Abstract
Aberrant Wnt signaling promotes oncogenesis by increasing cellular levels of β-catenin, which associates with DNA-bound transcription factors and activates Wnt target genes. However, the molecular mechanism by which β-catenin mediates gene expression is still poorly understood. Here, we show that cell cycle and apoptosis regulator 1 (CCAR1), which was recently shown to function as a transcriptional coactivator for nuclear receptors, also interacts with β-catenin and enhances the ability of β-catenin to activate expression of transiently transfected reporter genes. Furthermore, association of CCAR1 with the promoter of an endogenous Wnt/β-catenin target gene in a colon cancer cell line depends on the presence of β-catenin. Depletion of CCAR1 inhibits expression of several Wnt/β-catenin target genes and suppresses anchorage-independent growth of the colon cancer cell line. Thus, CCAR1 is a novel component of Wnt/β-catenin signaling that plays an important role in transcriptional regulation by β-catenin and that, therefore, may represent a novel target for therapeutic intervention in cancers involving aberrantly activated Wnt/β-catenin signaling.
The Wnt/β-catenin signaling cascade controls a variety of cell fate decisions during development and is important for cell proliferation and self-renewal of many types of stem cells, including intestinal epithelial and hematopoietic stem cells (1–5). Misregulation of this signaling has been recognized as a hallmark of many aggressive human cancers (2, 3, 5). Indeed, genetic alterations of genes involved in β-catenin degradation have been reported in various tumors (6–12). In addition, mutational analyses of clinical specimens and experiments with transgenic mice have proven the importance of β-catenin stabilization in adenoma formation, which is the earliest event of colorectal carcinogenesis. Therefore, the Wnt pathway has been causally linked to various cancers, most notably to colorectal cancers.
Secreted Wnt proteins bind to Frizzled receptors to initiate the signaling cascade (13). In the absence of Wnt signals, there is only a small pool of cytosolic β-catenin under normal physiological conditions, due to constitutive phosphorylation of β-catenin via a multiprotein complex composed of Axin, casein kinase I α, glycogen synthase kinase-3β, and tumor suppressor adenomatous polyposis coli (14–16). Phosphorylated β-catenin is then ubiquitinated by βTrCP and destroyed by proteasome-mediated proteolysis. Wnt signaling inhibits the function of this complex and thereby stabilizes β-catenin, resulting in increased cytoplasmic β-catenin, some of which then translocates into the nucleus. β-Catenin has been recognized as a pivotal factor for cancer development, because its interaction with various transcriptional activators such as lymphoid enhancer-binding factor (LEF)3/T cell factor (TCF) family members (17), NF-κB (18, 19), Prop1 (20), and nuclear receptors (21–24) are required for expression of a subset of target genes involved in regulation of cell proliferation, apoptosis, and tumor metastasis. For example, reported downstream targets of β-catenin-LEF/TCF-regulated transcription include genes involved in cell proliferation (e.g. c-myc (25) and c-jun (26)), inhibition of apoptosis (e.g. survivin (27, 28)), and tumor metastasis (e.g. MMP7 (29)). Hence, to better understand the contribution of Wnt/β-catenin deregulation in cancer, it is crucial to explore how β-catenin controls and regulates the transcription of these target genes.
As a potent primary coactivator for LEF/TCF transcription factors, β-catenin binds directly to DNA-bound LEF/TCF proteins and serves as a platform for recruiting additional secondary coactivators to promoters of a variety of LEF/TCF target genes. Generally, these secondary coactivators assist β-catenin in mediating transcriptional activation either through modulation of chromatin conformation or recruitment and activation of RNA polymerase II and its associated basal transcription machinery (17). To date, several coactivators have been reported to interact with β-catenin, including histone methyltransferases CARM1 (30) and MLL/Set1 (31); histone acetyltransferases p300 and CBP (32–35), and TRRAP/Tip60 (18, 36, 37); the Brg1 ATPase subunit of the SWI/SNF chromatin-remodeling complex (38); the MED12 component of the Mediator complex, which recruits in RNA polymerase II (39); pygopus (39, 40); casein kinase 2 (41); FLAP1 (42); Bcl9/Legless (43); GAC63 (44); GRIP1 (45); and CoCoA (46). Although a number of interacting proteins have been identified, the molecular details of their cooperation with β-catenin to control transcription are still poorly understood. To further explore the mechanism of coactivator function by β-catenin, it is important to define the specific functional relationships of β-catenin with the interacting proteins and to determine their molecular and physiological roles in Wnt signaling.
Cell cycle and apoptosis regulator 1 (CCAR1) is a regulator of apoptosis signaling as well as cell proliferation. For example, CCAR1 triggers apoptosis signaling in a retinoid-dependent manner (47). Additionally, cell growth-inhibitory and apoptosis-promoting effects elicited by inhibition of epidermal growth factor receptor involve CCAR1 (48). On the other hand, CCAR1 has also been shown to be important for estrogen-induced gene expression and estrogen-dependent growth of human breast cancer cells (49). Thus, previous studies suggest that CCAR1 can serve as a key intracellular transducer of either proliferation or apoptosis signaling pathways in response to different signals. Likewise, Wnt ligands are known as critical stimuli of the cellular communication network controlling multiple biological processes such as proliferation and cell fate determination. In addition, several coactivators for estrogen receptor and other nuclear receptors have also been found to cooperate with β-catenin in Wnt signaling. Hence, we speculated that CCAR1 may play a role in Wnt signaling. Here, we report that CCAR1 is a functional binding partner of β-catenin and assists β-catenin in transcriptional activation of Wnt target genes, some of which are implicated in proliferation and metastasis. In addition, depletion of CCAR1 inhibited the anchorage-independent growth of human colorectal cancer cells. Thus our results provided a novel insight into β-catenin-mediated tumorigenesis.
EXPERIMENTAL PROCEDURES
Plasmids
Vectors pGEX-4T1-β-catenin, pGEX-5X1-CCAR1 (with a BamHI-XhoI insert), and pGEX-5X1-LEF1 for bacterially expressed glutathione S-transferase (GST) fusion proteins, and luciferase reporter plasmids pGL3OT (for LEF1) and GK1-LUC (for Gal4) were described previously (45, 49). For expression of N-terminal Gal4 DNA binding domain (DBD) fusion proteins, pM-β-catenin and pM-LEF1 were constructed as described previously (45). The following mammalian expression vectors were described in previous publications as follows: pSG5.HA-LEF1, pSG5.HA-β-catenin, and the same vector encoding various HA-tagged fragments of β-catenin (45), pSG5.HA-CoCoA (50), pSG5.HA-CARM1 (51), pCMV-p300 (52), and pSG5.HA-CCAR1 (49). Plasmids encoding CCAR1 fragments (amino acids 290–630, 471–630, 631–1146, 670–900, and 955–1146) were created by PCR amplification and subcloning into EcoRI and XhoI sites of pSG5.HA (51); CCAR1 1–249 and CCAR1 1–289 were cloned into BamHI and XhoI sites of pSG5.HAb (gift from Martin A. Privalsky, University of California at Davis). For lentivirus production, the vesicular stomatitis virus envelope protein G expression construct pMD.G1, the packaging vector pCMV ΔR8.91 (53), and the transfer vector pHRCMVpuroSin8 (54) were used. The β-catenin short hairpin RNA (sh-β-catenin) transfer vectors were produced with PCR products containing the U6 promoter and sh-β-catenin coding sequence. The PCR products were inserted into SpeI and PstI sites of pHRCMVpuroSin8. Short hairpin RNA encoding sequences targeting β-catenin were as follows: 5′-AAAACTGCAGAAAAAGCTTCCAGACACGTATCATGCGTTTCTCTTGAAAACGCACGATAGCGCGTCTGGAAGCGGTGTTTCGTCCTTTCCACAAG-3′. The sh-CCAR1 transfer vector was described previously (49).
Cell Culture and Luciferase Reporter Gene Assay
HT29 cells were maintained in McCoy's 5a medium with 10% fetal bovine serum and penicillin and streptomycin. CV-1, COS-7, and 293T cell lines were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin and streptomycin. For transient reporter gene assay, CV-1 cells were plated at 105 cells per well in 12-well plates and transiently transfected by TargeFect F1 reagent (Targeting Systems) according to the manufacturer's protocol. Total DNA in every well was adjusted to a constant amount by adding empty expression vectors. Forty-eight hours after transfection, luciferase assays on cell extracts were performed with Promega Luciferase Assay kit. Data shown are the mean and range of variation of duplicate transfected cultures from a single experiment and are representative of at least two independent experiments. Luciferase activities were not normalized to internal controls, because expression of so-called constitutive reporter genes is affected by overexpression of many coactivators. Instead, multiple independent experiments with multiple plasmid preparations were used to demonstrate reproducibility.
GST Pulldown Assay
HA-β-catenin, HA-LEF1, and HA-CCAR1 were synthesized in vitro by using the TnT-Quick-coupled transcription/translation system (Promega, Madison, WI) according to the manufacturer's protocol. These in vitro synthesized proteins were then used for GST pulldown assay as described previously (50). Briefly, in vitro translated proteins and immobilized GST fusion proteins were mixed with NETN buffer (200 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, pH 7.6, and 0.01% Nonidet P-40) and incubated overnight at 4 °C. On the next day, beads were washed with NETN three times, and bound proteins were analyzed by immunoblot with anti-HA antibodies (Roche Applied Science). The amount of GST used as a negative control was always higher than that of GST fusion proteins.
Co-immunoprecipitation
COS-7 cells were grown in 100-mm-diameter dishes seeded with 106 cells and transfected with expression plasmids pSG5.HA-CCAR1 and pSG5.HA-β-catenin using TargeFect F2 reagent (Targeting Systems) according to the manufacturer's protocol. Two days after transfection cells were harvested with radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, and 0.1% SDS) supplemented with protease inhibitor mixture (Roche Applied Science), and cell extracts were used for immunoprecipitation with antibodies against β-catenin (BD Biosciences), normal mouse IgG (Santa Cruz Biotechnology), normal rabbit IgG (Bethyl Laboratories), or CCAR1 (Bethyl Laboratories). Protein A/G beads (Santa Cruz Biotechnology) were added, and the reaction was incubated for 3 h at 4 °C. Beads were then washed with radioimmune precipitation assay buffer, and bound proteins were analyzed by immunoblot with anti-HA antibody.
Chromatin Immunoprecipitation (ChIP) and Sequential ChIP Assays
ChIP assays were performed as described previously (50). Briefly, HT29 cells were grown in 150-mm cell culture dishes and treated with 1% formaldehyde for 20 min at room temperature. Cross-linking reactions were quenched with glycine, and cells were harvested and sonicated. After a pre-clearing step with Protein A/G beads (Santa Cruz Biotechnology), chromatin fractions were subjected to immunoprecipitation with 4 μg of β-catenin antibodies (Santa Cruz Biotechnology) or 8 μg of CCAR1 antibodies (Bethyl Laboratories, 435A unless specified otherwise). Protein A/G beads were added to capture immune complexes, which were then washed extensively, eluted, and heated to reverse formaldehyde cross-linking. Purified immunoprecipitated DNA and total input DNA were utilized as template in quantitative real-time PCR (qPCR) using a Stratagene Mx3000P instrument. The qPCRs contained SYBR Green QPCR Master Mix (Stratagene) and 150 nm of forward and reverse primers and were incubated through 40 cycles using the following conditions: 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 30 s. After the end of amplification, a melting curve analysis was carried out to confirm the homogeneity of products from each reaction. The primers used were as follows: hAxin2 3′-untranslated region (nucleotides +31681 to +31784 relative to transcription start site) (55); hAxin2 WRE (nucleotides −284 to −384 relative to transcription start site), 5′-TTTATAAAGTCCTCCAAGCC-3′ (forward) and 5′-AAGAACTGCAAGCAAGCAGATT-3′ (reverse); DKK1 Upstream Negative Control site (nucleotides −4462 to −4559 relative to transcription start site) (55); DKK1 WRE (nucleotides −827 to −931 relative to transcription start site) (55); c-Myc WRE (nucleotides −1223 to −1421 relative to transcription start site) (55); c-Myc open reading frame (nucleotides +2780 to +2866 relative to transcription start site (55). The relative standard curve method was utilized to determine the relative amount of immunoprecipitated DNA and input (unfractionated chromatin) signals. Signals from immunoprecipitated DNA were normalized to their respective input, and data were represented as the % of Input. In some experiments relative recruitment was calculated by dividing specific antibody signal by the signal from IgG_2 (Bethyl Laboratories). Data shown are mean and range of variation for two PCR reactions from a single experiment.
In sequential ChIP experiments (50), cross-linked DNA-protein complexes were eluted from primary immunoprecipitates by incubation with 10 mm dithiothreitol at 37 °C for 30 min. Eluates were then diluted 1:50 in immunoprecipitation dilution buffer and subjected to secondary immunoprecipitation with the indicated antibodies.
Real-time Reverse Transcriptase-PCR Assays
Total RNA from cell lines was extracted using TRIzol reagent (Invitrogen), and first-strand cDNA was synthesized by reverse transcribing 0.1 μg of total RNA using iScript cDNA Synthesis kit (Bio-Rad). 1 μl of the reverse transcription reaction was amplified by qPCR using the following primers: β-actin (49): α-tubulin, 5′-GATCTGGAGCCTACGGTCATTG-3′ (forward) and 5′-GAGCTGCTCTGGGTGGAAGA-3′ (reverse); MMP7 (55): BMP4, 5′-CACTGGTCCCTGGGATGTTC (forward) and 5′-GATCCACAGCACTGGTCTTGACTA-3′ (reverse); c-myc, 5′-CTCTCAACGACAGCAGCCCG-3′ (forward) and 5′-CAACATCGATTTCTTCCTCATCTTC-3′ (reverse); Axin2, 5′-CCACACCCTTCTCCAATCCA-3′ (forward) and 5′-TGGACACCTGCCAGTTTCTTT-3′ (reverse); DKK1, 5′-AAACGCTGCATGCGTCACGCTAT-3′ (forward) and 5′-AAAGCTTTCAGTGATGGTTT-3′ (reverse); and c-jun, 5′-GAAGTCGGCGAGCGGCTGCA-3′ (forward) and 5′-TTCTCTTGCGTGGCTCTC-3′ (reverse). Results shown are mean and range of variation for duplicate PCRs from a single cDNA preparation, and a minimum of two independent experiments were performed. Relative expression levels of target genes were determined by the standard curve method. All samples were corrected for total input RNA by normalizing to α-tubulin mRNA.
Cell Proliferation Assay
HT29 cells (1000 cells per well) were seeded in 24-well dishes in McCoy's 5a medium supplemented with 10% fetal bovine serum. Cell numbers were determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) assay (Promega CellTiter 96®) on the indicated days after plating. Results shown are the mean ± S.D. from triplicate cultures of a single experiment and are representative of two independent experiments.
Colony Formation in Soft Agar
0.8% of Noble agar (VWR International) in McCoy's 5a medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin was poured into 60-mm Petri dishes and allowed to solidify at room temperature. For each cell pool, 4000 cells were suspended in 0.3% molten agar and layered on top of the solidified 0.8% agar in each dish. These dishes were then placed at 37 °C and 5% CO2 in a cell culture incubator for 2–4 weeks until colonies were visible. During that period, 1 ml of medium was added approximately every 2 days to prevent drying of the agar gel. Right before colony counting, dishes were fixed with 100% ethanol for 15 min and then stained with 0.1% crystal violet (VWR International) in phosphate-buffered saline (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 1.3 mm KCl, 135 mm NaCl, pH 7.4). The colonies in each dish were counted by using the Molecular Imager Gel Doc XR System (Bio-Rad) and photographed by Leica microscopy (Plan-Apo 1×). Three dishes were set up for each cell line, and results shown are mean ± S.D. of the triplicate plates for a single experiment, which is representative of three independent experiments.
Lentivirus Packaging and Virus Transduction
Lentiviral particles were generated as described previously (56). Briefly, for each 100-mm plate of 293T cells, 8 μg of the transducing vector (pHRCMVpuroSin8-β-catenin or pHRCMVpuroSin8-CCAR1), 2.5 μg of the envelop plasmid pMD.G1, and 7.5 μg of the packaging vector pCMV ΔR8.91 were co-transfected by Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. The medium was changed the next day, and viruses were harvested by collecting the culture medium at 48 and 72 h post-transfection. Conditioned medium from 2 days was pooled, passed through a 0.45-μm filter, and stored at −80 °C. For lentiviral transduction, target cells were seeded in 12-well plates and reached 80% of confluency at the day of infection. Conditioned medium containing the virus was added to cells along with Polybrene (Millipore) at the final concentration of 6 μg/ml. Infection was allowed to proceed for 12–16 h before the addition of selection medium containing puromycin (5 μg/ml). Resistant cells were pooled and used for the indicated experiments.
RESULTS
Interaction between CCAR1 and β-Catenin
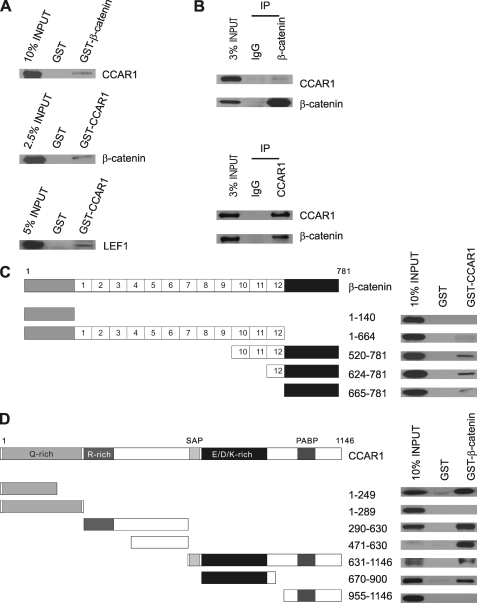
To investigate a possible physical interaction between β-catenin and CCAR1, CCAR1 or β-catenin synthesized in vitro was incubated with either GST, GST-β-catenin, or GST-CCAR1 bound to glutathione-Sepharose, and the bound proteins were analyzed by immunoblot. CCAR1 was bound by GST-β-catenin, but not by GST, and β-catenin was bound by GST-CCAR1 but not by GST alone (Fig. 1A, upper and middle panels). These data suggest that β-catenin may bind directly to CCAR1. LEF1 synthesized in vitro also bound specifically to GST-CCAR1, suggesting another possible direct interaction (lower panel).
FIGURE 1.
Binding of CCAR1 to β-catenin and LEF1. A, GST pulldown assays were performed as described under “Experimental Procedures,” using bacterially produced GST fusion proteins bound to glutathione-Sepharose beads and HA-tagged proteins translated in vitro. Bound proteins were detected by immunoblot analysis using antibodies against an HA tag. B, HA-tagged β-catenin and CCAR1 were expressed in COS-7 cells by transient transfection, and immunoprecipitation (IP) was performed on cell extracts, using the indicated antibodies against β-catenin or CCAR1, or normal IgG. Precipitated proteins were detected by immunoblot with antibodies against HA-tag. Uncropped images of the blots are shown in the supplemental materials (supplemental Fig. S1). C, HA-tagged fragments of β-catenin synthesized in vitro were incubated with GST-CCAR1 in GST pulldown assays, and bound proteins were detected by immunoblot using antibodies against the HA tag, as described in A. The diagram shows the 12 armadillo repeats of β-catenin flanked by N-terminal and C-terminal domains. D, GST pulldown assays were performed as in C using GST-β-catenin and HA-tagged fragments of CCAR1. The diagram shows the domains of CCAR1, including regions with a high content of specific amino acids, an SAF-Acinus-PIAS (SAP) domain, and a poly-A-binding protein (PABP) homology domain. GST pulldown assays were repeated at least twice, with results equivalent to those shown.
To examine whether CCAR1 associates with β-catenin in cultured cells, co-immunoprecipitation was carried out with extracts of COS-7 cells transiently transfected with plasmids encoding HA-β-catenin and HA-CCAR1. CCAR1 was specifically co-precipitated by antibodies against β-catenin, but not by normal IgG (Fig. 1B, upper panel; supplemental Fig. S1, upper panel). Conversely, β-catenin was precipitated by antibodies against CCAR1, but not by IgG (Fig. 1B, lower panel; supplemental Fig. S1, lower panel). Thus interaction between CCAR1 and β-catenin can occur in vitro and in vivo.
To further map the one or more regions of β-catenin that are responsible for its interaction with CCAR1, GST-CCAR1 was incubated in GST pulldown assays with five β-catenin fragments that were synthesized in vitro (amino acids 1–140, 1–664, 520–781, 624–781, and 665–781). Three overlapping C-terminal fragments of β-catenin (amino acids 520–781, 624–781, and 665–781) bound specifically to GST-CCAR1 (Fig. 1C), indicating that the C terminus of β-catenin is the major CCAR1 binding domain. Reciprocal mapping of the β-catenin-binding region(s) in CCAR1 (Fig. 1D) identified three separate domains of CCAR1 (amino acids 1–249, 471–630, and 670–900) capable of binding to β-catenin. Interestingly, addition of 40 amino acids to the 1–249 fragment of CCAR1 (amino acids 1–289) strongly inhibited the interaction between CCAR1 and β-catenin, suggesting that amino acids 250–289 of CCAR1 may regulate this interaction.
CCAR1 Cooperates with β-Catenin as a Coactivator for LEF1
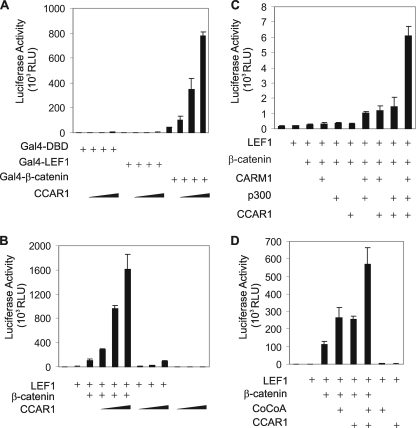
To address whether binding of CCAR1 to β-catenin can modulate β-catenin-directed transcriptional activity, we monitored the effect of overexpression of CCAR1 on transcriptional activation by β-catenin tethered to Gal4 DBD. Compared with Gal4 DBD alone, Gal4 DBD fused to β-catenin strongly activated expression from a luciferase reporter plasmid controlled by Gal4 response elements (Fig. 2A), and this activity was further enhanced by CCAR1 in a dose-dependent manner. However, CCAR1 overexpression had little or no effect on the activity of Gal4 DBD or Gal4 DBD fused to LEF1. Thus, even though CCAR1 can bind to LEF1 in vitro (Fig. 1A, lower panel), CCAR1 was unable to enhance transcriptional activation by LEF1. These results indicate that CCAR1 and β-catenin have a functional as well as a physical interaction.
FIGURE 2.
Cooperation of CCAR1 and β-catenin as coactivators for transcriptional activation by LEF1. A, CV-1 cells were transfected in 12-well plates with luciferase reporter plasmid GK1-Luc (300 ng) controlled by Gal4 response elements, pM plasmids encoding Gal4-DBD alone or fused to LEF1 or β-catenin (100 ng), and pSG5.HA-CCAR1 (300, 600, or 900 ng). Luciferase assays were conducted on cell extracts as described under “Experimental Procedures.” Results shown are from a single experiment and are representative of three independent experiments. B, CV1 cells were transfected with luciferase reporter plasmids pGL3OT (200 ng) containing LEF1-responsive elements, along with pSG5.HA-LEF1 (10 ng), pSG5.HA-β-catenin (200 ng), and pSG5.HA-CCAR1 (300, 600, or 900 ng). Results are from a single experiment, which is representative of three independent experiments. C and D, plasmids used for reporter gene assay were pGL3OT (200 ng) along with pSG5.HA-LEF1 (0.005 ng in C and 5 ng in D), pSG5.HA-β-catenin (200 ng), pSG5.HA-CARM1 (200 ng), pCMV.p300 (200 ng), pSG5.HA-CoCoA (200 ng), and pSG5.HA-CCAR1 (200 ng), as indicated. Luciferase activity was determined as in A.
Members of the LEF/TCF family of DNA-binding proteins recruit β-catenin, which then serves as a platform for recruiting additional transcriptional coactivators to activate transcription of LEF/β-catenin target genes. Because CCAR1 interacts with LEF1 and β-catenin, we tested whether CCAR1 can function as a coactivator for LEF1-mediated gene transcription, using the transiently transfected luciferase reporter plasmid pGL3OT, which is controlled by LEF/TCF-responsive elements. Overexpression of LEF1 alone produced a small enhancement of luciferase activity, and overexpression of β-catenin further enhanced luciferase activity (Fig. 2B). CCAR1 expression with LEF1 and β-catenin caused a dramatic additional enhancement. However, without overexpression of β-catenin, CCAR1 had very little effect on transcriptional activation by LEF1. The weak enhancement of LEF1 activity by CCAR1 in the absence of overexpressed β-catenin may result from endogenous β-catenin. Thus, although CCAR1 alone is a weak coactivator for LEF1, CCAR1 can cooperate synergistically with β-catenin to cause a dramatic enhancement of transcriptional activation by LEF1.
The very large number of transcriptional coactivators discovered to date suggests that initiation of transcription is a very complex process. We therefore tested whether CCAR1 can cooperate with several other coactivators that have been shown to associate with and enhance the coactivator activity of β-catenin: CoCoA (46), p300 (32), and CARM1 (30). For these transient transfection experiments, we used reduced levels of the plasmid encoding LEF1, because we have previously shown that these conditions are appropriate for observing synergistic cooperation among multiple coactivators (57). We observed synergistic cooperation of CCAR1 and β-catenin with CARM1 and p300 (Fig. 1C) and with CoCoA (Fig. 1D). Furthermore, this synergy was completely dependent on the co-expression of β-catenin (data not shown). These results are consistent with the conclusion that β-catenin associates directly with DNA-bound LEF1 and recruits additional coactivators, such as CCAR1, CARM1, p300, and CoCoA, each of which makes a specific and distinct contribution to the process of transcriptional activity.
β-Catenin Recruits CCAR1 to a Target Gene of Wnt Signaling
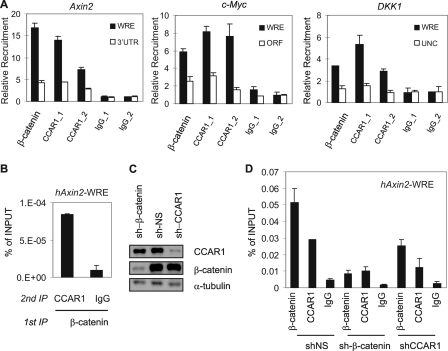
To investigate whether β-catenin recruits CCAR1 to target genes of LEF1 and the Wnt signaling pathway, we initially used ChIP to test whether CCAR1 associated with the endogenous Wnt-responsive enhancers (WREs) of the Axin2 gene (55, 58, 59), DKK1 gene (55, 60), and c-myc gene (31) in HT29 cells. HT29 is a human colon carcinoma cell line widely used for studying the Wnt/β-catenin signaling pathway, due to its high endogenous level of β-catenin and constitutively active intrinsic Wnt signaling. Both β-catenin and CCAR1 bound preferentially to the WREs of the Axin2 gene, DKK1 gene, and c-myc gene, compared with other sites within or near these genes, which lack WRE sequences (Fig. 3A). ChIP performed with two different normal IgG preparations showed equally low background recruitment signals at both WRE and negative control sites. Thus β-catenin and CCAR1 specifically occupy the WREs of the Axin2, DKK1, and c-myc promoters. To investigate whether β-catenin and CCAR1 occupy the Axin2 WRE site together as part of the same complex, we performed modified ChIP assays with two sequential immunoprecipitations. Sequential immunoprecipitations performed on HT29 chromatin with antibodies against β-catenin and CCAR1 indicated that these two proteins are found together in the same complex on the Axin2 WRE (Fig. 3B). Sequential ChIP performed by antibodies against β-catenin followed by normal IgG served as a negative control.
FIGURE 3.
Recruitment of CCAR1 to a Wnt target gene depends on β-catenin. A, ChIP assays were performed with HT29 cell chromatin, using antibodies against β-catenin or CCAR1 (CCAR_1 antibody, Bethyl 435A; CCAR_2 antibody, Bethyl 270A), or with normal IgG (IgG_1, Santa Cruz Biotechnology normal rabbit; IgG_2, Bethyl normal rabbit). Immunoprecipitated DNA was analyzed by qPCR with primers for the upstream WRE of the Axin2 c-myc, and DKK1 genes or with primers for the 3′-untranslated region (3′UTR) of the Axin2 gene, an upstream negative control (UNC) site near the DKK1 gene, and the open reading frame (ORF) of the c-myc gene. Relative recruitment was calculated by dividing specific antibody signal by the signal for IgG_2 (Bethyl Laboratories). B, sequential ChIP assays for hAxin2-WRE were performed with the indicated antibodies, and results are expressed relative to input DNA from the unfractionated chromatin. C, HT29 cells were infected with lentivirus encoding a puromycin resistance gene, and shRNAs against a nonspecific sequence (NS), β-catenin, or CCAR1, and puromycin-resistant cells were selected. At 7 days after the infection, cell extracts were analyzed by immunoblot using antibodies against CCAR1, β-catenin, or α-tubulin. D, the infected cells from C were analyzed by ChIP assay as in A. In A, B, and D, the mean and range of variation from duplicate PCR reactions are shown. ChIP results shown are from a single experiment, which is representative of two or more independent experiments.
Next, we explored how CCAR1 is recruited to the Axin2 promoter. Because CCAR1 can interact with both β-catenin and LEF1 (Fig. 1A), this raised a question as to whether CCAR1 binding to the Axin2 WRE is dependent upon β-catenin. Therefore, lentiviral vectors that express short-hairpin RNA (shRNA) against β-catenin or CCAR1 were introduced into HT29 cells to reduce endogenous levels of these proteins. Western blots confirmed that the protein levels of β-catenin and CCAR1 in HT29 cells were substantially reduced by their respective shRNAs, when compared with nonspecific shRNA (Fig. 3C). ChIP analysis of the Axin2 WRE demonstrated that shRNA against β-catenin (compared with the nonspecific shRNA) almost completely eliminated occupancy of the Axin2 WRE by both β-catenin and CCAR1 (Fig. 3D), indicating that CCAR1 is targeted to the Axin2 WRE by its interaction with β-catenin. Surprisingly, the depletion of CCAR1 also partially inhibited β-catenin binding to Axin2 promoter. Because the cellular level of β-catenin was not affected by reduction of the CCAR1 level (Fig. 3C), this observation suggests that CCAR1 may contribute to the stable occupancy of the Axin2 promoter by β-catenin.
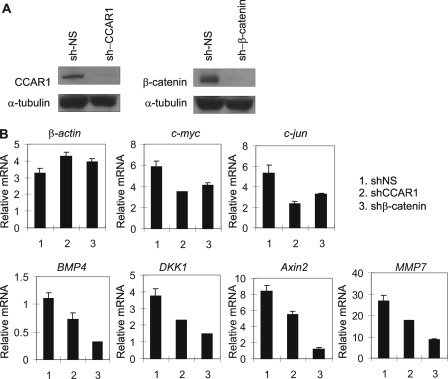
CCAR1 Is Required for Efficient Expression of Wnt Target Genes
The association of CCAR1 with β-catenin on the Axin2 WRE (Fig. 3) and the cooperative function of CCAR1 and β-catenin in the activation of transient reporter genes by LEF1 (Fig. 2) suggest that CCAR1 may be important for helping β-catenin to activate endogenous target genes of Wnt signaling, such as c-myc, c-jun, BMP4, MMP7, Axin2, and DKK1. Lentiviral vectors expressing shRNAs were used again to reduce endogenous levels of CCAR1 or β-catenin proteins in HT29 cells (Fig. 4A). Reduction of β-catenin levels had a moderate to dramatic impact on expression of the six Wnt/β-catenin target genes listed above. Thus, β-catenin appeared to play a major role in the expression of BMP4, MMP7, Axin2, and DKK1; however, the expression of c-myc and c-jun appeared to depend only partially on β-catenin and thus presumably is driven by other transcription factor complexes in addition to LEF1/β-catenin (Fig. 4B). Reduction of CCAR1 levels partially compromised the expression of all six of the Wnt target genes, indicating that CCAR1 is also important for efficient expression of these genes. In contrast, depletion of CCAR1 or β-catenin had no effect on the level of β-actin mRNA, demonstrating gene-specific requirement of CCAR1 and β-catenin. It is interesting to note that reducing the CCAR1 level had a larger effect than reducing the β-catenin level on expression of c-myc and c-jun, but had a lesser effect than reducing the β-catenin level on expression of the other four Wnt target genes. These results suggest somewhat different relative roles for CCAR1 and β-catenin in mediating expression of the two sets of Wnt target genes (see “Discussion”). Depletion of CCAR1 also reduced the Wnt3a-dependent expression of the Axin2 gene in RKO cells (data not shown). RKO is a colon cancer cell line with a normal adenomatous polyposis coli gene and normal Wnt3a-dependent regulation of β-catenin levels. Thus, CCAR1 is required for expression of the target genes of Wnt and β-catenin in cells with normal or abnormal regulation of β-catenin.
FIGURE 4.
Requirement of β-catenin and CCAR1 for expression of Wnt target genes. A, HT29 cells infected with lentivirus encoding shRNA against β-catenin, CCAR1, or a nonspecific sequence (NS) were analyzed by immunoblot as in Fig. 3C. B, total RNA from the lentivirus-infected cells in A was examined by quantitative reverse transcriptase-PCR analysis, using primers specific for the indicated Wnt target genes. Results shown are normalized to the level of α-tubulin mRNA, are the mean and range of variation for duplicate PCR reactions from a single experiment, and are representative of at least three independent experiments.
Role of CCAR1 and β-Catenin in Neoplastic Transformation
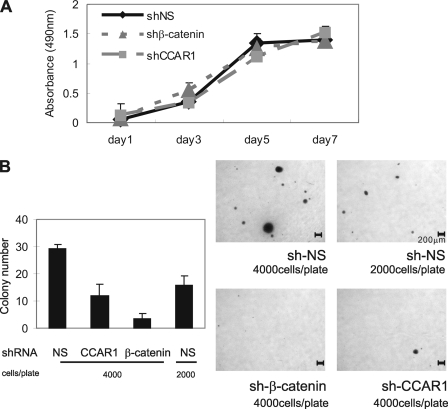
In colorectal cancers, c-myc, c-jun, and MMP7 are robustly expressed and play critical roles in tumor growth and progression (61–63). Because reduction of the endogenous CCAR1 level compromised the expression of these genes, we assessed the role of CCAR1 in colon cancer cell proliferation. Indistinguishable growth curves were obtained for HT29 cell populations stably infected with lentiviral vectors encoding shRNAs directed against a nonspecific sequence, β-catenin, or CCAR1 (Fig. 5A). To assess a possible role of CCAR1 and β-catenin in anchorage-independent growth, which is typically associated with a tumorigenic phenotype, the same HT29 cell populations were tested for their ability to form colonies in soft agar. HT29 cells expressing shRNA directed against β-catenin or CCAR1 formed fewer and smaller colonies than HT29 cells expressing the nonspecific shRNA (Fig. 5B and supplemental Figs. S2 and S3). Depletion of β-catenin nearly eliminated colony formation in soft agar, whereas colony formation was inhibited by >50% by depletion of CCAR1. As a control to test the linearity of the automated colony counting method, we plated two different amounts of HT29 nonspecific shRNA cells. As expected, reducing the number of cells plated by half also reduced by half the number of colonies detected (Fig. 5B). The result for depletion of β-catenin confirms previous conclusions that an aberrant elevated endogenous level of β-catenin is central to induction of neoplastic and morphological transformation of human colorectal cancer cells. The result for depletion of CCAR1 is consistent with the conclusion that CCAR1, along with other coactivators that are recruited to Wnt target genes by β-catenin, plays an important role in mediating transcriptional activation by β-catenin.
FIGURE 5.
Role of CCAR1 in anchorage-independent colony formation. A, HT29 cells were plated in standard tissue culture dishes, and cell proliferation was monitored by MTS assay. B, diluted HT29 cell suspensions containing the indicated number of cells were plated in soft agar, and colony formation was examined by staining after 2–4 weeks (right panels). Microscope images are shown with a scale bar representing 200 μm. Colonies in each dish were counted by Molecular Imager Gel Doc XR System (Bio-Rad). Results shown are mean ± S.D. from triplicate cultures from a single experiment and are representative of three independent experiments. The images from the triplicate plates used for the data shown in Fig. 5B are presented in the supplemental materials (supplemental Fig. S2). Results from an independent experiment are also presented in supplemental Fig. S3.
DISCUSSION
Defects in crucial components of the Wnt signaling pathway, including β-catenin, adenomatous polyposis coli, and the Axins, play a predominant role in the pathogenesis of human cancers. A proposed consequence of these Wnt pathway-related mutations is to elevate the levels of β-catenin both in the cytoplasm and nucleus, allowing more β-catenin to bind to LEF/TCF and promote transcription of LEF/TCF target genes. Proteins encoded by LEF/β-catenin target genes likely collaborate in executing a program leading to and/or maintaining neoplastic transformation. However, beyond the recruitment of β-catenin, the mechanism of activation of Wnt target gene expression is poorly understood. Here we extend the current understanding of Wnt signaling by identifying a novel β-catenin binding partner, CCAR1 (Fig. 1). Functionally, CCAR1 cooperated synergistically with β-catenin to cause robust enhancement of LEF1-mediated transcription of a transient reporter gene (Fig. 2). The coactivator function of CCAR1 in the transient reporter gene assays was almost completely dependent on co-expression of β-catenin. Thus, although CCAR1 can bind to LEF1 as well as β-catenin in vitro (Fig. 1), the requirement for β-catenin suggests that CCAR1 may be recruited to LEF1 target genes by β-catenin, not directly by LEF1. In agreement with this conclusion, depletion of endogenous β-catenin prevented recruitment of CCAR1 to the WRE associated with the Axin2 promoter (Fig. 3D). Thus, in the chromatin-based cellular environment, the LEF1-CCAR1 interaction, if it occurs, is not sufficient for stable recruitment of CCAR1 to the Axin2 WRE.
Wnt signaling is important for normal cell proliferation and differentiation. However, for precise control of Wnt signaling during development, normal tissue possesses an auto-regulatory mechanism to limit the duration or intensity of a Wnt-initiated signal. Axin2, which binds to β-catenin and induces its degradation, is a direct transcriptional target of LEF/β-catenin (59). In addition, the DKK1 gene encoding an extracellular inhibitor of Wnt signaling is also induced by Wnt signaling (60, 64, 65). By contrast, during tumorigenesis this negative feedback loop is disrupted by acquisition of oncogenic mutations of β-catenin or cellular components that normally cause regulated degradation of β-catenin, or by epigenetic silencing of components involved in the auto-regulatory loop. For example, hyper-methylation of the DKK1 gene promoter has been reported in human colorectal cancer cells (66). Here, our loss-of-function studies provide evidence for the involvement of CCAR1 in the auto-regulatory mechanism; depletion of CCAR1 moderately reduced expression of Axin2 and DKK1 (Fig. 4B). Additionally, depletion of β-catenin essentially abolished expression of these two genes, thus indicating that these two components of the Wnt/β-catenin autoinhibitory loop are regulated primarily by Wnt/β-catenin signaling. Loss of β-catenin at the Axin2 and DKK1 promoters would presumably prevent recruitment of multiple coactivators, including CCAR1, which are needed for transcriptional activation. In fact, the dismissal of β-catenin from the WRE of the Axin2 gene blocked recruitment of CCAR1 (Fig. 3D).
The importance of CCAR1 for Wnt targets is not limited to genes involved in the auto-regulatory mechanism but also extends to genes involved in neoplastic transformation. Depletion of CCAR1 caused a 30–40% suppression of the expression of the growth-promoting genes c-myc and c-jun (Fig. 4). It is interesting to note that depletion of β-catenin caused a stronger inhibition of Axin2 and DKK1 expression than depletion of CCAR1, whereas depletion of CCAR1 caused a slightly stronger inhibition of expression of the growth promoting genes c-myc and c-jun than depletion of β-catenin. These results suggest two different regulatory programs for the Wnt auto-regulatory target genes versus the growth-promoting Wnt target genes. The dramatic loss of expression of Axin2 and DKK1 upon depletion of β-catenin indicates that the Wnt/β-catenin pathway plays a dominant role in regulating these genes in HT29 cells. Because β-catenin is expected to recruit multiple secondary coactivators (including CCAR1) to the promoter, it is not surprising that depletion of CCAR1 caused less inhibition of these genes than depletion of β-catenin. In contrast, the more moderate loss of c-myc and c-jun expression observed upon depletion of β-catenin suggests that Wnt/β-catenin signaling is only one of multiple signaling pathways regulating expression of these genes. In fact, c-myc and c-jun are known to be regulated by signaling through a variety of transcription factors, including nuclear receptors (67). Because CCAR1 can function as a coactivator for LEF1/β-catenin (this report) as well as nuclear receptors and p53 (49), it would not be surprising if CCAR1 also functions as a coactivator for other classes of transcription factors. Thus, if CCAR1 is recruited to c-myc and c-jun promoters by multiple transcription factors, this could explain why depletion of CCAR1 had a stronger effect than depletion of β-catenin.
The involvement of CCAR1 as a mediator of signaling leading to apoptosis (47, 48) as well as proliferation (49) and aspects of oncogenic transformation (Fig. 5) appears contradictory at first glance. However, the finding that CCAR1 can serve as a coactivator for multiple classes of transcription factors indicates that the activity of CCAR1 can be directed by a variety of signaling pathways, which result in recruitment of CCAR1 to specific sets of target genes that depend upon the nature of the signal. Thus, interactions between two coactivators may control specific programs of gene expression with specific physiological outcomes. The fact, that depletion of CCAR1 or β-catenin suppressed one aspect of the neoplastic transformation of human colorectal cancer cells (i.e. colony formation in soft agar (Fig. 5 and supplemental Figs. S2 and S3), suggests the possibility that targeting the interaction between CCAR1 and β-catenin may be a potential strategy for therapeutic control of aberrant Wnt/β-catenin signaling in colorectal cancer.
Supplementary Material
Acknowledgments
We thank Kelly Chang, Daniel Gerke, and Irina Ianculescu for expert technical assistance, Dr. Robert Ladner (University of Southern California) for HT29 cells, and Dr. Didier Trono (University of Geneva) for pMD.G1 and pCMV ΔR8.91 vectors.
This work was supported, in whole or in part, by National Institutes of Health Grant DK43093 (to M. R. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- LEF
- lymphoid enhancer-binding factor
- TCF
- T-cell factor
- CCAR1
- cell cycle and apoptosis regulator 1
- GST
- glutathione S-transferase
- DBD
- DNA-binding domain
- HA
- hemagglutinin
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative PCR
- CMV
- cytomegalovirus
- WRE
- Wnt-responsive enhancer
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Nusse R. (2008) Cell Res. 18, 523–527 [DOI] [PubMed] [Google Scholar]
- 2.McDonald S. A., Preston S. L., Lovell M. J., Wright N. A., Jankowski J. A. (2006) Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 267–274 [DOI] [PubMed] [Google Scholar]
- 3.Reya T., Clevers H. (2005) Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 4.Hayward P., Kalmar T., Arias A. M. (2008) Development 135, 411–424 [DOI] [PubMed] [Google Scholar]
- 5.Fuerer C., Nusse R., Ten Berge D. (2008) EMBO Rep. 9, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienz M., Clevers H. (2000) Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 7.Dahmen R. P., Koch A., Denkhaus D., Tonn J. C., Sörensen N., Berthold F., Behrens J., Birchmeier W., Wiestler O. D., Pietsch T. (2001) Cancer Res. 61, 7039–7043 [PubMed] [Google Scholar]
- 8.Kinzler K. W., Vogelstein B. (1996) Cell 87, 159–170 [DOI] [PubMed] [Google Scholar]
- 9.Liu W., Dong X., Mai M., Seelan R. S., Taniguchi K., Krishnadath K. K., Halling K. C., Cunningham J. M., Boardman L. A., Qian C., Christensen E., Schmidt S. S., Roche P. C., Smith D. I., Thibodeau S. N. (2000) Nat. Genet. 26, 146–147 [DOI] [PubMed] [Google Scholar]
- 10.Polakis P. (2000) Genes Dev. 14, 1837–1851 [PubMed] [Google Scholar]
- 11.Satoh S., Daigo Y., Furukawa Y., Kato T., Miwa N., Nishiwaki T., Kawasoe T., Ishiguro H., Fujita M., Tokino T., Sasaki Y., Imaoka S., Murata M., Shimano T., Yamaoka Y., Nakamura Y. (2000) Nat. Genet. 24, 245–250 [DOI] [PubMed] [Google Scholar]
- 12.Wu R., Zhai Y., Fearon E. R., Cho K. R. (2001) Cancer Res. 61, 8247–8255 [PubMed] [Google Scholar]
- 13.Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 14.Ha N. C., Tonozuka T., Stamos J. L., Choi H. J., Weis W. I. (2004) Mol. Cell 15, 511–521 [DOI] [PubMed] [Google Scholar]
- 15.Xing Y., Clements W. K., Kimelman D., Xu W. (2003) Genes Dev. 17, 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y., Clements W. K., Le Trong I., Hinds T. R., Stenkamp R., Kimelman D., Xu W. (2004) Mol. Cell 15, 523–533 [DOI] [PubMed] [Google Scholar]
- 17.Städeli R., Hoffmans R., Basler K. (2006) Curr. Biol. 16, R378–R385 [DOI] [PubMed] [Google Scholar]
- 18.Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., Tran C., Chen C., Chung C. H., Huber O., Rose D. W., Sawyers C. L., Rosenfeld M. G., Baek S. H. (2005) Nature 434, 921–926 [DOI] [PubMed] [Google Scholar]
- 19.Deng J., Miller S. A., Wang H. Y., Xia W., Wen Y., Zhou B. P., Li Y., Lin S. Y., Hung M. C. (2002) Cancer Cell 2, 323–334 [DOI] [PubMed] [Google Scholar]
- 20.Olson L. E., Tollkuhn J., Scafoglio C., Krones A., Zhang J., Ohgi K. A., Wu W., Taketo M. M., Kemler R., Grosschedl R., Rose D., Li X., Rosenfeld M. G. (2006) Cell 125, 593–605 [DOI] [PubMed] [Google Scholar]
- 21.Easwaran V., Pishvaian M., Salimuddin, Byers S. (1999) Curr. Biol. 9, 1415–1418 [DOI] [PubMed] [Google Scholar]
- 22.Pálmer H. G., González-Sancho J. M., Espada J., Berciano M. T., Puig I., Baulida J., Quintanilla M., Cano A., de Herreros A. G., Lafarga M., Muñoz A. (2001) J. Cell Biol. 154, 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah S., Hecht A., Pestell R., Byers S. W. (2003) J. Biol. Chem. 278, 48137–48145 [DOI] [PubMed] [Google Scholar]
- 24.Song L. N., Herrell R., Byers S., Shah S., Wilson E. M., Gelmann E. P. (2003) Mol. Cell Biol. 23, 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 26.Mann B., Gelos M., Siedow A., Hanski M. L., Gratchev A., Ilyas M., Bodmer W. F., Moyer M. P., Riecken E. O., Buhr H. J., Hanski C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T., Otevrel T., Gao Z., Gao Z., Ehrlich S. M., Fields J. Z., Boman B. M. (2001) Cancer Res. 61, 8664–8667 [PubMed] [Google Scholar]
- 28.Kim P. J., Plescia J., Clevers H., Fearon E. R., Altieri D. C. (2003) Lancet 362, 205–209 [DOI] [PubMed] [Google Scholar]
- 29.Crawford H. C., Fingleton B. M., Rudolph-Owen L. A., Goss K. J., Rubinfeld B., Polakis P., Matrisian L. M. (1999) Oncogene 18, 2883–2891 [DOI] [PubMed] [Google Scholar]
- 30.Koh S. S., Li H., Lee Y. H., Widelitz R. B., Chuong C. M., Stallcup M. R. (2002) J. Biol. Chem. 277, 26031–26035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sierra J., Yoshida T., Joazeiro C. A., Jones K. A. (2006) Genes Dev. 20, 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Kolligs F. T., Hottiger M. O., Mosavin R., Fearon E. R., Nabel G. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12613–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagishi M., Fujii R., Hatta M., Yoshida E., Araya N., Nagafuchi A., Ishihara S., Nakajima T., Fukamizu A. (2000) J. Biol. Chem. 275, 35170–35175 [DOI] [PubMed] [Google Scholar]
- 34.Hecht A., Vleminckx K., Stemmler M. P., van Roy F., Kemler R. (2000) EMBO J. 19, 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takemaru K. I., Moon R. T. (2000) J. Cell Biol. 149, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer A., Chauvet S., Huber O., Usseglio F., Rothbächer U., Aragnol D., Kemler R., Pradel J. (2000) EMBO J. 19, 6121–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y., Lee N., Fearon E. R. (2003) Cancer Res. 63, 8726–8734 [PubMed] [Google Scholar]
- 38.Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. (2001) EMBO J. 20, 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrera I., Janody F., Leeds N., Duveau F., Treisman J. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6644–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker D. S., Jemison J., Cadigan K. M. (2002) Development 129, 2565–2576 [DOI] [PubMed] [Google Scholar]
- 41.Wang S., Jones K. A. (2006) Curr. Biol. 16, 2239–2244 [DOI] [PubMed] [Google Scholar]
- 42.Lee Y. H., Stallcup M. R. (2006) Nucleic Acids Res. 34, 5052–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramps T., Peter O., Brunner E., Nellen D., Froesch B., Chatterjee S., Murone M., Züllig S., Basler K. (2002) Cell 109, 47–60 [DOI] [PubMed] [Google Scholar]
- 44.Chen Y. H., Yang C. K., Xia M., Ou C. Y., Stallcup M. R. (2007) Nucleic Acids Res. 35, 2084–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Kim J. H., Koh S. S., Stallcup M. R. (2004) J. Biol. Chem. 279, 4212–4220 [DOI] [PubMed] [Google Scholar]
- 46.Yang C. K., Kim J. H., Li H., Stallcup M. R. (2006) J. Biol. Chem. 281, 3389–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rishi A. K., Zhang L., Boyanapalli M., Wali A., Mohammad R. M., Yu Y., Fontana J. A., Hatfield J. S., Dawson M. I., Majumdar A. P., Reichert U. (2003) J. Biol. Chem. 278, 33422–33435 [DOI] [PubMed] [Google Scholar]
- 48.Rishi A. K., Zhang L., Yu Y., Jiang Y., Nautiyal J., Wali A., Fontana J. A., Levi E., Majumdar A. P. (2006) J. Biol. Chem. 281, 13188–13198 [DOI] [PubMed] [Google Scholar]
- 49.Kim J. H., Yang C. K., Heo K., Roeder R. G., An W., Stallcup M. R. (2008) Mol. Cell 31, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J. H., Li H., Stallcup M. R. (2003) Mol. Cell 12, 1537–1549 [DOI] [PubMed] [Google Scholar]
- 51.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 52.Puri P. L., Sartorelli V., Yang X. J., Hamamori Y., Ogryzko V. V., Howard B. H., Kedes L., Wang J. Y., Graessmann A., Nakatani Y., Levrero M. (1997) Mol. Cell 1, 35–45 [DOI] [PubMed] [Google Scholar]
- 53.Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. (1997) Nat. Biotechnol. 15, 871–875 [DOI] [PubMed] [Google Scholar]
- 54.Zhao J., Pettigrew G. J., Thomas J., Vandenberg J. I., Delriviere L., Bolton E. M., Carmichael A., Martin J. L., Marber M. S., Lever A. M. (2002) Basic Res. Cardiol. 97, 348–358 [DOI] [PubMed] [Google Scholar]
- 55.Li J., Wang C. Y. (2008) Nat. Cell Biol. 10, 160–169 [DOI] [PubMed] [Google Scholar]
- 56.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 57.Lee Y. H., Koh S. S., Zhang X., Cheng X., Stallcup M. R. (2002) Mol. Cell Biol. 22, 3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung J. Y., Kolligs F. T., Wu R., Zhai Y., Kuick R., Hanash S., Cho K. R., Fearon E. R. (2002) J. Biol. Chem. 277, 21657–21665 [DOI] [PubMed] [Google Scholar]
- 59.Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Mol. Cell Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niida A., Hiroko T., Kasai M., Furukawa Y., Nakamura Y., Suzuki Y., Sugano S., Akiyama T. (2004) Oncogene 23, 8520–8526 [DOI] [PubMed] [Google Scholar]
- 61.Newell K. J., Witty J. P., Rodgers W. H., Matrisian L. M. (1994) Mol. Carcinog. 10, 199–206 [DOI] [PubMed] [Google Scholar]
- 62.Marcu K. B., Bossone S. A., Patel A. J. (1992) Annu. Rev. Biochem. 61, 809–860 [DOI] [PubMed] [Google Scholar]
- 63.Smith D. R., Myint T., Goh H. S. (1993) Br. J. Cancer 68, 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chamorro M. N., Schwartz D. R., Vonica A., Brivanlou A. H., Cho K. R., Varmus H. E. (2005) EMBO J. 24, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.González-Sancho J. M., Aguilera O., García J. M., Pendás-Franco N., Peña C., Cal S., García de Herreros A., Bonilla F., Muñoz A. (2005) Oncogene 24, 1098–1103 [DOI] [PubMed] [Google Scholar]
- 66.Aguilera O., Fraga M. F., Ballestar E., Paz M. F., Herranz M., Espada J., García J. M., Muñoz A., Esteller M., González-Sancho J. M. (2006) Oncogene 25, 4116–4121 [DOI] [PubMed] [Google Scholar]
- 67.Dubik D., Shiu R. P. (1988) J. Biol. Chem. 263, 12705–12708 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.