Abstract
The seven-pass transmembrane protein Smoothened (Smo) is an essential component of the Hedgehog (Hh) signaling pathway that is critically involved in normal animal development as well as pathological malignancies. In studying Hh-related biological processes, it would be highly desirable if Smo activity could be instantly switched between activation and inhibition. Using Gli1-dependent GFP transgenic zebrafish and in vitro biochemical assays, we identified and characterized two potent Smo inhibitors, SANT74 and 75 (Smoothened antagonist 74 and 75), by screening a small molecule library designed based on the scaffold of Smo agonist SAG. These compounds are structural analogs of SAG with the methyl group substituted by a propyl or allyl group in SANTs. We show that SANTs and SAG exert opposite effects on Smo activity by regulating protein conformation. Our study represents the first demonstration of conformational regulation of Smo by small molecule analogs, and the combinational use of these Smo modulators in a temporal controlled fashion should be useful for studying Hh biology.
The Hh signaling pathway is essential for embryonic development and adult tissue homeostasis in metazoans. Hh ligands function through three main components: a 12-transmembrane protein receptor Patched1 (Ptch1), a seven-transmembrane protein co-receptor Smo, and the transcription factors of Gli family (1). Binding of secreted Hh proteins to Ptch1 results in subcellular relocation and conformation changes of Smo that in turn promote the expression of Hh target genes through the Gli transcription factors (1).
Hh signaling regulates both cell proliferation and differentiation during normal embryogenesis. Recent studies indicate that Hh pathway activation is also essential for postembryonic tissue repair and stem cell regulation (2). Because of its critical roles in regulating cell fate and proliferation, overstimulation of Hh pathway has been implicated in several malignancies and cancers (3). Basal cell carcinoma, one of the most common cancers in the Western world, involves mutations in the Ptch1 and Smo (4–6). During brain development, Hh is a mitogen for cerebellar granule neuron progenitors and is down-regulated after postnatal development (7, 8). In contrast, more than 30% of human medulloblastoma exhibit high levels of Gli1 expression (9). Mutations in both Ptch1 and Smo have been identified in human medulloblastoma (10, 11).
In addition to its roles in embryonic patterning and cancer, the Hh signaling pathway is required for organogenesis such as pancreas formation and angiogenesis. Studies in zebrafish and mice indicate that Hh pathway deficiencies result in deceased vascular structures. Shh-deficient mice have abnormal pulmonary vasculature formation and diminished airway branching in lungs (12). Zebrafish embryos lacking Hh signaling display defects of the primary intersegmental vessel sprouting (13). Shh appears to promote the maturation of blood vessels by regulating the mRNA levels of angiopoietin-1 and -2 and vascular endothelial growth factor (14). Hh signaling also plays an important role in tumor-associated angiogenesis. When treated with cyclopamine, a plant-derived steroidal alkaloid antagonist of Hh signaling pathway, tumorgenic vascular structures become dramatically reduced (15, 16).
Because mutated Ptch1 or Smo proteins are mostly responsible for the abnormal activation of Hh related to human diseases, intense efforts have been invested to identify therapeutic inhibitors acting on or downstream of Smo. Cyclopamine is one of the compounds that specifically block Hh signaling pathway through direct interaction with Smo (17, 18). The effect of oncogenic mutations in Smo and Patched appears reversible by cyclopamine (19). A small scale clinic study has found that topical treatment with cyclopamine was effective in reducing the size of basal cell carcinoma lesions (20). Cyclopamine also causes the regression and apoptosis of human medulloblastomas in murine tumor allograft models (21, 22). Using cell-based screening platforms with a luciferase reporter under the control of multiple Gli1-binding sites, several additional synthetic Hh inhibitors with Smo binding affinity have been identified (23–26). Some of these Hh inhibitors are able to suppress basal cell carcinoma and medulloblastomas in animal models (25, 27).
With such a broad implication of Hh activity in normal biological processes and cancers, it is highly desirable to develop efficient screen platforms for identifying potent and specific inhibitors. To this end, we developed a Gli-GFP4 transgenic zebrafish model that allows in vivo detection of Hh activity in whole living embryos. Because genetic Hh-deficient zebrafish embryos have defined phenotypes, they can be used to characterize the efficiency and specificity of the inhibitors. For instance, treatment of zebrafish embryos with cyclopamine will induce phenotypes identical to those associated with Smo mutations in zebrafish, including U-shaped somites, shorter intersegmental blood vessels, and fewer pancreatic β cells.
Using zebrafish embryos, we screened a chemical library based on the structure of SAG, a synthetic Hh pathway agonist that directly targets Smo in a manner that antagonizes cyclopamine action (23, 24). Of the small molecule modulators of Smo that have been reported, SAG is particularly potent with efficacy in the low nanomolar range, and its effects on murine embryogenesis also suggest that this agonist is highly specific for the Hh pathway (23). Because agonists and antagonists of several transmembrane proteins are often structurally related (28), we reasoned that modification of SAG structure could generate additional inhibitors targeting Smo with high potency and specificity. Here we report two lead compounds identified from our studies and demonstrate their ability to inhibit Hh pathway activation in cultured cells and whole organisms through binding to Smo. Furthermore, we provide evidence that SAG and our compounds regulate Smo activity through conformation changes. These new inhibitors should be useful as reagents for interrogating Hh pathway-dependent physiology and as drug candidates for targeting dysregulated Hh pathway activity in pathological conditions.
EXPERIMENTAL PROCEDURES
Synthetic Protocols
The details are described in the supplemental materials and Ref. 29. Supplemental Fig. S1 illustrates our new synthetic route of SAG, and supplemental Fig. S2 demonstrates parallel syntheses of the initial SANT library. Accordingly, the scaffold of SANT could be constructed by combining five distinct modules. The design elements for SANT library included varying the substitution pattern of the biaryl fragment, replacing the cyclohexane-1,4-diamine with benzene-1,4-diamine, changing chloride on the benzo[b]thiophene ring to hydrogen, and displacing the methyl group with other alkyl groups. Synthetically, the biaryl subunit C1–4 was achieved by palladium-catalyzed coupling reaction of the commercially available aryl boronic acids A1–2 and aryl bromides B1–2 (see supplemental Fig. S2); C1–4 then underwent the reductive amination with amine D1–2 to form the secondary amines E1–8. After coupling with two types of acyl chloride F1–2, the formed products were then reacted with alkyl iodide, followed by treatment with trifluoroacetic acid to give a 48-membered SANT library in overall good yields (29).
Generation of Gli-GFP Transgenic Zebrafish
A DNA fragment containing the 8 Gli-binding sites with a minimal promoter was generated by PCR from Gli-dependent firefly luciferase plasmids (30) using primer 8gli (forward, CGAGCTAGCGGATCCCCGGGAACAGATTC, and reverse, GCGACGCGTTTTACCAACAGTACCGG). The PCR product digested with NheI and MluI was ligated to GFP and used to generate transgenic zebrafish by microinjection. We identified eight founder lines expressing GFP that reflect the pattern of Hh signaling activity in developing embryos. Four lines exhibiting stronger expression throughout development were maintained and used in this study.
Screening Methods
The primary screen was carried out in 96-well plates. 2 μl of each compound (8 mm in Me2SO) was diluted to a total volume of 200 μl with Holtfreter's buffer. The average concentration of each small molecule was ∼80 μm. The embryos were distributed to 96-well plates with three embryos placed in each well. Then diluted solution of each compound was added. The embryos were exposed to compound solution and incubated at 28.5 °C from 2 h post-fertilization (hpf) until 48 hpf. Phenotypes were observed for two times at 36 and 48 hpf, separately. Once active compounds were identified, each one was analyzed at 80, 40, 20, 10, 5, 2, and 0.5 μm to determine its lowest effective concentration.
Image Acquisition and Analysis
GFP-positive embryos or larvae were viewed under an Axioimager Z1 fluorescence microscope (Zeiss), equipped with 5, 10, and 20× lenses and filter set 10 for detection of GFP (excitation, 450–490 nm; barrier, 510 nm; emission, 515–565 nm).
Quantitative Real Time PCR
The larvae were exposed to different concentrations of small molecules from 2 hpf. Total RNA was extracted from the embryos at 36 hpf using the TRIzol kit (Ambion, Austin, TX) according to the manufacturer's protocols. Three groups/treatment, each composed of 20 embryos, were assayed using SYBRGreen qPCR kit by real time PCR (ABI Prism 7700 sequence detector system).
The primer sequences used were as follows: zgli1, forward, GGGGAACATCTACAGTCATC, and reverse, GTGGCAGTTCGTCTCATAAA; zptc1, forward, TGATTGTGACTCCTTTGG, and reverse, TCCTTATTAGGGGCACTG; and zbactin1, forward, CTATGAGCTGCCTGACGG, and reverse, TGGTGGAAGGAGCAAGAG.
Compound Activities in Shh-LIGHT2 Cells
Shh-LIGHT2 cells (ATCC CRL-2795), a NIH-3T3 cell line stably incorporating Gli-dependent firefly luciferase and thymidine kinase promoter-driven Renilla luciferase reporters (19), were cultured to confluency in 96-well plates using DMEM containing 10% calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were then treated with various concentrations of the compounds in DMEM containing 0.5% calf serum and a 1:20 dilution of Shh-N-conditioned medium (24). The cells were cultured for an additional 30 h under standard conditions. Firefly and Renilla luciferase activities were measured on a Veritas luminometer (Turner Biosystems) using a dual luciferase kit (Promega) according to the manufacturers' protocols.
Smo Binding Assay
HEK 293T cells were cultured in DMEM containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin to 50% confluency in 96-well plates and then transfected with a cytomegalovirus promoter-driven mouse Smoothened-Myc3 expression construct (100 ng of cDNA/well) with FuGENE 6 (Roche Applied Science) according to the manufacturer's protocols. After 12 h the cells were incubated with DMEM containing 0.5% fetal bovine serum, 10 μm Hoechst 33342, 5 nm BODIPY-labeled cyclopamine (17), and various concentrations of SANT 75 for 1 h. The cells were washed two times with phosphate-buffered saline buffer and then imaged in DMEM containing 0.5% fetal bovine serum using an ImageXpress 5000A microscopy system (Molecular Devices).
Constructs and mSmo Conformation Analysis by FRET Using Confocal Microscopy
To construct mSmo-CFPC/YFPC, CFP, or YFP was fused in frame to mSmo C terminus in pGE vector between NheI and SalI, respectively, with the following primer sequences: 5′-GTACGCTAGCATGGTGAGCAAGGGCGAGCTG-3′ and 5′-GTACGTCGACTCACTTGTACAGCTCGTCCATG-3′. To construct mSmo-CFPN/YFPN, CFP or YFP was inserted in frame into mSmo by SpeI site after signal peptide site (amino acid 31). The primer sequences are as follows: 5′-GTACACTAGTATGGTGAGCAAGGGCGAGCTG-3′, and 5′-GTACACTAGTCTTGTACAGCTCGTCCATG-3′. To construct mSmo-CFPL2YFPC, CFP was inserted into mSmoYFP construction by NotI after mSmo amino acid 355. The primer sequences are as follows: 5′-GTACGCGGCCGCTGGTGAGCAAGGGCGAGCTG-3′ and 5′-GTACGCGGCCGCACTTGTACAGCTCGTCCATG-3′.
The effect of SAG, SANT74, and SANT75 on mSmo conformation change was tested by FRET in NIH-3T3 cells that were co-transfected with mouse Smo CFP and Smo YFP reporter constructs. NIH-3T3 cells were cultured in DMEM containing 10% bovine calf serum and antibiotics PS at 5%CO2 in a humidified incubator. Transfection was carried out using FuGENE 6 (Roche Applied Science). For FRET analysis of cultured cells, CFP- and YFP-tagged constructs were transfected into NIH-3T3 cells. Transfected cells were treated in the presence of compounds with or without Shh-conditioned medium. The cells were washed with phosphate-buffered saline, fixed with 4% formaldehyde for 20 min, and mounted on slides in 80% glycerol. Fluorescence signals were acquired with the 100× objective of a Zeiss LSM510 confocal microscope. CFP was excited by 458-nm light, and the emission was collected through a BP 480–520-nm filter. YFP was excited by 514-nm light, and the emission was collected through a BP 535–590-nm filter. CFP signal was obtained once before and once after photobleaching YFP using the full power of the 514-nm laser line for 1–2 min at the top half of each cell, leaving the bottom half unbleached that serves as an internal control. The intensity of CFP was analyzed using the Metamorph software (Universal Imaging Corp). Energy transfer efficiency was calculated using the formula: FRET% = (CFP signal after photobleaching − CFP signal before photobleaching/CFP signal after photobleaching) × 100. Each data set was based on 10–15 individual cells. In each cell, four to five regions of interest in photobleached area were selected for analysis.
RESULTS
Chemical Synthesis, Gli-GFP Transgenic Zebrafish Generation and Library Screening
Our initial syntheses produced 48 compounds that were designed to survey the functional importance of the following structural elements in SAG: 1) position of nitrogen atom on pyridine, 2) direction of the two aryl rings, 3) chloro-substituent on benzo[b]thiophene, 4) the conformation and electric effect of the six-member ring connecting the amine and amide, and 5) size of the substituted group on the amine (supplemental Figs. S1 and S2).
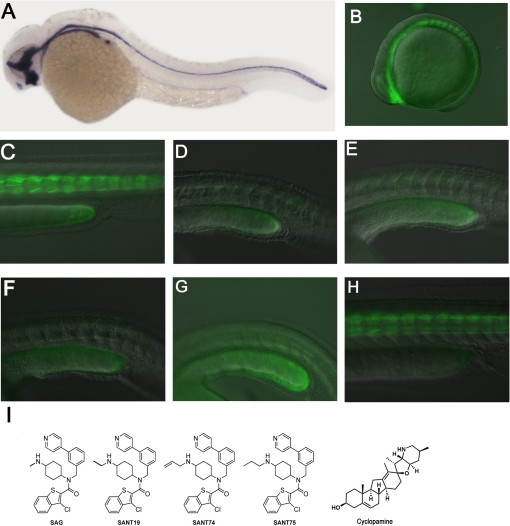
In zebrafish, Shh is expressed primarily in notochord during embryogenesis (Fig. 1A). We produced stable transgenic zebrafish carrying reporter constructs with eight Gli-binding sites linked to a minimal promoter and GFP. We obtained several lines in which the transgenic embryos exhibited the strongest GFP expression in the somite tissues immediately adjacent to the midline structure that has the highest Shh expression (Fig. 1, B and C). The GFP expression pattern is consistent with the expression of Gli1 and Ptc1 in zebrafish embryos at the stages analyzed. More importantly, this expression was abolished upon the treatment with 20 μm cyclopamine (Fig. 1D). These findings suggested that Gli-GFP expression in these transgenic fish was reflective of the endogenous activity pattern of Shh and was sensitive to Hh signaling level changes induced by chemical inhibitors.
FIGURE 1.
Identification of SANT74 and SANT75 using Gli-GFP transgenic zebrafish embryos. A, Shh expression in the midline structure of zebrafish embryo as detected by RNA whole mount in situ hybridization. B and C, expression of Gli-GFP in the somites near the midline of early embryos (13somite and 36 hpf). D, ablation of Gli-GFP by 20 μm cyclopamine in 36-hpf embryos. E. Ablation of Gli-GFP by 80 μm SANT19 in 36-hpf embryos. F and G, ablation of Gli-GFP by 20 μm SANT74 and 75 in 36-hpf embryos. H, Gli-GFP expression of 36-hpf embryos treated with 20 μm SAG. I, structures of SAG, SANT19, SANT74 SANT75, and cyclopamine.
Using these embryos as a tool, we screened the library mentioned above and identified one compound that could block Gli-GFP expression at ∼80 μm (Fig. 1E), producing a phenotype visually identical to that of 20 μm cyclopamine treatment. We named this compound SANT19 for Smoothened antagonist (Fig. 1I), which has a structure very similar to SAG but with an ethyl group on the terminal nitrogen. Under the same condition, embryos treated by SAG had no effect on Gli-GFP expression (Fig. 1H). This finding suggests that SAG derivatives with N-alkyl substituents larger than a methyl group might exhibit significant inhibitory activity. Based on this speculation, a second library containing 13 compounds was synthesized to explore the size and electric effect of the substituted group (supplemental Fig. S3).
Further screening of this new library identified two additional compounds that were able to abolish Gli-GFP expression in the zebrafish embryo assays at 20 μm (Fig. 1, F and G). We named these two compounds SANT74 and SANT75, respectively (Fig. 1I). In this transgenic zebrafish assay, action of SANT74 and SANT75 is dose-dependent, with an observable inhibition of Gli-GFP at 5 μm (supplemental Fig. S4) and induction of strong embryonic structural abnormality at 40 μm or above (data not shown). All three substitutions (allyl, propyl, and methyl) are hydrophobic; propyl and allyl groups are larger in size than methyl groups. (Fig. 1I).
Characterization of SANT74 and SANT75 in Zebrafish
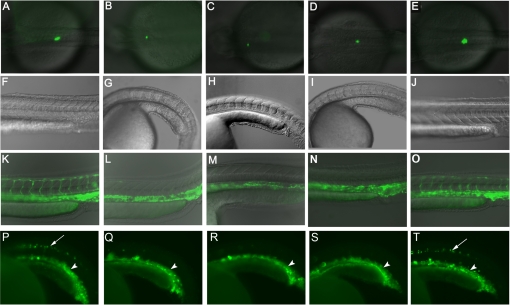
Genetic studies have demonstrated that characteristic phenotypes of zebrafish embryos with deficient Hh signaling activity include U-shaped somites, fewer pancreatic insulin positive β cells, and shorter intersegmental blood vessels (ISV). If SANT74 and SANT75 are specific inhibitors of Hh treating, zebrafish embryos should specifically produce these well characterized phenotypes. Other developmental defects such as general necrosis and abnormalities in unrelated organs would suggest off target effects of these compounds. We therefore selected these criteria to evaluate the specificity of SANT74 and SANT75 in zebrafish, using cyclopamine as an established control. To reveal changes in pancreas and blood vessels, transgenic zebrafish that have β cells labeled by insulin-GFP and vascular endothelial cells labeled by Flk-GFP were treated with either SANT74, SANT75, or cyclopamine. Similar to cyclopamine, treatment with 20 μm SANT74 and SANT75 from 2 to 36 hpf significantly reduced insulin-GFP cells and produced U-shaped somites (Fig. 2, A–J). The embryos treated with 40 μm SANT74, SANT75, and cyclopamine from 17 to 36 hpf completely lacked ISV (Fig. 2, L–N). These phenotypes were not observed in embryos treated with the same concentration of SAG (Fig. 2, E, J, and O), indicating that this inhibitory activity is specific to SANT74 and SANT75. We also treated transgenic zebrafish embryos expressing gata1-GFP, which labels the erythroid cell lineage that is in close contact with vascular endothelial cells and did not observe any changes (Fig. 2, P–T). This transgenic fish also showed some motor neurons labeled by GFP. The embryos treated with SANT74 SANT75 and cyclopamine lacked this expression in marked motor neurons compared with embryos treated with Me2SO and SAG (Fig. 2, P–T), which is consistent with the fact that Hh signaling is required for motor neuron development (31). The expression of Axin2, a Wnt signaling protein and target gene, is further detected by real time PCR, and the data showed no changes between embryos treated with 20 μm SANT74 and Me2SO (supplemental Fig. S5 C). These studies suggest that SANT74 and SANT75 have no obvious off target effects. In zebrafish assay, combining 10 μm of SANT74 or SANT75 and 10 μm of cyclopamine produced an inhibitory effect similar to 20 μm of each compound alone, suggesting additive action on the same protein target of Smo (data not shown).
FIGURE 2.
Characterization of SANT compounds in zebrafish. A, in untreated transgenic embryo (36 hpf), pancreatic islet cells revealed by insulin-GFP appear as green fluorescence. B–D, embryos treated with 20 μm SANT74 SANT75 and cyclopamine from 2 hpf display decreased number of islet cells in 36 hpf. E, embryos treated with 20 μm SAG from 2 hpf display normal number of islet cells in 36 hpf. F, the V-shaped somites in Me2SO-treated wild type embryo (36 hpf). G–I, embryos treated with 20 μm SANT74 SANT75 and cyclopamine from 2 hpf displayed ventrally curved bodies and U-shaped somites in 36 hpf. J, V-shaped somites of embryo treated with 20 μm SAG (36 hpf). K, in untreated transgenic embryo (36 hpf), blood vessels revealed by Flk-GFP appear as green fluorescence. L–N, embryos treated with 40 μm SANT74, SANT75 and cyclopamine from 17 hpf specifically lacks ISV but maintains normal V-shaped somites in 36 hpf. O, embryo treated with 40 μm SAG from 17 hpf display both normal ISV and V-shaped somites. P, in untreated transgenic embryo (21 hpf), red blood cells revealed by gata1-GFP appear as green fluorescence in the ventral trunk region (arrowhead). This transgenic embryo also has GFP expression in spinal motor neurons (arrow). Q–S, embryos treated with 20 μm SANT74, SANT75 and cyclopamin from 2 hpf show the same number of gata1-GFP labeled red blood cells (arrowhead) but reduced motor neurons (arrow) in 21 hpf. T, embryo treated with 20 μm SAG from 2 hpf appears same as the untreated control.
As a small molecule, SANT74 or SANT75 offers the temporal control of Hh activity. We tested the timing requirement of Hh for normal somite formation and sprouting of ISV by adding and washing away SANT74 at various developmental stages, followed by phenotypic analysis. Before 17 hpf, addition of SANT74 will induce U-shaped somites formation. When SANT74 was added to the embryos upon fertilization but washed away before 17 hpf, the sprouting of ISV could occur normally (supplemental Fig. S6). To affect ISV specifically without inducing U-shaped somites, SANT74 or SANT75 can be added between 17 and 24 hpf (Fig. 2, L and M). After 24 hpf, adding SANT74 or SANT75 no longer had any effect on the sprouting of ISV. These studies demonstrated that a critical stage for the sprouting of ISV is between 17 and 24 hpf and that development of ISV is independent of somites.
Ptc1 and Gli1 are two target genes of Hh signal pathway, and treatment of SANT74 or SANT75 should inhibit their expression. As measured by real time, quantitative PCR mRNA levels of Ptc1 and Gli1 in zebrafish embryos were reduced by SANT74 and SANT75 in a dose-dependent manner (supplemental Fig. S5; only SANT74 is shown).
Characterization of SANT74 and SANT75 in Mammalian Cells
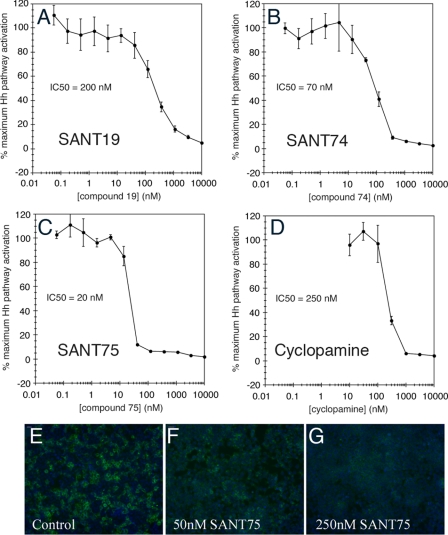
The data listed above demonstrated that SANT74 and SANT75 are potent and specific inhibitors of Hh in zebrafish. To address whether these compounds also inhibit Hh signaling in mammalian cells, Shh-LIGHT2 cells cultured in Shh-N-conditioned medium were treated with various concentrations of SANT19, SANT74, and SANT75, and luciferase reporter activity was measured as previously reported (1, 24). Under this condition, SANT19, SANT74, and SANT75 had an IC50 of 200, 70, and 20 nm, respectively (Fig. 3, A–C), all exhibiting inhibitory potency greater than that of cyclopamine (IC50 = 250 nm) (Fig. 3D). SAG has been shown as an agonist that competes with cyclopamine in binding to Smo protein at its heptahelical bundle (17, 24). To determine whether SANTs compete with cyclopamine in a similar fashion, Smo-overexpressing HEK 293T cells counterstained with Hoescht 33342 (blue) were incubated with 5 nm BODIPY-cyclopamine (green) and different concentrations of SANT75. As shown in Fig. 3 (E–G) and supplemental Fig. S7, SANT75 could effectively abrogate BODIPY-cyclopamine/Smo binding in a dose-dependent manner, and the concentrations of SANT75 correlated with its IC50 in the Shh-LIGHT2 cell assay. These findings confirmed that SANT75 is an inhibitor of cyclopamine that binds to Smo protein in mammalian cells.
FIGURE 3.
Inhibition of Hh activation by SANT compounds in Shh-LIGHT2 cells. A–C, Shh-LIGHT2 cells incubated with Shh-N-conditioned medium and various concentrations of SANTs were measured for Hh pathway-dependent firefly luciferase activity. SANT74 and SANT75 inhibited Hh pathway activation with potencies greater than that of cyclopamine (IC50 = 250 nm, D). E–G, SANT75 blocks the binding of BODIPY-cyclopamine to Smo. Smo-overexpressing HEK 293T cells were incubated with 5 nm BODIPY-cyclopamine (green) and then indicated with different concentrations of SANT75 (50 and 250 nm are shown here) and counterstained with Hoescht 33342 (blue). The concentrations at which SANT75 effectively abrogates BODIPY-cyclopamine/Smo binding correlate with the IC50 of this inhibitor in the Shh-LIGHT2 cell assay.
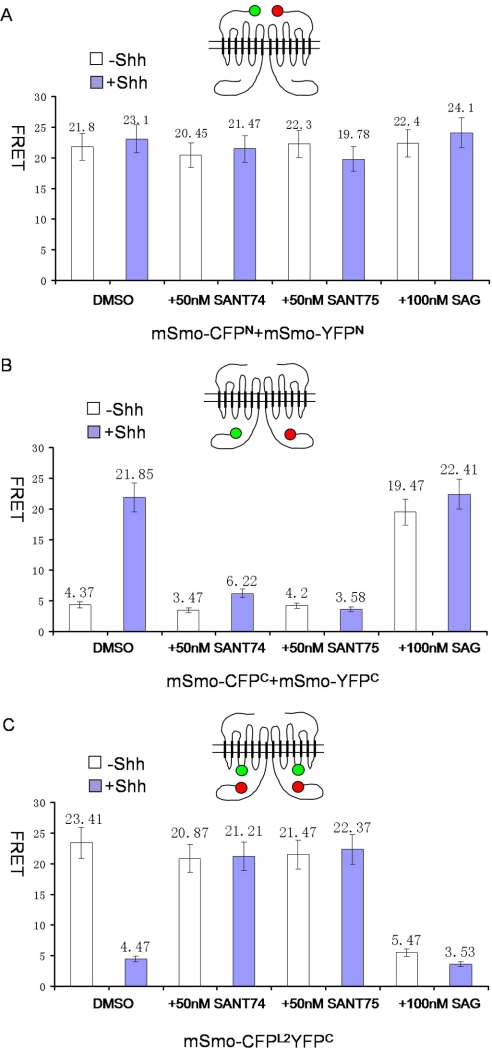
A recent study has suggested that Hh regulates Smo activity by inducing a conformational switch and resulting in increased proximity of its C-terminal cytoplasmic tails (C-tail) (32). SAG and SANT have very similar structures but modulate Smo activity in opposite directions. These two molecules therefore are likely candidates capable of inducing active or inactive conformations independent of Hh ligand. To test this, the conformation status of Smo was determined by FRET analyses using three pairs of fluorescently tagged Smo: 1) mSmo-CFPN and mSmoYFPN, which measures the distance between Smo N termini; 2) mSmo-CFPC and mSmo-YFPC, which measure the distance between Smo C termini; and 3) mSmo-CFPL2YFPC, which measures the distance between Smo C terminus and the second intracellular loop (Fig. 4) (32). NIH3T3 cells transfected with mSmo-CFPN and mSmoYFPN, mSmo-CFPC and mSmo-YFPC, or mSmo-CFPL2YFPC were treated with or without Shh and SANT74 (50 nm), or SANT75 (50 nm), or SAG (100 nm). mSmo-CFPN/mSmoYFPN exhibited high basal FRET (Fig. 4A), consistent with the previous observation that Smo exhibits as a constitutive dimer/oligomer (32). This high basal FRET value was not affected by either SAG or SANTs (Fig. 4A), suggesting that Smo dimer/oligomer does not dissociate or the distance between Smo N termini is not disturbed in the presence of SAG or the SANTs. In Fig. 4B, in the absence of Shh, the mSmo C-tails within the dimer/oligomer adopt an inhibitory conformation that prevents further aggregation. With Shh, mSmo is activated and increases the proximity between mSmo C-tails. The FRET value between mSmo C-tail CFP and mSmo C-tail YFP changed from ∼4.37 to ∼21.85% after Shh treatment. Shh-induced increase in FRET value between mSmo-CFPC and mSmo-YFPC (FRETC) was blocked by either SANT74 or SANT75 (Fig. 4B). In Fig. 4C, in the absence of Shh, the FRET value between the second internal loop CFP and C-tail YFP is high (∼23.41%) because mSmo keeps a “closed” conformation. Shh could activate mSmo and make mSmo C-tail conformation change to an “open” status, decreasing the FRET value. Similarly, Shh-induced decrease in FRET obtained from mSmo-CFPL2YFPC (FRETL2C) was also blocked by SANT74 and SANT75 (Fig. 4C), suggesting that both SANT74 and SANT75 can override Shh-induced Smo conformational change. Thus, SANT appears to lock mSmo in the “closed” inactive conformation. In contrast, SAG induced an increase in FRETC and decrease in FRETL2C independent of Shh (Fig. 4, B and C), suggesting that SAG binding to Smo promotes its “open” active conformation.
FIGURE 4.
Conformational changes of Smo induced by SAG and SANT compounds. Mouse Smo proteins were fused to CFP and YFP at the N terminus, the C terminus, and intramolecularly. FRET was assayed in the absence or presence of Shh with SANT 74, SANT75 and SAG. A, Shh-independent FRET observed at the extracellular ends is not affected by these compounds. B, Shh-induced FRET at the cytoplasmic tails is reduced by SANT 74 and SANT75 but increased by SAG. C, Shh inhibited FRET between one cytoplasmic tail, and the intramolecular domain is increased by SANT 74 and SANT 75 but reduced by SAG.
DISCUSSION
Here we developed an effective phenotype-based transgenic zebrafish embryo assay for Hh signaling pathway inhibitors. This system, using Gli-GFP as a reporter to link activity of a compound to Hh and using other characteristic phenotypes to measure specificity, allows rapid selection of highly potent small molecule inhibitors of Hh. This complements the cell-based Hh modulator screen systems that are less effective to eliminate nonspecific compounds. Because the GFP expression in the embryos is specific and reflective of Hh activity, it is possible to use this fish line in an automated system that detects a simple change of the GFP fluorescence level.
Using zebrafish coupled with mammalian cell assays, our screen identified two highly specific and potent inhibitors of Smo. The IC50 values of these two compounds are in the range of 20–50 nm, which are the best among all reported small molecule Hh pathway inhibitors. It is worth mentioning that the zebrafish assay and Shh-Light2 cell assay have different values in evaluating the effective concentrations. The minimal active concentrations of SANT74 and SANT75 in Shh-Light2 cell assay are 50–100 times more effective than that of zebrafish assay. This is consistent with the previous observations for cyclopamine analyzed by Shh-Light2 cell assay and zebrafish assay. It is likely due to drug metabolism and distribution difference between single cell suspension in vitro and whole embryo body in vivo.
According to the screening results, the following observations about chemistry and structure activity of SANT74 and SANT75 on Smo are apparent. 1) Considering the pyridine-based biaryl fragment, the pyridine ring should be on the meta-position to the entire scaffold, and the nitrogen of pyridine located at the para-position gave the best result as compared with the substrates with meta-substitution. 2) The “Cl” group on the benzo[b]thiophene is necessary to maintain the activity because the replacement of “Cl” group with “H” leads to much lower activity. 3) In comparison with the substitution effects of alkyl group of cyclohexane-1,4-diamine on the outcomes of activity, the substitution group should be hydrophobic, and the activity was increased as the size of substitution group becomes bigger ( H <
H <  CH3 <
CH3 <  CH2CH3 <
CH2CH3 <  CH2CH2CH3 or
CH2CH2CH3 or  CH2CH = CH2). However, when bulky groups, such as
CH2CH = CH2). However, when bulky groups, such as  CH2CH2CH2CH3 and
CH2CH2CH2CH3 and  CH2Ph, were employed as the substitutions, no activity was observed, presumably because the steric hindrance of those bulky groups prevents them from binding to their target (supplemental Fig. S3 and Table S1).
CH2Ph, were employed as the substitutions, no activity was observed, presumably because the steric hindrance of those bulky groups prevents them from binding to their target (supplemental Fig. S3 and Table S1).
How Smo is regulated to transmit Hh signal is still not well understood. It is proposed that secreted Hh ligands act directly on Ptch1 and change the subcellular distribution as well as conformation of Smo protein in the primary cilium (1, 33) (32). In the absence of Hh, Ptch1 is thought to pump out an unknown endogenous agonist or pump in an endogenous inhibitor of Smo so that Smo is kept in an inactive status. Binding of Hh to Ptch1 stops this pumping action, allowing transfer of Smo into primary cilium and changing it to an active status. In Drosophila, Smo cytoplasmic tails stay as an inactive conformation through intramolecular electrostatic interactions mediated by multiple arginine residues. Hh-induced phosphorylation disrupts the interaction and induces a conformational switch, allowing dimerization of Smo cytoplasmic tails, eventually leading to activation of Hh pathway (32). The cytoplasmic tail of mammalian Smo is essential for its signaling activity too. Although the phosphorylation clusters in Drosophila Smo are not conserved in mSmo, a recent study has suggested that mSmo is phosphorylated in response to Shh, and GPRK2 is a likely Smo kinase, whereas the phosphorylation sites in mSmo have yet to be defined (34). So it is conceivable that mSmo phosphorylation by GPRK2 or other kinases induced by Hh may control a conformational change in mSmo that leads to its activation. Our studies established that SAG and SANT74/SANT75 induce opposite conformational changes involving the cytoplasmic tail but have no effect at the extracellular end of Smo, in a manner corresponding to changes induced by Hh ligand. The structure of SANT75 only differs from SAG by containing a propyl substitution, which is larger than methyl group in SAG. Because SAG and cyclopamine both bind to heptahelical bundles of Smo (17, 24), SANT74 and SANT75 should likely bind to the same site. This argues that induction of conformation change at the cytoplasmic tail may not result from direct binding of these small molecules to this part of the protein. It is possible that SAG and SANT74/75 exert an opposing effect on Smo phosphorylation that could regulate its conformation. Although the mechanism of action by SAG and SANT74/75 remains to be elucidated, our studies provide a good example of structurally related analogs inducing drastic activity change of a protein through regulation of its conformation. The findings here also favor that the endogenous regulator of Smo likely acts on conformation of Smo protein. Oxysterols have recently been identified as an endogenous agonist to activate Hh signal pathway by helping Smo to reach the primary cilium (35, 36). It would be interesting to determine whether oxysterols and SANT74/75 have any interactive roles in regulating Smo conformation.
SANT74 and SANT75 are stronger inhibitors than cyclopamine in mammalian cells and should be useful reagents for studying Hh biology. Cyclopamine is a relatively weak compound, with an IC50 of about 250 nm in cultured cells, and has a structure that is less amendable for modification (Fig. 1I). In addition, cyclopamine is acid-sensitive and therefore is not ideal for oral delivery as a drug candidate. In our studies, SANT74 and SANT75 are relatively stable and have small structures. Their effects in whole zebrafish embryos assays are highly specific, showing no obvious off target toxicity. These observations suggest that SANT74 and SANT75 have good potential as therapeutic agents.
Supplementary Material
Acknowledgments
We thank Prof. Jianguo Chen for sharing the real time PCR instrument. We are grateful to Prof. Bo Zhang and Prof. Jiahua Chen for support and discussion.
This work was supported by National Science Foundation of China Grants 20325208, 20225318, 20521202, and 20672004, Ministry of Education of China Program 985, the Shenzhen Science and Technology Key Laboratory Promotion Fund, the Instrumental Analysis Fund of Peking University, and funds from the Sidney Kimmel Foundation for Cancer Research (to J. K. C.), the Astellas USA Foundation (to J. K. C.), and the Brain Tumor Society/Rachel Molly Markoff Foundation (to J. K. C.).

The on-line version of this article (available at http://www.jbc.org) contains Supplemental Figs. S1–S7 and Table S1.
- GFP
- green fluorescent protein
- hpf
- hours post-fertilization
- DMEM
- Dulbecco's modified Eagle's medium
- FRET
- fluorescence resonance energy transfer
- ISV
- intersegmental blood vessel(s).
REFERENCES
- 1.Rohatgi R., Scott M. P. (2007) Nat. Cell Biol. 9, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 2.Karhadkar S. S., Bova G. S., Abdallah N., Dhara S., Gardner D., Maitra A., Isaacs J. T., Berman D. M., Beachy P. A. (2004) Nature 431, 707–712 [DOI] [PubMed] [Google Scholar]
- 3.Jacob L., Lum L. (2007) Science 318, 66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie J., Murone M., Luoh S. M., Ryan A., Gu Q., Zhang C., Bonifas J. M., Lam C. W., Hynes M., Goddard A., Rosenthal A., Epstein E. H., Jr., de Sauvage F. J. (1998) Nature 391, 90–92 [DOI] [PubMed] [Google Scholar]
- 5.Gailani M. R., Ståhle-Bäckdahl M., Leffell D. J., Glynn M., Zaphiropoulos P. G., Pressman C., Undén A. B., Dean M., Brash D. E., Bale A. E., Toftgård R. (1996) Nat. Genet. 14, 78–81 [DOI] [PubMed] [Google Scholar]
- 6.Johnson R. L., Rothman A. L., Xie J., Goodrich L. V., Bare J. W., Bonifas J. M., Quinn A. G., Myers R. M., Cox D. R., Epstein E. H., Jr., Scott M. P. (1996) Science 272, 1668–1671 [DOI] [PubMed] [Google Scholar]
- 7.Tostar U., Malm C. J., Meis-Kindblom J. M., Kindblom L. G., Toftgård R., Undén A. B. (2006) J. Pathol. 208, 17–25 [DOI] [PubMed] [Google Scholar]
- 8.Wechsler-Reya R. J., Scott M. P. (1999) Neuron 22, 103–114 [DOI] [PubMed] [Google Scholar]
- 9.Lee Y., Miller H. L., Jensen P., Hernan R., Connelly M., Wetmore C., Zindy F., Roussel M. F., Curran T., Gilbertson R. J., McKinnon P. J. (2003) Cancer Res. 63, 5428–5437 [PubMed] [Google Scholar]
- 10.Hahn H., Wojnowski L., Miller G., Zimmer A. (1999) J. Mol. Med. 77, 459–468 [DOI] [PubMed] [Google Scholar]
- 11.Wechsler-Reya R., Scott M. P. (2001) Annu. Rev. Neurosci. 24, 385–428 [DOI] [PubMed] [Google Scholar]
- 12.Pepicelli C. V., Lewis P. M., McMahon A. P. (1998) Curr. Biol. 8, 1083–1086 [DOI] [PubMed] [Google Scholar]
- 13.Gering M., Patient R. (2005) Dev. Cell 8, 389–400 [DOI] [PubMed] [Google Scholar]
- 14.Pola R., Ling L. E., Silver M., Corbley M. J., Kearney M., Blake Pepinsky R., Shapiro R., Taylor F. R., Baker D. P., Asahara T., Isner J. M. (2001) Nat. Med. 7, 706–711 [DOI] [PubMed] [Google Scholar]
- 15.Nagase M., Nagase T., Koshima I., Fujita T. (2006) Microvasc. Res. 71, 85–90 [DOI] [PubMed] [Google Scholar]
- 16.Geng L., Cuneo K. C., Cooper M. K., Wang H., Sekhar K., Fu A., Hallahan D. E. (2007) Angiogenesis 10, 259–267 [DOI] [PubMed] [Google Scholar]
- 17.Chen J. K., Taipale J., Cooper M. K., Beachy P. A. (2002) Genes Dev. 16, 2743–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper M. K., Porter J. A., Young K. E., Beachy P. A. (1998) Science 280, 1603–1607 [DOI] [PubMed] [Google Scholar]
- 19.Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. (2000) Nature 406, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 20.Tabs S., Avci O. (2004) Eur. J. Dermatol. 14, 96–102 [PubMed] [Google Scholar]
- 21.Sanchez P., Ruiz i, Altaba A. (2005) Mech. Dev. 122, 223–230 [DOI] [PubMed] [Google Scholar]
- 22.Berman D. M., Karhadkar S. S., Hallahan A. R., Pritchard J. I., Eberhart C. G., Watkins D. N., Chen J. K., Cooper M. K., Taipale J., Olson J. M., Beachy P. A. (2002) Science 297, 1559–1561 [DOI] [PubMed] [Google Scholar]
- 23.Frank-Kamenetsky M., Zhang X. M., Bottega S., Guicherit O., Wichterle H., Dudek H., Bumcrot D., Wang F. Y., Jones S., Shulok J., Rubin L. L., Porter J. A. (2002) J. Biol. 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J. K., Taipale J., Young K. E., Maiti T., Beachy P. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 14071–14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams J. A., Guicherit O. M., Zaharian B. I., Xu Y., Chai L., Wichterle H., Kon C., Gatchalian C., Porter J. A., Rubin L. L., Wang F. Y. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 4616–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J., Wu X., Pasca di Magliano M., Peters E. C., Wang Y., Hong J., Hebrok M., Ding S., Cho C. Y., Schultz P. G. (2007) Chembiochem. 8, 1916–1919 [DOI] [PubMed] [Google Scholar]
- 27.Romer J. T., Kimura H., Magdaleno S., Sasai K., Fuller C., Baines H., Connelly M., Stewart C. F., Gould S., Rubin L. L., Curran T. (2004) Cancer Cell 6, 229–240 [DOI] [PubMed] [Google Scholar]
- 28.Fischer J., Ganellin C. R.International Union of Pure and Applied Chemistry (2006) Analogue-based Drug Discovery, pp. xxxi and 575, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 29.Wang N., Xiang J., Ma Z., Quan J., Chen J., Yang Z. (2008) J. Comb. Chem. 10, 825–834 [DOI] [PubMed] [Google Scholar]
- 30.Sasaki H., Hui C., Nakafuku M., Kondoh H. (1997) Development 124, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 31.Roelink H., Augsburger A., Heemskerk J., Korzh V., Norlin S., Ruiz i, Altaba A., Tanabe Y., Placzek M., Edlund T., Jessell T. M., Dodd J. (1994) Cell 76, 761–775 [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y., Tong C., Jiang J. (2007) Nature 450, 252–258 [DOI] [PubMed] [Google Scholar]
- 33.Denef N., Neubüser D., Perez L., Cohen S. M. (2000) Cell 102, 521–531 [DOI] [PubMed] [Google Scholar]
- 34.Chen W., Ren X. R., Nelson C. D., Barak L. S., Chen J. K., Beachy P. A., de Sauvage F., Lefkowitz R. J. (2004) Science 306, 2257- 2260 [DOI] [PubMed] [Google Scholar]
- 35.Dwyer J. R., Sever N., Carlson M., Nelson S. F., Beachy P. A., Parhami F. (2007) J. Biol. Chem. 282, 8959–8968 [DOI] [PubMed] [Google Scholar]
- 36.Corcoran R. B., Scott M. P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 8408–8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.