Abstract
In plants, a family of more than 20 heat stress transcription factors (Hsf) controls the expression of heat stress (hs) genes. There is increasing evidence for the functional diversification between individual members of the Hsf family fulfilling distinct roles in response to various environmental stress conditions and developmental signals. In response to hs, accumulation of both heat stress proteins (Hsp) and Hsfs is induced. In tomato, the physical interaction between the constitutively expressed HsfA1 and the hs-inducible HsfA2 results in synergistic transcriptional activation (superactivation) of hs gene expression. Here, we show that the interaction is strikingly specific and not observed with other class A Hsfs. Hetero-oligomerization of the two-component Hsfs is preferred to homo-oligomerization, and each Hsf in the HsfA1/HsfA2 hetero-oligomeric complex has its characteristic contribution to its function as superactivator. Distinct regions of the oligomerization domain are responsible for specific homo- and hetero-oligomeric interactions leading to the formation of hexameric complexes. The results are summarized in a model of assembly and function of HsfA1/A2 superactivator complexes in hs gene regulation.
Heat stress transcription factors (Hsfs)4 are the terminal components of signal transduction chains mediating the activation of genes responsive to heat stress (hs) and a large number of other environmental or chemical stressors (1–4). Besides their central role in response to stress-related stimuli, Hsfs are also involved in the regulation of cell growth and survival under normal physiological and developmental conditions (for review see Refs. 4–6). Remarkably, the Hsf-controlled stress response system is conserved throughout the eukaryotic kingdom. Hsfs are maintained in an inactive state under normal growth conditions controlled by interaction with molecular chaperones, but they are rapidly activated under stress conditions (1, 2, 7–10). As a result, heat stress proteins (Hsps) are synthesized, many of which act as molecular chaperones protecting proteins against stress damage or assisting in their folding, intracellular distribution, and degradation (11–16).
Heat stress-inducible genes in eukaryotes share conserved promoter elements (heat shock elements, HSE) with the consensus motif (AGAAn)(nTTCT) (17, 18). Similar to other transcriptional activators, Hsfs have a modular structure (4, 19) with an N-terminal conserved DNA binding domain with a central helix-turn-helix motif and a domain with extended heptad repeats of hydrophobic amino acid residues (HR-A/B region) required for oligomerization. The C-terminal domains of Hsfs are more divergent, and many of them include localization signals for nuclear import (NLS) (20) and export (NES) (21) as well as short peptide motifs with aromatic, large hydrophobic, and acidic amino acid residues (AHA motifs) for the activator function (22, 23).
In yeast, Drosophila and nematodes Hsfs are encoded by unique genes, whereas multiple Hsfs exist in vertebrates. Hsf1, one of 3–4 Hsfs identified in vertebrates has been shown to function as ubiquitous regulator of the stress response, while the other members are assumed to have tissue- or development-specific functions (6, 19).
In contrast to all other organisms investigated so far, plants possess an extraordinarily complex Hsf family both in terms of total number of representatives (usually more than 20) as well as of their structural and functional diversification. Three classes of plant Hsfs (classes A, B, and C) are defined by peculiarities of their HR-A/B regions (9). Evidently, the Hsf mixture changes in a tissue-specific manner and in response to stress treatments (see reviews in Refs. 3, 4, 24).
Remarkably, despite many similarities, our current knowledge indicates plant-specific variations of the Hsf network. (i) In tomato, HsfA1 functions as master regulator of the hs response (25), whereas in Arabidopsis such a master regulator function could not be identified so far. Even the simultaneous knock-down of two HsfA1 isogenes (Hsfs A1a and A1b) had no marked effect on the hs response, and only a subset of hs genes was affected (26). (ii) In contrast to the situation in Arabidopsis, tomato HsfA1 operates upstream of hs-induced expression of chaperones and HsfA2. Hence, HsfA2 accumulates during repeated cycles of heat stress and recovery and becomes the dominant Hsf in thermotolerant cells. Its activity is further controlled by a network of proteins involving HsfA1 and chaperones of the sHsp family influencing its solubility, intracellular localization, and activator function (21, 27, 28). Arabidopsis HsfA2 is also hs-inducible and highly expressed after heat stress treatment, but it is different in its physicochemical properties when compared with tomato HsfA2. The analysis of HsfA2 overexpression and knock-out plants of Arabidopsis indicate that HsfA2 functions as enhancer of hs gene expression with specific activity on a subset of Hsf-dependent genes (29–31).
Plant Hsfs interact with each other physically and/or functionally with the result of mutual enhancement or repression of their activities (21, 27, 32, 33). Although only a few examples of Hsf cooperation have been investigated in greater detail, remarkable differences in the underlying molecular mechanisms are apparent. The coactivator function of tomato HsfB1 depends on a histone-like motif in its C-terminal domain required for recruitment of the histone acetyltransferase-like protein HAC1, a plant ortholog of the p300/CBP (CREB-binding protein) family. Formation of ternary complexes of HsfA1, HsfB1, and HAC1 with enhanced DNA binding activities does not require physical interaction between the two Hsfs (32). In contrast, cooperation between class A Hsfs depends on their oligomerization domains and the formation of hetero-oligomeric complexes. However, despite a similar basic structure of the HR-A/B regions (Fig. 1), interactions between class A Hsfs are highly selective. The interaction of HsfA1 and HsfA2 leads to nuclear retention of HsfA2 and formation of superactivator complexes with enhanced transcriptional activities (21, 27). Interestingly, the opposite functional consequence was observed for the HsfA4b/HsfA5 pair. By binding to HsfA4b, HsfA5 impairs DNA binding and consequently the activator function of Hsf4b (33).
FIGURE 1.
Domain structure of tomato heat stress transcription factors. Numbers indicate amino acid residues. OD, oligomerization domain corresponding to the hydrophobic heptad repeat region (HR-A/B). The term HsfA1 is used synonymously throughout this report and corresponds to the master regulator HsfA1a in Lycopersicon esculentum (3, 25). The patterns of AHA motifs are Hsf-specific (33, 36, 37).
Here we challenge the current model of hetero-oligomerization dependence of the synergistic action of tomato HsfA1 and HsfA2. We demonstrate that multimeric complexes can be formed in the absence of DNA and that transcriptional activity strongly depends on hetero-complex formation. Besides nuclear retention of HsfA2, the combination of specific functional properties of both Hsfs in A1/A2 hetero-complexes is essential for the superactivator function.
EXPERIMENTAL PROCEDURES
General Reagents and Procedures
Standard protocols were used for cloning and nucleic acid analysis (Refs. 34, 35 and supplemental Table S1). PCR fragments for subcloning were generated using the High Fidelity PCR Enzyme Mix (Fermentas, St. Leon-Rot, Germany). Protein extraction, SDS-polyacrylamide gel electrophoresis (PAGE), and protein blotting analysis were performed as described (25, 28). The loading of equal amounts of total protein extracts corresponding to 20,000 protoplasts and the efficiency of protein transfer were controlled by staining of the protein blots with Ponceau before performing consecutive cycles of immunodetection to monitor the protein levels of transiently expressed Hsfs and endogenous Hsp17-CI on the same blot. The generation and use of specific antisera against individual tomato (Lycopersicon pervianum) Hsfs (HsfA1, A2, A3) and Hsp17-CI were described before (20, 28, 36). Primary antibodies for immunodetection of HA-tagged proteins were obtained from Hiss Diagnostics (Freiburg, Germany). Horseradish peroxidase-conjugated secondary antibodies were obtained from Sigma-Aldrich. For transient gene expression studies, tobacco (Nicotiana plumbaginifolia) leaf mesophyll protoplasts were used. Polyethylene glycol-mediated co-transformation of reporter and Hsf expression plasmids was carried out as described previously (22, 27, 37).
Plasmid Constructs for Transient Expression Studies in Protoplasts
The Hsf-dependent reporter plasmid pGmhsp17.3B- CI::GUS was described before (22). Plasmid constructs for Hsf expression in plant cells are based on the pRT series of vectors (38). Constructs for Hsfs A1, A2, A3, and A4b were described before (20, 33, 36). Further deletions or modifications were done on the basis of these parental expression vectors. Primer sequences (supplemental Table S1) and mutant constructs are given (supplemental Table S2). For subcellular localization studies, GFP-HsfA2 fusion constructs were generated by subcloning of corresponding PCR fragments into p35dS::GFP (39) and analyzed by fluorescence microscopy in protoplasts according to procedures described before (21, 33). For microscopic analysis, a Zeiss Axiophot microscope (Zeiss, Oberkochen, Germany) combined with a Color View F12 System (Olympus, Hamburg, Germany) was used. Captured images were resized and combined using Photoshop 5.5 Software (Adobe Systems, La Jolla, CA).
GUS Reporter Assay
The promoter-GUS activity was determined as described (20). The synergism was calculated by Equation 1,
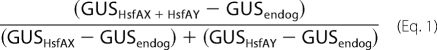 |
where GUSHsfAX+HsfAY is the activity determined for the combination of the indicated Hsfs or Hsf mutant forms, GUSHsfAX and GUSHsfAY are the activities of the individual Hsfs themselves, and GUSendog corresponds to the endogenous Hsf activity on the GUS reporter in protoplasts after mock transformation with the empty pRT vector. For data presentation in Fig. 5B, the additional stimulation (AS) of GUS activity by the combination of two transcription factors above additive activities proportional to the combination of wt HsfA1 and HsfA2 was calculated by Equation 2.
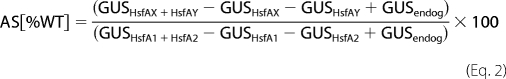 |
GUSHsfA1+HsfA2 is the activity determined for the combination of wild type HsfA1 and HsfA2, while GUSHsfA1 and GUSHsfA2 are the activities of the individual wild type Hsfs, respectively.
FIGURE 5.
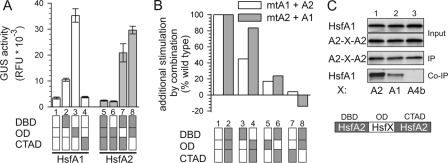
The synergistic function of HsfA1/A2 complexes depends on the heterologous combination of the oligomerization and the activator domains. A, individual activities of domain-swapping mutants of HsfA1 (white bars) or HsfA2 (gray bars) in the GUS reporter assay. The domain structure of wt HsfA1 (1, white block diagram) and HsfA2 (5, gray block diagram) is compared with the corresponding domain-swapping mutants (2–4 and 6–8). B, ratio of additional stimulation of GUS activity (AS) by combination of the mutants of HsfA1 (samples 3, 5, 7) with wt HsfA2 (white bars) or the mutants of HsfA2 (samples 4, 6, 8) combined with wt HsfA1 (gray bars) compared with the wt combination (samples 1, 2) was calculated as described under “Experimental Procedures.” The corresponding domain-swapping mutants are indicated at the bottom. C, HsfA1 and HsfA2 (wt, lane 1) or mutant forms of HsfA2 containing the HR-A/B domain of HsfA1 (lane 2) or HsfA4b (negative control, lane 3) were co-expressed in tobacco protoplasts and complexes immunoprecipitated with HsfA2 antibodies. The levels of HsfA1 and HsfA2 forms (A2-X-A2) before precipitation (Input), the amount of HsfA2 forms immunoprecipitated (IP), and the amount of HsfA1 co-immunoprecipitated (Co-IP) are shown as indicated. The domain structure of the HsfA2 mutants is illustrated below. For abbreviations see legend to Fig. 1.
Co-immunoprecipitation
Protein extracts from 4 samples each of 1 × 105 protoplasts transformed with 5 μg of indicated Hsf expression plasmids were prepared in 100 μl of NEB500 lysis buffer containing 25 mm HEPES, pH 7.5, 500 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 10 mm NaF, 10 mm β-mercaptoethanol, 0.2% (w/v) Nonidet P-40, and 10% (w/v) glycerol and Complete protease inhibitor mixture tablets (Roche Applied Science). 5 μl were taken as input control; the residual extract was supplemented with NEB0 (i.e. NEB500 without NaCl) to a total volume of 400 μl. HsfA2 antiserum was coupled to NHS-activated Sepharose beads (GE Healthcare, Freiburg, Germany) according to the manufacturer's protocol and added as 1:10 slurry to the sample (40 μl). After incubation at 4 °C for 1 h with gentle agitation, beads were pelleted and washed three times with 500 μl of 10 mm Tris buffer (pH 8.0) supplied with 140 mm NaCl and 500 mm LiCl and once with 10 mm Tris buffer (pH 8.0) containing 140 mm NaCl. Bound proteins were eluted with 30 μl of 2× SDS sample buffer. Samples of 10 μl were separated by SDS-PAGE and processed for immunoblotting.
Size Exclusion Chromatography (SEC)
Protein extracts in NEB500 corresponding to ∼500,000 protoplasts were separated by SEC on Superdex 200 HR30/10 filtration column (GE-Healthcare) at 4 °C by eluting with 25 mm HEPES, pH 7.5, 500 mm NaCl, 1 mm EDTA, 0.2% (w/v) Nonidet P-40, and 1 mg ml−1 pefabloc at a flow rate of 0.4 ml min−1. Fractions of 0.8 ml were collected, precipitated with acetone, and redissolved in SDS sample buffer for immunoblot analyses. As molecular size standards, thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and bovine serum albumin (67 kDa) were used.
Cross-linking of Hsf Complexes
Cross-linking reactions were performed with protein extracts in NEB500 buffer corresponding to 1 × 105 protoplasts by incubation in the presence of 2 mm glutaraldehyde at room temperature. The reaction was stopped after 10 min by the addition of 200 mm glycine and precipitation with acetone. After washing the precipitate with acetone and ethanol, proteins were dissolved in SDS sample buffer, separated on linear 4–12% SDS-PAGE gradient gels, and processed for immunoblotting of Hsf cross-linking products.
RESULTS
Synergistic Gene Expression by HsfA1 and HsfA2
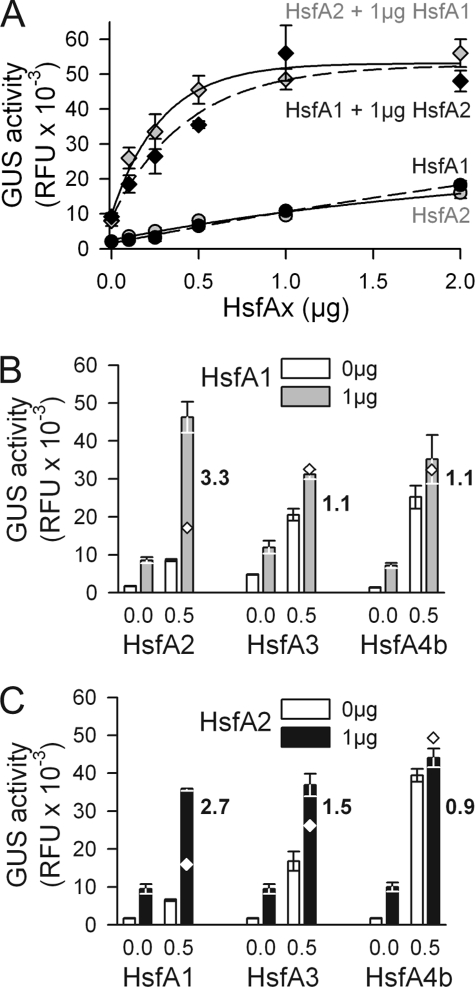
To investigate the specificity of interactions of tomato class A Hsfs, we performed transient reporter assays using an Hsf-dependent hsp17-promoter::GUS reporter construct in tobacco mesophyll protoplasts (22). GUS activity was measured upon coexpression of different combinations of HsfA1, HsfA2, HsfA3, and HsfA4b (Fig. 1). First we examined the influence of increasing amounts of expression constructs in combinations of HsfA1 and HsfA2 by titration of either HsfA1 expression against a constant amount of HsfA2 or vice versa. An increasing stimulation of GUS activity was observed for all combinations of HsfA1 and HsfA2 (Fig. 2A, diamonds), which was ∼2.5–3.5-fold higher than expected for additive effects as determined from the individual activities (Fig. 2A, circles). Evidently, the interaction of HsfA1 and HsfA2 is synergistic with maximal activities at a stoichiometry close to 1:1 (HsfA1/HsfA2) (see also supplemental Fig. S1). In contrast to the HsfA1/HsfA2 combination, neither HsfA1 (Fig. 2B) nor HsfA2 (Fig. 2C) showed comparable effects with HsfA3 or HsfA4b. Based on these results, we conclude that HsfA1 and HsfA2 form a pair of specifically interacting Hsfs, which collaborate synergistically as a type of superactivator of hs gene expression.
FIGURE 2.
Cooperative action between tomato Hsfs. A–C, Hsf activity was monitored by expression of the Hsf-dependent GUS reporter construct PGmhsp17. 3B-CI::GUS (supplemental Fig. S1) in tobacco protoplasts. A, co-transformation with the indicated amounts of plasmids encoding HsfA1 or HsfA2, either alone (gray and black dots, respectively) or in combination with 1 μg of plasmid encoding the corresponding partner Hsf (diamonds). B and C, same as in A with 0 or 0.5 μg of plasmids encoding the indicated Hsfs, either alone (white bars) or in combination with 1 μg of HsfA1 (B, gray bars) or HsfA2 (C, black bars). Diamonds indicate the additive values of GUS activities determined by transformation of protoplasts with the individual partner Hsfs alone. The synergism of GUS activity for the corresponding Hsf combinations is given as numbers and was calculated as described under “Experimental Procedures.”
Synergism Depends on Specific Features of the Oligomerization and Activation Domains
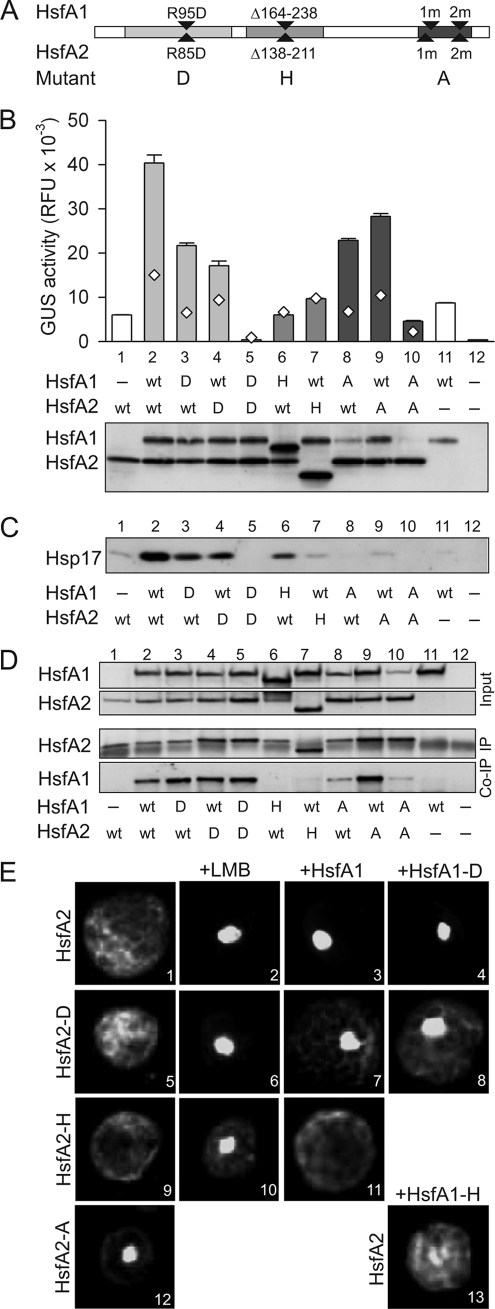
Hsfs have a modular structure with distinct domains performing specific functions such as DNA binding (DBD) and oligomerization (OD). The C-terminal activator domain (CTAD) contains motifs required for nuclear import and export and transcriptional activation (Fig. 1). To characterize their roles in synergistic gene activation, we constructed HsfA1 and HsfA2 mutants defective either in DNA binding, oligomerization, or activator functions (Fig. 3A). The DNA binding mutants of HsfA1 and HsfA2 were generated by replacing an invariant and functionally essential arginine residue in the DNA recognition helices (40). The oligomerization mutants bear a deletion of the entire OD, and the transcriptional activation mutants are characterized by replacements of aromatic and hydrophobic amino acid residues in the C-terminal activator motifs (AHA motifs, Ref. 37). In reporter assays, all HsfA1 and HsfA2 mutants showed strongly reduced activities, i.e. usually <15% of their wild type forms (supplemental Fig. S2).
FIGURE 3.
Importance of functional domains for interaction and synergistic activity of HsfA1 and HsfA2. A, mutations introduced into HsfA1 and HsfA2 as well as the abbreviations are given (for further details of AHA mutations (mutant forms A) see also Fig. 4A). B and C, analysis of HsfA1/A2 cooperation in reporter gene expression assays in tobacco protoplasts after coexpression of the indicated wild type (wt) and mutant forms (samples 1–12). B, GUS reporter activities are presented as bars and diamonds and indicate the ratio of GUS activities contributed by additive effects (see legend to Fig. 2). Below, expression controls for the transformed Hsf constructs determined by immunoblot analysis are shown. C, expression levels of endogenous Hsp17-CI proteins were determined by immunodecoration performed on the same protein blot used in B. D, co-immunoprecipitation of HsfA1 with anti-HsfA2 antibodies. Expression levels (10% input) for both Hsfs (upper panels) as well as results of immunoprecipitation (IP) of HsfA2 and co-immunoprecipitation (Co-IP) of HsfA1 (lower panels) are shown. The additional band appearing in all lanes of the HsfA2-IP panel results from co-elution of IgG heavy chains. E, expression constructs for HsfA2 (samples 1–4 and 13) and its mutant forms D (samples 5–8), H (samples 9–11), and A (sample 12) with N-terminally fused GFP were transformed alone (images 1, 2, 5, 6, 9, 10, 12) or co-transformed with wt HsfA1 (+HsfA1, samples 3, 7, 11) or its mutant forms D (+HsfA1-D, samples 4, 8) and H (+HsfA1-H, sample 13), respectively. For inhibition of nuclear export, leptomycin B (+LMB, samples 2, 6, 10) was added 2 h before harvesting.
Next, the influence of these mutations on the synergistic activation in the presence of the corresponding partner Hsf was analyzed. In addition to the plasmid-borne GUS reporter, we used the activation of the endogenous chromatin-embedded Hsp17-CI gene expression as an alternative reporter system (32, 33). Loss of the DNA binding function (mutants D) had only moderate effects as long as the partner Hsf was wild type (Fig. 3, B and C, lanes 3–5). Similarly, in the GUS reporter assay, the activator domain mutants (A) also showed synergism (even though at a reduced level) as long as the activator domain of one Hsf was wild type (Fig. 3B, lanes 8–10). In contrast, OD deletion mutants showed only the basal reporter activity, because of the activity of the wild type partner Hsf (Fig. 3B; compare samples 1 and 6, samples 11 and 7). Remarkably, the results were similar for both types of reporters, i.e. plasmid-borne GUS and endogenous Hsp17-CI expression. Exceptions were observed with the CTAD mutants where a much more pronounced reduction of synergism was found in the Hsp17-CI-based reporter assay (Fig. 3C, lanes 8 and 9) compared with the GUS activity (Fig. 3B, samples 8 and 9). Evidently, expression of the chromatin-embedded Hsp17-CI genes appears to be more sensitive to the composition of the activator complex. Based on these results, we conclude that the activator and the oligomerization domains are essential for the synergistic gene activation by HsfA1 and HsfA2, whereas an intact DBD is only required for either one of the two partner Hsfs.
We next examined the formation of hetero-oligomeric complexes between wild type and mutant forms of HsfA1 and HsfA2. In co-immunoprecipitation assays, HsfA1 was detectable in immunoprecipitates of wild type HsfA2 (Fig. 3D, lane 2), as well as in combinations including DBD (lanes 3–5) and CTAD mutants (lanes 8–10) from both Hsfs. However, no HsfA1 was detectable in immunoprecipitates of the OD deletion mutants (lanes 6 and 7). As expected, hetero-oligomeric complexes were only formed by HsfA1 and HsfA2 forms with intact ODs.
An alternative test to show the direct interaction between the two Hsfs in vivo is the nuclear retention assay of HsfA2. HsfA2 shuttles between the nucleus and cytoplasm; but due to its relatively strong NES, the equilibrium is shifted toward a dominant cytoplasmic localization (21). Only for HsfA2-A, but not for the other mutants, a nuclear localization was observed (Fig. 3E, samples 1, 5, 9, and 12). Most likely, in the HsfA2-A mutant, exchange of Leu-341 to Ala in the AHA2 motif (see Fig. 4A) also influences the NES function. To confirm the nucleo-cytoplasmic shuttling, we used leptomycin B (LMB) as inhibitor of nuclear export. In the presence of LMB, wild type HsfA2 and all its mutants showed a dominant nuclear localization (samples 2, 6, and 10). However, nuclear retention of HsfA2 can also be achieved by co-expression of HsfA1 (Ref. 21, and sample 3 in Fig. 3E). As expected, the nuclear retention of HsfA2 was only slightly affected by mutation of the DNA binding domain even when both Hsf binding mutants, HsfA1-D and HsfA2-D were co-expressed (samples 7 and 8). This confirms that interaction of the two Hsfs is sufficient for nuclear retention, and DNA binding is not required for the formation of HsfA1/A2 complexes. In contrast, when the OD was deleted in either HsfA2 or HsfA1, almost no nuclear retention of HsfA2 was observed (samples 11 and 13). So far, we cannot explain the evident discrepancies for the combination of wt HsfA2 with the HsfA1-H mutant. The observed lack of synergism in GUS expression (Fig. 3B, sample 6), of co-immunoprecipitation (Fig. 3D, lane 6) and nuclear retention (Fig. 3E, image 13) contrasts to a significant signal in the Hsp17 expression (Fig. 3C, lane 6).
FIGURE 4.
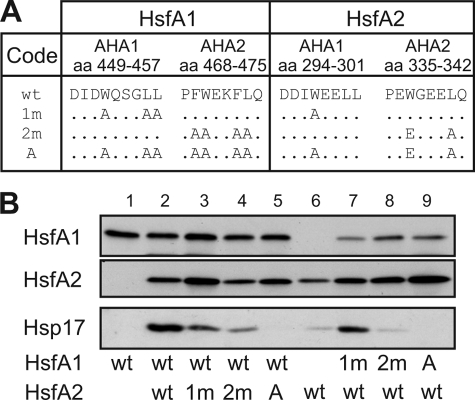
Role of the AHA motifs for the synergistic activity of HsfA1/A2 hetero-oligomers. A, mutations in the AHA1 and AHA2 motifs in the CTADs of HsfA1 and HsfA2, respectively, and annotation of the mutants. As shown previously (32, 37), the complete functional knock-out of HsfA1was only achieved by exchanging all seven indicated amino acid residues to alanine. B, expression levels of wild type (wt) and mutant forms of HsfA1 and HsfA2 after co-expression in combinations as indicated below (upper panels), and stimulation of endogenous Hsp17-CI gene expression in the corresponding samples 1–9 (lower panel).
Contribution of the AHA Motifs in the C-terminal Activator Domain
The CTADs of HsfA1 and HsfA2 contain each of two peptide motifs assigned as AHA1 and AHA2, which are essential for the activator function (Fig. 1 and Refs. 22, 32, 37). Because these motifs are potential interaction sites for components of the transcription machinery, it is tempting to speculate that functional complementation between the four AHA motifs in the HsfA1/A2 hetero-oligomers are crucial for synergistic activation of gene expression. To challenge this assumption, we tested different AHA mutants of HsfA1 and HsfA2 (Fig. 4A). We analyzed the endogenous Hsp17-CI expression because of the higher sensitivity compared with the GUS reporter. The strong synergistic expression of endogenous Hsp17-CI in the presence of the two wild type Hsfs (Fig. 4B, lane 2) was significantly reduced with the AHA1 mutants (lanes 3 and 7), but even more with the AHA2 mutants (lanes 4 and 8). Mutation of both AHA motifs in either HsfA1 or HsfA2 abolished the Hsp17-CI expression (lanes 5 and 9). Hence, although both activator motifs contribute to the synergistic effects, the function of the AHA2 motifs is more important.
Domain-swapping Experiments
To underscore the differential contribution of the functional domains to the activity of HsfA1/A2 hetero-oligomers, we generated hybrid constructs by domain-swapping (Fig. 5). Interestingly, the hybrid forms with exchanged OD domains of both HsfA1 and HsfA2 (Fig. 5A, hybrid forms 3 and 7) have a higher individual transcriptional activity than wild type. This holds true for the HsfA2 hybrid form with the CTAD of HsfA1 as well (Fig. 5A, sample 8). It is tempting to speculate that HsfA1 and HsfA2 are kept in an attenuated form because of intramolecular interactions between the OD and the CTAD, as suggested earlier for the yeast, Drosophila, and human Hsf1 (41–43). Thus, the intramolecular shielding could be released in the hybrid Hsfs 3, 7, and 8 (Fig. 5A).
In combination with the corresponding partner Hsf, these hybrid constructs allow analysis of the contribution of individual domains to the synergistic activity, by formation of HsfA1/A2 complexes, which carry one domain changed to a homo-oligomeric constellation while all other parts are identical to wild type HsfA1/A2 complexes. As expected, exchange of the DNA binding domains has only a moderate effect on the synergistic activity, especially when the DNA binding domain of HsfA1 was placed into the HsfA2 background (Fig. 5B, 3 and 4). In contrast, exchange of the ODs strongly reduces the synergistic effects (Fig. 5B, 5 and 6), and exchange of the CTADs even abolishes any synergism (Fig. 5B, 7 and 8), irrespectively, whether the hetero-oligomers contained only the activation domain of HsfA1 or of HsfA2. These results were confirmed by analysis of Hsp17-CI expression (see supplemental Fig. S3). This confirms the essential function of the combination of the CTADs from both partners in the HsfA1/A2 co-complexes, usually mediated or reinforced by interaction of the two heterologous ODs.
The results obtained by replacement of the oligomerization domains prompt the question whether hetero-oligomerization is preferred to homo-oligomerization. To answer this, we co-immunoprecipitated HsfA1 co-expressed with wild type HsfA2 or HsfA2 mutants with the OD replaced by that of HsfA1 or HsfA4b. The latter was used as negative control because no interaction of HsfA1 with HsfA4b was previously observed (33). Indeed, comparing the co-immunoprecipitation efficiency, we observed a strong preference for the wild type HsfA1/A2 hetero-oligomers as compared with the combination of HsfA1 with the HsfA2xA1OD hybrid, i.e. the combination of homologous ODs (Fig. 5C, lanes 1 and 2). As expected, the use of the HsfA2xA4bOD hybrid as bait could not co-immunoprecipitate HsfA1 (Fig. 5C, lane 3). These results are in agreement with yeast two-hybrid interaction data reported earlier (27) and provide further evidence that the oligomerization domains mediate the preferential formation of hetero-oligomeric HsfA1/A2 complexes.
The Dual Function of the Oligomerization Domain
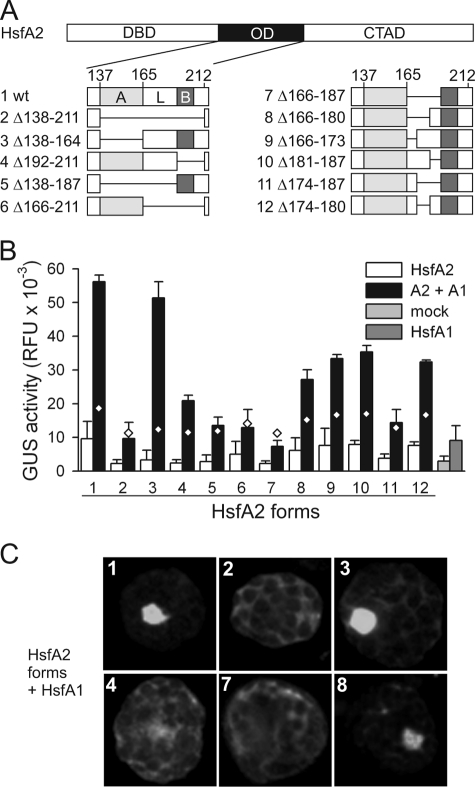
Plant class A Hsfs differ from all non-plant Hsfs by an extension of the OD by insertion of 21 amino acid residues between the HR-A and HR-B region. To investigate the contribution of different parts of the OD to hetero-oligomerization, a series of HsfA2 mutants with deletions in the OD region was created (Fig. 6A) and tested for synergistic interaction with HsfA1 (Fig. 6B). As already shown for the HsfA2-H mutant in Fig. 3, deletion of the OD abolished synergistic interaction and activity of HsfA2 (Fig. 6B, lane 2). However, deletion of parts of the OD revealed differential effects. The HR-A part seems to be less important for the synergistic function (sample 3) than HR-B (sample 4). The synergism was 4.2-fold for the wild type combination and 7.4-fold for the combination with HsfA2 mutant form 3, but reduced to 3.2-fold for mutant form 4. In contrast, a mutant HsfA2 with deletion of the central linker region L (sample 7) showed the lowest GUS activity, and no synergistic effect was observed. Similar results were obtained with other HsfA2 mutants lacking the linker (samples 5 and 6).
FIGURE 6.
Functional dissection of the oligomerization domain. A, deletions in the HsfA2 oligomerization domain are illustrated as bar diagrams and the numbers of deleted amino acid residues are indicated. B, GUS reporter expression in tobacco protoplasts transformed with the indicated HsfA2 forms (constructs 1–12) either alone (white bars) or in combination with HsfA1 (black bars). Bars on the right show the mock result (gray) and the activity of HsfA1 alone (dark gray). Diamonds indicate the additive GUS activity values for each of the indicated HsfA2 mutant form in combination with HsfA1. For control of Hsf expression and induction of endogenous Hsp17-CI expression, see supplemental Fig. S4. C, intracellular localization of the indicated forms of HsfA2 (numbers corresponding to A) with N-terminally fused GFP in protoplasts co-transformed with HsfA1.
The linker region appears to be essential for the synergistic interaction of HsfA2 with HsfA1. Hence, we generated mutants of HsfA2 with partial deletions of the linker region (Fig. 6A, mutants 8–12). The only mutant without any stimulatory or synergistic activity was mutant 11 where the C-terminal part of the linker was removed (Fig. 6B, sample 11). At the same time, preserving the C-terminal 7-amino acid residues of the linker was sufficient to maintain the capability of HsfA2 for interaction with HsfA1 (Fig. 6B, sample 8). Interestingly, mutant 10, not containing this C-terminal heptapeptide also shows a significant synergistic action. This might be explained by the similarity between the two adjacent amino acid heptads forming the middle and the C-terminal part of the linker (IIAMGEK and IETQERK, respectively). Basically similar results were obtained, when the synergistic activity of the HsfA2 mutants was analyzed by expression of endogenous Hsp17-CI (supplemental Fig. S4; also presenting the Hsf expression controls for Fig. 6B).
Finally, the strength of the interaction of HsfA2 mutants with HsfA1 was confirmed by nuclear retention assays (Fig. 6C). In agreement with the results from the reporter assays, nuclear retention was abolished with mutants 2, 4, and 7 but preserved with mutants 3 and 8. Taken together, results indicate that the region mediating hetero-oligomerization is formed essentially by HR-B and parts of the preceding linker sequence.
Formation of Homo- and Hetero-oligomeric Hsf Complexes
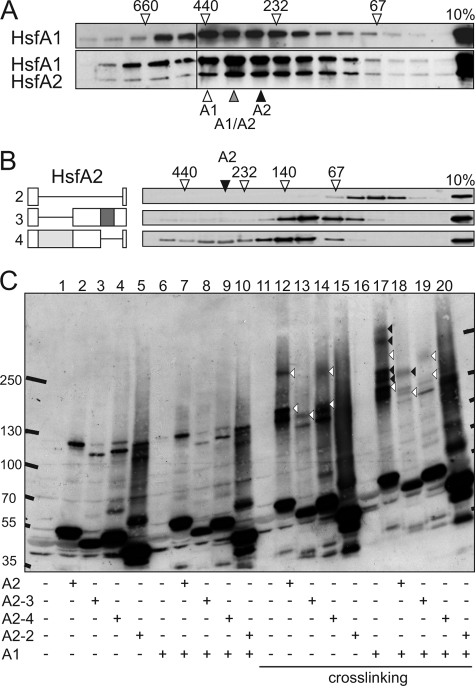
To investigate the formation and size of Hsf complexes in more detail, we used the protoplast system for Hsf expression and characterization of oligomeric complexes by SEC. In an earlier study, we had shown that HsfA2 alone forms complexes eluting in a broad peak with a maximum corresponding to an apparent molecular size in the range of 320–350 kDa indicating the formation of hexamers (28). Here we find that expression of HsfA1 alone leads to similar complexes with an apparent molecular size in the range of 400–440 kDa (Fig. 7A, upper panel). When both, HsfA1 and HsfA2 were coexpressed, the peak fractions for both proteins coincided, and they migrated at an intermediate molecular size of ∼350–400 kDa (Fig. 7A, lower panel). Hence, HsfA1 and HsfA2 can form homo- and hetero-oligomeric hexameric complexes.
FIGURE 7.
SEC and cross-linking of homo- and hetero-oligomeric Hsf complexes. A, protein complexes formed by expression of HsfA1 (top panel) in combination with HsfA2 (bottom panel) in transformed tobacco protoplasts were analyzed by SEC on Superdex 200. Proteins of the consecutive fractions were analyzed by SDS-PAGE and immunoblotting. On the top, migration and molecular sizes of marker proteins are indicated. On the bottom, triangles indicate peak fractions of HsfA1 complexes (white) and HsfA1/A2 co-complexes (gray). The black triangle indicates the corresponding peak fraction of complexes formed by HsfA2 alone (deduced from Fig. 5A in Ref. 28). B, SEC of complexes formed by HsfA2 mutants with deletions in the HR-A/B region as illustrated on left (numbering as in Fig. 6). The peak fraction corresponding to the size of wt HsfA2 complexes is indicated as in A. C, HsfA1, HsfA2, and its mutants were expressed either alone or in combination in tobacco protoplasts as indicated on the bottom. Aliquots of protein extracts were incubated before (lanes 1–10) or after chemical cross-linking (lanes 11–20) as described under “Experimental Procedures.” Shown is the immunoblot processed with antibodies against HsfA2. Extract of protoplasts transformed with empty vector was loaded for control (lanes 1 and 11). The position of marker proteins is indicated on the left and right margin. Estimation of relative molecular weights was done after compensation of bending (not shown). White arrowheads indicate cross-linking products formed by HsfA2 or its mutants alone; black arrowheads indicate cross-linking products formed by HsfA1/A2.
Next, we analyzed complex formation of HsfA2 mutants carrying deletions in the HR-A/B region (Fig. 6A). As expected from the results presented above (Figs. 3–6), deletion of the entire oligomerization domain led to an exclusively monomeric form (Fig. 7B, mutant 2). In contrast, the mutant protein that lacks the HR-B region and is unable to interact with HsfA1 (Fig. 6C) migrated at a molecular size of about 120–150 kDa (Fig. 7B, mutant 4) corresponding to trimeric complexes. Interestingly, the mutant lacking the HR-A region and still interacting with HsfA1 (Fig. 6C) formed only dimeric complexes with a molecular size of about 80–110 kDa (Fig. 7B, mutant 3). Together, these results indicate the existence of trimeric subcomplexes in the hexameric wild type HsfA2, which are formed by interaction between the HR-A regions. These trimers can be further assembled to hexamers by dimerization at the interaction surface provided by the HR-B and preceding linker regions. Probably, the same holds true for the HsfA1/A2 hetero-hexamer representing a dimeric homotrimer.
To challenge this observation, we performed glutaraldehyde cross-linking experiments in protein extracts from protoplasts expressing HsfA2 or its HR-A/B mutants either alone or in combination with HsfA1. Formation of cross-linking products of HsfA2 was analyzed after separation on a SDS polyacrylamide gradient gel and immunodecoration (Fig. 7). Specific cross-linking products became visible only after glutaraldehyde treatment (lanes 11–20), with exception of some SDS resistant artifacts in the range of 120 kDa detectable for all HsfA2 forms (lanes 2–5 and 7–10). Two major cross-linking products at about 130 and 200 kDa are detectable for wild type HsfA2 (lane 12). The migration corresponds to a dimeric and trimeric composition, respectively. Higher order complexes as deduced from the SEC results (Fig. 7A) could not be detected. This could be explained by limitations of the method used, because the immunodetectability might be compromised by either low abundances of complexes with higher molecular weight or by extensive crosslinking of proteins within such complexes. The results obtained with the mutant forms of HsfA2 (lanes 13–15, white arrowheads) are in line with the SEC results (Fig. 7B). While products with molecular weights corresponding to dimeric and trimeric compositions are detectable for the mutant form lacking HR-B (A2-4, lane 14), only cross-linking products corresponding to a dimer are observed for the mutant with deletion of the HR-A region (A2-3, lane 13). For the mutant lacking the whole OD no corresponding cross-linking products are observed (A2-2, lane 15).
In the presence of HsfA1 additional crosslinking products appeared indicating heteromeric HsfA1/A2 interactions. As expected, these products (marked by black arrowheads) are only observed in combination with wild type HsfA2 (Fig. 7C, lane 17) or the mutant A2-3 (lane 18), but not with the mutant lacking the HR-B region (lanes 19 and 20). This confirms the requirement of the HR-B region for the formation of hetero-oligomeric HsfA1/A2 complexes (Fig. 6C). Furthermore, the detection of only one additional heteromeric cross-linking product in the combination of HsfA1 with the A2-3 mutant lacking HR-A indicates that the linker/HR-B region provides the surface for interactions contributing to the formation of homo- and heterodimeric complexes (Fig. 7, B and C, lanes 13 and 18). The formation of higher order complexes depends on the HR-A region, which, however, does not provide the surface for heteromeric interactions (Figs. 6C and 7C, lane 14 versus 19). Taken together, the results show that multiple interactions between the oligomerization domains contribute to the formation and stabilization of higher order Hsf complexes and confirm the postulated existence of similar substoichiometric complexes in both HsfA2 homo-oligomers and HsfA1/A2 hetero-oligomers.
DISCUSSION
Specificity and Synergistic Activity of the HsfA1/A2 Superactivator
Plants have evolved a highly efficient and flexible stress response system to ensure survival and normal development under versatile environmental conditions which often involve hs (3, 24, 44, 45). During hs, three consecutive phases of gene expression control can be discriminated: (i) Rapid reprogramming of gene expression at the onset of stress connected with a block of housekeeping gene expression, (ii) induction and maintenance of stress gene expression at high levels ensuring the necessary accumulation of chaperones and other protective proteins, and (iii) after return to normal conditions, efficient recovery of housekeeping gene expression is a prerequisite to allow resumption of normal plant development.
In tomato, three transcription factors HsfA1, HsfA2, and HsfB1 form a functional triad connected to the regulation of the hs response (4). At the onset of hs, the constitutively expressed master regulator HsfA1 triggers the expression of not only Hsp-encoding genes but also of the genes encoding HsfA2 and HsfB1 (25). Both, HsfA2 and HsfB1 subsequently cooperate with HsfA1 to ensure efficient production of Hsps, maintenance of thermotolerance, and return to the expression of housekeeping genes during recovery (27, 32).
Although HsfA2 strongly accumulates in thermotolerant tomato cells, it is largely inactive on its own and needs interaction with HsfA1 for nuclear localization and prevention of aggregation in the cytoplasm (21, 28). In addition, formation of HsfA1/A2 hetero-oligomeric complexes leads to synergistic effects on the activation of expression of Hsf-dependent genes, which is usually 3–5-fold higher than the additive activities of the individual partners (Fig. 2 and supplemental Fig. S1). As shown by transcriptional stimulation in reporter assays, co-immunoprecipitation, and nuclear retention of HsfA2, the interaction between HsfA1 and HsfA2 is very specific and depends on peculiarities of their ODs (Figs. 2 and 3). These findings are in line with observations on the interaction of HsfA4b and HsfA5, which form another pair of specifically interacting Hsfs (33). Despite considerable similarity in sequence and basic domain organization of the ODs (9), there are evidently subtle but decisive differences between the HR-A/B regions of class A Hsfs leading to the formation of these pairs of specifically interacting Hsfs. Replacement of the OD in HsfA2 by that of HsfA4b abolishes the interaction of the hybrid protein with HsfA1 (Fig. 5), while a HsfA1 hybrid protein carrying the OD of HsfA4b is susceptible to repression by HsfA5, an effect not observed with wild type HsfA1 (33). Although only few examples are investigated so far, these findings support the idea that the large family of plant Hsfs is organized in a network of specifically interacting members with distinct functions to ensure tight regulation of Hsf-dependent gene expression, not only under hs but also other conditions of environmental stress.
Considering our earlier findings and the data presented in this report, the synergistic activity of the HsfA1/A2 co-activator results from the contribution of cooperative effects on multiple functional levels.
(i) Nuclear retention of cytoplasmic HsfA2 in the presence of HsfA1 (21, 27).
(ii) Stabilization of both Hsfs in the hetero-oligomeric complexes, especially if either of the two partners is expressed at substoichiometric levels (Fig. 3 and supplemental Fig. S1). Similar effects were observed for the stabilization of the HsfA4b/A5 pair in tomato (33) and have been reported as well for hetero-oligomeric complexes formed by human Hsf1 and Hsf2 (46).
(iii) Although indispensable for the superactivator function, DNA binding is not required for the formation of HsfA1/A2 complexes themselves, which evidently precedes the recognition of HSEs in the promoter region of target genes (Fig. 3).
(iv) The differential interaction of individual AHA motifs with components of the transcriptional machinery, i.e. TFIID, SAGA, and SWF/SNF complexes, was reported recently for several Arabidopsis class A Hsfs (23). Our reporter assays with AHA mutants of tomato HsfA1 and HsfA2 demonstrate that the special combination of different AHA motifs in the HsfA1/A2 complex is important for the synergistic activation. The functional complementation of the CTADs of both Hsfs is particularly striking when the expression of endogenous, chromatin-embedded sHsp genes is analyzed (Fig. 4).
(v) Formation of HsfA1/A2 complexes stimulates the activity of HsfA1 by releasing its repressed CTAD (Fig. 5). It is tempting to speculate that this type of derepression depends on conformational changes and contributes to the hs inducibility of HsfA1 activity in situ as detected by band shift assays and hs gene transcription (27, 47). A frequently discussed model used to explain the inactive state of Hsfs involves shielding of functional domains by intramolecular interactions between C-terminal parts and the N-terminal DBD and HR-A/B (OD) region (41–43, 48–50). Disturbing the molecular context in some of the domain-swapping mutants of HsfA1 and HsfA2 evidently compromises such an intramolecular shielding (Fig. 5A).
The Function of the HR-A/B Region as Homo- and Hetero-oligomerization Domain
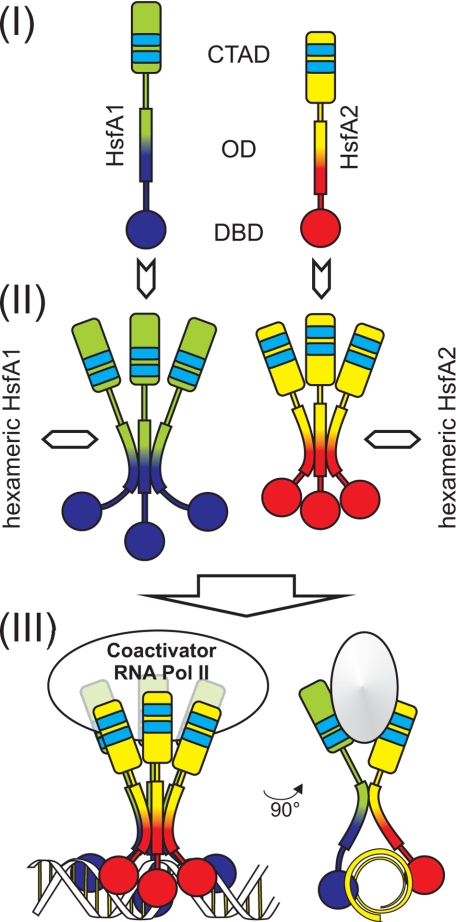
The oligomerization domain is conserved in all Hsfs and consists of a heptad repeat pattern of hydrophobic amino acid residues (HR-A/B region), which is predicted to form a trimeric, α-helical coiled-coil structure of the leucine zipper-type (51). Formally, a tripartite structure can be assigned. The N-terminal portion (HR-A region) is usually formed by 4 to 5 heptad repeats, which are separated by a short glutamine-rich linker of 6 amino acid residues from the C-terminal short hydrophobic heptad region (HR-B). The importance of the HR-A/B region for Hsf oligomerization and activity was frequently reported, and for most non-plant Hsfs the formation of homotrimers is assumed (1, 7). In contrast to most other Hsfs including plant class B Hsfs, the linker region of class A Hsfs is extended by an insertion of 21 amino acid residues (9), and it was intriguing to ask, whether this extension is a structural prerequisite for the formation of hetero-oligomeric HsfA1/A2 complexes and whether the more separated HR-A and B regions may have distinct functions in the formation of homo- and hetero-oligomeric Hsf complexes. The analysis of HsfA2 mutants harboring deletions in the HR-A/B region with respect to functionality (Fig. 6) and complex formation (Figs. 6 and 7) provide interesting new insights into particular functions of the OD of HsfA2 and implicate consequences for assembly and structural properties of oligomeric Hsf complexes. The major conclusions that can be drawn from the data are summarized in the model illustrated in Fig. 8. Newly synthesized Hsf molecules are rapidly assembled in homotrimeric subcomplexes supported by interactions between their HR-A regions (Fig. 7). The subsequent formation of hexameric complexes is mediated by providing an interface for dimerization formed by C-terminal parts of the linker and the adjacent HR-B region (Figs. 6 and 7). In the presence of both HsfA1 and HsfA2, the dimerization of the trimeric subcomplexes to the hexameric HsfA1/A2 superactivator complex is strongly favored over the formation of homo-oligomeric hexamers. Although the linker and the HR-B regions are essential and sufficient to form synergistically active HsfA1/A2 complexes and the HR-A regions from both Hsfs do not interact with each other, all three parts of the OD are required for the stabilization of the hexameric complex.
FIGURE 8.
Model of plant Hsf assembly. Monomeric Hsfs (i) assemble to a trimeric homo-oligomer (ii) by interaction via HR-A. The trimer forms a hexamer through heterodimeric interactions (iii) mediated by the linker and HR-B. To illustrate the two types of interactions, a 90° rotation of the model is shown on the right. For further details see “Discussion.”
In summary, the existence of homo-oligomeric complexes of HsfA1 and HsfA2 with moderate activities contrasts the strong tendency to form highly active hexameric HsfA1/A2 complexes by dimerization of homotrimeric subcomplexes. These results lead to the intriguing question whether in the native surrounding of heat-stressed tomato cells homo- and hetero-oligomeric complexes of the two Hsfs coexist in a dynamic equilibrium, which is controlled by members of the chaperone network (Hsp17, Hsp70, Hsp90) previously shown to be involved in the regulation of Hsf activity in plants (28, 52, 53).
Supplementary Material
Acknowledgments
We thank Enrico Schleiff for helpful discussions and support during preparation of the manuscript and Markus Bohnsack for critical proofreading during revision.
This work was supported by the Deutsche Forschungsgemeinschaft (to L. N. (No 249/4) and K. D. S. (Scha 577/6)).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- Hsf
- heat stress transcription factor
- Hsps
- heat stress proteins
- hs
- heat stress
- AHA
- activator motifs: peptide motifs consisting of aromatic, large hydrophobic and acidic amino acid residues
- GFP
- green fluorescent protein
- OD
- oligomerization domain
- DBD
- DNA binding domain
- CTAD
- C-terminal activator domain
- SEC
- size exclusion chromatography
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- HR
- heptad repeat
- wt
- wild type.
REFERENCES
- 1.Morimoto R. I. (1998) Genes Dev. 12, 3788–3796 [DOI] [PubMed] [Google Scholar]
- 2.Schöffl F., Prändl R., Reindl A. (1998) Plant Physiol. 117, 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baniwal S. K., Bharti K., Chan K. Y., Fauth M., Ganguli A., Kotak S., Mishra S. K., Nover L., Port M., Scharf K. D., Tripp J., Weber C., Zielinski D., von Koskull-Döring P. (2004) J. Biosci. 29, 471–487 [DOI] [PubMed] [Google Scholar]
- 4.von Koskull-Döring P., Scharf K. D., Nover L. (2007) Trends Plant Sci. 12, 452–457 [DOI] [PubMed] [Google Scholar]
- 5.Akerfelt M., Trouillet D., Mezger V., Sistonen L. (2007) Ann. N.Y. Acad. Sci. 1113, 15–27 [DOI] [PubMed] [Google Scholar]
- 6.Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C. (1995) Annu. Rev. Cell Dev. Biol. 11, 441–469 [DOI] [PubMed] [Google Scholar]
- 8.Nakai A. (1999) Cell Stress Chaperones 4, 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nover L., Bharti K., Döring P., Mishra S. K., Ganguli A., Scharf K. D. (2001) Cell Stress Chaperones 6, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voellmy R., Boellmann F. (2007) Adv. Exp. Med. Biol. 594, 89–99 [DOI] [PubMed] [Google Scholar]
- 11.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 12.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 13.Millar A. H., Whelan J., Small I. (2006) Curr. Opin. Plant Biol. 9, 610–615 [DOI] [PubMed] [Google Scholar]
- 14.Han J. H., Batey S., Nickson A. A., Teichmann S. A., Clarke J. (2007) Nat. Rev. Mol. Cell Biol. 8, 319–330 [DOI] [PubMed] [Google Scholar]
- 15.Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 16.Oreb M., Tews I., Schleiff E. (2008) Trends Cell Biol. 18, 19–27 [DOI] [PubMed] [Google Scholar]
- 17.Pelham H. R., Bienz M. (1982) EMBO J. 1, 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nover L. (ed) (1991) Heat Shock Response, CRC Press, Boca Raton, Florida [Google Scholar]
- 19.Pirkkala L., Nykänen P., Sistonen L. (2001) FASEB J. 15, 1118–1131 [DOI] [PubMed] [Google Scholar]
- 20.Lyck R., Harmening U., Höhfeld I., Scharf K. D., Nover L. (1997) Planta 202, 117–125 [DOI] [PubMed] [Google Scholar]
- 21.Heerklotz D., Döring P., Bonzelius F., Winkelhaus S., Nover L. (2001) Mol. Cell. Biol. 21, 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treuter E., Nover L., Ohme K., Scharf K. D. (1993) Mol. Gen. Genet. 240, 113–125 [DOI] [PubMed] [Google Scholar]
- 23.Kotak S., Port M., Ganguli A., Bicker F., von Koskull-Döring P. (2004) Plant J. 39, 98–112 [DOI] [PubMed] [Google Scholar]
- 24.Miller G., Mittler R. (2006) Ann. Bot. 98, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra S. K., Tripp J., Winkelhaus S., Tschiersch B., Theres K., Nover L., Scharf K. D. (2002) Genes Dev. 16, 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann C., Eggers-Schumacher G., Wunderlich M., Schöffl F. (2004) Mol. Genet. Genomics 271, 11–21 [DOI] [PubMed] [Google Scholar]
- 27.Scharf K. D., Heider H., Höhfeld I., Lyck R., Schmidt E., Nover L. (1998) Mol. Cell. Biol. 18, 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Port M., Tripp J., Zielinski D., Weber C., Heerklotz D., Winkelhaus S., Bublak D., Scharf K. D. (2004) Plant Physiol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006) Plant J. 48, 535–547 [DOI] [PubMed] [Google Scholar]
- 30.Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., von Koskull-Döring P. (2006) Plant Mol. Biol. 60, 759–772 [DOI] [PubMed] [Google Scholar]
- 31.Charng Y. Y., Liu H. C., Liu N. Y., Chi W. T., Wang C. N., Chang S. H., Wang T. T. (2007) Plant Physiol. 143, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bharti K., von Koskull-Döring P., Bharti S., Kumar P., Tintschl-Körbitzer A., Treuter E., Nover L. (2004) Plant Cell 16, 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baniwal S. K., Chan K. Y., Scharf K. D., Nover L. (2007) J. Biol. Chem. 282, 3605–3613 [DOI] [PubMed] [Google Scholar]
- 34.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (eds) (1993) Current Protocols in Molecular Biology, John Wiley & Sons Inc., Indianapolis [Google Scholar]
- 35.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Bharti K., Schmidt E., Lyck R., Heerklotz D., Bublak D., Scharf K. D. (2000) Plant J. 22, 355–365 [DOI] [PubMed] [Google Scholar]
- 37.Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000) Plant Cell 12, 265–278 [PMC free article] [PubMed] [Google Scholar]
- 38.Töpfer R., Schell J., Steinbiss H. H. (1988) Nucleic Acids Res. 16, 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reichel C., Mathur J., Eckes P., Langenkemper K., Koncz C., Schell J., Reiss B., Maas C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5888–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damberger F. F., Pelton J. G., Liu C., Cho H., Harrison C. J., Nelson H. C., Wemmer D. E. (1995) J. Mol. Biol. 254, 704–719 [DOI] [PubMed] [Google Scholar]
- 41.Nieto-Sotelo J., Wiederrecht G., Okuda A., Parker C. S. (1990) Cell 62, 807–817 [DOI] [PubMed] [Google Scholar]
- 42.Rabindran S. K., Haroun R. I., Clos J., Wisniewski J., Wu C. (1993) Science 259, 230–234 [DOI] [PubMed] [Google Scholar]
- 43.Zuo J., Baler R., Dahl G., Voellmy R. (1994) Mol. Cell. Biol. 14, 7557–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K. D. (2007) Curr. Opin. Plant Biol. 10, 310–316 [DOI] [PubMed] [Google Scholar]
- 45.Larkindale J., Vierling E. (2008) Plant Physiol. 146, 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandqvist A., Björk J. K., Akerfelt M., Chitikova Z., Grichine A., Vourc'h C., Jolly C., Salminen T. A., Nymalm Y., Sistonen L. (2009) Mol. Biol. Cell 20, 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scharf K. D., Rose S., Zott W., Schöffl F., Nover L., Schöff F. (1990) EMBO J. 9, 4495–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakai A., Morimoto R. I. (1993) Mol. Cell. Biol. 13, 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P. C., Thiele D. J. (1999) J. Biol. Chem. 274, 17219–17225 [DOI] [PubMed] [Google Scholar]
- 50.Chen T., Parker C. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peteranderl R., Rabenstein M., Shin Y. K., Liu C. W., Wemmer D. E., King D. S., Nelson H. C. (1999) Biochemistry 38, 3559–3569 [DOI] [PubMed] [Google Scholar]
- 52.Kim B. H., Schöffl F. (2002) J. Exp. Bot. 53, 371–375 [DOI] [PubMed] [Google Scholar]
- 53.Yamada K., Fukao Y., Hayashi M., Fukazawa M., Suzuki I., Nishimura M. (2007) J. Biol. Chem. 282, 37794–37804 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.