Abstract
Light converts rhodopsin, the prototypical G protein-coupled receptor, into a form capable of activating G proteins. Recent work has shown that the light-activated state of different rhodopsins can possess different molecular properties, especially different abilities to activate G protein. For example, bovine rhodopsin is ∼20-fold more effective at activating G protein than parapinopsin, a non-visual rhodopsin, although these rhodopsins share relatively high sequence similarity. Here we have investigated possible structural aspects that might underlie this difference. Using a site-directed fluorescence labeling approach, we attached the fluorescent probe bimane to cysteine residues introduced in the cytoplasmic ends of transmembrane helices V and VI in both rhodopsins. The fluorescence spectra of these probes as well as their accessibility to aqueous quenching agents changed dramatically upon photoactivation in bovine rhodopsin but only moderately so in parapinopsin. We also compared the relative movement of helices V and VI upon photoactivation of both rhodopsins by introducing a bimane label and the bimane-quenching residue tryptophan into helices VI and V, respectively. Both receptors showed movement in this region upon activation, although the movement appears much greater in bovine rhodopsin than in parapinopsin. Together, these data suggest that a larger conformational change in helices V and VI of bovine rhodopsin explains why it has greater G protein activation ability than other rhodopsins. The different amplitude of the helix movement may also be responsible for functional diversity of G protein-coupled receptors.
Rhodopsin, the photosensitive G protein-coupled receptor (GPCR),3 is responsible for transmitting a light signal into an intracellular signaling cascade through activation of G protein in visual and non-visual photoreceptor cells. Rhodopsin consists of a protein moiety (opsin, comprising seven transmembrane α-helical segments) combined with a chromophore (11-cis retinal) that acts as the light-sensitive ligand. Photoisomerization of the 11-cis retinal to the all-trans form induces structural changes in the protein moiety that then enable it to couple with and activate the G protein.
The crystal structure of inactive bovine rhodopsin has been extensively investigated (1–3). Recently, a crystal structure of inactive invertebrate squid rhodopsin was also solved (4), and crystal structures of the inactive form of β-adrenergic receptors and A2 adenosine receptor have been reported (5–7). Remarkably, all of these crystal structures exhibit a very similar arrangement for the seven transmembrane helices (4, 8). Together, these facts suggest that the architecture for the inactive form is conserved among rhodopsin-like GPCRs.
The structural features of an activated GPCR are much less defined. Thus, a variety of biochemical and biophysical methods, including cross-linking methods (9, 10) and site-directed spin and fluorescence labeling methods (10–13), have been employed to identify the dynamic and structural changes involved in forming the activated state. The data from these studies consistently suggest that some kind of movement of helix VI is involved in the formation of the active state of the rhodopsins. In particular, the cytoplasmic end of helix VI has been proposed to rotate and/or tilt toward helix V (10–13). Remarkably, the recent crystal structures of bovine opsin are consistent with the widely accepted helix motion model. Both the structures of opsin (the ligand-free form of rhodopsin that has partial G protein activation ability) and a complex of opsin with a peptide derived from the G protein C terminus show a movement of helix VI toward helix V, compared with the dark state rhodopsin structure (14, 15). Studies of β-adrenergic and muscarinic receptors also show that agonist binding promotes movement of helix VI toward helix V in these receptors (16, 17). Because the region between the cytoplasmic ends of helices V and VI in various GPCRs is a main site of interaction with G proteins (18), it is possible that movement of helices V and VI leads to formation of a conformation capable of interacting with G protein (19).
Together, these studies imply that the active state conformation of GPCRs may be similar. However, a detailed comparison of the active-state conformation for two different GPCRs has never been precisely undertaken in the same laboratory using the same methods.
In this context we have been investigating rhodopsins with different functional properties to determine whether their active states have different conformations. Our goal was to determine whether any functional or structural differences in the active states of these GPCRs could be detected under the exact same experimental conditions.
Previously, we have found that several rhodopsins, such as an invertebrate rhodopsin and a vertebrate non-visual rhodopsin parapinopsin (20, 21), can be activated not only by light but also by exogenous all-trans retinal acting as a full agonist (22). This is in contrast to vertebrate visual rhodopsins, including bovine rhodopsin, which cannot fully form the active state by direct binding of all-trans retinal (23), although all-trans retinal can fully activate some rhodopsin mutants (24). Other invertebrate rhodopsin (25) and the circadian photoreceptor melanopsin (26) can also bind all-trans retinal directly.
Interestingly, the active form of the all-trans retinal-activated rhodopsins exhibit some striking differences in their spectroscopic and biochemical properties compared with vertebrate visual rhodopsins (27). In particular, the efficiency of bovine rhodopsin for activating G protein is ∼20∼50-fold higher than that of parapinopsin and invertebrate rhodopsin. This difference could be related to the difference in position of a specific amino acid residue counterion that is essential for rhodopsin to absorb visible light, namely one at position 113 or 181 (28).4 Further biochemical analyses using chimeric mutants combining rhodopsins with lower and higher G protein activation abilities suggested that the difference in G protein activation ability was because of a structural difference in transmembrane helices in the active states but not because of difference in amino acid sequence of G protein interaction site (29) (Fig. 1, A–C). In addition, the active states of parapinopsin and the invertebrate rhodopsin are thermally stable and can be reconverted to the inactive state by subsequent light absorption, showing photo-regenerable or bistable nature (21, 28), unlike the active state of bovine rhodopsin, which is thermally unstable and cannot revert to the inactive state by subsequent light absorption (30).
FIGURE 1.
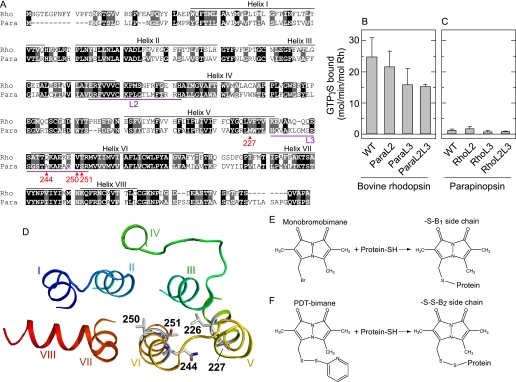
Molecular properties and sites of fluorescent probe attachment for bovine rhodopsin and parapinopsin. A, sequence alignment of bovine rhodopsin and parapinopsin. Amino acid residues to which cysteine and fluorescence label were introduced are marked with red. The amino acid residues identical and similar between bovine rhodopsin and parapinopsin are shown with white characters with black and gray background, respectively. Bovine rhodopsin and parapinopsin show 41% sequence identity and 61% similarity. In this paper the residue number of parapinopsin is described by using the bovine rhodopsin numbering system. B and C, comparison of G protein activation ability of rhodopsin and parapinopsin wild type (WT) proteins and loop-replaced mutants. In these mutants the second and/or third cytoplasmic loop was swapped between the two receptors. ParaL2 and ParaL3 indicate mutants of bovine rhodopsin in which second and third loops were replaced with the corresponding loop of parapinopsin, respectively. RhoL2 and RhoL3 indicate mutants of parapinopsin in which the second and third loops were replaced with the corresponding loops of bovine rhodopsin, respectively. ParaL2L3 and RhoL2L3 are mutants of bovine rhodopsin and parapinopsin in which both the second and third loops were swapped, respectively. See Terakita et al. (29) for more details. Data are presented as the means ± S.E. of three separate experiments except for mutants RhoL3, RhoL2L3, and ParaL2L3 (n = 2). D, model of bovine rhodopsin. Amino acid residues which were mutated to cysteine to enable attachment of the fluorescent probe bimane or mutated to tryptophan are indicated. Positions 226, 227, 244, 250, and 251 in the crystal structure of the dark state of bovine rhodopsin (PDB code 1GZM) are shown. E, reaction of the mBBr label with a sulfhydryl group. The mutants labeled with mBBr are named by the number of the residue and the suffix B1. F, reaction of the PDT-bimane with a sulfhydryl group. The mutants labeled with PDT-bimane are named by the number of the residue and the suffix B2. The disulfide linkage between the label and protein can be cleaved using Tris(2-carboxyethyl)phosphine (32).
In this study we used site-directed fluorescence labeling (13, 31) to compare the structural features of active states of bovine rhodopsin with lamprey parapinopsin, a UV-sensitive non-visual pigment in the pineal organs (21). Parapinopsin shows relatively high sequence similarity (∼60%) to bovine rhodopsin, yet it has a greatly reduced ability to activate G protein (see Fig. 1, A–C) (21, 28). Using established protocols, we introduced cysteine residues into the cytoplasmic ends of helices V and VI, the region proposed to rearrange upon activation in GPCRs (11, 12, 14, 18). We then site-specifically labeled these cysteines with the small, non-polar fluorescent probe, bimane, and used the spectral properties of these bimane probes to act as reporter groups for environmental changes around their site of attachment upon formation of the photoactivated state for both rhodopsins.
In addition, we measured changes in the relative proximity of the cytoplasmic ends of helix VI to helix V in both rhodopsin and parapinopsin using the tryptophan-induced-quenching of bimane (TrIQ-bimane) fluorescence method (31, 32). TrIQ-bimane measures the efficiency of intramolecular fluorescence quenching of bimane caused by tryptophan (Trp), which occurs in a distance-dependent manner. The goal of this study was to determine whether the helices in both receptors moved in the same way during formation of the active state. Our results show that whereas movement of helix VI relative to helix V occurs during formation of the active state for both parapinopsin and bovine rhodopsin, the “amplitude” of the movement is markedly different between the two rhodopsins.
EXPERIMENTAL PROCEDURES
Construction of Rhodopsins
The coding region of bovine rhodopsin and parapinopsin tagged with the monoclonal antibody rho 1D4 epitope sequence were inserted into plasmid vectors SRα and pcDNA3.1 (Invitrogen), respectively (21). Bovine rhodopsin mutant C140S/C316S/C322S/C323S and parapinopsin mutant V138F/C140A/C316A/C323A were used as respective “background” mutants in which reactive cysteine residues are substituted according to a previous report (13). It should be noted that in parapinopsin, the V138F mutation recovered expression levels that were decreased by the C140A mutation, and the background mutant of parapinopsin showed normal photoreaction and light-dependent G protein activation (supplemental Fig. S1).
Purification and Labeling of Rhodopsins with Bimane
Expression of the bovine rhodopsin and parapinopsin wild types and mutants in human embryonic kidney 293S cells were carried out as described (21, 29). The pigments reconstituted with 11-cis retinal were extracted with buffer A (1% n-dodecyl-β-maltoside, 50 mm HEPES, 140 mm NaCl, pH 6.5) and mixed with 1D4-agarose for 6 h. Labeling of rhodopsins with monobromobimane or PDT-bimane and subsequent purification was carried out when the protein was bound to the 1D4 matrix as previously described (13, 19). It should be noted that labeling procedures were done at pH 7.8 (bovine rhodopsin) or 6.8 (parapinopsin) for 18 h at 4 °C.
Label incorporation in each mutant of bovine rhodopsin was roughly estimated from difference spectra between the bimane-labeled mutant and wild type without label. Difference absorbance at around 380 nm was assumed to reflect absorbance of attached label, and the labeling efficiency was roughly estimated using excitation coefficients ϵ380 = 5000 for bimane (31). The labeling efficiencies of parapinopsin mutants were estimated by comparison of the fluorescence intensity after denaturation of protein using SDS and removal of the retinal chromophore using hydroxylamine with the intensity of labeled bovine rhodopsin 250C mutant, of which labeling efficiency was already estimated, after the same treatment.
For bovine rhodopsin, the labeling efficiencies for mutants 227C, 244C, 250C, and 251C were estimated as ∼0.4, ∼0.8, ∼1.1, and ∼1.6 label per the rhodopsin, respectively. The small increase in the number of labels with the 251C mutant may be because of the contribution of labels bound to cysteine residues in the population of non-functional rhodopsin proteins, which do not possess a retinal chromophore, as we routinely noticed a slightly lower efficiency of reconstitution with retinal for the mutant. In the case of parapinopsin, the labeling efficiencies for mutants 227C, 244C, 250C, and 251C were estimated as ∼0.7, ∼1.0, ∼0.9, and ∼1.1 label per parapinopsin, respectively. The background mutants of bovine rhodopsin and parapinopsin were labeled with bimane at less than 0.1 and 0.2 label per rhodopsin, respectively. Importantly, the labels did not appear to inhibit formation of the active state, as all of the labeled mutants showed almost the same absorption spectra as respective wild types before and after photoactivation. In particular, all bovine rhodopsin and parapinopsin mutants in this study were almost completely converted to their active states.
Spectrophotometry
Absorption spectra of purified samples were recorded with the Shimadzu UV2400 spectrophotometer, and irradiations of the samples were carried out as previously described (21, 28). UV and yellow lights were supplied by the light source with a UTVAF-50S-36U glass filter (Sigma-Koki) and a Y-52 glass filter (Toshiba), respectively.
Steady-state Fluorescence Assays
Steady-state fluorescence measurements were carried out using the Jasco FP-6300 fluorescence spectrophotometer. Measurement temperature was 10 °C. Excitation was set at 380 nm using a 2.5-nm bandpass setting. Excitation light was attenuated to ∼1/50 by a neutral density filter (Sigma-Koki) to avoid photoactivation of the pigments by excitation light. The fluorescence spectra of buffer were subtracted. In the KI quenching assay (Fig. 3) the salt concentration was kept at 25 mm by the addition of a corresponding amount of KCl, and Na2S2O3 was added to 0.1 mm to inhibit formation of I3−.
FIGURE 3.
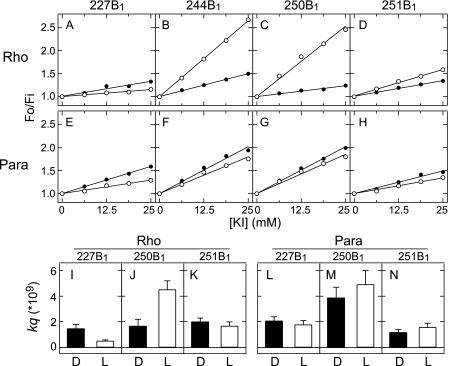
Accessibility of mBBr labeled bovine rhodopsin and parapinopsin mutants for fluorescence quenching agent KI. Top panels, A–H, Stern-Volmer plots of bovine rhodopsin mutants 227B1 (A), 244B1 (B), 250B1 (C), and 251B1 (D) and parapinopsin mutants 227B1 (E), 244B1 (F), 250B1 (G), and 251B1 (H) are shown. Stern-Volmer plots report the ratio of fluorescence intensity in the absence (Fo) and presence (Fi) of quencher as a function of quencher concentration. All of the mutants were labeled with mBBr. The closed and open circles indicated Fo/Fi values in the dark state and after photoactivation, respectively. Bottom panels, I–N, bimolecular quenching constant (kq) of respective mutants. The kq value was determined as the ratio of the slop of Stern-Volmer plot Ksv over the fluorescent lifetime τ (kq = Ksv/τ) (see supplemental Table S1). Black and white bars indicate kq values before (D) and after (L) photoactivation, respectively. Data are presented as the means ± S.E. of at least three separate experiments.
Fluorescence Lifetime Measurements
Fluorescence lifetime measurements were carried out using a PTI Laserstrobe fluorescence lifetime instrument (13) and a PTI EasyLife LED fluorescence lifetime instrument. Measurements were taken at 10 °C using 380-nm excitation pulses while monitoring the emission through long pass filters. Measurements used 100 μl of 250∼500 nm samples placed in 2-mm cuvette and represented two averages of 5 shots (Laserstrobe) or three averages of 0.3-s integration (EasyLife), collected in 150 channels. Each measurement took less than 5 min to minimize a time-dependent decay of photoproducts. The fluorescence decays were fit to a double exponential using the commercial PTI program FeliX32, and average fluorescence lifetimes 〈τ〉 were determined as previously described (〈τ〉 = α1τ1 + α2τ2, where α1 and α2 are the fractional amplitudes of each lifetime τ1 and τ2, respectively) (33).
Cleavage of Bimane from PDT-bimane-labeled Rhodopsins to Enable Normalization of Comparison of Spectra
Reduction of the disulfide bond between PDT-bimane and rhodopsin and parapinopsin was carried out as previously described (32).
G Protein Activation Assays of Loop-replaced Mutants
Bovine rhodopsin and parapinopsin mutants in which the second and/or third cytoplasmic loop were swapped between each other was prepared as previously reported (29). The second cytoplasmic loop is the region from position 134 to position 152 in bovine rhodopsin numbering system, and the third loop is the region from position 231 to position 252 (see Fig. 1A).
Functionality was measured using a radionucleotide filter binding assay, which measures GDP/guanosine GTPγS exchange by Gi (29). The assay mixture consisted of 50 mm HEPES, pH 6.5, 140 mm NaCl, 8 mm MgCl2, 1 mm dithiothreitol, 1 μm [35S]GTPγS, and 4 μm GDP. The samples were either irradiated or kept in the dark and then mixed immediately with Gi solution (final concentration of 600 nm).
RESULTS
A cysteine residue was introduced at position 227, 244, 250, or 251 of bovine rhodopsin (Rho) and parapinopsin (Para) (Fig. 1, A and D) in a background mutant in which reactive cysteines were mutated to alanine or serine. These sites were chosen because spin labels at these positions in bovine rhodopsin were previously shown to detect conformational changes upon light activation (11). Each cysteine was labeled with the fluorescent probe monobromobimane (mBBr), as this probe has previously been used to detect conformational changes in bovine rhodopsin (13) and the β-adrenergic receptor (5). As described under “Experimental Procedures,” these cysteine mutants were expressed and purified, and mBBr was attached to the cysteine residue (Fig. 1E). The labeling efficiency of each mutant indicated specific bimane labeling (see “Experimental Procedures”). Hereafter, the mutants labeled with mBBr are named by the number of the residue and the suffix B1.
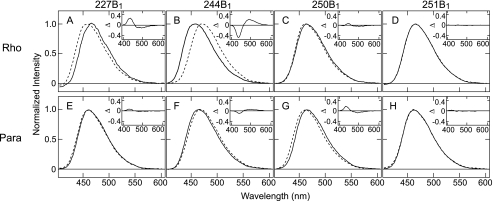
We first analyzed light-dependent changes in the wavelength of maximum emission intensity (λmax) for the bimane-labeled mutants, as the λmax value of bimane reflects the local polarity surrounding its attachment site on the surface of a protein (32, 34). Photoactivation of the bovine rhodopsin mutants produced an ∼14-nm red-shift in the λmax for mutant 244B1 and an ∼7-nm blue-shift in the λmax for mutant 227B1 (Fig. 2, A and B). Essentially no shift was observed for mutants 250B1 and 251B1 (Fig. 2, C and D). A qualitatively similar pattern of shifts was observed upon photoactivation of the parapinopsin mutants, except that wavelength shifts were smaller (Fig. 2, E–H), with the parapinopsin mutant 244B1 showing an ∼4-nm red-shift and mutant 227B1 showing an ∼1-nm blue-shift. These results suggest a similar, but not identical light-induced conformational change occurs in both rhodopsins, with the magnitude of change appearing smaller for the parapinopsin mutant. Interestingly, upon activation, a relatively small red-shift (∼5 nm) was observed for a bimane probe at the equivalent 244 site on the β-adrenergic receptor under similar conditions (5). There are subtle differences in absolute λmax values of the respective mutants between two rhodopsins, which might either be because of differences in environment around the introduced fluorescent probe or may be partially caused by the differences in the conditions of the ionic lock between helices III and VI (see “Discussion”). Because our data cannot rule out either interpretation, here we focus on light-induced changes of λmax.
FIGURE 2.
Fluorescence emission spectra of the mBBr labeled mutants of bovine rhodopsin and parapinopsin. Emission spectra of mBBr labeled mutants 227C (A), 244C (B), 250C (C), and 251C (D) of bovine rhodopsin and parapinopsin mutants 227C (E), 244C (F), 250C (G), and 251C (H) are shown. Spectra are normalized by maximal fluorescence intensity as 1.0. Solid and broken lines indicate spectra before and after photoactivation, respectively. Experimental conditions are described under “Experimental Procedures.” λmax values for the labeled bovine rhodopsin mutants are 466 nm (227B1 in the dark), 459 nm (227B1 after photoactivation), 457 nm (244B1 in the dark), 471 nm (244B1 after photoactivation), 464 nm (250B1 in the dark), 462 nm (250B1 after photoactivation, 464 nm (251B1 in the dark), and 464 nm (251B1 after photoactivation). The emission λmax values for the labeled parapinopsin mutants are 462 nm (227B1 in the dark), 461 nm (227B1 after photoactivation), 464 nm (244B1 in the dark), 468 nm (244B1 after photoactivation), 466 nm (250B1 in the dark), 462 nm (250B1 after photoactivation, 463 nm (251B1 in the dark), and 463 nm (251B1 after photoactivation). The insets show the difference in the emission spectra of before versus after photoactivation. The spectral changes in 244B1 mutants upon photoactivation of bovine rhodopsin and parapinopsin were not changed between pH 6 and 8 (see supplemental Fig. S5), suggesting that differences of conformational changes between the two rhodopsins are not because of differences in pH effects.
We further investigated these light-induced structural changes by measuring the susceptibility of each probe to collision with the water-soluble fluorescence quenching agent KI. Probe accessibility is often analyzed using a Stern-Volmer plot, which shows the ratio of fluorescence intensity in the absence (Fo) and presence (Fi) of quencher as a function of quencher concentration. The slope of the plot is named Ksv. Indeed, changes in the Ksv upon photoactivation of bovine rhodopsin and parapinopsin were detected (Fig. 3, A–H). In rhodopsin studies Ksv values provide only limited information on probe accessibility as Ksv depends not only on the collision rate of the probe with quenching agent but also on the fluorescence lifetime of the probe. In rhodopsin samples the fluorescence lifetime of bimane can vary depending on the amounts of spectral overlap between the emission spectrum of the bimane probe and retinal absorption spectrum (see supplemental Fig. S2, A–D for further explanation). For rhodopsin studies, a better measure of probe accessibility to quencher is to determine the bimolecular quenching constant kq. The kq value is simply the ratio of Ksv over the fluorescent lifetime τ (kq = Ksv/τ), and the kq value reflects the true collision rate of quencher with the probe. For these reasons we measured fluorescence lifetimes in addition to Ksv values for each bimane-labeled mutant to calculate kq and, therefore, directly compare probe accessibility for the different rhodopsin mutants.
The kq values obtained from these Ksv values and fluorescence lifetimes are reported in supplemental Table S1 and are plotted in Fig. 3, I–N.5 For bovine rhodopsin mutants 227B1 and 250B1 the kq values changed 0.35- and 2.6-fold upon photoactivation, respectively (Fig. 3, I and J, and supplemental Table S1). In contrast, the kq values at mutants 227B1 and 250B1 of parapinopsin changed only 0.9- and 1.3-fold, respectively (Fig. 3, L and M, and supplemental Table S1). The kq values of 251B1 mutants of bovine rhodopsin and parapinopsin did not change significantly upon photoactivation (Fig. 3, K and N). The kq values clearly show that whereas large light-induced changes in KI accessibility occur at several sites on bovine rhodopsin, more moderate changes occur in parapinopsin. Together, these results indicate that the environment around the labeled residues in helices V and VI changes more dramatically in bovine rhodopsin than parapinopsin upon photoactivation.
We next set out to determine whether these differences are because of differences in the rearrangement of helices V and VI upon photoactivation. To do this we employed an intramolecular fluorescence quenching approach. This approach exploits TrIQ-bimane technique to assess the proximity between the bimane-label and Trp residue on proteins, based on the ability of the Trp residue to quench bimane fluorescence in a distance dependent manner (31, 32, 35).
For these studies we labeled a cysteine residue at position 251 in helix VI with bimane and introduced a Trp residue at position 226 or 227 in helix V (see Fig. 1D). For these measurements it was critical to develop a procedure to enable comparison of the fluorescence intensity between different samples as well as to take into account differences in background energy transfer to the retinal chromophore. Thus, rather than using mBBr, we labeled the samples using the bimane derivate PDT-bimane (32). Hereafter these mutants are named by the residue number with B2. We used PDT-bimane because it attaches through a reducible disulfide linkage that can be cleaved to release free bimane from the protein (Fig. 1F). After reduction, one can directly compare the intensity of the released free label from two different samples to easily and precisely obtain a normalizing factor that can be used to subsequently compare the total original starting fluorescence intensity for the probe bound to each protein sample (32). In our analysis we employed this approach to reliably compare the extent of fluorescence decrease by normalizing the sample spectra, correcting for any differences in the amount of attached fluorescent probe between the two tryptophan mutants or differences in energy transfer to the retinal chromophore.
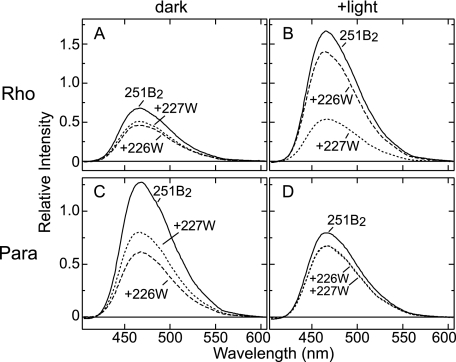
The results of the TrIQ-bimane studies are shown in Fig. 4. In the dark state of bovine rhodopsin, the bimane probe at position 251 was quenched slightly more effectively by Trp-226 than by Trp-227 (Fig. 4A). This small but clear difference is reproduced in separate experiments and likely reflects the closer distance between positions 226 and 251 (Cα-Cα distance is ∼9 Å) than between positions 227 and 251 (Cα-Cα distance is ∼11 Å) in the bovine rhodopsin crystal structure (2, 3) (see Fig. 1D). As already noted, the general arrangement of transmembrane helices are almost identical among all of the GPCRs whose inactive structure have so far been determined (4, 7, 8). If one assumes the structure of inactive (dark-state) parapinopsin follows this trend, then position 251 should be closer to position 226 than to position 227. Indeed, we find in dark-state parapinopsin the bimane at position 251 is more efficiently quenched by Trp-226 than Trp-227 (Fig. 4C). Taken together, these results suggest a similar relative arrangement of helices V and VI in dark-state bovine rhodopsin and parapinopsin.6
FIGURE 4.
Fluorescence emission spectra of the mutants containing 251B2 of bovine rhodopsin and parapinopsin. A and B, emission spectra of PDT-bimane labeled bovine rhodopsin mutants in the dark (A) and after photoactivation (B). C and D, emission spectra of PDT-bimane labeled parapinopsin mutants in the dark (A) and after photoactivation (B). Spectra are normalized to the fluorescence intensity after reductive cleavage of the disulfide linkage between bimane label and rhodopsin using the reducing agent Tris(2-carboxyethyl)phosphine (see “Experimental Procedures”). The +226W and +227W indicate the introduction of a Trp residue at positions 226 and 227, respectively, to act as a local quencher group.
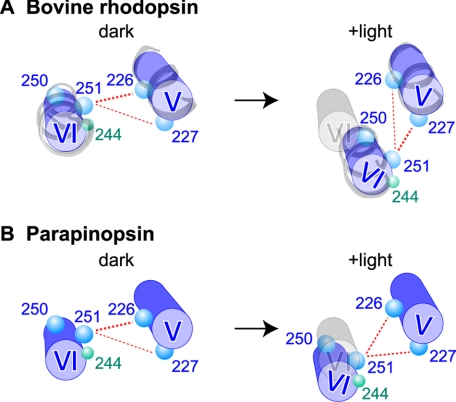
In contrast to their dark-state similarities, the bovine rhodopsin and parapinopsin mutants showed different TrIQ-bimane quenching profiles upon photoactivation. For the bovine rhodopsin mutants, photoactivation caused the bimane at position 251 to be more effectively quenched by Trp-227 than by Trp-226 (Fig. 4B), consistent with previous models proposing a tilting and/or rotation of helix VI relative to helix V during formation of the active state (see Fig. 5A). In contrast, photoactivation of the parapinopsin mutants produced a different pattern of TrIQ-bimane quenching in which both Trp-226 and Trp-227 showed about the same ability to quench 251B2 (Fig. 4D). Thus, these results suggest that in parapinopsin a different extent of helix V/VI movement occurs. In particular, the amino acid residue at position 251 moves to a position equidistant from positions 226 and 227 (Fig. 5B). This arrangement differs from the arrangement in bovine rhodopsin, in which position 251 is farther from position 226 and closer to position 227 (Fig. 5A). We also attempted to test the model in Fig. 5 by studying changes in a bimane probe placed at position 250. Unfortunately, parapinopsin mutant 250C does not efficiently label with PDT-bimane, and we, thus, had to use mBBr, which cannot be reduced off the protein to enable quantification. However, the studies with mBBr at position 250 (shown in supplemental Fig. S3) are in complete agreement with the model proposed in Fig. 5 (see the supplemental text) and previous studies (35).
FIGURE 5.
Model of conformational changes in helices V and VI upon photoactivation of bovine rhodopsin and parapinopsin. The models (blue) represent the proposed arrangement of helices V and VI in the inactive and active forms as viewed from the cytoplasmic surface of bovine rhodopsin (A) and parapinopsin (B), based on the intramolecular quenching data on amino acid residues at positions 226, 227, 250, and 251 (blue circles). The presumed original position of helix VI in the dark models is also shown (gray) in the +light models. For the bovine rhodopsin model, the actual arrangement of helices V and VI based on the crystal structures of bovine rhodopsin (PDB code 1GZM) and bovine opsin (PDB code = 3CAP) has also been included for both the dark and +light models of bovine rhodopsin, respectively (gray ribbon models). It should be noted that the relative arrangement of the helices and residues shown in the right panel of A are consistent with other experimental data. These include the crystal structures of bovine opsin, which show residues at positions 250 and 251, forming van del Waals contacts with residues at positions 226 and 227, respectively (14, 15), and recent models based on double electron-electron resonance spectroscopy (42). The models also explain the differences we observe in environmental changes around residue at position 244 (green circles) upon photoactivation of bovine rhodopsin and parapinopsin (Fig. 2, B and F).
Together, these results strongly suggest that the active state is different for parapinopsin compared with bovine rhodopsin. Our data suggest that for both rhodopsins, movement of helices V and VI does occur during photoactivation, but the movement is smaller for parapinopsin. In other words, we propose that a similar relative movement of the helices V and VI occurs for both, but the amplitude of movement is larger in bovine rhodopsin than in parapinopsin. This interpretation is also consistent with our data on the emission maximum and fluorescence quenching. Both studies showed the polarity and quenching for labeled residues in helices V and VI changed upon photoactivation, and the extent of the changes was larger in bovine rhodopsin than in parapinopsin (Figs. 2 and 3).
DISCUSSION
In this study we set out to determine whether protein dynamics might explain why two similar GPCRs would possess different abilities to activate the same G protein. Specifically, we tested bovine rhodopsin and parapinopsin to determine whether their marked difference in ability to activate G protein is related to a difference in conformational change between helix V and helix VI upon photoactivation. Our approach was to use a site-directed fluorescence labeling in which we introduced bimane residues at specific sites on both proteins and then compared the fluorescence properties before and after light activation. Several lines of evidence indicate that upon photoactivation both rhodopsins undergo a conformational change that is similar but different in magnitude. At several positions we observed clear differences in the λmax of the fluorescence emission spectra as well as differences in accessibility to water-soluble quencher agents (Figs. 2 and 3). We further defined the differences in movement by using TrIQ-bimane method (Fig. 4). A model consistent with our results is schematically summarized in Fig. 5.
One possibility for these differences could be a partial breaking of the ionic lock between helices III and VI in the inactive state, as has been observed for the crystal structures of β-adrenergic receptors (36). In this line of thinking, if the status of the ionic lock differed for the inactive state of bovine rhodopsin versus parapinopsin, it might result in differences in their relative arrangement of helices V and VI. However, we feel this possibility is unlikely. All of the main amino acid residues participating to the ionic lock in both bovine and squid rhodopsin (Arg-135 and Glu-247 in bovine rhodopsin) are conserved in parapinopsin. And for both bovine rhodopsin and squid rhodopsins (an invertebrate rhodopsin which shares similarities in photoreaction to parapinopsin), the crystal structures show an intact ionic lock in the inactive (dark) form (2–4). Thus, based on these observations, we feel it is reasonable to assume the ionic lock is likely present in the dark state of parapinopsin, with perhaps a slight difference in its strength.
The ionic lock in parapinopsin may contribute to keeping the relative arrangement of the residues at positions 226, 227, and 251 similar to the arrangement in bovine rhodopsin. In fact, the arrangement of helices V and VI in the inactive (dark) state are highly conserved among the crystal structures of bovine and squid rhodopsins. The slight differences we observed in the TrIQ-bimane profiles between bovine rhodopsin and parapinopsin in the dark (Fig. 4, A and C) may be because of subtle differences of relative arrangement of helices V and VI and/or subtle differences of local environment around the bimane-label and the tryptophan.
In the case of the active state, differences are much more dramatic; the quenching efficiency of 227W on 251B2 fluorescence is much greater in bovine rhodopsin than in parapinopsin (Fig. 4, B and D). Because we experimentally eliminated the possibility that this difference could be because of differences in polarity between the two positions (see supplemental Fig. S4), the only explanation left is that the difference in the quenching efficiency is caused by a different relative arrangement of amino acid residues at positions 227 and 251 between the active states of bovine rhodopsin and parapinopsin. Thus, based on our data, we propose that during photoactivation of both rhodopsins, a similar relative movement of helix VI toward helix V with tilting and/or rotation takes place, but the amount or amplitude of these helical movements differ, with the amount of movement being larger for bovine rhodopsin. One intriguing possibility is that this difference in the helical movement could be because of a difference in the status of the ionic lock between helices III and VI in the active state, because the status of the ionic lock is reported to be important in the active state formation based on structural studies of bovine rhodopsin (14, 37).
Interestingly, the region between the cytoplasmic ends of helices V and VI is important for interacting with G protein (15, 18, 29, 38). It has been proposed that one key aspect of receptor activation is to expose a “hydrophobic patch” in this region which provides a critical site for high affinity binding of the G protein α-subunit C terminus (19). It is, thus, possible that the larger amplitude of light-induced conformational changes we detected for bovine rhodopsin makes this binding site more accessible for G protein binding, thus explaining the ∼20–50-fold greater ability of bovine rhodopsin to activate G protein compared with parapinopsin and invertebrate rhodopsin (see Fig. 1, B and C) (28).
The difference in the amplitude of the light-induced conformational change may also be related to another difference between the two rhodopsins, namely the difference in their photoreversibility. The larger conformational change observed in bovine rhodopsin might enable so much internal rearrangement within the protein that it cannot be converted back to a dark state structure by further light-induced photoisomerization of retinal. Therefore, it is tempting to speculate that during molecular evolution of rhodopsins, vertebrate visual pigments including bovine rhodopsin acquired higher G protein activation ability through acquisition of the larger conformational change, a byproduct of which also resulted in abolishment of photoreversibility in the rhodopsins at the same time. Interestingly, metarhodopsin I, the precursor of the active state of bovine rhodopsin (metarhodopsin II), can be converted back to the dark state by subsequent light absorption (39). Comparison of conformation between metarhodopsin I and the active state of parapinopsin would provide a further evolutionary suggestion.
It has been suggested that invertebrate rhodopsins have the counterion glutamic acid at position 181 (Glu-181) and vertebrate rhodopsins acquired Glu-113 counterion during the molecular evolution (28, 40). Parapinopsin has both Glu-113 and Glu-181, but the Glu-181 serves as the counterion. Therefore, the difference in the amplitude of the light-induced conformational change may be related to differences in the position of counterion, although it is still unclear how the large conformational change is generated. Detailed mechanisms of the large conformational change will be revealed by future studies.
Rhodopsin is the best understood member of the GPCR family. A detailed analysis of the human genome reveals more than 800 unique GPCRs, of which ∼700 are rhodopsin-like GPCRs (41). These receptors receive various extracellular signals and drive the intracellular signaling cascade through various G proteins. Diversity of GPCRs is considered to be generated by diversity in ligand binding sites and G protein interacting sites. On the other hand, there are different receptors that bind the same ligand and activate the same G protein. Our findings open the possibility that G protein activation efficiency (signal transmission efficiency) may be regulated by amplitude of conformational changes in the multiple receptors, leading to generation of functional diversity of GPCRs.
Supplementary Material
Acknowledgments
We thank Yoshinori Shichida (Kyoto University) for helpful discussions at the initial stage of this work and Robert S. Molday (University of British Columbia) for the kind supply of rho 1D4-producing hybridoma.
This work was supported, in whole or in part, by National Institutes of Health Grants DA018169 and EY015436 (to D. L. F.). This work was also supported in part by grants-in-aid for Scientific Research from the Japanese Ministry of Education, Science, Sports, and Culture (to A. T. and M. K.) and by the Yamada Science Foundation (to A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
In this paper the residue number of parapinopsin is described by using the bovine rhodopsin numbering system.
The lifetime measurements were not carried out for the 244B1 mutants because of low yields of the parapinopsin 244B1 mutant. It should be noted that differences of kq values between the dark states of bovine rhodopsin and parapinopsin may be partially because of differences in the conditions of the ionic lock between helices III and VI or because of differences in the polarity of the surrounding amino acid sequence or differences in accessibility to solvent.
The slight differences in the quenching profiles between bovine rhodopsin and parapinopsin in the dark may be because of subtle differences of the relative arrangement of helices V and VI and/or subtle differences of the local environment around the bimane-label and the tryptophan (see “Discussion”).
- GPCR
- G protein-coupled receptor
- GTPγS
- guanosine 5′-3-O-(thio) triphosphate
- λmax
- wavelength of maximum emission intensity
- mBBr
- monobromobimane
- PDT-bimane
- (2-pyridyl)dithiobimane
- TrIQ-bimane
- tryptophan-induced quenching of bimane.
REFERENCES
- 1.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 2.Okada T., Sugihara M., Bondar A. N., Elstner M., Entel P., Buss V. (2004) J. Mol. Biol. 342, 571–583 [DOI] [PubMed] [Google Scholar]
- 3.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. (2004) J. Mol. Biol. 343, 1409–1438 [DOI] [PubMed] [Google Scholar]
- 4.Murakami M., Kouyama T. (2008) Nature 453, 363–367 [DOI] [PubMed] [Google Scholar]
- 5.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 6.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefkowitz R. J., Sun J. P., Shukla A. K. (2008) Nat. Biotechnol. 26, 189–191 [DOI] [PubMed] [Google Scholar]
- 9.Sheikh S. P., Zvyaga T. A., Lichtarge O., Sakmar T. P., Bourne H. R. (1996) Nature 383, 347–350 [DOI] [PubMed] [Google Scholar]
- 10.Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 11.Altenbach C., Yang K., Farrens D. L., Farahbakhsh Z. T., Khorana H. G., Hubbell W. L. (1996) Biochemistry 35, 12470–12478 [DOI] [PubMed] [Google Scholar]
- 12.Hubbell W. L., Altenbach C., Hubbell C. M., Khorana H. G. (2003) Adv. Protein Chem. 63, 243–290 [DOI] [PubMed] [Google Scholar]
- 13.Dunham T. D., Farrens D. L. (1999) J. Biol. Chem. 274, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 14.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. (2008) Nature 454, 183–187 [DOI] [PubMed] [Google Scholar]
- 15.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 16.Ghanouni P., Steenhuis J. J., Farrens D. L., Kobilka B. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5997–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward S. D., Hamdan F. F., Bloodworth L. M., Wess J. (2002) J. Biol. Chem. 277, 2247–2257 [DOI] [PubMed] [Google Scholar]
- 18.Gether U. (2000) Endocr. Rev. 21, 90–113 [DOI] [PubMed] [Google Scholar]
- 19.Janz J. M., Farrens D. L. (2004) J. Biol. Chem. 279, 29767–29773 [DOI] [PubMed] [Google Scholar]
- 20.Blackshaw S., Snyder S. H. (1997) J. Neurosci. 17, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koyanagi M., Kawano E., Kinugawa Y., Oishi T., Shichida Y., Tamotsu S., Terakita A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6687–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukamoto H., Terakita A., Shichida Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jäger S., Palczewski K., Hofmann K. P. (1996) Biochemistry 35, 2901–2908 [DOI] [PubMed] [Google Scholar]
- 24.Han M., Smith S. O., Sakmar T. P. (1998) Biochemistry 37, 8253–8261 [DOI] [PubMed] [Google Scholar]
- 25.Koutalos Y., Ebrey T. G., Tsuda M., Odashima K., Lien T., Park M. H., Shimizu N., Derguini F., Nakanishi K., Gilson H. R. (1989) Biochemistry 28, 2732–2739 [DOI] [PubMed] [Google Scholar]
- 26.Peirson S., Foster R. G. (2006) Neuron 49, 331–339 [DOI] [PubMed] [Google Scholar]
- 27.Terakita A. (2005) Genome Biology 6, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terakita A., Koyanagi M., Tsukamoto H., Yamashita T., Miyata T., Shichida Y. (2004) Nat. Struct. Mol. Biol. 11, 284–289 [DOI] [PubMed] [Google Scholar]
- 29.Terakita A., Yamashita T., Nimbari N., Kojima D., Shichida Y. (2002) J. Biol. Chem. 277, 40–46 [DOI] [PubMed] [Google Scholar]
- 30.Bartl F. J., Ritter E., Hofmann K. P. (2001) J. Biol. Chem. 276, 30161–30166 [DOI] [PubMed] [Google Scholar]
- 31.Mansoor S. E., McHaourab H. S., Farrens D. L. (2002) Biochemistry 41, 2475–2784 [DOI] [PubMed] [Google Scholar]
- 32.Mansoor S. E., Farrens D. L. (2004) Biochemistry 43, 9426–9438 [DOI] [PubMed] [Google Scholar]
- 33.Sommer M. E., Smith W. C., Farrens D. L. (2005) J. Biol. Chem. 280, 6861–6871 [DOI] [PubMed] [Google Scholar]
- 34.Mansoor S. E., McHaourab H. S., Farrens D. L. (1999) Biochemistry 38, 16383–16393 [DOI] [PubMed] [Google Scholar]
- 35.Janz J. M. (2004) Structural Dynamics of Rhodopsin: Relationships between Retinal Schiff Base Integrity and Receptor Signaling States. Ph.D. thesis, Oregon Health and Science University [Google Scholar]
- 36.Weis W. I., Kobilka B. K. (2008) Curr. Opin. Struct. Biol. 18, 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salom D., Lodowski D. T., Stenkamp R. E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J. A., Palczewski K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.König B., Arendt A., McDowell J. H., Kahlert M., Hargrave P. A., Hofmann K. P. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6878–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritter E., Zimmermann K., Heck M., Hofmann K. P., Bartl F. J. (2004) J. Biol. Chem. 279, 48102–48111 [DOI] [PubMed] [Google Scholar]
- 40.Terakita A., Yamashita T., Shichida Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14263–14267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B. (2003) Mol. Pharmacol. 63, 1256–1272 [DOI] [PubMed] [Google Scholar]
- 42.Altenbach C., Kusnetzow A. K., Ernst O. P., Hofmann K. P., Hubbell W. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7439–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.