbit antisera to recombinant wheat eIF1, eIF2α, and eIF5 as described under “Experimental Procedures.” C, similar set of Ni-NTA pulldown experiments was performed in the presence of His-eIF3c-NTD, eIF1, eIF2, and eIF5. CK2 and ATP/AMP-PCP are as indicated.
In addition to binding recombinant wheat eIF1 and eIF5, His-eIF3c-NTD unexpectedly bound to native wheat eIF2 in the Ni-NTA pulldown experiments. Based on interaction of recombinant eIF2β with His-eIF3c (Fig. 6A), it is likely that the interaction of eIF3c with native eIF2 occurs primarily through the β-subunit.
FIGURE 6.
MFC interactions with eIF2. A, His-eIF3c (250 pmol) was mixed with ∼250 pmol of recombinant wheat eIF2β. 10 pmol of recombinant A. thaliana CK2α1 or 10 pmol of recombinant A. thaliana CK2α1 plus 10 pmol of CK2β1 were used to phosphorylate initiation factors as indicated. Control reactions were conducted in the absence of the His6-tagged protein to evaluate the interaction between non-His6-tagged proteins and Ni-NTA matrix. The elution was analyzed by SDS-PAGE, and the gel was cut such that bands could be visualized by Coomassie Blue staining or transferred to PVDF and analyzed with rabbit antisera to recombinant wheat eIF2β. B, similar analysis was performed using His6-eIF5 as described in A. C, native wheat eIF2 (200 pmol) was incubated with 200 pmol of recombinant wheat eIF1. Factors were phosphorylated as described under “Experimental Procedures,” and affinity-purified antibodies to eIF2β and protein A-Sepharose beads (Sigma) were added to each reaction and analyzed with a One-step Western Complete kit (GenScript) using a 1:5000 dilution of rabbit antisera to recombinant wheat eIF1. A loading and Western control lane (1*) is shown that contained only eIF1 (∼10 pmol).
Effect of CK2 Phosphorylation on eIF3c-NTD Pulldowns
The N-terminal domain of wheat eIF3c is highly phosphorylated by CK2 in vitro, and the presence of CK2α significantly increased the interaction of His-eIF3c-NTD with eIF1, eIF2, and eIF5 in Ni-NTA pull downs (Fig. 4B, compare lanes 1, 3, and 5 with 2, 4, and 6, respectively). When eIF1, eIF2, and eIF5 are all present, the binding of individual subunits to His-eIF3c-NTD is increased as observed previously (Fig. 4B, lane 7); however, the presence of CK2α provides a further enhancement in the binding of eIF1, eIF2, and eIF5 to the His-eIF3c-NTD (Fig. 4B, compare lanes 7 and 8). The increased interaction that occurs in the presence of CK2α is ATP-dependent, as no similar increase occurs in the presence of the nonhydrolyzable analogue AMP-PCP (Fig. 4C). CK2α phosphorylates eIF2α and eIF5 in vitro, and thus the increased binding in the presence of CK2α may not be solely attributed to the phosphorylation of eIF3c but may represent the effects of multiple phosphorylation events. eIF2β is only phosphorylated in the presence of the CK2 holoenzyme (41); thus in the absence of CK2β subunits only eIF3c, eIF2α, and eIF5 are phosphorylated.
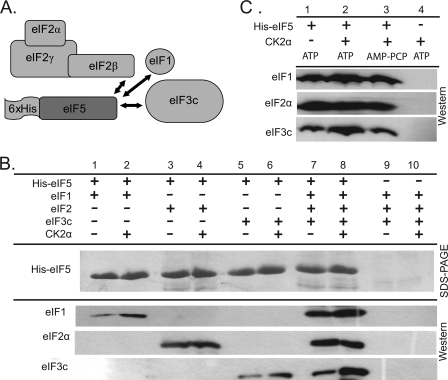
Interaction of Wheat Initiation Factors with eIF5 in Vitro
The interaction of eIF1, eIF2, and eIF3c with wheat eIF5 was also evaluated using a series of pulldown experiments (Fig. 5A). As shown in Fig. 5B, the interaction with native eIF2 is robust (lane 3); however, the interaction of recombinant wheat eIF1 (Fig. 5B, lane 1) and full-length eIF3c (lane 5) with His-eIF5 occurs modestly under the in vitro binding conditions used in this study. However, the amount of these factors pulled down by His-eIF5 is enhanced in the presence of all three factors involved in MFC formation (native wheat eIF2, recombinant wheat eIF1, and recombinant full-length wheat eIF3c; Fig. 5B compare lanes 1, 3, and 5 with lane 7). This enhanced interaction is likely because of the cooperative binding of factors, as demonstrated between eIF3c-NTD and both eIF1, eIF2, and eIF5 (Fig. 4B). Assays conducted in the presence of native wheat eIF3 were inconclusive, as the eIF3 complex bound to the Ni-NTA matrix in the absence of any His-tagged proteins. The interaction of eIF3 with Ni-NTA was detected even when reactions were conducted in the presence of 50 mm imidazole. The cause of this nonspecific interaction is unknown, as a sequence analysis of wheat eIF3 subunits revealed no more than two consecutive histidine residues.
FIGURE 5.
CK2 enhances interaction of MFC components with His-eIF5 in vitro. A, potential interactions of wheat initiation factors with His-eIF5 were evaluated by a series of Ni-NTA pulldowns. B, His-eIF5 (400 pmol) was mixed with ∼400 pmol of the following initiation factors as indicated: recombinant wheat eIF1, native wheat eIF2 (22), and/or recombinant wheat eIF3c. 10 pmol of recombinant A. thaliana CK2α1 was used to phosphorylate initiation factors. Control reactions (lanes 9 and 10) were conducted in the absence of the His6-tagged protein to evaluate the interaction between wheat initiation factors/CK2α and the Ni-NTA beads. The eluted proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining or transferred to PVDF and analyzed with rabbit antisera to recombinant wheat eIF1, eIF2α, and eIF3c as described under “Experimental Procedures.” C, similar set of Ni-NTA pulldown experiments was performed in the presence of eIF1, eIF2, eIF3c, and His-eIF5. CK2 and ATP/AMP-PCP are as indicated.
Effect of CK2 Phosphorylation on His-eIF5 Pulldowns
In the presence of CK2α, the binding of eIF1 and eIF3c to His-eIF5 is enhanced in pulldown assays (Fig. 5B, compare lanes 1 and 5 with lanes 2 and 6). The increased interaction of eIF1 and eIF3c with His-eIF5 that occurs in the presence of CK2α is phosphorylation-dependent, as no similar increase in binding occurs in the presence of AMP-PCP (Fig. 5C). A similar increase in the interaction of eIF2 with eIF5 was not observed in the presence of CK2α (Fig. 5B, compare lanes 3 and 4). The interaction of the eIF5 C terminus with the N-terminal domain of eIF2β has been previously demonstrated in yeast (12). Studies in mammalian cells suggest that the eIF2/eIF5 interaction may be dependent on CK2 phosphorylation of eIF5 (19).
The binding of recombinant eIF2β to His-eIF3c is increased by the presence of either CK2α1 or CK2α1β1 holoenzyme (Fig. 6A). It appears that the phosphorylation of eIF3c is responsible for the increased binding to eIF2β, because no further enhancement in eIF2β-eIF3c binding is achieved by the addition of the CK2β subunit, which is required for eIF2β phosphorylation (Fig. 6A, compare lanes 5 and 6).
CK2 does not appear to have any effect on the interaction of wheat eIF2 (Fig. 5B) or recombinant wheat eIF2β (Fig. 6B) with His-eIF5. It was initially suspected that this was the result of in vitro binding conditions being highly favorable, and thus any increase in interaction that was mediated by CK2 was negligible. However, even in the presence of higher salt concentrations (150 mm KCl) and lower protein concentrations (∼20 μm), no clear effect of CK2 phosphorylation could be demonstrated on eIF2/eIF5 binding. In separate experiments with recombinant wheat eIF2β, an increase in the salt concentration during binding inhibited the interaction of wheat eIF5 with eIF2β, suggesting an ionic interaction occurs between these proteins in plants (results not shown).
This study provides the first evidence that CK2 phosphorylation of wheat eIF5 influences its interaction with eIF1. CK2 phosphorylation enhances the interaction of the His-eIF3c-NTD and eIF5 (Fig. 4B) and full-length wheat eIF3c with His-eIF5 (Fig. 5B). In addition, CK2 phosphorylation of wheat eIF5 did not directly affect its in vitro interaction with eIF2; however, it cannot be ruled out that this interaction is influenced by CK2 phosphorylation in vivo.
It is likely that the interaction of eIF1 with eIF3c stabilizes the eIF1/eIF5 interaction, as the His-eIF3c-NTD was found to bind both eIF1 and eIF5. Yeast eIF1 has been shown to interact with all three factors involved in the MFC (4), and thus it is conceivable that eIF2 may enhance the eIF1/eIF5 interaction through a similar bridging interaction. However, the direct interaction of native wheat eIF2 and recombinant wheat eIF1 could not be demonstrated in this study in either the presence or the absence of CK2 subunits (Fig. 6C).
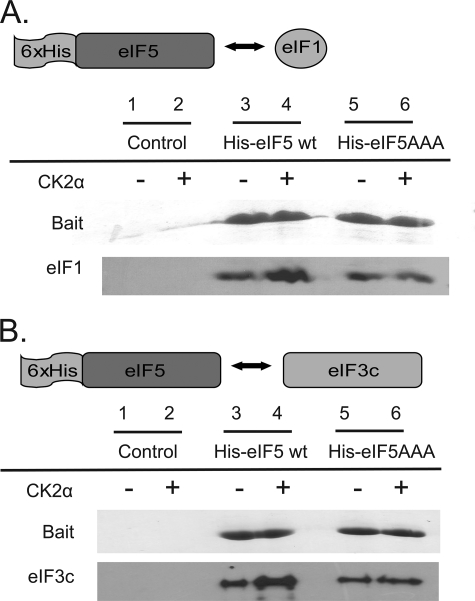
Effect of eIF5 Mutagenesis on MFC Formation
Mutation of the three eIF5 CK2 phosphorylation sites to alanine prevents the CK2-mediated increase in eIF1 binding (Fig. 7A, compare lanes 4 and 6). Similarly, the presence of CK2 subunits enhances the interaction of wheat His-eIF5 with eIF3c. However, in the presence of CK2 the amount of eIF3c pulled down by His-eIF5AAA is not increased, suggesting that the phosphorylation of eIF3c alone does not mediate the enhanced interaction. This suggests that the interaction of wheat eIF5 with both eIF1 and eIF3c is enhanced by CK2 phosphorylation of eIF5. Both recombinant wheat His-eIF5 and His-eIF5AAA appear to be folded in one-dimensional NMR spectra, suggesting that the change in binding is not because of misfolding of the mutant protein.3 It was recently reported that a basic/hydrophobic surface of yeast eIF1 participates in binding to the eIF5 C terminus, and mutations that alter basic residues in this domain of eIF1 were lethal (4). It is likely that phosphorylation of wheat eIF5 enhances interactions with positively charged domains in wheat eIF1.
FIGURE 7.
Increased in vitro interaction with eIF1 and eIF3c upon phosphorylation of eIF5. Alanine mutations at CK2 phosphorylation sites prevent the CK2-mediated increase in eIF1 and eIF3c binding to eIF5. His-eIF5 (400 pmol) or His-eIF5AAA was mixed with ∼400 pmol of recombinant wheat eIF1 (A) or eIF3c (B). 10 pmol of recombinant A. thaliana CK2α1 was used to phosphorylate initiation factors. Control reactions were conducted in the absence of the His-tagged protein to evaluate the interaction between non-His-tagged proteins and 10 μl of Ni-NTA matrix. The elution was analyzed by SDS-PAGE, and the gel was cut such that bands could be visualized by Coomassie Blue staining or transferred to PVDF and analyzed with a 1:5000 dilution of rabbit antisera to recombinant wheat eIF1 (A) or eIF3c (B) as described under “Experimental Procedures.” wt, wild type.
eIF5 was immunoprecipitated from wheat germ extracts to further analyze the formation of the MFC and the effect of CK2 phosphorylation on eIF5. eIF1, eIF2, and eIF3 in addition to eIF5 were all detected in the immunoprecipitation elution (see supplemental Fig. D). The interaction of native factors with eIF5 was modestly increased by supplementing reactions with the phosphatase inhibitor calyculin A and CK2. In additional experiments, eIF5-depleted extracts were supplemented with recombinant eIF5 or eIF5AAA. The amount of eIF1, eIF2, and eIF3 that interacts with wild-type eIF5 is increased by the presence of both calyculin A and CK2; however, a similar increase was not detected when extracts were supplemented with eIF5AAA suggesting that phosphorylation was necessary for the increase. These results are similar to those obtained with the purified factors that showed phosphorylation of eIF5 enhances its interaction with eIF3c and eIF1 in the MFC.
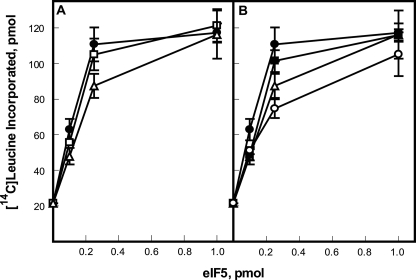
Effect of eIF5 Mutagenesis on in Vitro Translation
The ability of recombinant eIF5 to reactivate translational activity of eIF5-depleted extracts was evaluated by the incorporation of [14C]leucine into protein in the presence of A. thaliana capped heat shock protein (HSP) 21 mRNA.4 The recombinant wild-type and mutant eIF5 preparations were analyzed by both Bradford and SDS-PAGE to ensure equivalent levels of protein were tested in in vitro translation. No significant difference was observed between the ability of wild-type eIF5 and eIF5 S451E, a phosphomimic, to stimulate translation in vitro (Fig. 8A). However, mutation of S451A reduced the activity ∼20% at low levels of eIF5 (Fig. 8A). The double mutation of S209A/T240A has a modest effect on activity compared with the single mutant S451A (Fig. 8B) suggesting that phosphorylation of Ser-209/Thr-240 are not as critical for optimal activity as Ser-451. The ability of the triple mutant S209A/T240A/S451A (eIF5AAA) to stimulate in vitro translation in eIF5-depleted wheat germ extracts is reduced by ∼40% at low levels compared with wild-type eIF5 (Fig. 8B). These results suggest that the phosphorylation of wheat eIF5 by CK2, particularly at Ser-451, plays an important role in mediating eIF5 activity in the formation of the MFC.
FIGURE 8.
eIF5-dependant translation assay. The ability of recombinant wheat eIF5 to stimulate translation in eIF5-depleted wheat germ S30 extracts was analyzed. Each 50-μl reaction contained 20 μl of eIF5-depleted wheat germ extract, 5 pmol of HSP21 mRNA template, and recombinant eIF5 as indicated. A, comparison of Ser-451 single mutants; wheat eIF5 (●), eIF5 S451E (□), or eIF5 S451A (Δ). B, comparison of single, double, and triple mutants; wheat eIF5 (●), eIF5 S451A (Δ), eIF5 S209A/T240A (■), or eIF5 S209A/T240A/S451A (○). Reactions were incubated for 30 min at 27 °C, and the amount of [14C]leucine incorporated into polypeptide was determined as described previously (22, 26).
DISCUSSION
The differential phosphorylation of recombinant wheat initiation factors by plant CK2 isoforms has been demonstrated in vitro (41). Numerous CK2 phosphorylation sites were identified in recombinant wheat and A. thaliana translation initiation factors (Table 1). Evidence of CK2 phosphorylation was also found in native preparations of wheat eIF2 and eIF5. This is the first study to extensively identify the CK2 phosphorylation sites in plant initiation factors. Identification of the specific residues modified by CK2 phosphorylation initially provided some insight into the possible role CK2 plays in translation. A majority of these sites were located in the N and C termini of proteins, which generally have less structure, and are therefore good targets for phosphorylation and/or participating in protein/protein interactions. If phosphorylation sites were critical to function, one would expect the residues to be conserved. This is indeed the case for a number of the CK2 phosphorylation sites identified by this study (Fig. 2).
This study is also the first to evaluate the role CK2 phosphorylation plays in the interaction of factors within the MFC, which consists of eIF1/eIF3/eIF5/eIF2/GTP/Met-tRNAiMet. It is striking that three of the four factors involved are all substrates for CK2 and that a number of the key domains involved in factor interaction were the targets of CK2. Wheat eIF1 does not appear to be phosphorylated by CK2, nor does it contain any predicted phosphorylation sites for CK2 or any other protein kinases. Of particular interest is the heavily phosphorylated N terminus of eIF3c and the C terminus of eIF5, which both serve as hubs for yeast MFC assembly (6, 9). Binding studies from yeast suggest that acidic residues in the N terminus of eIF3c interact with basic residues in eIF1 and eIF5 (6), and an acidic/aromatic domain in the C terminus of eIF5 interacts with the K-boxes of eIF2β (10). The results of this study showed that CK2 phosphorylation of wheat initiation factors enhanced the interaction of several components of the MFC.
In this study, wheat eIF2α was found to be phosphorylated only on Ser-318 by CK2 in vitro. This CK2 phosphorylation site (Ser-318) is distinct from the Ser-51 involved in regulation of mammalian eIF2 guanine nucleotide exchange. In addition, native wheat eIF2α purified in the presence of calyculin A was observed to be at least partially phosphorylated on this residue. A. thaliana eIF2α is also phosphorylated by CK2 in vitro at two sites; however the phospho-peptide(s) were not detected by mass spectrometry. A. thaliana eIF2α is likely phosphorylated on Ser-322 and Ser-317, as truncation of A. thaliana eIF2α C terminus prevents phosphorylation by CK2. Phosphorylation of the plant eIF2α C terminus by CK2 is not unique, as three CK2 phosphorylation sites have been previously identified in the yeast eIF2α (SUI2) C terminus (33). Mutation of the yeast eIF2α to alter its three CK2 sites to alanine did not cause a detectable phenotype; however, when combined with other mutations that reduced the efficiency of nucleotide exchange (GCN2, GCN3, and GDC7), the alanine mutations exacerbated growth defects. This would suggest that the phosphorylation of eIF2α in yeast may be required for optimum eIF2 activity, and it is possible that Ser-318 phosphorylation in wheat may play a similar role. In addition, Ser-318 is conserved across plants, suggesting its functional relevance. There is no site similar to Ser-318 in mammalian eIF2α, and mammalian CK2 does not appear to phosphorylate eIF2α (34).
Wheat eIF2β was phosphorylated in vitro in the presence of the CK2 holoenzyme on Thr-52 and Thr-85, whereas A. thaliana eIF2β was phosphorylated in a similar region on Ser-42, Ser-80, and Ser-112. CK2 phosphorylation of the eIF2β N-terminal domain has been reported in mammalian cells (35), yet this phosphorylation does not appear to be conserved in yeast (34). In mammalian cells, the proper phosphorylation of eIF2β by CK2 is thought to be required for its function, as overexpression of a nonphosphorylatable mutant of eIF2β leads to a reduction in the rate of protein synthesis stimulated by serum (35). The primary site of eIF2β phosphorylation in mammalian cells was identified as S2, with a secondary (partial) phosphorylation occurring on Ser-67 (35). It is not known if the wheat eIF2β phosphorylation sites are functionally comparable. The S2 site in mammalian eIF2β does not appear to be conserved in plants or yeast. The N-terminal domain of mammalian and yeast eIF2β contains a series of three lysine blocks that participate in binding to eIF5, eIF2Bϵ, and mRNA (12). Interestingly, the N-terminal domain of plant eIF2β only contains two lysine blocks; thus some variance in the function and regulation of this domain is expected.
The in vitro phosphorylation of eIF3c by CK2 is extensive, and 12 sites were identified in this study. A large number of these sites are conserved across plants and with other eukaryotes. This is the first study to show the phosphorylation of eIF3c enhances its interaction with eIF1, -2, and -5. A growing body of evidence suggests that eIF3 may function as the central hub in the coordination of translation initiation (36). The interaction of residues in the N-terminal domain of yeast eIF3c (NIP1) with eIF1 and eIF5 has been shown to play an important role in AUG selection; these interactions appear to link the function of eIF1 in recognizing proper codon-anticodon base pairing with the both the GTPase-activating protein activity of eIF5 and the release of the Pi generated by eIF2-GTP hydrolysis (6). In this study, a similar binding event has been demonstrated with recombinant wheat eIF3c, and this binding was enhanced by the presence of CK2. Multiple sequence alignment of eIF3c sequences aligns a number of the CK2 phosphorylation sites identified in wheat with key domains in yeast that have been shown to mediate binding to eIF1 and eIF5 (10). It is likely that the phosphorylation of the eIF3c-NTD by CK2 enhances its interaction with positively charged binding domains in its MFC partners, as basic domains containing high levels of lysine and arginine residues in yeast eIF1 (4) and eIF5 (9) play a critical role in their association with yeast eIF3c. A hexaphosphorylated tryptic peptide has been identified in vivo from the N-terminal region of the mammalian eIF3c (37). The potential kinases responsible for their phosphorylation were predicted by Scansite to be the acidophilic serine kinases, CK1 or CK2. It is likely that the phosphorylation sites identified here represent a similar and possibly more dramatic phosphorylation event in plants. Although large scale phosphoproteomic analyses in planta are only beginning to gain momentum, the Arabidopsis Protein Phosphorylation Site data base (PhosPhAt 2.1) contains spectra identifying the phosphorylation of a consensus CK2 site (Ser-40) in the N terminus of A. thaliana eIF3c (28). Alignment of CK2 phosphorylation sites in the N-terminal sequences of eIF3c suggests that the phosphorylation of A. thaliana at Ser-40 corresponds to the in vitro phosphorylation of wheat eIF3c Ser-53 that was observed in this study. Based on the findings of this study, it is likely that future analysis of eIF3c in planta will provide further evidence of in vivo phosphorylation.
In vitro CK2 phosphorylation sites were identified in wheat eIF5 at Ser-209, Thr-240, and Ser-451 and in A. thaliana eIF5 at Ser-201, Thr-230, Ser-428, Ser-431, and Ser-433. Our analysis of native wheat eIF5 revealed that it is multiply phosphorylated and Ser-451 is almost entirely phosphorylated on two distinct isoforms of eIF5 (Fig. 3). This analysis also revealed phosphopeptides representing the partial phosphorylation of tryptic fragments containing Ser-209 and Thr-240. All three of the CK2 phosphorylation sites in eIF5 are conserved across plants, and similar CK2 phosphorylation sites exist in the C terminus of yeast and mammalian protein sequences (Fig. 2). An interaction between eIF1 and eIF5 has been previously established with yeast factors (4); however, it was not clear that this interaction was conserved across other systems as an interaction of human eIF1 with recombinant human eIF5 could not be demonstrated (11). Wheat eIF5 interacts with eIF1 in vitro, and this is the first report that the interaction of eIF5 with eIF1 is enhanced by CK2 phosphorylation of eIF5. Site-directed mutagenesis of eIF5 to remove CK2 phosphorylation sites not only prevented the CK2-mediated increase in its interaction with eIF1 and eIF3c but also resulted in reduced stimulation of translation initiation by eIF5 in vitro. This supports findings with mammalian cells, where the expression of mutants that prevented CK2 phosphorylation disrupted synchronous cell cycle progression and significantly reduced growth rates (19).
The results of this study suggest that plant CK2 phosphorylation, unlike mammalian CK2, does not influence the interaction of wheat eIF5 with either native wheat eIF2 or recombinant wheat eIF2β. The C terminus of human eIF5 not only contains two sequential CK2 phosphorylation sites, but it is also ∼33 additional amino acids longer than plant and yeast eIF5. In addition, the N-terminal domain of plant eIF2β is significantly shorter than other forms of eIF2β and contains only two lysine-rich K-boxes; thus some variation in the regulation/binding of these domains is not unexpected between plant and other systems.
This is the first study to observe a direct interaction between eIF3c and eIF2β. It is possible that this interaction is plant-specific. Yeast eIF5 binds to eIF2β through an AA-box (acidic/aromatic-box) in its C terminus, and this interaction is required for MFC formation (38). It has been observed previously that the physical association of yeast eIF5 with the eIF2β N terminus enhances the eIF3/eIF5 interactions (39). An indirect interaction of these subunits has been observed in yeast, as eIF3a (TIF32), eIF1, and eIF5 all serve as intermediaries, binding simultaneously to both eIF3c (NIP1) and the β-subunit of eIF2 (4, 10). The findings presented here indicate that wheat eIF2 and eIF2β are both capable of binding to the eIF3c-NTD in the absence of other factors, and this interaction is enhanced by CK2 phosphorylation of eIF3c.
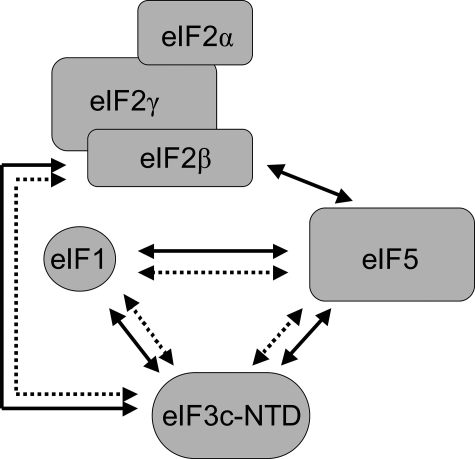
These data suggest that plant initiation factors interact in a manner similar to the yeast MFC (3). A series of in vitro interactions were clearly observed between recombinant wheat eIF1, eIF3c, eIF5, and eIF2β/native wheat eIF2 in the absence of 40 S ribosomal subunits (Fig. 9, solid lines). Similar interactions were also observed in native extracts. Because of the presence of multiple forms of CK2 subunits in plants, it is tempting to speculate a role for multiple forms of CK2 subunits in the regulation of plant translation initiation (41). The results of this study demonstrate that the phosphorylation of initiation factors by CK2 enhances the interaction of plant MFC components in vitro (Fig. 9, dotted lines) and suggest a potential role for CK2 phosphorylation in regulating the activity of plant translation initiation factors through their MFC interactions.
FIGURE 9.
Model for in vitro plant multifactor complex interactions and the influence of CK2. A number of in vitro interactions have been observed with plant initiation factors in the absence of 40 S ribosomal subunits. A solid line indicates interactions of plant factors detected in this study. The findings of this study suggest plant initiation factors are capable of forming multifactor complexes similar to those previously reported in yeast consisting of eIF1-eIF3-eIF5-eIF2-GTP-Met-tRNAiMet (3). In addition a number of these interactions (dotted line) were enhanced by CK2 phosphorylation in vitro.
Supplementary Material
Acknowledgments
We thank Dr. David Hoffman (Department of Chemistry and Biochemistry, University of Texas, Austin) for NMR analysis of recombinant proteins; Laura Mayberry and Dr. Andrew Lellis for technical advice; and Tanya Trynosky (undergraduate) and Isaac Hernandez (National Science Foundation Research Experience for Undergraduate Students CHE0552422) for assistance with preparation of CK2 proteins. We also thank Asma Sharif and Jacqulyn Tolson of the Analytical Instrumentation Facility Core for performing IMAC enrichment of phosphopeptides. Their successful separations allowed the detection of numerous phosphopeptides by MALDI-TOF/TOF analysis.
This work was supported by Grant DE-FG02-04ER15575 from the United States Department of Energy, Grant MCB0214996 from the National Science Foundation, and Grant F1339 from The Welch Foundation (to K. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table A and Figs. A–D.
D. Hoffman, personal communication.
Mayberry, L. K., Allen, M. L., Dennis, M. D., and Browning, K. S., (2009) Plant Physiol., in press.
- MFC
- multifactor complex
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- DTT
- dithiothreitol
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MOPS
- 4-morpholinepropanesulfonic acid
- Ni-NTA
- nickel-nitrilotriacetic acid
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- IMAC
- immobilized metal affinity chromatography
- AMP-PCP
- adenosine 5′-(β,γ-methylenetriphosphate)
- PVDF
- polyvinylidene difluoride.
REFERENCES
- 1.Hershey J. W., Merrick W. C. (2000) in Translational Control of Gene Expression ( Sonenberg N., Hershey J. W., Mathews M. B. eds) pp. 33–88, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2.Mathews M. B., Sonenberg N., Hershey J. W. (2007) in Translational Control in Biology and Medicine ( Mathews M. B., Sonenberg N., Hershey J. W. eds) pp. 1–40, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 3.Asano K., Clayton J., Shalev A., Hinnebusch A. G. (2000) Genes Dev. 14, 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reibarkh M., Yamamoto Y., Singh C. R., del Rio F., Fahmy A., Lee B., Luna R. E., Ii M., Wagner G., Asano K. (2008) J. Biol. Chem. 283, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch A. G., Dever T. E., Asano K. (2007) in Translational Control in Biology and Medicine ( Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 225–269, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 6.Valásek L., Nielsen K. H., Zhang F., Fekete C. A., Hinnebusch A. G. (2004) Mol. Cell. Biol. 24, 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asano K., Phan L., Anderson J., Hinnebusch A. G. (1998) J. Biol. Chem. 273, 18573–18585 [DOI] [PubMed] [Google Scholar]
- 8.Phan L., Zhang X., Asano K., Anderson J., Vornlocher H. P., Greenberg J. R., Qin J., Hinnebusch A. G. (1998) Mol. Cell. Biol. 18, 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., Wagner G., Asano K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valásek L., Nielsen K. H., Hinnebusch A. G. (2002) EMBO J. 21, 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher C. M., Pestova T. V., Hellen C. U., Wagner G. (1999) EMBO J. 18, 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asano K., Krishnamoorthy T., Phan L., Pavitt G. D., Hinnebusch A. G. (1999) EMBO J. 18, 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinna L. A. (2002) J. Cell Sci. 115, 3873–3878 [DOI] [PubMed] [Google Scholar]
- 14.Litchfield D. W. (2003) Biochem. J. 369, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meggio F., Pinna L. A. (2003) FASEB J. 17, 349–368 [DOI] [PubMed] [Google Scholar]
- 16.Olsten M. E., Weber J. E., Litchfield D. W. (2005) Mol. Cell. Biochem. 274, 115–124 [DOI] [PubMed] [Google Scholar]
- 17.Homma M. K., Homma Y. (2005) Mol. Cell. Biochem. 274, 47–52 [DOI] [PubMed] [Google Scholar]
- 18.Maiti T., Bandyopadhyay A., Maitra U. (2003) Yeast 20, 97–108 [DOI] [PubMed] [Google Scholar]
- 19.Homma M. K., Wada I., Suzuki T., Yamaki J., Krebs E. G., Homma Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15688–15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metz A. M., Browning K. S. (1997) Arch. Biochem. Biophys. 342, 187–189 [DOI] [PubMed] [Google Scholar]
- 21.Mayberry L. K., Dennis M. D., Leah Allen M., Ruud Nitka K., Murphy P. A., Campbell L., Browning K. S. (2007) Methods Enzymol. 430, 397–408 [DOI] [PubMed] [Google Scholar]
- 22.Lax S. R., Lauer S. J., Browning K. S., Ravel J. M. (1986) Methods Enzymol. 118, 109–128 [DOI] [PubMed] [Google Scholar]
- 23.Monzingo A. F., Dhaliwal S., Dutt-Chaudhuri A., Lyon A., Sadow J. H., Hoffman D. W., Robertus J. D., Browning K. S. (2007) Plant Physiol. 143, 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Person M. D., Monks T. J., Lau S. S. (2003) Chem. Res. Toxicol. 16, 598–608 [DOI] [PubMed] [Google Scholar]
- 25.Browning K. S., Humphreys J., Hobbs W., Smith G. B., Ravel J. M. (1990) J. Biol. Chem. 265, 17967–17973 [PubMed] [Google Scholar]
- 26.Mayberry L. K., Browning K. S. (2006) Curr. Protocols Microbiol. 16K.1.1–16K.1.13 [DOI] [PubMed] [Google Scholar]
- 27.Browning K. S., Yan T. F., Lauer S. J., Aquino L. A., Tao M., Ravel J. M. (1985) Plant Physiol. 77, 370–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heazlewood J. L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W. X. (2008) Nucleic Acids Res. 36, D1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterhout J. J., Lax S. R., Ravel J. M. (1983) J. Biol. Chem. 258, 8285–8289 [PubMed] [Google Scholar]
- 30.Lax S. R., Osterhout J. J., Ravel J. M. (1982) J. Biol. Chem. 257, 8233–8237 [PubMed] [Google Scholar]
- 31.Bonnet H., Filhol O., Truchet I., Brethenou P., Cochet C., Amalric F., Bouche G. (1996) J. Biol. Chem. 271, 24781–24787 [DOI] [PubMed] [Google Scholar]
- 32.Jiang X., Wang Y. (2004) Biochemistry 43, 15567–15576 [DOI] [PubMed] [Google Scholar]
- 33.Feng L., Yoon H., Donahue T. F. (1994) Mol. Cell. Biol. 14, 5139–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llorens F., Sarno S., Sarró E., Duarri A., Roher N., Meggio F., Plana M., Pinna L. A., Itarte E. (2005) Mol. Cell. Biochem. 274, 53–61 [DOI] [PubMed] [Google Scholar]
- 35.Llorens F., Duarri A., Sarró E., Roher N., Plana M., Itarte E. (2006) Biochem. J. 394, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch A. G. (2006) Trends Biochem. Sci. 31, 553–562 [DOI] [PubMed] [Google Scholar]
- 37.Damoc E., Fraser C. S., Zhou M., Videler H., Mayeur G. L., Hershey J. W., Doudna J. A., Robinson C. V., Leary J. A. (2007) Mol. Cell. Proteomics 6, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 38.Das S., Maitra U. (2000) Mol. Cell. Biol. 20, 3942–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh C. R., Yamamoto Y., Asano K. (2004) J. Biol. Chem. 279, 49644–49655 [DOI] [PubMed] [Google Scholar]
- 40.Majumdar R., Bandyopadhyay A., Deng H., Maitra U. (2002) Nucleic Acids Res. 30, 1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis M. D., Browning K. S. (2009) J. Biol. Chem. 284, 20602–20614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.