Abstract
Close packing of three chains in a standard collagen triple helix requires Gly as every third residue. Missense mutations replacing one Gly by a larger residue in the tripeptide repeating sequence in type I collagen are common molecular causes of osteogenesis imperfecta. The structural and dynamic consequences of such mutations are addressed here by NMR studies on a peptide with a Gly-to-Ser substitution within an α1(I) sequence. Distances derived from nuclear Overhauser effects indicate that the three Ser residues are still packed in the center of the triple helix and that the standard 1-residue stagger is maintained. NMR dynamics using H-exchange and temperature-dependent amide chemical shifts indicate a greater disruption of hydrogen bonding and/or increased conformational flexibility C-terminal to the Ser site when compared with N terminal. This is consistent with recent suggestions relating clinical severity with an asymmetric effect of residues N- versus C-terminal to a mutation site. Dynamic studies also indicate that the relative position between a Gly in one chain and the mutation site in a neighboring staggered chain influences the disruption of the standard hydrogen-bonding pattern. The structural and dynamic alterations reported here may play a role in the etiology of osteogenesis imperfecta by affecting collagen secretion or interactions with other matrix molecules.
Mutations in collagen result in a variety of connective tissue diseases (1, 2), with the clinical phenotype depending on the location and function of the collagen type. For instance, mutations in type I collagen, the major collagen in bone, lead to a bone disorder, osteogenesis imperfecta (OI),3 whereas mutations in type III collagen, which is present in high amounts in blood vessels, lead to aortic rupture in Ehlers-Danlos syndrome type IV (1, 2). All collagens have a triple helix motif composed of three polyproline II-like chains that are staggered by 1 residue and supercoiled about a common axis. The smallest residue Gly is typically present as every 3rd residue in each chain because of the tight packing of the chains, which generates the characteristic (Gly-Xaa-Yaa)n repeating sequence. The Gly residues are all buried in the center, and the structure is stabilized by interchain N–H (Gly) … C O (Xaa) hydrogen bonds (3–5). The most common type of mutation leading to collagen disorders is a missense mutation that replaces 1 Gly in the repeating sequence by a larger residue.
O (Xaa) hydrogen bonds (3–5). The most common type of mutation leading to collagen disorders is a missense mutation that replaces 1 Gly in the repeating sequence by a larger residue.
The best characterized collagen disease is OI, or brittle bone disease, which is distinguished by fragile bones due to mutations in type I collagen (2, 6). More than 400 Gly substitution missense mutations in the α1(I) and α2(I) chains of type I collagen have been reported to lead to OI (7). The severity of the disease varies widely from mild cases with multiple fractures to perinatal lethal cases (2, 6, 7). A single base change in a Gly codon can lead to one of 8 residues (Ser, Ala, Cys, Val, Arg, Asp, Glu, Trp) or a missense mutation. The smallest residue Ala is underrepresented in OI, suggesting that it may not always lead to pathology, whereas Ser mutations are overrepresented, corresponding to the most common substitutions observed. The 152 mutations leading to a Gly to Ser substitution account for ∼39% of all missense mutations in the α1(I) of type I collagen (7), with 115 associated with mild phenotypes and 37 associated with lethal phenotypes.
The identity of the residue replacing Gly may be a determinant in the clinical severity of OI. Model peptide studies indicate that the degree of triple helix destabilization depends on the residue replacing Gly, with a ranking of the least destabilizing to the most destabilizing Ala,Ser<Cys<Arg<Val<Glu,Asp<Trp (8). There is some correlation between clinical severity of OI cases and this destabilization scale, with the strongly destabilizing residues Val, Arg, Asp, and Glu associated largely with lethal phenotypes (8). However, as cited above, a Gly to Ser mutation can lead to a mild, a severe, or a lethal OI case, with no obvious molecular explanation. Other factors suggested to contribute to clinical phenotype include the rigidity of its immediate sequence environment; its location with respect to the C terminus; its proximity to salt bridges; and its presence at an interaction site, such as the binding site for proteoglycans on collagen fibrils (7, 9). A recent study of the stability of OI collagens supported the importance of the domain location of the mutation (10), whereas a network analysis of the mutations suggested the importance of a destabilizing tripeptide sequence C-terminal to the mutation site (11).
The standard triple helix conformation must undergo some structural perturbation as a result of a Gly replacement that is likely to relate to the development of the disorder. Thus it is important to define the structural consequences of a Gly substitution. It has not proved possible to obtain molecular information for the long collagen molecules themselves, but model collagen peptides have proved amenable to x-ray crystallography and NMR techniques (12, 13). The structure of a peptide containing a Gly to Ala substitution near the center of the peptide (Pro-Hyp-Gly)10 has been solved by x-ray crystallography (5). This structure shows an overall straight molecule with standard triple helical structures at both ends and a localized conformational deformation at the Ala replacement site. The direct N–H (Gly) … C O (Xaa) hydrogen bond is replaced by a water-mediated hydrogen bond N–H (Ala) … H2O … C
O (Xaa) hydrogen bond is replaced by a water-mediated hydrogen bond N–H (Ala) … H2O … C O (Xaa).
O (Xaa).
Here, NMR spectroscopy is used to define the structural and dynamic effect of a Gly to Ser replacement through the application of recently developed NMR methodology on selectively 13C/15N doubly labeled collagen peptides (14). This strategy includes chain assignments, measurement of NOEs, and scalar J-couplings to define the conformation of the peptide. These results combined with NMR hydrogen exchange experiments and temperature-dependent chemical shift data demonstrate the disturbed dynamic features and hydrogen bonding around the Ser substitution site. The NMR data of the Gly to Ser peptide are compared with the NMR and x-ray high resolution structure of the peptide containing a Gly to Ala substitution (5).
EXPERIMENTAL PROCEDURES
Peptide Design
Previous studies reported the design and characterization of a collagen peptide that could fold completely around a Gly to Ala substitution site in the context of an α1(I) sequence (15). The peptide design contains a GPO(GAO)3 renucleation region on the N terminus and a (GPO)4 nucleation sequence at the C terminus (15). However, introduction of a Gly to Ser substitution in this peptide design showed a population of partially disordered equilibrium states in addition to fully folded trimer and unfolded monomer, making this peptide not ideal for NMR structure analysis.4 In this study, the renucleation sequence at the N terminus was redesigned from GPO(GAO)3 to (GPO)4 to increase the stability of the peptide and to ensure that all trimeric NMR signals arise from the fully folded species. The new peptide Ac-(GPO)4GPVSPAGAR(GPO)4GY-CONH2 is designated as the T1–898(G901S) peptide.
Sample Preparation
The T1–898(G901S) peptide was synthesized by the Tufts University Core Facility (Boston, MA). The peptide was made with selectively 13C/15N doubly labeled residues: residues Gly13, Pro14, Val15, Ser16, Pro17, Ala18, Gly19, and Gly28. The peptide was purified using a Waters XTerra Prep C18 column and then an Amersham Biosciences Superdex 75 Prep column on an Amersham Biosciences fast protein liquid chromatography system. The identity of the peptide was confirmed by matrix-assisted laser desorption ionization mass spectrometry. Peptide (POG)4POA(POG)5, designated as the Gly → Ala peptide, was synthesized by the Tufts University Core Facility and purified using a Waters XTerra Prep C18 column on an Amersham Biosciences fast protein liquid chromatography system. The Gly → Ala peptide was made with selectively 15N labeled residues at Ala15 and Gly24 positions; labeling was limited by the repetitive sequence. Samples for T1–898(G901S) and the Gly → Ala peptide were prepared in 10% D2O/90% H2O at pH 2 with concentrations of 4 and 3 mm, respectively.
NMR Experiments
NMR experiments for the T1-898(G901S) peptide were performed on a Varian INOVA 600-MHz spectrometer equipped with a cold probe. For the sequential assignment, 1H-15N heteronuclear single quantum coherence (HSQC) and HNCA experiments (16) were obtained at 15 °C. Three-dimensional 15N-edited TOCSY-HSQC experiments (17, 18) with a mixing time of 45 ms and three-dimensional 15N-edited NOESY-HSQC experiments (17–19) with mixing times of 30–50 ms were performed at 10, 15, and 20 °C as described before (14). A three-dimensional H(CCO)NH experiment was performed at 20 °C (20) and paired with NOESY-HSQC to help assign NOE peaks. The three-dimensional H(CCO)NH experiment comprised 30 (t1) × 70 (t2) × 512 (t3) complex points and were recorded with spectral widths of 2000.0 (F1), 7000.0 (F2), and 7022.5 (F3) Hz. Three-dimensional HNHA experiments (21) were performed to measure homonuclear 3JHNHα coupling constants at 15 °C, with an H–H coupling period of 25 ms. The correction factor for the 3JHNHα coupling constants was obtained as described before (14). Hydrogen exchange experiments were carried out at 10 °C, pDcorrect 2.4, as described earlier (22). For measurements of amide proton temperature gradients, 1H-15N HSQC spectra were obtained at 0–25 °C with an interval of 5 °C. The sample was equilibrated at each temperature for at least 3 h. Amide proton temperature gradients were obtained by linear regression analysis of the amide proton chemical shifts versus temperature.
NMR experiments for the Gly → Ala peptide were performed on a Varian INOVA 500-MHz spectrometer. The HSQC experiment was performed at 15 °C. The 3JHNHα coupling constants were measured and corrected, and the amide proton temperature gradients were measured similarly as for the T1–898(G901S) peptide.
All data were processed using the FELIX 2004 software package (Felix NMR, Inc., San Diego, CA) and/or NMRPipe (23) and analyzed with FELIX 2004 or NMRView (24) as described before (14). For HC(CO)NH experiments, a solvent suppression filter was applied in the acquisition dimension to the data prior to apodization with a 90° sine-bell window function. The data were subsequently zero-filled to 1024 complex points and Fourier-transformed. The t1 and t2 dimensions were increased 1.5 times by forward-backward linear prediction (25), multiplied by a sine-bell window function, zero-filled to 256 complex points, and Fourier-transformed. The final three-dimensional data for HC(CO)NH experiment included 256 × 256 × 512 real points.
Generation of NOE Contact Map and Molecular Modeling
The predicted background map was generated as described before from a classic triple helical conformation (14). The NOE contact map for peptide T1–898(G901S) was made from observed NH–H NOEs in the three-dimensional 1H-15N NOESY-HSQC experiment and classified as NH–NH, NH–Hα, and NH side chain (Hβ, Hγ, Hδ).
A computer model structure of the T1–898(G901S) peptide was generated based on the x-ray crystal structure of Gly → Ala (Protein Data Bank (PDB) ID: 1CAG) (5) using the Molecular Operating Environment 2006.08 (Chemical Computing Group Inc., Montreal, Canada). Residues GPO-APO-GPO were substituted to GPV-SPA-GAR. A different starting structure of T3–785 (PDB ID: 1BKV) (26) was also tried, and residues GIT-GAR-GLA were substituted to GPV-SPA-GAR. The models were energy-minimized with dihedral angle ϕ and NOE distance restraints similarly as described before (14). The structures were solvated with a standard Molecular Operating Environment water soak procedure. The input NMR restraints included 15 ϕ angle restraints (residues of Gly13, Val15, Ser16, Ala18, and Gly19 in three chains) and four NOE distances (1Ser16NH–3Ser16NH, 1Ser16NH–3Gly13NH, 3Ser16NH–1Gly19NH, 1Ser16NH–1Val15NH). Multiple solutions of dihedral angle ϕ from 3JHNHα couplings are incorporated by starting with very loose restraints to cover all ϕ solutions and following with iterative procedure of back calculation and structure validation. For four NOE restraints, a highly precise distance cannot be derived from the NOE peak intensity because there are inherent errors associated with the accuracy of volume measurement, spin diffusion, and dynamics. Therefore a lower limit of 1.8 Å and a higher limit of 5.0 Å were set for these four distance restraints. Back calculation of 3JHNHα values and back calculation of an NOE map were used to eliminate models that were not consistent with the experimental data. Three representative structures consistent with experimental 3JHNHα values and all 1H-1H NOEs were selected for the T1–898(G901S) peptide.
RESULTS
NMR Characterization of the Gly → Ala Peptide
NMR studies were carried out on the peptide with a Gly to Ala substitution, (POG)4POA(POG)5 (designated the Gly → Ala peptide) whose high resolution structure has been solved by x-ray crystallography (PDB ID: 1CAG) (5). Because of the repetitive nature of this peptide, the potential for studying labeled residues is limited. Specific 15N labels were placed at the Gly24 residue and at the unique Ala15 residue to obtain local conformation and dynamic information that could be compared with features in the crystal structure.
HSQC spectra were determined for the Gly → Ala peptide. The Gly24 residues showed a single trimer peak, due to the overlapping of the 3 Gly residues in the triple helix in the repetitive Pro-Hyp-Gly triple helix environment, and a set of monomer peaks, due to cis-trans isomers, as reported for other triple helical peptides (27). Three well resolved trimer peaks of Ala15 are downfield shifted in the proton dimension relative to the Ala15 monomer, with one chain shifted to 9.4 ppm (data not shown). This supports the non-equivalence of the 3 Ala residues within the triple helix. The large downfield shift may reflect an altered conformation or hydrogen-bonding behavior of Ala15 when compared with standard triple helical peptides. Due to the lack of stagger information from the labeling of the isolated Ala15 residues, chain assignments could not be obtained.
3JHNHα coupling measurements were carried out on the Gly → Ala peptide (Table 1) and show that one of the Ala15 trimer peaks has a very high J-coupling constant (8.6 Hz) relative to the other two and relative to the J-coupling constant value seen for Gly24. Multiple ϕ angle solutions were calculated for Ala15 and compared with the ϕ angles in the x-ray crystal structure (Table 1). In the crystal structure, the ϕ angle for chain 2 is significantly different from those of chains 1 and 3 and from the value for the Gly24 residue, consistent with the unusually high J-coupling constant for one Ala15 peak. A disruption of the triple helical PPII conformation at the Ala15 in one chain at the substitution site is thus observed both in solution and in the crystal.
TABLE 1.
3JHNHα of the Gly → Ala and the T1–898(G901S) peptide and the corresponding multiple dihedral angle solutions from the Karplus equation and from the x-ray crystal structure, and the NH temperature gradients of Gly → Ala and the T1–898(G901S) peptide
| Peptide name | Residue | 3JHNHα | ϕ(1) | ϕ(2) | ϕ(3) | ϕ(4) | ϕ in x-ray structure | NH temperature gradientsa |
|---|---|---|---|---|---|---|---|---|
| Hz | ppb/°C | |||||||
| Gly → Ala | 1Ala15b | 5.6 ± 0.8 | 76 ± 16 | −168 ± 6 | 44 ± 16 | −72 ± 6 | −81 | −12.2 |
| 2Ala15 | 8.6 ± 0.5 | −144 ± 5 | −96 ± 5 | −104 | −10.7 | |||
| 3Ala15 | 5.6 ± 0.6 | 78 ± 11 | −168 ± 5 | 42 ± 11 | −72 ± 5 | −61 | −4.3 | |
| Gly24 | 4.7 ± 0.3 | 93 ± 3 | −175 ± 2 | 27 ± 3 | −65 ± 2 | −72c | −3.5 | |
| T1–898(G901S) | 1Ser16 | 6.1 ± 0.9 | 72 ± 15 | −165 ± 7 | 47 ± 15 | −75 ± 7 | NA | −5.3 |
| 2Ser16 | 6.4 ± 0.3 | 66 ± 6 | −163 ± 2 | 53 ± 6 | −77 ± 2 | NA | −4.0 | |
| 3Ser16 | 5.1 ± 0.3 | 88 ± 4 | −172 ± 2 | 32 ± 4 | −68 ± 2 | NA | −7.8 | |
| Gly28 | 5.3 ± 0.2 | 86 ± 3 | −171 ± 2 | 34 ± 3 | −69 ± 2 | NA | −3.1 |
a NH temperature gradients are used to distinguish hydrogen-bonded (NH gradient > −4.5ppb/°C) from non-bonded (NH gradient < −4.5ppb/°C) NH groups (28).
b Tentative chain numbers are shown in superscript. Chains 1, 2, and 3 may not indicate leading, middle and trailing chains because the chain assignment cannot be obtained from the NMR data of isolated Ala15.
c Average ϕ angle of Gly24 in three chains in x-ray structure was used.
Amide temperature gradient measurements were performed on the Gly → Ala peptide to characterize whether the amides of Gly24 and Ala15 are hydrogen-bonded as indicated by an NH gradient > −4.5 ppb/°C (28). Only one of the three Ala15 trimers has a temperature gradient that is > −4.5 ppb/°C, indicating the formation of a single hydrogen bond at the Gly to Ala mutation site (Table 1). Gly24 in the (GPO) region shows a chemical shift temperature gradient typical of hydrogen bonding as expected. Previous hydrogen exchange experiments of the Gly → Ala peptide showed that all three Ala15 have a very low degree of protection from solvent when compared with the control Gly24 (29).
Peptide Design of the T1–898(G901S) Peptide
The features of the Gly → Ala peptide were compared with the effect of a Gly to Ser replacement in a triple helical peptide T1–898(G901S). A stable triple helical collagen model peptide containing the immediate sequence surrounding a natural Gly to Ser OI mutation at site 901 in the α1(I) chain was studied by NMR. The peptide design is based on and modified from previous studies on the renucleation of model peptide (15) (see “Experimental Procedures” for details). A renucleation sequence (GPO)4 was put at the N terminus to increase the stability of the peptide and to ensure no partially disordered intermediates in solution as measured by NMR diffusion experiments.4 This new peptide Ac-(GPO)4-GPV-SPA-GAR-(GPO)4GY-CONH2 (designated as T1–898(G901S)) includes the collagen sequence (GPV-SPA-GAR) from position 898–906 in the α1(1) chain and a Gly to Ser substitution at the 901 position associated with a mild case of OI. A Tyr is added in the C terminus of the peptide to allow the determination of concentration by UV absorbance. Circular dichroism (CD) and differential scanning calorimetry studies on this peptide showed a thermal stability with Tm of 28 °C and a high calorimetric enthalpy of 413 kJ/mol.5 The peptide was synthesized with consecutive 13C/15N doubly labeled residues at the Gly13, Pro14, Val15, Ser16, Pro17, Ala18, Gly19, and Gly28 positions to allow chain assignment (14).
NMR Chain Assignments and Chemical Shifts of the T1–898(G901S) Peptide
The HSQC spectrum of the T1–898(G901S) peptide shows one or more trimer peaks as well as monomer peaks for each doubly labeled residue that has an amide proton (Fig. 1, A and B). Gly28, within the Gly-Pro-Hyp repeating region at the C terminus, shows only a single trimer resonance at the typical position for Gly in standard triple helical peptides (14, 30). For the labeled residues Gly13, Val15, Ser16, Ala18, and Gly19, three trimer peaks are observed. It indicates that a well defined structure with three non-equivalent chains is present in the substitution region. Most notably, one of the Ser16 trimers has shifted downfield significantly in the 1H dimension (9.1 ppm).
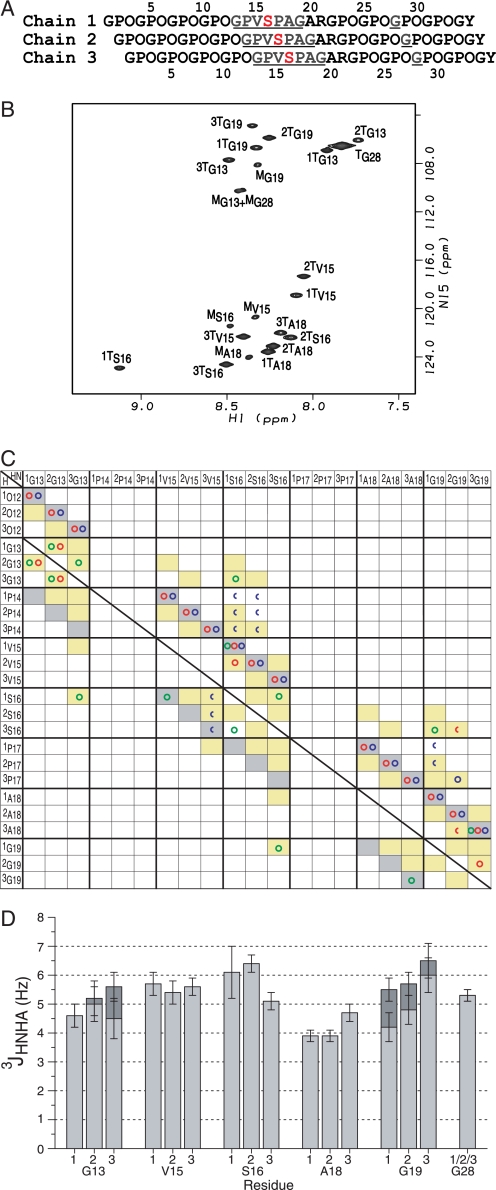
FIGURE 1.
NMR conformational characterization of the T1–898(G901S) peptide. A, the sequence diagram of the peptide shows the characteristic 1-residue stagger. Leading, middle, or trailing chain stagger assignment is indicated as chain 1, 2, or 3, respectively. The 13C/15N doubly labeled residues are underlined and colored in blue, and the Gly to Ser substation site is shown in red. B, the HSQC spectrum of the T1–898(G901S) peptide at 15 °C. The peaks corresponding to the monomer and trimer state are denoted with a superscript M or T, respectively. Chain stagger is indicated as a number in front of the superscript T. C, comparison of experimental NOEs of T1–898(G901S) peptide with a standard triple helical conformation. A predicted contact map obtained from a standard triple helix model structure is shown in shaded boxes as background (14). Contacts are shaded in gray for intrachain distances less than 5 Å and in yellow for interchain distances less than 5 Å. Experimental NOEs for T1–898(G901S) peptide are represented by circles (HN-HN (green circles), HN-Hα (red circles), and HN side chain protons (blue circles)) and are overlaid on the shaded contacts, showing the expected intrachain and interchain NOEs and one new contact (1Ser16 NH to 3Ser16 NH) consistent with the 1-residue stagger between chains and the packing of the Ser residues at the substitution site. Half circles represent overlapped NOEs. The superimposition of the experimental NOEs and the background indicates the 1-residue stagger of the triple helix throughout the substitution site. The diagnostic interchain NH–NH and NH–Hα NOEs between the three chains of the Gly13 residues (1Gly13NH to 2Gly13NH and 2Gly13Hα, 2Gly13NH to 1Gly13NH and 1Gly13Hα, 2Gly13NH to 3Gly13NH and 3Gly13Hα, 3Gly13NH to 2Gly13NH) and between Gly19 residues (3Gly19NH to 2Gly19Hα) are seen as well as interchain NOEs from 2Gly19NH to 3Pro17Hβ/γ supporting the 1-residue stagger. In addition, the NOEs normally unique between Gly are observed between the Ser16 residues and residues Gly13 and Gly19 (1Ser16NH to 3Gly13NH, 3Ser16NH to 1Gly19NH), suggesting that the Ser16 residues are packed into the center of the triple helix at the substitution site, in a behavior similar to a Gly at the same position. D, the histogram of the 3JHNHα coupling constants for the T1–898(G901S) peptide. 3JHNHα coupling constants from the two Hα Gly residues are shown in dark gray and light gray bars.
Although the three chains of the triple helix are identical in amino acid sequence, they are structurally distinct due to the 1-residue stagger between the chains (Fig. 1A). To perform detailed conformational characterization, it is necessary to identify the leading, middle, and trailing individual chains. The chain assignment of trimer resonances can be divided into two steps: sequential assignment by triple resonance experiments and chain stagger identification by NOE distances (14). HNCA and HCAN experiments were used to establish sequential assignments. However, the sensitivity of the HCAN experiment used to correlate Pro with the next residue (31) was too low for the trimer resonances and could not be used. Therefore assignments were obtained for short segments via a single HNCA experiment. The segments Gly13, Val15–Ser16, and Ala18–Gly19 were connected via NOE experiments by assuming a 1-residue stagger (See the next section for details). All observed trimer resonances in the T1–898(G901S) peptide could be assigned to specific chains of the triple helix as indicated in the HSQC spectrum by the superscripted number (Fig. 1B).
NMR Conformational Characterization of the T1–898(G901S) Peptide through NOE Contacts and Scalar J-Coupling Constants
NMR conformational parameters were measured on the T1–898(G901S) peptide, including NOEs to obtain distance information and 3JHNHα constants to obtain the dihedral angle ϕ. Examination of an NOE contact map summarizing the NH–H distance information provides a way to identify leading, middle, and trailing chains and to detect any deviation from a standard triple helix conformation (14). An NOE contact map diagram (Fig. 1C) was constructed for the experimental NOESY-HSQC data for the T1–898(G901S) peptide. Experimental NOEs (represented as circles) corresponded to intermolecular distances (yellow squares) of a standard triple helix showing the 1-residue stagger is maintained throughout the Ser16 substitution site. In the normal triple helix, 3Gly13 in the leading strand will be within 5 Å of 1Gly16 in the trailing strand, and similarly, 3Gly16 will come close to 1Gly19 (Fig. 1C). When the Ser replaces Gly, the analogous 1Ser16NH to 3Gly13NH and 3Ser16NH to 1Gly19NH are observed. One NOE observed between 1Ser16NH and 3Ser16NH is not predicted from the standard triple helix and is weak relative to the 1Ser16NH to 3Gly13NH and 3Ser16NH to 1Gly19NH NOEs.
3JHNHα coupling constants that can be related to the dihedral angle ϕ were obtained for the T1–898(G901S) peptide from the HNHA experiment (Fig. 1D). Many of the 3JHNHα coupling constants are similar to one another in the range of 4–6 Hz, but the 3JHNHα of 2Ser16 is slightly higher than the other residues. It is not possible to establish a definite value for the angle ϕ because there are multiple solutions from the parameterized Karplus equation (21). Instead the multiple possible ϕ solutions are taken as restraints during the molecular modeling and used to eliminate incorrect models.
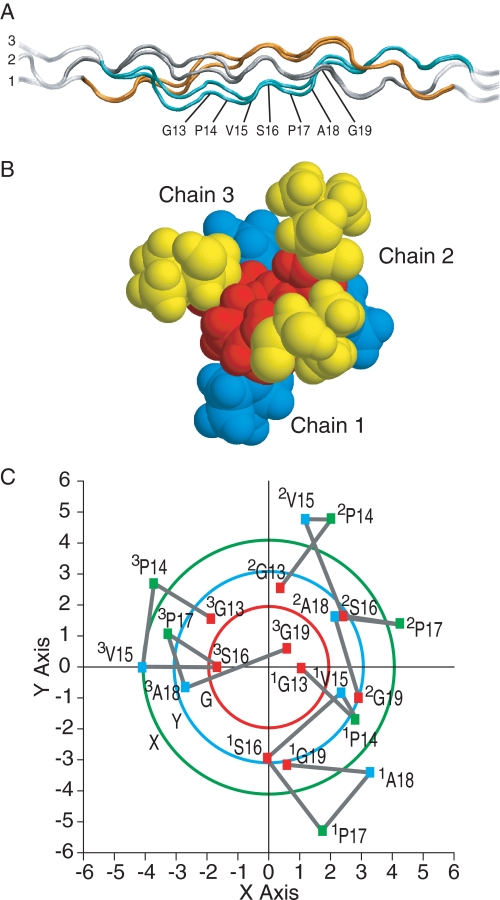
To visualize the conformation in solution for the T1–898(G901S) peptide, molecular modeling was performed with the incorporation of NMR ϕ angle restraints from 3JHNHα measurements and distance restraints from NOESY experiments using the strategy described previously (14). Back calculations of NMR parameters from the energy-minimized structures were performed to eliminate many structures that were not consistent with the experimental data. The resulting representative models are not unique but are examples that gave good agreement with NMR data (see Fig. 3A). The small differences between the model structure and the x-ray structures are consistent with the small differences in NOEs and J-coupling data.
FIGURE 3.
Model structure of the T1–898(G901S) peptide. A, ribbon diagram of T1–898(G901S) peptide models represented with the N-terminal at the left and chain numbers. The NMR parameters are used as the restraints in the energy minimization, including the ϕ angle restraints from J-coupling constants and distance restraints from NOEs. Three model structures that are consistent with all NMR experimental data are obtained and show similar features. A set of two models from the Gly → Ala starting structure is shown for illustration. Leading, middle, and trailing chains are represented as 1, 2, and 3 and are colored in gray, orange, and cyan, respectively, for the energy-minimized segment of residues 6–26. The Gly13 to Gly19 residues in chain 3 of one structure are indicated. B, space-filling model of the cross-section view from the N terminus to the C terminus of one T1–898(G901S) peptide model. Views show that the Ser16 residues are closely packed at the center. The VSP segment is colored as Val15 (yellow squares), Ser16 (red squares), and Pro17 (blue squares). C, the cross-view of the Cα of residues around the substitution region in the T1–898(G901S) peptide model. The structure was aligned to the central axis of the triple helix molecule using the pdbinertia program. The x and y axes indicate the coordinates in the structure. The projection of Cα atoms of residues Gly13 to Gly19 is shown in squares. Residues at the Gly, Xaa, and Yaa positions are colored in red, green, and blue, respectively. The substituting residues are colored in red. The Cα traces are shown in gray lines. In a standard triple helical conformation, the Cα of Gly, Xaa, and Yaa residues are an equal distance from the central axis of the triple helix, respectively. Three circles indicate the Cα traces of Gly, Xaa, and Yaa residues in the standard triple helical conformation from the (PPG)10 x-ray crystal structure (38). Traces of residues at Gly, Xaa, and Yaa positions are colored in red, green, and blue, respectively.
Hydrogen Exchange and NH Temperature Gradient Studies of the T1–898(G901S) Peptide
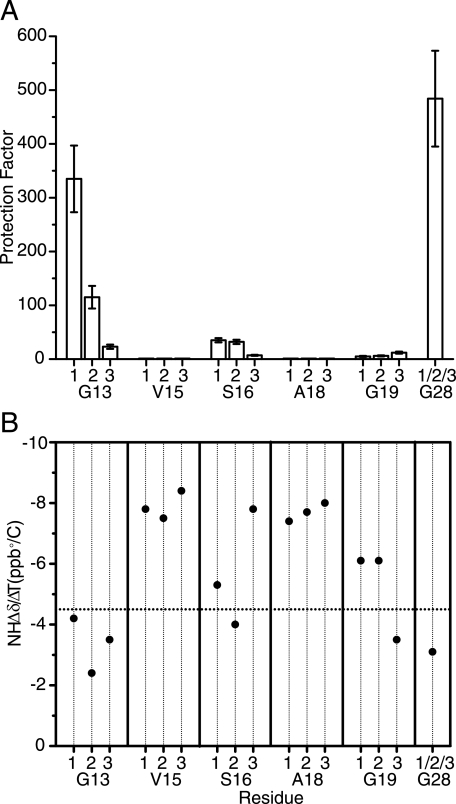
Hydrogen exchange and amide temperature gradient measurements were performed to characterize the hydrogen-bonding features and dynamic consequences of the Gly to Ser substitution in the T1–898(G901S) peptide. Hydrogen exchange experiments conducted to study the protection of amide protons from solvent showed that Gly28 in the stable C-terminal GPO repeating region has a very high degree of protection from exchange (Fig. 2A). The residues located at the Gly positions surrounding the substitution site (Gly13 and Gly19) and the Ser position are less protected than Gly28 (p = 484). The protection factors of the Gly13 residues are variable across the three chains with chain 1 (p = 335) almost as fully protected as Gly28 followed by chain 2 (p = 115), with an intermediate protection (three times less than 1Gly13), and then chain 3 (p = 23), which shows little protection (∼300 less than 1Gly13). All 3 Ser16 residues show low protection factors on the order of 10–40, with chain 3 again the least protected. The 3 Gly19 residues (p = 5–12) in the triplet C-terminal to the Ser substitution site show lower protection factors relative to Gly13 and Ser16, with Gly19 in chain 3 showing a slightly larger protection factor (p = 12). In the central GPV-SPA-GAR sequence, the 2 residues Val15 and Ala18 showed almost no protection, as expected for residues in the Yaa position.
FIGURE 2.
Hydrogen exchange and NH temperature gradient studies on the T1–898(G901S) peptide. A, histogram of hydrogen/deuterium protection factors for the labeled residues in the T1–898(G901S) peptide. B, amide NH Δδ/ΔT plot for T1–898(G901S) peptide. The dashed horizontal line corresponds to Δδ/ΔT = −4.5 ppb/°C, which provided the cutoff line of hydrogen bonding.
Amide temperature gradient studies have been used in peptides to distinguish hydrogen-bonded (NH gradient > −4.5 ppb/°C) from non-bonded (NH gradient < −4.5 ppb/°C) NH groups (Fig. 2B) (28). They are performed here on the T1–898(G901S) peptide to complement the hydrogen exchange studies and to evaluate the existence of hydrogen bonds at the different labeled positions. As expected, the NH temperature gradient for Gly28, which is located in the rigid (GPO)4 region, is more positive than −4.5 ppb/°C, indicating the existence of hydrogen bonds in this C-terminal GPO-rich end (28). Val15 and Ala18 have very negative values of NH temperature gradient for all residues, suggesting that they are not involved in hydrogen bonding, as expected for residues in the Yaa positions. Residue Gly13, located one triplet N-terminal to the mutation site, and residue Gly19, located one triplet C-terminal to the mutation site, differ in their NH temperature dependence. All three chains of Gly13 have more positive temperature gradients than the cutoff, suggesting the formation of hydrogen bonds at all three positions. For Gly19 residues, the NH temperature gradients are non-equivalent, with the indication of a hydrogen bond for chain 3 only, the residue farthest from the substitution site due to the stagger. At the mutation site, the Ser16 residues show non-equivalence in the temperature gradients of the three chains, with only chain 2 having a value greater than −4.5 ppb/°C. This suggests that at the mutation site, chain 2 is involved in hydrogen bonding, whereas the other two are not. This is similar to what is observed for the Gly → Ala peptide where a single hydrogen bond at the mutation site is indicated from temperature gradients (Table 1).
DISCUSSION
It is important to understand how the structural perturbation of the collagen triple helix by OI mutations is affected by the residue that replaces Gly and by the flanking tripeptide sequences (13). The issues are addressed here by NMR studies on a peptide with a Gly to Ser substitution within a 9-residue α1(I) sequence and its comparison with the NMR and x-ray studies of a peptide with a Gly to Ala sequence within a repeating Pro-Hyp-Gly sequence.
NMR conformational studies on labeled residues in the central region indicate that the 1-residue stagger is maintained throughout the region including the Ser substitution and that most of the residues have ϕ angles expected for the triple helix. NOEs between backbone amide protons that are normally unique to Gly residues in the standard triple helix are observed for the Ser16 residues in the substituted peptide. These NOEs indicate that despite the larger size of Ser relative to Gly, the 3 Ser residues are trying to pack into the center of the triple helix in a manner similar to a Gly at the same position. However, residues larger than Gly cannot fit into all three chains and pack in this helical structure. Unlike Gly, the 3 Ser residues are non-equivalent, and a new weak NOE indicates that two Ser16 residues from different chains are packing closer than expected. The conformational distortion due to the mutation is highly localized and absorbed by a small number of residues. A similar distortion appears to be present in the Gly → Ala peptide as predicted from the x-ray structure. Interestingly an unusual downfield shift is seen for 1 Ser residue in the T1–898(G901S) peptide and 1 Ala residue in the Gly → Ala peptide structure, supporting an unusual conformational or hydrogen-bonding feature (Fig. 3).
Although the replacement of a Gly by an Ala or by a Ser residue leads to a similar local perturbation at the substitution site, there are subtle differences between the Gly → Ala and T1–898(G901S) peptides as indicated by some distinctive NOE contacts. In addition, the 3JHNHα of 1 Ala residue in the Gly → Ala peptide is very high, indicating a non-polyproline II conformation, whereas the coupling constants of all 3 Ser16 residues in the T1–898(G901S) peptide are consistent with polyproline II (Table 1). These subtle differences could relate to the additional hydroxyl group in Ser when compared with Ala or to the presence of a real collagen α1(I) sequence, GPVSPAGAR, for the Ser peptide when compared with the Gly to Ala substitution in an all Gly-Pro-Hyp environment GPOAPOGPO (5).
Although conformational changes caused by a Gly to Ser substitution are subtle, more significant and specific perturbations are observed by NMR dynamics experiments including H-exchange and temperature-dependent amide chemical shifts. Interestingly the dynamics studies reflect the effect of the stagger on the relative position between a Gly in one chain and the mutation site Ser in a different chain within a triple helix. The 3 Gly13 residues show hydrogen bonding, but the degree of protection depends on whether they are in the leading, middle, or trailing chain. Because of the staggering of the three chains, the leading chain 1 is surrounded by an almost normal triple helix environment, whereas the Gly13 in the trailing chain 3 is falling heavily under the influence of the Ser substitution site. The larger degree of disruption of hydrogen bonding when a Gly is closer in its axial position relative to a Ser may arise from increased flexibility or breathing at that site. Only 1 Gly19 residue shows a temperature-dependent amide shift, indicating H-bonding, and it is located in the leading strand, which is farthest away from the Ser residues. All 3 Gly19 residues show fast exchange, but Gly19 in chain 3 appears a little more protected because that chain is the farthest in axial position from the Ser perturbation. A similar asymmetric disruption is also suggested by the x-ray structure of the Gly → Ala peptide, where an indirect water-mediated hydrogen bond rather than a direct hydrogen bond is observed for the Gly of the triplet C-terminal to the mutation in the trailing strand, which is closest to the Ala mutation site in the leading strand.
The NMR dynamics experiments that include H-exchange and temperature-dependent chemical shift experiments suggest increased disruption of hydrogen bonding in the C- versus the N-terminal triplets flanking the Gly to Ser mutation. A recent computational study has focused on the asymmetric influence of tripeptides flanking a Gly to Ser mutation site, noting that a destabilizing triplet C-terminal to a Gly to Ser substitution is correlated with a lethal OI phenotype, whereas no correlation is seen for an N-terminal destabilizing triplet (11). Consistent with this result, peptide studies showed a greater destabilization effect when a tripeptide of low triple helix propensity is put on the C-terminal side when compared with the N-terminal side of a Gly to Ser substitution (11). The NMR dynamic studies presented here on the T1–898(G901S) peptide suggest an inherent asymmetry surrounding a mutation site with less regular hydrogen bonding or increased conformational flexibility on the C-terminal side, and this inherent effect could be synergistic with the sequence-dependent stability of neighboring triplets.
At the mutation site, the normal backbone N–H (Gly) … C O (Pro) hydrogen bond is replaced by indirect water-mediated N–H (Ala) … H2O … C
O (Pro) hydrogen bond is replaced by indirect water-mediated N–H (Ala) … H2O … C O (Pro) bonds in the Gly → Ala structure. NMR hydrogen exchange data of the 3 Ser16 residues all show low protection from solvent, suggesting either that there are no direct N–H (Ser) … C
O (Pro) bonds in the Gly → Ala structure. NMR hydrogen exchange data of the 3 Ser16 residues all show low protection from solvent, suggesting either that there are no direct N–H (Ser) … C O (Xaa) bonds present or that there is increased conformational flexibility at the mutation site. However, the replacement of a Gly by a Ser residue introduces the possibility of additional hydrogen bonding through the hydroxyl side chain. Mooney and Klein (32) carried out molecular dynamics studies on a range of Gly to Ser OI mutation sites and found the Ser OH hydrogen-bonded to the backbone of an adjacent chain for a significant fraction of the time. Several types of interchain hydrogen bonds involving the hydroxyl group of Ser are possible. The presence of a defined hydrogen bond, e.g. between the Ser and a backbone carbonyl, or the possibility of an ensemble of different hydrogen bonds is consistent with the high calorimetric enthalpy observed for this peptide. Such hydrogen bonding may play a role in stabilizing the triple helix assembly and compensating for the destabilizing loss of direct backbone hydrogen bonds.
O (Xaa) bonds present or that there is increased conformational flexibility at the mutation site. However, the replacement of a Gly by a Ser residue introduces the possibility of additional hydrogen bonding through the hydroxyl side chain. Mooney and Klein (32) carried out molecular dynamics studies on a range of Gly to Ser OI mutation sites and found the Ser OH hydrogen-bonded to the backbone of an adjacent chain for a significant fraction of the time. Several types of interchain hydrogen bonds involving the hydroxyl group of Ser are possible. The presence of a defined hydrogen bond, e.g. between the Ser and a backbone carbonyl, or the possibility of an ensemble of different hydrogen bonds is consistent with the high calorimetric enthalpy observed for this peptide. Such hydrogen bonding may play a role in stabilizing the triple helix assembly and compensating for the destabilizing loss of direct backbone hydrogen bonds.
The structural and dynamic alterations reported here may play a role in the etiology of OI by affecting collagen secretion or interactions with other matrix molecules. There is increasing evidence that lethal mutations may occur at sites important for binding proteoglycans (7, 37). The conformational and hydrogen-bonding alterations indicated by the NMR data would lead to an altered appearance of the exterior of the triple helix molecule as seen by binding partners. Plotting the cross-section of the T1–898(G901S) peptide indicates that the Cα atoms of residues in the Xaa and Yaa positions surrounding the Ser are located farther out from center than a standard triple helix (Fig. 3C). This altered molecular cross-section could generate defective fibril formation or an abnormal collagen fibril exterior and thus affect ligand recognition and binding.
The peptides studied here are homotrimers containing three Gly replacements in each molecule. In contrast, type I collagen is a heterotrimer consisting of two α1(I) chains and one α2(I) chain, and the dominant disorder OI is caused by a Gly mutation in only one or two of the chains in a trimer (2). Gauba and Hartgerink (33) recently used electrostatic interactions to form heterotrimer model peptides with one, two, or three Gly to Ser mutations (34). Their results show that the incorporation of a first Gly to Ser mutation in one chain leads a large drop in thermal stability, whereas the addition of the second and then the third mutation leads to only a small further decrease in thermal stability, as predicted from computational studies (35). More heterotrimer studies are being reported recently (33, 36), and such studies will ascertain whether asymmetric perturbation and hydrogen-bonding properties seen for the homotrimers are present in triple helical molecules with only one or two mutants chains.
Acknowledgment
We thank Dr. Seho Kim for help with implementation of triple resonance experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants GM45302 (to J. B.) and GM60048 (to B. B.). This work was also supported by National Science Foundation Grants DBI-0320746 (to J. B.) and DBI-0403062 (to J. B.).
- OI
- osteogenesis imperfecta
- Hyp
- hydroxyproline (three-letter code)
- O
- hydroxyproline (single-letter code)
- HSQC
- heteronuclear single quantum coherence
- NOE
- nuclear Overhauser effect
- NOESY
- nuclear Overhauser effect spectroscopy.
REFERENCES
- 1.Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 2.Kuivaniemi H., Tromp G., Prockop D. J. (1997) Hum. Mutat. 9, 300–315 [DOI] [PubMed] [Google Scholar]
- 3.Rich A., Crick F. H. (1961) J. Mol. Biol. 3, 483–506 [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran G. N. ed (1967) Treatise on Collagen, pp 103–184, Academic Press, New York [Google Scholar]
- 5.Bella J., Eaton M., Brodsky B., Berman H. M. (1994) Science 266, 75–81 [DOI] [PubMed] [Google Scholar]
- 6.Byers P. H., Cole W. G. (2002) in Connective Tissue and Its Hereditable Disorders ( Royce P. M., Steinmann B. eds) pp 385–430, Wiley-Liss, New York [Google Scholar]
- 7.Marini J. C., Forlino A., Cabral W. A., Barnes A. M., San Antonio J. D., Milgrom S., Hyland J. C., Körkkö J., Prockop D. J., De Paepe A., Coucke P., Symoens S., Glorieux F. H., Roughley P. J., Lund A. M., Kuurila-Svahn K., Hartikka H., Cohn D. H., Krakow D., Mottes M., Schwarze U., Chen D., Yang K., Kuslich C., Troendle J., Dalgleish R., Byers P. H. (2007) Hum. Mutat. 28, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck K., Chan V. C., Shenoy N., Kirkpatrick A., Ramshaw J. A., Brodsky B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K., Nowak I., Kirchner M., Xu Y. (2008) J. Biol. Chem. 283, 34337–34344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makareeva E., Mertz E. L., Kuznetsova N. V., Sutter M. B., DeRidder A. M., Cabral W. A., Barnes A. M., McBride D. J., Marini J. C., Leikin S. (2008) J. Biol. Chem. 283, 4787–4798 [DOI] [PubMed] [Google Scholar]
- 11.Bodian D. L., Madhan B., Brodsky B., Klein T. E. (2008) Biochemistry 47, 5424–5432 [DOI] [PubMed] [Google Scholar]
- 12.Baum J., Brodsky B. (1999) Curr. Opin. Struct. Biol. 9, 122–128 [DOI] [PubMed] [Google Scholar]
- 13.Brodsky B., Persikov A. V. (2005) Adv. Protein Chem. 70, 301–339 [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Brodsky B., Baum J. (2007) J. Biol. Chem. 282, 22699–22706 [DOI] [PubMed] [Google Scholar]
- 15.Hyde T. J., Bryan M. A., Brodsky B., Baum J. (2006) J. Biol. Chem. 281, 36937–36943 [DOI] [PubMed] [Google Scholar]
- 16.Ikura M., Kay L. E., Bax A. (1990) Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 17.Fesik S. W., Zuiderweg E. R. (1988) J. Magn. Reson. 78, 588–593 [Google Scholar]
- 18.Messerle B. A., Wider G., Otting G., Weber C., Wüthrich K. (1989) J. Magn. Reson. 85, 608–613 [Google Scholar]
- 19.Marion D., Kay L. E., Sparks S. W., Torchia D. A., Bax A. (1989) J. Am. Chem. Soc. 111, 1515–1517 [Google Scholar]
- 20.Grzesiek S., Anglister J., Bax A. (1993) J. Magn. Reson. B 101, 114–119 [Google Scholar]
- 21.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 22.Mohs A., Popiel M., Li Y., Baum J., Brodsky B. (2006) J. Biol. Chem. 281, 17197–17202 [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 25.Zhu G., Bax A. (1992) J. Magn. Reson. 98, 192–199 [Google Scholar]
- 26.Kramer R. Z., Bella J., Mayville P., Brodsky B., Berman H. M. (1999) Nat. Struct. Biol. 6, 454–457 [DOI] [PubMed] [Google Scholar]
- 27.Buevich A., Baum J. (2001) Philos. Trans. R Soc. Lond. B Biol. Sci. 356, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baxter N. J., Williamson M. P. (1997) J. Biomol. NMR 9, 359–369 [DOI] [PubMed] [Google Scholar]
- 29.Liu X. (1997) Real-time NMR Investigations of Triple Helical Peptide Folding and Collagen Folding Diseases. Ph.D. dissertation, p. 101, Rutgers, the State University of New Jersey [Google Scholar]
- 30.Mohs A., Li Y., Doss-Pepe E., Baum J., Brodsky B. (2005) Biochemistry 44, 1793–1799 [DOI] [PubMed] [Google Scholar]
- 31.Powers R., Gronenborn A. M., Clore G. M., Bax A. (1991) J. Magn. Reson. 94, 209–213 [Google Scholar]
- 32.Mooney S. D., Klein T. E. (2002) Mol. Cell Proteomics 1, 868–875 [DOI] [PubMed] [Google Scholar]
- 33.Gauba V., Hartgerink J. D. (2008) J. Am. Chem. Soc. 130, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 34.Brodsky B., Baum J. (2008) Nature 453, 998–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mooney S. D., Huang C. C., Kollman P. A., Klein T. E. (2001) Biopolymers 58, 347–353 [DOI] [PubMed] [Google Scholar]
- 36.Madhan B., Xiao J., Thiagarajan G., Baum J., Brodsky B. (2008) J. Am. Chem. Soc 130, 13520–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Lullo G. A., Sweeney S. M., Korkko J., Ala-Kokko L., San Antonio J. D. (2002) J. Biol. Chem. 277, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 38.Berisio R., Vitagliano L., Mazzarella L., Zagari A. (2002) Protein Sci. 11, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]