Abstract
Fibrosis is characterized by elevated transforming growth factor β (TGFβ) signaling, resulting in extracellular matrix accumulation and increased PAI-1 (plasminogen activator inhibitor) expression. PAI-1 induces the internalization of urokinase plasminogen activator/receptor and integrin αvβ3 from the cell surface. Since increased αvβ3 expression correlates with increased TGFβ signaling, we hypothesized that aberrant PAI-1-mediated αvβ3 endocytosis could initiate an autocrine loop of TGFβ activity. We found that in PAI-1 knock-out (KO) mouse embryonic fibroblasts), αvβ3 endocytosis was reduced by ∼75%, leaving αvβ3 in enlarged focal adhesions, similar to wild type cells transfected with PAI-1 small interfering RNA. TGFβ signaling was significantly enhanced in PAI-1 KO cells, as demonstrated by a 3-fold increase in SMAD2/3-containing nuclei and a 2.9-fold increase in TGFβ activity that correlated with an increase in αvβ3 and TGFβ receptor II expression. As expected, PAI-1 KO cells had unregulated plasmin activity, which was only partially responsible for TGFβ activation, as evidenced by a mere 25% reduction in TGFβ activity when plasmin was inhibited. Treatment of cells with an αvβ3-specific cyclic RGD peptide (GpenGRGD) led to a more profound (59%) TGFβ inhibition; a nonspecific RGD peptide (GRGDNP) inhibited TGFβ by only 23%. Human primary fibroblasts were used to confirm that PAI-1 inhibition and β3 overexpression led to an increase in TGFβ activity. Consistent with a fibrotic phenotype, PAI-1 KO cells were constitutively myofibroblasts that had a 1.6-fold increase in collagen deposition over wild type cells. These data suggest that PAI-1-mediated regulation of αvβ3 integrin is critical for the control of TGFβ signaling and the prevention of fibrotic disease.
Fibrotic disorders can result from environmental toxins, persistent infection, autoimmune disease, or mechanical injury, leading to the hardening and scarring of tissues. In fibrotic diseases, such as liver cirrhosis, renal fibrosis, and idiopathic lung fibrosis, or in pathological wound healing, such as hypertrophic scarring, scleroderma, and Dupuytren disease, the persistence of myofibroblasts contributes to disease progression by overproduction of extracellular matrix (ECM)2 and by excessive contraction (1–3). A shift in the balance of growth factors and cytokines that promote ECM deposition and proteases that degrade matrix often contributes to fibrotic disease (4, 5). Plasmin, a broad spectrum protease that is generated from plasminogen by uPA, is one of the proteases that degrades matrix and activates growth factors and other proteases (6). Since uPA activity is inhibited by PAI-1, the overexpression of PAI-1 results in matrix accumulation. For this reason, PAI-1 is a key prognostic marker for fibrotic disease. PAI-1 exerts its inhibitory activity on uPA by stimulating the endocytosis of the cell surface uPA·uPAR complex through the low density lipoprotein receptor-related protein (7). Integrin αvβ3 is also internalized with the uPA·uPAR·low density lipoprotein receptor-related protein complex (8). After endocytosis, uPAR and integrins are recycled back to the cell surface for another round of binding (8, 9). uPAR and αvβ3 promote cellular attachment and spreading, since they are receptors for the extracellular matrix molecule, vitronectin (10). Thus, cycling of the complex is thought to stimulate the attachment and detachment that is necessary for cell migration (8). Consequently, a shift in the expression of any of these components (PAI-1/uPA/uPAR/αvβ3) can result in either aggressive migration, as seen in cancer invasion, or a persistent increase in cell adhesion and cell tension, as seen in myofibroblasts in fibrotic tissue.
The family of TGFβ growth factors has been intensively studied for their role in fibrotic wound healing. Up-regulation of TGFβ results in amplified and persistent overproduction of molecules, such as integrins and PAI-1 and other protease inhibitors (e.g. TIMPs) (2, 3). Up-regulated integrins continue the cycle of TGFβ signaling by participating in the sustained activation of TGFβ from its latent form. To date, studies have found that various αv integrins participate in the activation of TGFβ (αvβ3, αvβ5, αvβ6, and αvβ8), but the mechanism differs (11–15). Integrins can serve as docking proteins to localize proteases that cleave and activate latent TGFβ in the ECM, or they can directly activate latent TGFβ in a protease-independent manner. Recently, it was discovered that latent TGFβ is also activated by mechanical stress generated from an integrin-mediated interaction between myofibroblasts and the ECM, primarily involving αvβ5. The mechanical stress promotes a conformational change that activates the latent TGFβ complex (15). αv integrins also modulate TGFβ signaling through the binding of αvβ3 to TGFβ receptor II (TGFβRII) in the presence of TGFβ. This interaction was shown to promote a dramatic increase in the proliferation of lung fibroblasts and induce invasion of epithelial breast cancer cells (16, 17).
Our data establish a role for the PAI-1-mediated control of αvβ3 expression and support a significant role for αvβ3 in TGFβ signaling. Using PAI-1 KO cells, we tested the hypothesis that the absence of PAI-1 would result in the accumulation of αvβ3 on the cell surface, since PAI-1 promotes the endocytosis of uPA·uPAR·αvβ3. PAI-1-mediated endocytosis of β3 was significantly reduced in the PAI-1 KO cells. Correspondingly, we report that β3 accumulated at the cell surface in enlarged β3-containing focal adhesions. Thus, we explored whether the accumulation of αvβ3 on the cell surface had fibrogenic effects even in the absence of profibrotic PAI-1. Our results demonstrate dramatically increased TGFβ activity and an increase in collagen expression in PAI-1 KO cells. Together, these findings suggest that PAI-1 modulates β3 expression and localization and, in turn, TGFβ signaling. Our data reveal that maintaining precise levels of PAI-1 is a key to preventing fibrosis. Understanding the consequence of regulating PAI-1 activity is critical in light of the many clinical therapies currently under development that target PAI-1 (18, 19).
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Cells
Transformed mink lung epithelial cells (TMLCs) containing the PAI-1 promoter fused to the luciferase gene were a generous gift of Dr. Daniel Rifkin (New York University). Antibodies to integrin β3 (R36) used for immunofluorescence were a generous gift of Dr. Barry Coller (Rockefeller University, New York, NY). Anti-vimentin antibody was a generous gift of Dr. Paul FitzGerald (University of California, Davis, CA). Antibodies against mouse PAI-1 were from American Diagnostica (sheep) and R&D Systems (rat), and TGFβRII (ab28382) was from Abcam (Cambridge, MA). β3 antibody used for immunoprecipitation and Western blotting (ab1932) was from Chemicon (Billerica, MA); β3 antibody (16-0611) and hamster IgG (16-4888) used for the endocytosis assay were from eBioscience (San Diego, CA); anti-early endosome antigen 1 (EEA1) antibody was from Abcam (ab15846-200); SMAD2/3 antibody was from BD Biosciences; SMAD2, SMAD3, phospho-SMAD2, and phospho-SMAD3 antibodies were from Cell Signaling (Danvers, MA); and vinculin antibody was from Sigma. Anti-α-SMA antibody was from Sigma. The FITC labeling kit was from Novus Biologicals (Littleton, CO). Secondary Alexa-488 and Alexa-568 were from Molecular Probes, Inc. (Eugene, OR). HRP-conjugated anti-Streptavidin antibodies and all HRP-conjugated secondary antibodies were from Jackson Laboratories (Bar Harbor, ME). Mouse PAI-1 siRNAs were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and the non-targeting fluorescent nucleotide control (siGlo) was from Dharmacon (Lafayette, CO). Human β3 cDNA (pcDNA3) was a generous gift from Dr. Peter Newman (Blood Research Institute, Blood Center of Wisconsin, Milwaukee, WI), and the GFP construct (EGFP-C1) was from Clontech (Mountain View, CA). Aprotonin was from Calbiochem. The control RGD peptide (GRGDNP) was from Biomol (Plymouth Meeting, PA), and the cyclic RGD peptide (GpenGRGD) was from Bachem (Torrance, CA). Collagen (PureCol) was from Inamed (Fremon, CA), and vitronectin was from Sigma.
Cells and Media
PAI-1 MEFs were maintained in PAI-1 complete media: DMEM (Invitrogen) with 10% fetal calf serum (Hyclone, Logan, UT), 1 mm l-glutamine, 1× non-essential amino acids (Invitrogen), and penicillin/streptomycin (Sigma). For experiments except where noted, cells were plated on 10 μg/ml collagen in supplemented serum-free media (SSFM): DMEM, 1× RPMI 1640 vitamin mix, 1× ITS liquid media supplement, 1 μg/ml glutathione (all from Sigma) and 2 mm l-glutamine, 1 mm sodium pyruvate, 1× non-essential amino acids with penicillin/streptomycin.
Human primary corneal fibroblasts (HCFs) were derived from the stroma of human corneas that were not suitable for transplantation (obtained from NDRI, Pittsburgh, PA). Stromal fibroblasts were isolated as previously described (20).
Immunocytochemistry
Cells were fixed with 3% p-formaldehyde (Fisher) and permeabilized with 0.1% Triton X-100 (Sigma). After blocking with 3% normal mouse serum (Jackson ImmunoResearch), cells were incubated with primary antibody (anti-αvβ3 (R36), anti-vinculin, anti-SMAD2/3, anti-SMAD3, or anti-α-SMA), washed in PBS, and incubated with secondary antibodies (Alexa-488 or Alexa-568). Coverslips were viewed with a Zeiss Axioplan 2 microscope, and images were captured using a SPOT-2 CCD camera (Diagnostic Instruments, Sterling Heights, MI) and processed by Adobe PhotoShop software.
Western Blots
24 h after seeding, cells were detached with detachment buffer (20 mm Tris, pH 7.5, 250 mm sucrose), pelleted, and lysed in 1% SDS plus complete protease inhibitor tablet (Roche Applied Science), phenylmethylsulfonyl fluoride (Sigma), Na3VP4 (Fisher), and benzonase (Sigma). 30 μg of protein was separated on 4–12% NuPAGE gradient gels and transferred to nitrocellulose membranes (gel for αvβ3 was non-reducing, gels for TGFβRII, vimentin, SMAD2 and -3, and pSMAD2 and -3 were reducing). For anti-αvβ3, streptavidin-HRP, and pSMAD2 and -3, blots were processed with 5% bovine serum albumin in Tris-buffered saline. TGFβRII, PAI-1, vimentin, SMAD2, and SMAD3 blots were blocked with 5% milk in Tris-buffered saline. Primary and secondary antibodies conjugated to HRP were diluted in bovine serum albumin or milk, correspondingly. Bands were visualized with ECL (Pierce).
Immunoprecipitation (IP)
For cell surface expression, cells were biotinylated for 30 min with 0.5 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Pierce) prior to detachment with detachment buffer. For αvβ3 IP, cell pellets were lysed in Triton Buffer (1% Triton, 150 mm NaCl, 20 mm Tris, 1 mm MgCl2, 1 mm CaCl2, 10% glycerol, pH 7.5) plus protease inhibitors (as above). For TGFβRII, cell pellets were lysed in 1% SDS (Fisher) plus protease inhibitors. SDS lysates were diluted to 0.1% SDS for protein determination and IPs. For each IP, protein A or G beads (Upstate Biotechnology, Inc., Lake Placid, NY) and 5 μg of antibody were added to 0.5 mg of total protein and incubated overnight at 4 °C. Eluted proteins were separated under reducing conditions and transferred to nitrocellulose before blocking with bovine serum albumin and incubation with streptavidin-HRP. Bands were visualized with ECL (Pierce). For PAI-1 IP, Triton lysates were immunoprecipitated with anti-PAI-1 (American Diagnostica) and detected with anti-PAI-1 (R&D Systems).
Endocytosis Assay
PAI-1 WT and KO cells were seeded in PAI-1 complete media at 1 × 104 cells/ml. FITC-labeled anti-β3 antibody and FITC-labeled control IgG (both without azide) (eBioscience) were incubated in 0.5% fetal bovine serum in DMEM for 30 min at 37 °C before fixation with p-formaldehyde. Cells were then co-immunostained for EEA1 to identify early endosomes. Autofluorescence was not quenched in this procedure to maintain the endocytosed FITC signal. MetaMorph® image analysis software was used to quantify overlap between β3 and EEA1.
Transfections
Transient transfection was performed using the Amaxa Nucleofection® system (Gaithersburg, MD). PAI-1 MEFs were transfected using the MEF2 kit with either 1 μm siRNA and 1 μg of EGFP plasmid or EGFP alone, and cells were seeded in PAI-1 complete media, without antibiotics. HCFs were co-transfected with 1 μm human PAI-1 siRNA, 1 μm siGlo, 2.5 μg of human β3 cDNA with 2.5 μg EGFP plasmid, or EGFP alone, using the normal human dermal fibroblasts kit and seeded in HCF complete media without antibiotics. Axiovert images were captured with an Axiocam MRm CCD camera.
RNA Extraction and qPCR
Total RNA was extracted from cell lysates using the RNeasy kit (Qiagen). First stranded cDNA was generated from 200 ng of total RNA using the Omniscript RT kit (Qiagen) and random primers (Invitrogen) according to the manufacturer's instructions. Triplicate determinations were analyzed by qPCR using the RT2 Real time detection system (SuperArray, Frederick, MD) and ABI 7900 sequence detection system. Annealing temperature was 55 °C for all reactions. Primers used were mouse PAI-1 forward (5′-GGGGTAGGTTGGGAGAATGT-3′) and reverse (5′-TCTGGGACAAAGGCTAAGGA-3′), human β3 forward (5′-TCACCAGTAACCTGCGGATTG-3′) and reverse (5′-GTAGCCAAACATGGGCAAGC-3′). Primers for human PAI-1, human β-actin, and mouse β-actin were obtained from Realtimeprimers.com (Elkins Park, PA).
Plasminogen Activator (PA) Activity Assay
This assay was performed as previously described (20). Briefly, cells were plated onto collagen or vitronectin in SSFM alone or with 1 ng/ml FGF-2 (Invitrogen) and 1 μg/ml heparin (Fisher) or with 1 ng/ml TGFβ1 (R&D Systems) and grown for 24 h. After cell lysis, uPA activity was determined using a chromogenic substrate for plasmin (Spectrozyme PL; American Diagnostica). The reaction product was measured on a Biotek spectrophotometer (A405 nm) at 1 h.
TGFβ Activity Assay
This protocol was slightly modified from previously published assays (12, 21). PAI-1 cells were co-cultured with TMLCs, which contain the PAI-1 promoter fused to the luciferase gene. PAI-1 WT or KO MEFS and TMLCs were plated at 1 × 105 cells/well (each) in 24-well dishes in DMEM, 10% fetal bovine serum, 1 mm l-glutamine, and penicillin/streptomycin. After 24 h, the media were replaced with (0.1% bovine serum albumin in DMEM, penicillin/streptomycin) and incubated for 24 h. Luciferase activity was measured in triplicate using the Bright-Glo detection system (Promega), and luminescence was determined using a Synergy 2 Biotek microplate reader (Winooski, VT). To determine the contribution of plasmin to TGFβ activity, 10 μg/ml aprotinin was added when cells were seeded and readded when the medium was changed. Similarly, for the peptide inhibition experiment, a 100 nm concentration of either control GRGDNP or cyclic-RGD peptide, GpenGRGD, was readded when the medium was changed. In addition, cells were plated on 10 μg/ml collagen to prevent cell detachment when peptides were added. PAI-1 siRNA and β3-transfected HCFs were co-cultured with TMLCs and assayed for luciferase activity as described above.
Quantitative Assay for Collagen Synthesis
This assay was performed as per Ref. 22. Briefly, 1 × 105 PAI-1 WT and KO cells were seeded in PAI-1 complete media into 24-well dishes and grown for 2 days before fixation in Boulin's fixative (75% picric acid (Sigma), 20% formaldehyde, 5% glacial acetic acid (Fisher)). Cells were washed and dried overnight at room temperature. To stain for collagen synthesis, Sirius Red stain (0.25 g of Direct Red (Sigma), 250 ml of picric acid) was added for 1 h. Wells were washed with 0.01 n HCl (Fisher), and dye was eluted with 0.5 ml of 0.1 n NaOH (Fisher). Duplicate points (150 μl) from triplicate wells were read at A550 on a Synergy 2 Biotek Reader in a 96-well plate. After elution, wells were washed and air-dried. Cell count was determined with Crystal Violet (Fisher). To determine the amount of collagen synthesis per cell, the ratio of Sirus Red to Crystal Violet was calculated.
Statistical Analysis
Each experiment was repeated 3–6 times. Numerical data are expressed as the mean ± S.D.
RESULTS
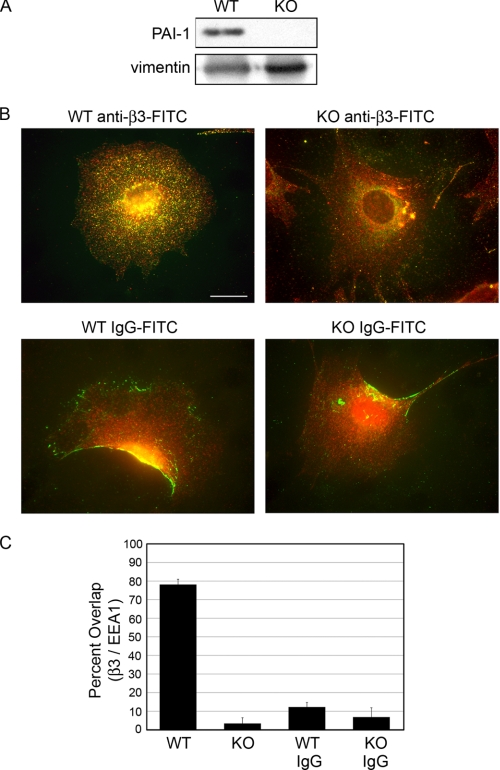
To confirm that PAI-1 KO MEFs lacked PAI-1, lysates of WT and KO cells were immunoprecipitated with anti-PAI-1 antibody. These data confirm the presence of PAI-1 in WT cells and its absence from KO cells (Fig. 1A). As expected, PAI-1 KO lysate did not yield a signal for PAI-1 despite equal loading of protein.
FIGURE 1.
In PAI-1 KO cells, β3 endocytosis is impaired. A, to confirm that PAI-1 was absent from PAI-1 KO cells, lysates were immunoprecipitated for PAI-1 and Western blotted with a second PAI-1 antibody. Loading controls demonstrate that equal quantities of the fibroblast protein, vimentin, are present in each lane. B, β3 and EEA1 co-localize in WT cells, but not in KO cells. PAI-1 WT and KO cells were incubated with FITC-labeled anti-β3 antibody (green) (B, top) and FITC-labeled control IgG (green) (B, bottom) for 30 min at 37 °C before fixation. Cells were immunostained for EEA1 to identify early endosomes (red). Co-localization of FITC-β3 and EEA1 is shown in WT (top left) only (yellow), demonstrating overlap. Bar, 20 μm. C, percentage overlap of β3 with EEA1 in each condition as determined using MetaMorph image analysis software. Approximately 30 cells were analyzed for each condition in three independent experiments.
Endocytosis of β3 in PAI-1 KO Cells Is Significantly Reduced
To determine if PAI-1 KO cells had reduced endocytosis of β3, PAI-1 WT and KO cells were incubated with FITC-labeled anti-β3 antibody that binds to a cell surface epitope of β3. After 30 min at 37 °C, cells were fixed and co-stained with an antibody to the early endosome protein (EEA1). Whereas WT cells have significant co-localization between β3 and EEA1 (Fig. 1B, top left, yellow punctate dots), KO cells have very few co-localizing endocytic vesicles (Fig. 1B, top right). FITC-labeled IgG was used as control (bottom panels). Neither the WT nor KO cells showed a dramatic uptake of the control IgG. Overlap between β3 and control IgG with EEA1 was quantified using Metamorph analysis (Fig. 1C). Of note is that the FITC-β3 does not accumulate in focal adhesions under these conditions (see Fig. 3 for focal adhesion staining). Instead, the antibody-integrin complex is internalized into endocytic vesicles. These data suggest that PAI-1 KO cells have impaired endocytosis of β3. Next, we determined if impaired endocytosis of β3 resulted in an accumulation of β3 on the cell surface of PAI-1 KO cells.
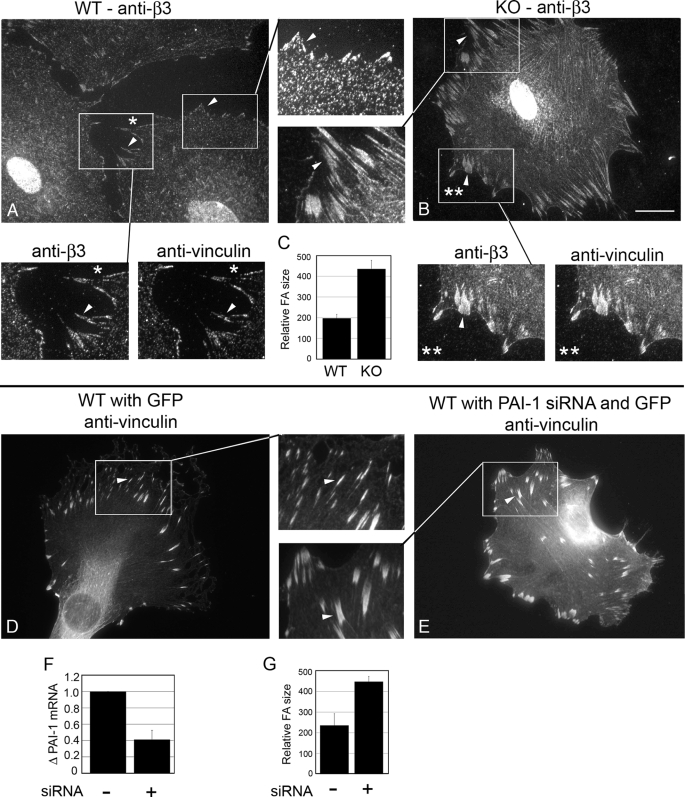
FIGURE 3.
PAI-1 KO β3-containing focal adhesions were enlarged and extended. WT and KO cells were immunostained for β3 (A and B). Insets, enlarged images of focal adhesions (arrows). Relative focal adhesion size between WT and KO cells was quantified using MetaMorph image analysis software (C). To demonstrate the co-localization of β3 and vinculin (a focal adhesion marker), WT and KO cells were co-immunostained for β3 and vinculin (A* and B**). To down-regulate PAI-1 in WT cells, WT cells were co-transfected with PAI-1 siRNA and GFP and then immunostained for vinculin (E). D, GFP alone. PAI-1 knockdown was confirmed using Real time PCR (F). PAI-1 knockdown led to a similar enlargement of focal adhesions compared with non-targeting siRNA controls (G). Bar, 20 μm. Approximately 300 focal adhesions were analyzed in each of three experiments.
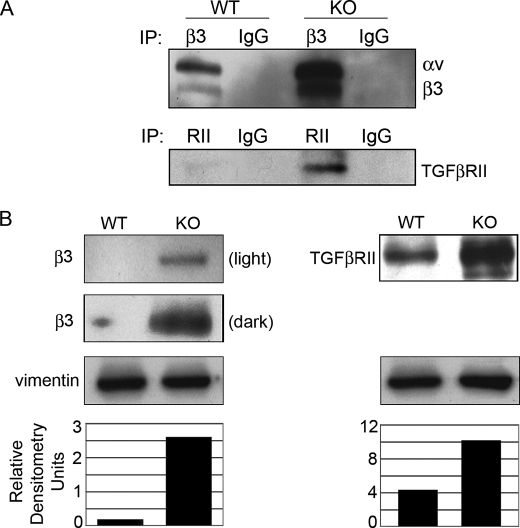
PAI-1 KO Cells Displayed Significantly Increased Cell Surface Expression of β3 and TGFβRII
Previously, PAI-1 was shown to promote the endocytosis of αvβ3 through a uPA·uPAR-mediated mechanism (8). To determine if there was an increase of αvβ3 on the cell surface of PAI-1 KO cells, WT and KO cells were cell surface-biotinylated before lysing and IP with antibodies to β3. Resultant Western blots were probed with streptavidin-HRP, revealing specific bands whose integrin identity was determined by molecular weight. Fig. 2A shows that αvβ3 expression was increased on the cell surface. Since β3 participates in the activation of TGFβ (11, 12), and increased TGFβ activity is associated with overexpression of TGFβRII in models of fibrosis (23, 24), we investigated if TGFβRII was up-regulated on the cell surface of PAI-1 KO cells. Indeed, as with αvβ3, TGFβRII cell surface expression was increased in KO cells. Similar to the cell surface expression of αvβ3 and TGFβRII, Western blots showed that total expression was greatly enhanced in lysates of PAI-1 KO cells (Fig. 2B). Mechanistically, β3 cell surface overexpression could increase TGFβ signaling by directly activating latent TGFβ in the matrix and/or by constitutively binding to the TGFβRII, and overexpression of TGFβRII would enhance this binding. Both pathways would sustain TGFβ signaling, trapping the cells in an autocrine loop of TGFβ activation. Many attempts were made by IP to detect endogenous binding between αvβ3 and TGFβRII; however, this interaction could not be captured using available reagents. Next we determined if impaired endocytosis of β3 changed not only the expression levels of β3 but also the localization of β3 in focal adhesions.
FIGURE 2.
β3 and TGFβRII expression is increased in PAI-1 KO cells. A, cell surface expression. WT and KO cells were biotinylated on the cell surface before lysis and immunoprecipitated for β3 or TGFβRII. Western blots were probed with streptavidin-HRP (SA-HRP) to detect biotinylated proteins. Bands were identified by molecular weight. B, total expression. WT and KO cell lysates were Western blotted for β3 or TGFβRII. Vimentin controls confirm equal loading (densitometry).
β3-containing Focal Adhesions in PAI-1 KO Cells Are Larger and More Extensive
β3-containing focal adhesions in PAI-1 WT and KO cells were compared by epifluorescence microscopy. Fig. 3 shows that PAI-1 WT cells (A) have smaller β3-containing focal adhesions compared with KO cells (B), which have large and extended focal adhesions. Since vinculin is a basic component of focal adhesions (25), we used anti-vinculin antibody to confirm the altered organization of focal adhesions in the WT and KO cells (Fig. 3, denoted with * and **, respectively). Relative focal adhesion size was quantified, demonstrating that focal adhesions in KO cells were ∼2-fold larger than in WT cells (C). Further, PAI-1 WT cells transfected with PAI-1 siRNA produced focal adhesions that were similar to those in PAI-1 KO cells (E) compared with control (D), confirming the specificity of the result. qPCR for PAI-1 showed a 60% decrease in PAI-1 RNA, confirming the effect of PAI-1 siRNA transfection in WT cells (F). Again, focal adhesion size in the PAI-1 siRNA-containing cells showed a 2-fold increase in size compared with WT cells with GFP alone (G). Together, these data show that the absence of PAI-1 produces enlarged focal adhesions containing β3.
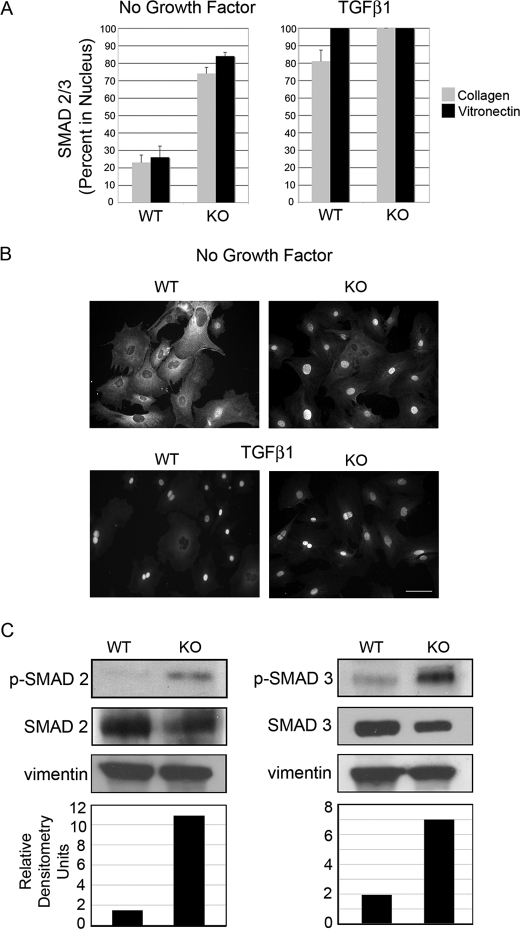
SMAD2/3 Is Localized to the Nucleus in PAI-1 KO Cells
TGFβ1 elicits its biological effects by interacting with TGFβRII, which recruits and activates TGFβRI. Activated TGFβRI phosphorylates SMAD2 and SMAD3, which forms a complex with SMAD4 and translocates to the nucleus to elicit gene transcription (26). Thus, the localization of SMAD2/3 to the nucleus is an indicator of TGFβ signaling. In WT and KO cells, the localization of SMAD2/3 to the nucleus was compared. Previous studies demonstrating the binding of β3 with TGFβRII in the presence of TGFβ1 also showed that TGFβ signaling was enhanced in the presence of vitronectin (16). Thus, to determine if vitronectin played a role in SMAD2/3 localization, cells plated on collagen were compared with cells plated on vitronectin. The results demonstrate that PAI-1 KO cells had significantly higher endogenous TGFβ activity than WT cells, ∼3-fold more nuclear SMAD2/3 on collagen and 3.5-fold more nuclear SMAD2/3 on vitronectin (Fig. 4A, left).
FIGURE 4.
SMAD2/3 is constitutively active in PAI-1 KO cells. A, PAI-1 WT and KO cells were seeded on collagen (gray) or vitronectin (black) in SSFM for 24 h (left) or SSFM plus 1 ng/ml TGFβ1 for 1 h (right). Cells were fixed and immunostained with antibodies for SMAD2/3. The percentage of nuclear localization was determined by counting ∼100 cells/condition in each of three experiments. B, representative images of cells seeded on collagen and quantified in A. Bar, 40 μm. C, Western blots for SMAD2 and -3 and phosphorylated SMAD2 and -3. Densitometry is represented by the ratio of pSMAD to SMAD that was first equalized to vinculin (pSMAD/(SMAD/vinculin)).
To determine the impact of exogenous TGFβ on SMAD2/3 localization, after 24 h in SSFM, cells were treated with TGFβ1 for 1 h prior to cell fixation (Fig. 4A, right). As expected, at 1 h, both WT and KO cells responded to exogenous TGFβ addition with 80% nuclear localization of SMAD2/3 on collagen and 100% on vitronectin for WT cells. Because of the enhanced basal level of SMAD2/3 signaling in the KO cells, cells grown on either matrix have 100% nuclear SMAD2/3. The increased TGFβ signaling on vitronectin suggests that the interaction between β3 and TGFβRII may contribute to the TGFβ signaling in these cells. Experiments were repeated with anti-SMAD3 antibody with similar results (data not shown). Representative images of SMAD2/3 localization are shown in Fig. 4B. To confirm the SMAD2/3 localization data, Western blots were performed using antibodies to SMAD2 and SMAD3 and their activated forms, phospho-SMAD2 and phospho-SMAD3. Results support those reported in Fig. 4, A and B. Both phospho-SMAD2 and -3 were significantly increased in PAI-1 KO cells (Fig. 4C).
PA Activity Is Unregulated in PAI-1 KO Cells
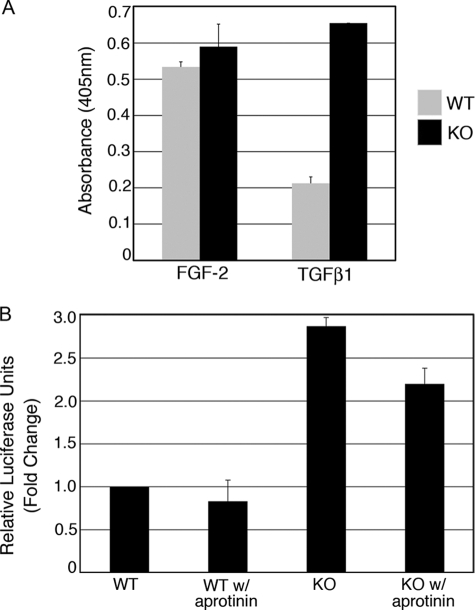
Plasmin activates proteases and growth factors, including TGFβ from the latent TGFβ complex. Since PAI-1 is the primary inhibitor of uPA activity, it follows that the absence of PAI-1 would result in unregulated (increased) uPA activity and, in turn, unregulated plasmin. To confirm this, a standard assay for PA activity was performed. This colorimetric assay quantifies the generation of plasmin. Previous work from our laboratory and others have shown that in fibroblasts, FGF-2 increases uPA activity, and TGFβ decreases this activity, due to the induction of PAI-1 by TGFβ (20, 27, 28). In WT cells, FGF-2 stimulated PA activity, whereas TGFβ reduced it by ∼50%. In contrast, in PAI-1 KO cells, PA activity was equally high in cells grown with FGF-2 or with TGFβ1 (Fig. 5A).
FIGURE 5.
PA is constitutively active in PAI-1 KO cells, but plasmin generation only partially accounts for the observed increase in TGFβ activity. PA activity is not regulated by FGF-2 or TGFβ1 in KO cells. A, PA activity was measured colorimetrically by adding a chromogenic substrate for plasmin to cell lysates. PAI-1 WT (gray) and KO cells (black) were seeded on collagen in SSFM with 1 ng/ml FGF-2 plus heparin or with 1 ng/ml TGFβ and grown for 24 h. B, TMLC assay for TGFβ activity. PAI-1 MEFs were co-cultured with TMLCs containing the luciferase gene fused to the PAI-1 promoter. Luciferase activity was measured using the Bright-Glo detection system. Where indicated, 10 μg/ml aprotinin was added to the culturing medium.
Unregulated Plasmin Activity Is Only Partially Responsible for the TGFβ Activity in PAI-1 KO Cells
To investigate if high levels of plasmin were principally responsible for the increase in TGFβ activity in KO cells, we performed a TGFβ activity bioassay that utilizes the induction of the luciferase gene fused to a PAI-1 promoter in TMLCs (12, 21). Active TGFβ was quantified by co-culturing PAI-1 WT and KO cells with TMLCs containing the lucerifase-PAI-1 construct. We found a 2.9-fold increase in TGFβ activity in KO over WT cells. When we inhibited plasmin activity by performing the assay in the presence of aprotinin, we found only a 25% decrease in TGFβ activity in both WT and KO cells (Fig. 5B). (Aprotinin was shown to abolish plasmin activity in the PA activity assay (data not shown).) These data demonstrate that plasmin is playing a role in TGFβ activation but that the majority of the TGFβ signaling can be attributed to other factors. Next, we investigated the importance of increased β3 expression to TGFβ activity.
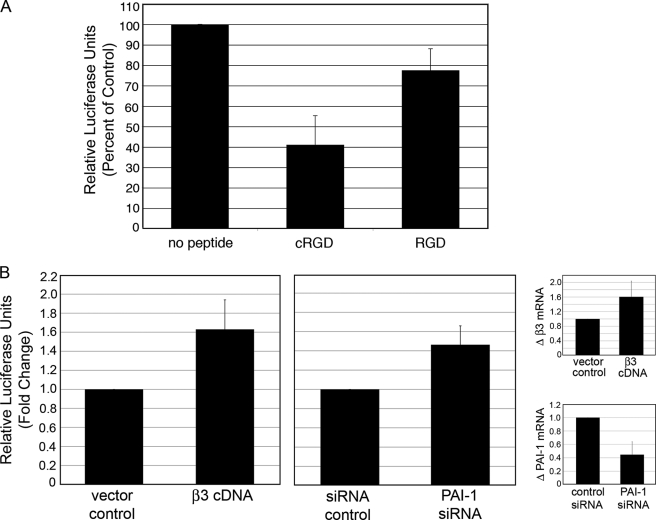
Inhibition of αvβ3 Significantly Decreases TGFβ Activity
To determine if disrupting β3 interactions would affect TGFβ activity, a cyclic peptide that specifically interferes with β3/RGD binding was utilized in the TGFβ activity assay (29). Although the RGD motif is contained in the ligands for both β1 and β3 integrins, such as latent TGFβ and ECM, this cyclic RGD peptide (GpenGRGD) preferentially disrupts β3/RGD binding (29), whereas the general RGD peptide (GRGDNP) preferentially disrupts β1/RGD binding (30, 31). In our assay, the cyclic peptide reduced TGFβ activity by 59%, whereas the RGD peptide reduced activity by only 23% (Fig. 6A). These data provide evidence that β3 accumulation preferentially promotes TGFβ signaling. Furthermore, these data are consistent with a previous report that the cyclic RGD peptide but not the RGD peptide inhibits TGFβ signaling resulting from αvβ3/TGFβRII binding (16).
FIGURE 6.
Modulation of β3 function regulates TGFβ activity in PAI-1 KO MEFs and human corneal fibroblasts. A, αvβ3 function was disrupted by adding 100 nm β3-specific cyclic RGD peptide (GpenGRGD) compared with 100 nm general RGD peptide (GRGDNP) to co-cultured PAI-1 KO and TMLCs (see “Experimental Procedures”). TGFβ activity was quantified using the Bright-Glo luciferase detection system. B, HCFs transfected with β3 or PAI-1 siRNAs showed enhanced TGFβ activity compared with GFP or non-targeting siRNAs, respectively. Quantitative qPCR confirmed β3 overexpression and PAI-1 knockdown.
Overexpression of β3 cDNA and Inhibition of PAI-1 Promotes TGFβ in Human Primary Fibroblasts
To confirm our findings in the PAI-1 MEFs and to show that similar results can be obtained with human cells, we utilized HCFs to show that up-regulation of β3 and knockdown of PAI-1 increase TGFβ activity. HCF cells were transfected with human β3 cDNA or siRNA to human PAI-1 and co-cultured with TMLCs. Results show that when β3 is overexpressed, TGFβ activity increased by 1.63-fold compared with vector control and that when PAI-1 synthesis is inhibited, TGFβ activity increased by 1.45-fold (Fig. 6B). Overexpression of β3 (1.6-fold) and knockdown of PAI-1 (56%) was confirmed by qPCR (small panels in Fig. 6B). These data confirm our findings in the PAI-1 WT and KO cells and extend our data to suggest that PAI-1 regulation of β3 to affect TGFβ activity is a general mechanism.
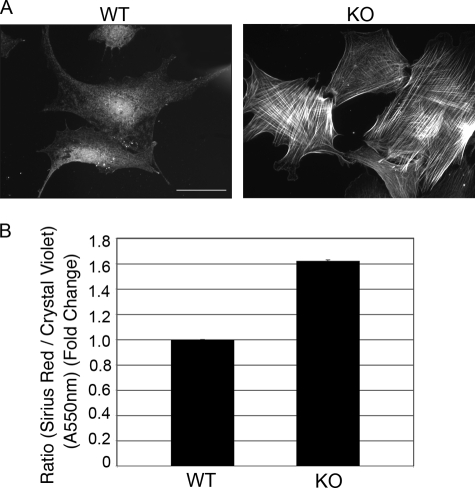
PAI-1 KO Cells Are Myofibroblasts That Synthesize Increased Collagen
Two markers of fibrotic disease and wound healing with scarring are the persistence of myofibroblasts and the excessive synthesis and accumulation of extracellular matrix, including collagen. To determine if PAI-1 KO cells are constitutively myofibroblasts, PAI-1 WT and KO cells were immunostained for α-SMA actin, a marker for myofibroblast differentiation. In Fig. 7A, we show that the KO cells have a propensity to become myofibroblasts. To quantify collagen accumulation, the Sirius Red colorimetric assay for collagen detection was used (22). In this assay, Sirius Red dye binds to newly synthesized as well as deposited collagen and is normalized to cell number provided by Crystal Violet staining. Our results demonstrate that PAI-1 KO cells consistently produce 1.6-fold more collagen than WT cells (Fig. 7B). These data support the finding that TGFβ signaling is increased in PAI-1 KO cells and thus these cells are constitutively myofibroblasts that produce more ECM, in accordance with established models of fibrotic disease.
FIGURE 7.
α-Smooth muscle actin expression and collagen synthesis is enhanced in PAI-1 KO cells. A, PAI-1 WT and KO were immunostained for α-SMA. Bar, 40 μm. B, collagen accumulation was determined by staining with Sirius Red dye (550 nm). Cell numbers were determined by staining with Crystal Violet (550 nm). The ratio of Sirus Red to Crystal violet was graphed.
DISCUSSION
Fibrotic myofibroblasts participate in an autocrine loop of TGFβ activation (32). Toward the goal of elucidating the mechanisms that promote enhanced TGFβ activity, we investigated the role of profibrotic PAI-1 in regulating integrin αvβ3 and TGFβ activity. We discovered that the absence of PAI-1 resulted in a dramatic up-regulation of total and cell surface expression of integrin αvβ3 and TGFβRII. In addition, the β3-containing focal adhesions were greatly enlarged compared with WT and resembled what is seen in myofibroblasts (2). The overexpression of endogenous β3 contributed to a 2.9-fold increase in TGFβ activity, since specifically inhibiting β3 function significantly reduced TGFβ activity. Further, overexpression of integrin β3 and knockdown of PAI-1 in human primary fibroblasts led to a 1.63- and 1.45-fold increase in TGFβ activity, respectively, suggesting that PAI-1 regulation of β3 and resultant activation of TGFβ is a general mechanism.
αvβ3 overexpression on the cell surface may induce TGFβ activity by activating the latent TGFβ complex and/or by enhanced signaling through the TGFβRII. αvβ3 binds to the RGD sequence in latent TGFβ (11), and the suggestion that αvβ3 activates latent TGFβ on the cell surface was derived from 1) the need to co-culture sclerodermal fibroblasts with the TMLC reporter cells to generate a TGFβ signal (suggesting that the TGFβ activity is localized to the plasma membrane and not freely diffusible) and 2) the fact that in the presence of anti-β3 blocking antibodies the TGFβ activity was reduced by ∼50% (12). We also needed to co-culture the PAI-1 WT and KO cells with the TMLCs to generate a TGFβ signal. Furthermore, a β3-specific cRGD peptide reduced this activity by 59%, suggesting that αvβ3 directly activates latent TGFβ. However, the β3 integrin may also be signaling through the TGFβRII. Because αvβ3 is not endocytosed in PAI-1 KO cells and thus remains on the cell surface, its binding to TGFβRII may be prolonged, leading to enhanced signaling. The increase in TGFβ signaling would in turn up-regulate synthesis of αvβ3 (12, 16), resulting in an autocrine loop of TGFβ activity. Both mechanisms may work in tandem to produce a fibrotic response, such as the generation of myofibroblasts and an increase in collagen synthesis, as was demonstrated in Fig. 7. These data are in agreement with the finding that overexpression of αvβ3 in dermal fibroblasts produced an increase in collagen α2(I) promoter activity and that a blocking antibody to αvβ3 reduced synthesis of type I procollagen in sclerodermal fibroblasts (12). The TGFβRII was also up-regulated in PAI-1 KO cells, which is consistent with an up-regulation of this receptor in models of fibrosis when PAI-1 is overexpressed (23, 24). The up-regulation may be a result of enhanced TGFβ signaling or a secondary consequence of the PAI-1 KO model. Further study is needed to distinguish between these mechanisms; however, the finding that overexpression of PAI-1 and elimination of PAI-1 both correlate with an increase in TGFβRII suggests that PAI-1 mediates TGFβRII expression levels.
Our data suggest that “normal” levels of PAI-1 are needed to avoid fibrosis. Overexpression of PAI-1 reduces PA activity, leading to the accumulation of matrix and scarring. We have found that eliminating PAI-1 also promotes a fibrotic phenotype. The in vivo data for PAI-KO models are mixed. Some studies have demonstrated that fibrosis is ameliorated in PAI-1 KO models of kidney, liver, and lung, whereas other studies using PAI-1 KO mice have shown an increase in fibrosis in these same tissues (reviewed in Refs. 19, 33, and 34). Furthermore, in models of cardiac disease, overexpression of uPA and PAI-KO has consistently resulted in macrophage accumulation and cardiac fibrosis (35).3 Interestingly, one study reported that PAI-1 null mice initially had reduced renal fibrosis, but older mice (30 weeks) that were either null for PAI-1 or overexpressing PAI-1 (but not WT), had dramatically increased renal tubulointerstitial area, TGFβ protein, and α-SMA expression, all of which are associated with renal fibrosis. The authors concluded that normal levels of PAI-1 were necessary to maintain healthy renal architecture and function in aging (36). This study suggests that the antifibrotic effects of PAI-1 deficiency may be time-dependent. Perhaps the initial increase in PA activity dominates in controlling fibrosis, whereas over time the overexpression of β3 and TGFβRII and the resultant increase in TGFβ produce a fibrotic phenotype.
Fibrotic healing is marked by the persistence of myofibroblasts in the wounded tissue (2, 32). Myofibroblast regulation is likely to be tissue-specific, since myofibroblasts in fibrotic tissue can be derived from stromal fibroblasts, epithelial-mesenchymal transition, or fibrocytes. These differences in myofibroblast origin may contribute to the differential results reported with the PAI-KO models. The data in this study suggest that PAI-1 regulation of αvβ3 is important for maintaining normal levels of TGFβ activity and that aberrant regulation of αvβ3 by a PAI-1-mediated mechanism can result in a dramatic increase in TGFβ signaling that is associated with fibrosis. This is an important finding, since several clinical therapies that target PAI-1 activity are currently under development (18, 19). Our study suggests that levels of PAI-1 are critical to modulating its function, and therefore, drugs in development that reduce but not eliminate PAI-1 may demonstrate longer term success in preventing fibrosis.
Acknowledgments
We are grateful to Dr. Sandra Masur and Dr. Lilliana Ossowski for insightful comments about the manuscript, to Dr. MaryAnne Stepp for technical advice on the Sirius Red assay, and to Dr. Ralf-Peter Czekay for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NEI, Grant R01 EYO17030 (to A. M. B.), Core Grant P30-EY01867, and a Research to Prevent Blindness grant. Microscopy was performed at the Mount Sinai School of Medicine-Microscopy Shared Research Facility, supported, in part, with funding from NIH, NCI, Shared Resources Grant 5R24 CA095823-04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and NIH Shared Instrumentation Grant 1 S10 RR0 9145-01.
Z. Xu, personal communication.
- ECM
- extracellular matrix
- uPA
- urokinase-type plasminogen activator
- uPAR
- uPA receptor
- TGF
- transforming growth factor
- EEA1
- early endosome antigen 1
- FITC
- fluorescein isothiocyanate
- HRP
- horseradish peroxidase
- GFP
- green fluorescent protein
- MEF
- mouse embryo fibroblast
- DMEM
- Dulbecco's modified Eagle's medium
- SSFM
- supplemented serum-free medium
- HCF
- human primary corneal fibroblast
- IP
- immunoprecipitation
- WT
- wild type
- KO
- knock-out
- EGFP
- enhanced GFP
- qPCR
- quantitative PCR
- PA
- plasminogen activator
- TMLC
- transformed mink lung epithelial cell
- siRNA
- small interfering RNA
- TGFβRII
- TGFβ receptor II
- FGF
- fibroblast growth factor.
REFERENCES
- 1.Desmoulière A., Darby I. A., Gabbiani G. (2003) Lab. Invest. 83, 1689–1707 [DOI] [PubMed] [Google Scholar]
- 2.Hinz B. (2007) J. Invest. Dermatol. 127, 526–537 [DOI] [PubMed] [Google Scholar]
- 3.Wynn T. A. (2008) J. Pathol. 214, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sisson T. H., Simon R. H. (2007) Curr. Drug Targets 8, 1016–1029 [DOI] [PubMed] [Google Scholar]
- 5.Huang Y., Noble N. A. (2007) Curr. Drug Targets 8, 1007–1015 [DOI] [PubMed] [Google Scholar]
- 6.Ragno P. (2006) Cell Mol. Life Sci. 63, 1028–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czekay R. P., Kuemmel T. A., Orlando R. A., Farquhar M. G. (2001) Mol. Biol. Cell 12, 1467–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czekay R. P., Aertgeerts K., Curriden S. A., Loskutoff D. J. (2003) J. Cell Biol. 160, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nykjaer A., Conese M., Christensen E. I., Olson D., Cremona O., Gliemann J., Blasi F. (1997) EMBO J. 16, 2610–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blasi F., Carmeliet P. (2002) Nat. Rev. Mol. Cell Biol. 3, 932–943 [DOI] [PubMed] [Google Scholar]
- 11.Ludbrook S. B., Barry S. T., Delves C. J., Horgan C. M. (2003) Biochem. J. 369, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asano Y., Ihn H., Yamane K., Jinnin M., Mimura Y., Tamaki K. (2005) J. Immunol. 175, 7708–7718 [DOI] [PubMed] [Google Scholar]
- 13.Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 14.Mu D., Cambier S., Fjellbirkeland L., Baron J. L., Munger J. S., Kawakatsu H., Sheppard D., Broaddus V. C., Nishimura S. L. (2002) J. Cell Biol. 157, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wipff P. J., Rifkin D. B., Meister J. J., Hinz B. (2007) J. Cell Biol. 179, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaffidi A. K., Petrovic N., Moodley Y. P., Fogel-Petrovic M., Kroeger K. M., Seeber R. M., Eidne K. A., Thompson P. J., Knight D. A. (2004) J. Biol. Chem. 279, 37726–37733 [DOI] [PubMed] [Google Scholar]
- 17.Galliher A. J., Schiemann W. P. (2006) Breast Cancer Res. 8, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaughan D. E., De Taeye B. M., Eren M. (2007) Curr. Drug Targets 8, 962–970 [DOI] [PubMed] [Google Scholar]
- 19.Ha H., Oh E. Y., Lee H. B. (2009) Nat. Rev. Nephrol. 5, 203–211 [DOI] [PubMed] [Google Scholar]
- 20.Bernstein A. M., Twining S. S., Warejcka D. J., Tall E., Masur S. K. (2007) Mol. Biol. Cell 18, 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe M., Harpel J. G., Metz C. N., Nunes I., Loskutoff D. J., Rifkin D. B. (1994) Anal. Biochem. 216, 276–284 [DOI] [PubMed] [Google Scholar]
- 22.Heng E. C., Huang Y., Black S. A., Jr., Trackman P. C. (2006) J. Cell. Biochem. 98, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubo M., Ihn H., Yamane K., Tamaki K. (2002) J. Rheumatol. 29, 2558–2564 [PubMed] [Google Scholar]
- 24.Hill C., Flyvbjerg A., Grønbaek H., Petrik J., Hill D. J., Thomas C. R., Sheppard M. C., Logan A. (2000) Endocrinology 141, 1196–1208 [DOI] [PubMed] [Google Scholar]
- 25.David-Pfeuty T., Singer S. J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 6687–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts A. B., Piek E., Böttinger E. P., Ashcroft G., Mitchell J. B., Flanders K. C. (2001) Chest 120, 43S–47S [DOI] [PubMed] [Google Scholar]
- 27.Lund L. R., Riccio A., Andreasen P. A., Nielsen L. S., Kristensen P., Laiho M., Saksela O., Blasi F., Danø K. (1987) EMBO J. 6, 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieuwerts A. M., Martens J. W., Dorssers L. C., Klijn J. G., Foekens J. A. (2002) Thromb. Haemost. 87, 674–683 [PubMed] [Google Scholar]
- 29.Srivatsa S. S., Fitzpatrick L. A., Tsao P. W., Reilly T. M., Holmes D. R., Jr., Schwartz R. S., Mousa S. A. (1997) Cardiovasc. Res. 36, 408–428 [DOI] [PubMed] [Google Scholar]
- 30.Pierschbacher M. D., Ruoslahti E. (1987) J. Biol. Chem. 262, 17294–17298 [PubMed] [Google Scholar]
- 31.Mogford J. E., Davis G. E., Meininger G. A. (1997) J. Clin. Invest. 100, 1647–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., Brown R. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 349–363 [DOI] [PubMed] [Google Scholar]
- 33.Arteel G. E. (2008) J. Gastroenterol. Hepatol. 23, S54–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gharaee-Kermani M., Hu B., Phan S. H., Gyetko M. R. (2008) Expert Opin. Investig. Drugs 17, 905–916 [DOI] [PubMed] [Google Scholar]
- 35.Moriwaki H., Stempien-Otero A., Kremen M., Cozen A. E., Dichek D. A. (2004) Circ. Res. 95, 637–644 [DOI] [PubMed] [Google Scholar]
- 36.Lassila M., Fukami K., Jandeleit-Dahm K., Semple T., Carmeliet P., Cooper M. E., Kitching A. R. (2007) Diabetologia 50, 1315–1326 [DOI] [PubMed] [Google Scholar]