Abstract
In the current post-genomic era, large scale efforts are underway to functionally explore the proteome by assembling large antibody libraries. However, because many proteins are modified post-translationally to regulate their function, collections of modification-specific sensors are also needed. Here we applied a novel approach to select monoclonal phosphospecific antibodies directly from the full-length protein and without up-front phosphoamino acid identification. We chose as antigen GRASP65, a well studied Golgi phosphoprotein. Bacterially produced full-length protein was first incubated with mitotic cytosol, thus allowing modification by naturally occurring kinases, and then used directly for affinity-based antibody selection using a single chain variable fragment phagemid library. In less than 1 week, three distinct and highly functional monoclonal phosphospecific antibodies against two GRASP65 epitopes were obtained and subsequently characterized. The presented approach is carried out fully in vitro, requires no prior knowledge of the phosphoamino acid identity, and is fast and inexpensive. It therefore has great potential to be an attractive alternative to classic animal-based protocols for the selection of post-translation modification sensors and thus to become an invaluable tool in our quest to understand the proteome in all its complexity.
In the current post-genomic era, large scale efforts are underway to study the emerging proteome. Because of their ability to bind their respective targets with extremely high specificity, antibodies are essential tools in this endeavor. In fact, non-profit organizations like the European “ProteomeBinders” Consortium (1) and the “Clinical Proteomic Technologies for Cancer” network (2) as well as commercial enterprises are currently sponsoring large projects to assemble all-inclusive antibody libraries.
To regulate their function, many proteins are altered after their initial synthesis. Because such post-translational modifications change the physicochemical properties of a given protein, this further increases the complexity of the proteome. Therefore, the true challenge is to have not only all-inclusive antibody libraries but also to assemble collections of sensors, which specifically detect post-translational changes.
One of the best studied modifications after polypeptide synthesis is reversible protein phosphorylation through the action of specific kinases and phosphatases. A plethora of cellular processes hinges upon the correct phosphorylation state of key regulators (3), and errors may result in cell death or malignancy (4). It is therefore not surprising that protein phosphorylation represents a very active area of research in the quest for new therapeutic cancer targets (5). Phosphospecific antibodies, first described over 25 years ago (6–8), represent key tools in the study of cellular processes regulated by phosphorylation. Their production, however, is relatively costly and time consuming.
In our laboratory we have succeeded in obtaining highly functional, conformation-specific recombinant antibodies using the method of phage display (9, 10).5 This not only allowed us to follow the fate of proteins upon activation but also demonstrated that the diversity of available recombinant antibody libraries is sufficiently broad. We thus decided to use a similar approach for the selection of phosphospecific antibodies. We chose GRASP65 as the antigen, the predominant phosphoprotein of mitotic Golgi membranes (11). Careful studies have mapped several phospho-residues of this structural Golgi protein, and phosphospecific antibodies have been generated using classical methods (12–14). This study describes a novel approach, which allowed the direct selection and characterization of three distinct and highly functional monoclonal phosphospecific antibodies.
EXPERIMENTAL PROCEDURES
Antigen Preparation
Poly-His-tagged full-length rat GRASP65 was expressed in Escherichia coli and purified on Ni-NTA6 beads as described (12). The still bound protein was enzymatically phosphorylated by incubating it at 37 °C with mitotic cell extract prepared as described before (15). After washing six times, the protein was eluted with a 200 mm imidazole-containing buffer followed by dialysis in phosphate-buffered saline, 20 mm β-glycerol phosphate.
Successful modification was confirmed using the mobility shift assay described before (11). Prior to phage display, the antigen was biotinylated using EZ-Link maleimide PEO2-biotin (Pierce).
Antibody Selection Using Phage Display
The Griffin.1 library protocol was used with modifications as published before (16, 17). After three rounds of affinity selection, 80 and 96 clones, respectively, were randomly selected and analyzed using two approaches. First, clones were analyzed directly by immunofluorescence using scFv-containing bacterial supernatants and HeLa cells transfected with rat GRASP65-GFP or nontransfected NRK cells. From all positive clones (34 in total) plasmids were extracted, and the scFv-coding region was sequenced. In the second approach, clones were first screened for the presence of antibodies by PCR using the primers 5′-CA GGA AAC AGC TAT GAC-3′ and 5′-TGA ATT TTC TGT ATG AGG-3′. Amplified products were cleaned using a 96-well format kit (Machery-Nagel) and then sequenced. Known and duplicate scFvs were discarded. The remaining unique positive clones were subcloned into an Fc-containing mammalian expression vector (see above). Subsequently produced full antibodies were characterized by immunofluorescence using NRK cells.
Characterization of Selected Antibodies
For the postfixation treatment with λ-phosphatase (New England Biolabs), cells were fixed in paraformaldehyde and permeabilized with Triton X-100. After washing in phosphate-buffered saline, cells were subsequently incubated for 1 h at 30 °C in the presence of the enzyme in TET buffer (50 mm Tris, pH 7.5, 0.1 mm EDTA, 0.01% Triton X-100, 2 mm MnCl2) before immunofluorescence staining.
For epitope mapping, purified His-tagged GRASP65 full-length proteins (wild type and various mutants mA, mB, mC, mD, mE, mF, mG, mH, mI, and mJ, where known/potential phosphorylation sites had been mutated to alanines) were incubated with mitotic or interphase cytosol at 37 °C for 1 h and then were further subjected to one round of purification with Ni-NTA beads. The eluates were resolved with SDS-PAGE and probed with the indicated antibodies on Western blot.
For more detailed “Experimental Procedures,” see supplemental material.
RESULTS
Full-length Antigen Is Synthesized in Bacteria and Directly Phosphorylated
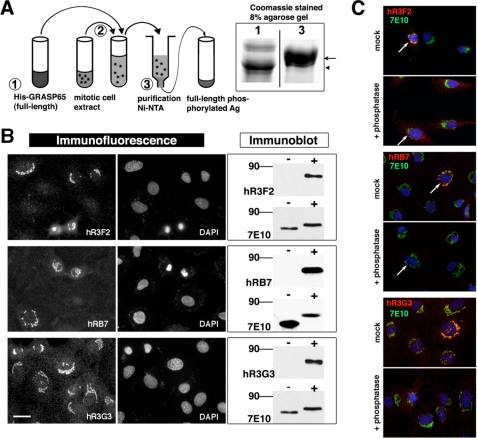
Classical methods to obtain phosphospecific antibodies require the design and subsequent synthesis of highly specific short phosphopeptides, which are ultimately used for animal immunization. In stark contrast, in our in vitro approach, the full-length protein can be used. In a first step, rodent GRASP65 fused to a polyhistidine tag was produced in bacteria. It was next purified on Ni-NTA resin and then directly phosphorylated by exposing it to kinase-containing mitotic cell extract, prepared as described before (15). Finally, the now enzymatically phosphorylated full-length antigen was eluted. Prior to antibody selection, successful modification was confirmed using a mobility shift assay (11) as shown in Fig. 1A.
FIGURE 1.
A simple antigen preparation protocol followed by in vitro affinity selection yields functional phosphospecific antibodies. A, antigen preparation. 1) Full-length GRASP65 is produced bacterially and purified with a poly-His tag. 2) The still bound antigen is exposed to mitotic cell lysate. 3) The modified antigen is re-purified and concentrated. Mobility shift assay (Coomassie-stained gel, right) shows slower migration of phospho-GRASP65 (arrowhead, nonmodified GRASP65, lane 1; arrow, phospho-GRASP65, lane 3). Ag, antigen. B, three antibodies functional in immunofluorescence and on immunoblot are selected. Note that hR3F2 and hRB7 only stain a subset of cells. Bar, 20 μm. DAPI, 4′,6-diamidino-2-phenylindole. For immunoblot, GRASP65 was produced bacterially, purified, and incubated in buffer (−) or with mitotic cell extract (+). The selected antibodies only bind GRASP65 when pretreated with mitotic extracts, although 7E10, a nonphosphospecific anti-GRASP65 antibody, also binds to nonmodified GRASP65. C, selected antibodies are phosphospecific. Post-fixation λ-phosphatase treatment abrogates staining, whereas that of total GRASP65 (7E10) is unaffected. Arrows mark cells in early mitosis. (For more detailed images of the experiment shown in C see supplemental Fig. 4.).
Three Distinct scFvs Are Identified after Affinity-based in Vitro Selection of Antibodies
Using the phosphorylated full-length GRASP65 protein as antigen (immobilized on magnetic beads), scFv antibodies were selected from a semi-synthetic phagemid library by using a protocol essentially as described before (17). Briefly, several rounds of affinity selection and amplification were performed. After the third round, randomly picked clones were analyzed by testing bacterially produced scFvs through immunofluorescence on GRASP65-overexpressing mammalian cells. Additionally, unique scFvs were identified using up-front DNA sequencing directly from bacterial colonies followed by immunofluorescence.
Combining the two approaches, three distinct antibodies, R3F2, RB7, and R3G3, were retained for further analysis. R3F2 was the most abundant scFv antibody, representing ∼15% of all clones, whereas the other two were each retrieved only once. Prior to further characterization, they were subcloned into a human Fc domain containing vector as described before (18), thus generating dimeric antibodies (hR3F2, hRB7, and hR3G3) with enhanced avidity and greater ease of use. In this format, all three antibodies clearly labeled perinuclear Golgi-like structures also of nontransfected cells (Fig. 1B). Furthermore, they easily detected bacterially produced rat GRASP65 on immunoblot. However, this was the case only if the antigen had first been exposed to mammalian cell extract (Fig. 1B).
We noticed that hR3F2 and hRB7, when used for immunofluorescence, labeled only a subset of cells. Detailed analysis revealed Golgi membrane staining exclusively in dividing cells (supplemental Figs. 1 and 2). Because phosphorylation of GRASP65 mostly takes place at the onset of mitosis (11, 13), this strongly suggested recognition of an epitope modified by a mitotic kinase. In contrast, hR3G3 labeled intracellular structures of all cells, although staining of mitotic cells was more pronounced. This implied binding of this third scFv to either a nonmodified residue or to an epitope modified by both interphase and mitotic kinases.
All Three Selected scFvs Specifically and Exclusively Bind Rodent GRASP65
In co-immunofluorescence experiments, the antibody labeling of all three scFvs strongly overlapped with that of other Golgi markers, including GRASP65, detected by a monoclonal antibody named 7E10 (19). All three selected antibodies readily recognized overexpressed GRASP65, although none bound to overexpressed GRASP55, which is closely related to GRASP65. Protein expression knockdown with GRASP65-specific small interfering RNA led to a loss of immunofluorescence signal (data not shown). All three antibodies, although readily staining cells of rodent origin, never decorated human cells. Species specificity was confirmed through immunofluorescence analysis of a mixed HeLa cell population, where cells stably expressing rat GRASP65 were mixed with wild type HeLa cells. All three antibodies bound the recombinant rodent protein, whereas nontransfected cells were never stained (supplemental Fig. 3).
All Three Selected Antibodies Are Phosphospecific
Our immunofluorescence and immunoblot data (Fig. 1B) suggested that all three antibodies would specifically recognize phospho-epitopes. To confirm this, FR3T3 cells were fixed, permeabilized, and incubated with a broad-acting λ-phosphatase to dephosphorylate cellular proteins. Subsequent immunofluorescence analysis showed loss of labeling by all three selected antibodies, whereas staining with nonphosphospecific antibodies was not affected (Fig. 1C and supplemental Fig. 4). Furthermore, a GRASP65 mutant, in which eight known/putative phospho-residues had been replaced by alanines, was no longer recognized by any of the three antibodies. This not only confirmed the λ-phosphatase experiment but also suggested that the antibody epitopes would be found among those eight residues.
Specific Target for Each of the Three Antibodies Is Determined
In a last set of experiments, epitope mapping for all three phosphospecific antibodies was performed through immunoblotting and the use of a set of GRASP65 mutants with substitutions of known/putative phosphoamino acids (Fig. 2A). For hR3F2, phosphoserine at position 376 was found as the epitope because the antibody failed to recognize any construct in which this residue had been mutated (Fig. 2, B–E, mC, mE, mF, mG, and mI). The second mitosis-specific antibody, hRB7, displayed an identical pattern (Fig. 2, D and E), thus representing a distinct second phosphorylation sensor against the same epitope. Previously, it has been shown that Ser(P)-376 is modified by the mitotic kinase Cdk1-cyclin B, and polyclonal phosphospecific antibodies have generated a staining pattern very similar to that seen with our selected hR3F2 and hRB7 (13).
FIGURE 2.
Epitope mapping reveals Ser(P)-376 (for hR3F2 and hRB7) and Ser(P)-277 (for R3G3) as phospho-targets. A, schematic depiction of known and putative phosphorylation sites and used His-tagged mutants. B–E, phospho-epitope mapping. Purified full-length His-tagged GRASP65 constructs (either wild type or phosphorylation-deficient mutants) were incubated with interphase (B) or mitotic (C–E) cell extracts, loaded in equal amounts onto separate gels, resolved with SDS-PAGE, and transferred onto nitrocellulose. Loss of signal for a given construct indicates that at least one of the mutated phospho-residues must be present as wild type and be phosphorylated for antibody binding.
Finally, epitope mapping was also performed for the third antibody, hR3G3. Analysis of binding patterns of this scFv to the different mutant constructs enabled identification of Ser(P)-277 as the target of the antibody (Fig. 2, B and C, mB). This finding is again in agreement with published data; it has been shown that this serine residue at position 277 is phosphorylated in both interphase and during mitosis by the action of two distinct kinases (14).
DISCUSSION
This work describes a novel antibody selection approach where the antigen is prepared by enzymatically phosphorylating the full-length target protein and where the antibodies are then subsequently obtained by utilizing a fully in vitro affinity-based selection process. Using the example of GRASP65, a well studied Golgi phosphoprotein, we show that highly functional conformation-specific sensors can be obtained; in one screen three distinct scFvs against two different phospho-epitopes were found and subsequently characterized.
Our method has several inherent advantages over animal-based techniques (schematically depicted in Fig. 3). First, the in vitro approach requires less time and resources. It allows skipping the steps of selection, synthesis, and injection of short synthetic phosphopeptides as antigen usually needed for the production of phosphospecific antibodies and uses the full-length protein directly during the antibody selection process. Classical methods depend on a robust immune response by the mammalian host and entail a fair amount of animal experimentation, including the need for repeated immunizations often with multiple different antigens. Thus, routinely milligram quantities of several different phosphopeptides and a significant time investment are required because either purification of the obtained serum to remove nonspecific binders or, in case of classic monoclonal antibodies, screening of a large number of hybridoma clones are necessary. In our animal-free method, microgram amounts of the bacterially produced antigen are sufficient, and high quality scFv antibodies can be obtained in a few days fully in vitro. Successful antibody selection is independent of the recognition of the target protein by the immune system, and the selected antibodies can be analyzed at the molecular level because their encoding nucleotide sequences are readily available.
FIGURE 3.
Schematic comparison of the classic approach with the in vitro approaches to obtain phosphospecific antibodies. For both approaches the antigen (Ag) is produced in full length and then phosphorylated (top portion, shaded gray). In the classic in vivo method (left, shaded blue), phospho-epitope mapping through mass spectrometry (mass spec), selection of the phospho-epitope to be targeted, and synthesis of a short phosphopeptide to be injected are all performed before immunization of animals. Additionally, downstream affinity purification or, alternatively, screening of a large number of hybridoma clones must be performed. Overall, this process takes months and yields one serum or one monoclonal antibody (monocl. Ab) directed against one predefined epitope. In the in vitro approach (right, shaded red), up-front phosphoamino acid identification is omitted, and the full-length antigen is used directly for antibody selection. Random clones are analyzed, and eventually Fc portion-containing antibodies are produced. In total, this approach takes only weeks, allows direct access to the DNA of the antibody, and has the potential of leading to several distinct monoclonal antibodies in endless supply directed against multiple epitopes.
An even more distinct advantage of our method, however, lies in the fact that there is no necessity to determine the phospho-residue composition of the protein of interest prior to antibody production. Because the entire selection process occurs in vitro, events activated after the injection of animals, such as protein degradation, dephosphorylation, and antigen processing, are all circumvented. The entire full-length protein prepared and enzymatically modified under quasi-physiologic conditions can thus be used directly as the antigen. This way, in a single phage display screen, multiple phosphospecific antibodies targeted at more than one phospho-epitope can be selected as evidenced by the present study. Furthermore, our approach may favor the selection of antibodies against the major phosphorylation site(s) of a given antigen. Indeed, two of the selected scFvs were found to bind one and the same residue, and one of them (R3F2) was by far the most abundant antibody found in the screen. Their target residue, Ser(P)-376, represents one of the key phospho-residues on GRASP65 (13).
Intriguingly, no nonphosphospecific antibodies were obtained using our method. Although this seems to be a paradox, previous work in our laboratory has similarly led to the selection of scFvs, which exclusively bind active domains. For instance, only antibodies specifically recognizing the GTP-bound form were obtained when the small GTPases Rab6 (10) and Rab15 were used as antigens. Upon activation through GTP loading (or analogously through phosphorylation), new protein-protein interaction domains are formed on the surface of the molecule. In contrast to animal-based approaches, the in vitro selection process relies on the recognition of three-dimensional rather than linear epitopes and is thus biased toward these interaction domains because they represent ideal binding sites for antibodies. Altogether this is yet another argument in favor of our approach because it may more efficiently yield highly functional conformation sensors and help determine the major phosphorylation site of a given protein.
Finally, our approach benefits from inherent advantages of the recombinant approach. Selected antibody clones can be stored as plasmids and at a fraction of the cost, which all but eliminates the risk of losing a clone. Antibodies can be produced in an endless supply and with high reproducibility, including in the form of fully humanized antibodies synthesized by mammalian cells. Because the Fc portion to which a given scFv would be fused can be chosen freely, various versions of the antibody, with the same variable regions but differing Fc fragments (mouse, rabbit, human, etc.), can be produced, as recently described by our laboratory (18). Finally, additional modifications, including fusions to fluorescent tags, can be easily made.
In summary, we have described a novel in vitro approach that allows the selection of high quality phosphospecific antibodies directly from full-length phosphorylated antigens, thus obviating the need for up-front phospho-residue mapping. To meet the challenges of understanding the proteome in all its complexity, collections of such modification-specific sensors will have to be assembled in addition to already existing antibody libraries. We strongly believe that our in vitro approach, by providing a simple and robust alternative to classical methods and at the same time offering savings in both time and resources, will be a reliable tool in this endeavor.
Supplementary Material
Acknowledgments
We thank G. Warren (Max F. Perutz Laboratories, Vienna, Austria) for support throughout this study. We also thank all members of the Perez laboratory for their discussions and constant support, especially N. Camper and A.-J. Jimenez.
This work was supported, in whole or in part, by National Institutes of Health Grant GM087364 (to Y. W.). This work was also supported by the Pardee Cancer Research Foundation, the Curie Institute, CNRS, and by grants from the “Agence National de la Recherche” and the “Association pour la Recherche Contre le Cancer” (to F. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. 1–4, and additional references.
- Ni-NTA
- nickel-nitrilotriacetic acid
- scFv
- single chain variable fragment
- h
- human.
REFERENCES
- 1.Taussig M. J., Stoevesandt O., Borrebaeck C. A., Bradbury A. R., Cahill D., Cambillau C., de Daruvar A., Dübel S., Eichler J., Frank R., Gibson T. J., Gloriam D., Gold L., Herberg F. W., Hermjakob H., Hoheisel J. D., Joos T. O., Kallioniemi O., Koegl M., Koegll M., Konthur Z., Korn B., Kremmer E., Krobitsch S., Landegren U., van der Maarel S., McCafferty J., Muyldermans S., Nygren P. A., Palcy S., Plückthun A., Polic B., Przybylski M., Saviranta P., Sawyer A., Sherman D. J., Skerra A., Templin M., Ueffing M., Uhlén M. (2007) Nat. Methods 4, 13–17 [DOI] [PubMed] [Google Scholar]
- 2.Haab B. B., Paulovich A. G., Anderson N. L., Clark A. M., Downing G. J., Hermjakob H., Labaer J., Uhlen M. (2006) Mol. Cell. Proteomics 5, 1996–2007 [DOI] [PubMed] [Google Scholar]
- 3.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 4.Johnson L. (2007) Biochem. Soc. Trans. 35, 7–11 [DOI] [PubMed] [Google Scholar]
- 5.Ventura J. J., Nebreda A. R. (2006) Clin. Transl. Oncol. 8, 153–160 [DOI] [PubMed] [Google Scholar]
- 6.Ross A. H., Baltimore D., Eisen H. N. (1981) Nature 294, 654–656 [DOI] [PubMed] [Google Scholar]
- 7.Nairn A. C., Detre J. A., Casnellie J. E., Greengard P. (1982) Nature 299, 734–736 [DOI] [PubMed] [Google Scholar]
- 8.Davis F. M., Tsao T. Y., Fowler S. K., Rao P. N. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2926–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrov A., Quesnoit M., Moutel S., Cantaloube I., Poüs C., Perez F. (2008) Science 322, 1353–1356 [DOI] [PubMed] [Google Scholar]
- 10.Nizak C., Monier S., del Nery E., Moutel S., Goud B., Perez F. (2003) Science 300, 984–987 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. (2003) EMBO J. 22, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Satoh A., Warren G. (2005) J. Biol. Chem. 280, 4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preisinger C., Körner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. (2005) EMBO J. 24, 753–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura S., Yoshioka K., Barr F. A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. (2005) J. Biol. Chem. 280, 23048–23056 [DOI] [PubMed] [Google Scholar]
- 15.Rabouille C., Misteli T., Watson R., Warren G. (1995) J. Cell Biol. 129, 605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths A. D., Williams S. C., Hartley O., Tomlinson I. M., Waterhouse P., Crosby W. L., Kontermann R. E., Jones P. T., Low N. M., Allison T. J., et al. (1994) EMBO J. 13, 3245–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nizak C., Martin-Lluesma S., Moutel S., Roux A., Kreis T. E., Goud B., Perez F. (2003) Traffic 4, 739–753 [DOI] [PubMed] [Google Scholar]
- 18.Moutel S., El Marjou A., Vielemeyer O., Nizak C., Benaroch P., Dübel S., Perez F. (2009) BMC Biotechnol. 9, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., Barr F. A. (1999) EMBO J. 18, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.