Abstract
The tumor suppressor gene HIC1 (Hypermethylated in Cancer 1) that is epigenetically silenced in many human tumors and is essential for mammalian development encodes a sequence-specific transcriptional repressor. The few genes that have been reported to be directly regulated by HIC1 include ATOH1, FGFBP1, SIRT1, and E2F1. HIC1 is thus involved in the complex regulatory loops modulating p53-dependent and E2F1-dependent cell survival and stress responses. We performed genome-wide expression profiling analyses to identify new HIC1 target genes, using HIC1-deficient U2OS human osteosarcoma cells infected with adenoviruses expressing either HIC1 or GFP as a negative control. These studies identified several putative direct target genes, including CXCR7, a G-protein-coupled receptor recently identified as a scavenger receptor for the chemokine SDF-1/CXCL12. CXCR7 is highly expressed in human breast, lung, and prostate cancers. Using quantitative reverse transcription-PCR analyses, we demonstrated that CXCR7 was repressed in U2OS cells overexpressing HIC1. Inversely, inactivation of endogenous HIC1 by RNA interference in normal human WI38 fibroblasts results in up-regulation of CXCR7 and SIRT1. In silico analyses followed by deletion studies and luciferase reporter assays identified a functional and phylogenetically conserved HIC1-responsive element in the human CXCR7 promoter. Moreover, chromatin immunoprecipitation (ChIP) and ChIP upon ChIP experiments demonstrated that endogenous HIC1 proteins are bound together with the C-terminal binding protein corepressor to the CXCR7 and SIRT1 promoters in WI38 cells. Taken together, our results implicate the tumor suppressor HIC1 in the transcriptional regulation of the chemokine receptor CXCR7, a key player in the promotion of tumorigenesis in a wide variety of cell types.
HIC1 (Hypermethylated in Cancer 1) is a tumor suppressor gene that resides on the short arm of chromosome 17, a region that is frequently deleted and epigenetically silenced in human cancers (1–4). HIC1 encodes a transcriptional repressor with five Krüppel-like C2H2 zinc fingers mediating DNA binding via its consensus binding site consisting of a 5′-(C/G)NG(C/G)GGGCA(C/A)CC-3′ sequence centered on a GGCA motif (2, 5). It also contains a central region that recruits CtBP5 co-repressor complexes (6) as well as an N-terminal BTB-POZ domain capable of autonomous transcriptional repression (7).
HIC1 is a direct target gene of p53 transactivation through a p53-responsive element (2, 8, 9). A regulatory feedback loop between HIC1 and p53 has been deciphered in which HIC1 directly represses the transcription of SIRT1, which deacetylates and thereby inactivates p53 (10, 11). Therefore, inactivation of one allele of HIC1 results in the de-repression of SIRT1 causing decreased p53-mediated transactivation of the remaining HIC1 allele. In addition, SIRT1 also deacetylates HIC1 and thereby favors its SUMOylation, thus establishing optimal transcriptional repression (12). Recently, HIC1, SIRT1, and E2F1 have also been implicated in a regulatory feedback loop because HIC1 represses the E2F1 promoter (13) and because E2F1 is a transcriptional activator of HIC1 (40). Furthermore, E2F1 is a crucial activator of SIRT1 transcription in response to DNA damage, but SIRT1 binds E2F1 and deacetylates it thus inhibiting E2F1-mediated gene activation (14, 15).
Current evidence places HIC1 inactivation as an initiating event in tumorigenesis because of the propensity of Hic1+/− mice to form spontaneous tumors (16) and the presence of HIC1 silencing events in pre-neoplastic conditions such as smoker's lung, colonic polyps, and cirrhotic liver (17). Elucidation of the tumorigenic mechanisms initiated by HIC1 inactivation is dependent upon the identification of the genes targeted by HIC1-mediated transcriptional repression. To identify these target genes, we created an adenoviral vector encoding a FLAG-HIC1 fusion protein and infected U2OS osteosarcoma cells, a cell line known to have lost HIC1 expression. Gene expression profiling was used to identify putative target genes, and confirmatory studies were then performed. Collectively, these studies identified CXCR7 as a direct transcriptional target of HIC1.
EXPERIMENTAL PROCEDURES
Construction of Replication-defective Recombinant Adenoviral Vectors
A HindIII-XbaI fragment containing the coding sequence of human HIC1 fused to an in-frame N-terminal FLAG epitope was prepared from the pcDNA3-FLAG-HIC1 vector (6) and cloned into the pAdCMV2 vector. Recombinant adenovirus vectors (Ad-FLAG-HIC1) were obtained as described previously (18). The recombinant adenovirus encoding green fluorescent protein (Ad-GFP) has been described previously (8).
Expression Profiling
U2OS cells were infected by adding virus stocks directly to the culture medium at an input multiplicity of 100 viral particles/cell (18). At nine time points after adenoviral infection (from 8 to 26 h), total RNA was isolated from each sample using TRIzol reagent (Invitrogen). 6.0 μg of total RNA from each sample was then converted to double-stranded cDNA using the SuperScript Choice System (Invitrogen). cDNA was purified using a phenol/chloroform/isoamyl alcohol extraction. Clean cDNA was used for the in vitro synthesis of biotin-labeled cRNA using the BioArray RNA transcript labeling kit (Enzo Diagnostics, Farmingdale, NY). cRNA was cleaned using RNeasy mini kits (Qiagen, Valencia, CA) and fragmented randomly to ∼200 bp. Labeled cRNAs were hybridized to human HG-U133A chips (Affymetrix) for 16 h. Each chip was scanned using a confocal laser scanner after staining with streptavidin phycoerythrin followed by a signal-amplifying second antibody. Data analysis was performed using the Affymetrix Microarray Suite 5.0 software to generate an absolute analysis for each chip. Each chip was scaled to a target intensity value of 150 to allow for inter-array comparisons. Per gene normalizations were performed by normalizing the genes in each HIC1 chip to their corresponding gene on the control GFP chip for each of the time points. Genes that were flagged as absent across all time points for both cell types were removed from the analysis. Self-organizing maps were used to identify major trends in expression. Raw data can be obtained at NCBI GEO (www.ncbi.nlm.nih.gov), accession number GSE9854, or the Children's National Medical Center Public Expression Profiling Resource site.
Western Blot and Antibodies
Proteins were fractionated by SDS-PAGE and transferred onto nitrocellulose membranes that were treated as described previously (6). Antibodies against HIC1 (325 and 2563) have been described previously (6). Anti-FLAG M2 (F3165, Sigma) is a monoclonal antibody directed against the epitope tag. We have also used monoclonal antibodies against Hsp60 (sc-13115, Santa Cruz Biotechnology) and polyclonal antibodies against CtBP1 (C8741, Sigma). The secondary antibodies were anti-rabbit and anti-mouse immunoglobulins and a horseradish peroxidase-linked whole antibody from Amersham Biosciences.
Generation of a Stable HIC1 Knockdown Cell Line
pLKO1 lentiviral vectors expressing short hairpin (sh) RNAs targeting HIC1 were purchased from Sigma. Two shRNAs (clone ID; NM_006487.1-1763s1c1 and 1-1982s1c1) that have been shown to efficiently knock down HIC1 mRNA (19) as well as a control shRNA (nontargeting shRNA vector, SHC002, Sigma) were used in these experiments. These vectors contain a puromycin resistance gene for selection of the transduced cells. Lentivirus production, transduction of cells, and selection of the transduced cells for 1 week using 2.0 μg/ml puromycin was performed as described (19).
Construction of Plasmids, Transient Transfection, and Luciferase Repression Assays
The CXCR7 promoter region was PCR-amplified from genomic DNA extracted from peripheral blood lymphocytes of healthy donors using forward and reverse primers containing KpnI and XhoI restriction sites (supplemental Table S1). After restriction digestion, the fragment was cloned in the pGL3 basic reporter gene vector to generate the CXCR7 promoter construct, pGL3 CXCR7 −813/+168. The −386/+164, −191/+164, and −26/+164 CXCR7 promoter constructs were prepared from this construct by PCR (supplemental Table S1). The −191/+164 and −26/+164 ΔXI mutants were generated by the two-round PCR strategy with the following mutant oligonucleotides: sense CAAAGCCATCATCTAGAGGGC and antisense CCTCTAGATGATGGCTTTGTAACCC, in which TGC in the HiRE site is replaced by CAT, a mutation shown to abolish HIC1 binding (5). The −863/+168 ΔXI construct has been obtained through restriction fragment swapping using a unique SpeI site (position −75 to −70). Briefly, the KpnI-SpeI fragment from the −863/+168 construct was exchanged with the small KpnI-SpeI fragment from the −26/+164 ΔXI mutant. All constructs were verified by nucleotide sequencing.
U2OS cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and transfected in Opti-MEM (Invitrogen) by the polyethyleneimine (Euromedex) method in 12-well plates with 500 ng of DNA (20). Cells were transfected for 6 h and then were incubated in fresh complete medium. They were rinsed in cold phosphate-buffered saline 48 h after transfection and lysed with the luciferase assay buffer. Luciferase and β-galactosidase activities were measured by using, respectively, beetle luciferin (Promega) and the Galacto-light kit (Tropix) with a Berthold chemiluminometer. After normalization to β-galactosidase activity, the data were expressed as fold activation relative to the empty pGL3 basic control vector. Results represented are the mean values and standard deviations from at least two independent transfections in triplicate.
Quantitative RT-PCR
Total RNA was reverse-transcribed using random primers and MultiScribeTM reverse transcriptase (Applied Biosystems). Real time PCR analysis was performed by FastStart DNA Master SYBR Green I PCR (Roche Applied Science) in a LightCycler fluorescence temperature cycler (Roche Applied Science) according to the manufacturer's instructions. The primers for SIRT1 and CXCR7 as well as control primers for actin and GAPDH are summarized in supplemental Table S2. Primers were used at a concentration of 0.5 μm. According to a melting point analysis, only one PCR product was amplified under these conditions. RNAs extracted from U2OS Ad-GFP were used to generate a standard curve for each gene. Results were normalized with respect to the internal controls and are expressed relative to the levels found in Ad-GFP-infected U2OS cells. Similar experiments were performed with cells transduced by the control shRNA or the shRNA targeting HIC1.
Chromatin Immunoprecipitation
ChIP was performed according to published protocols with slight modifications. Briefly, formaldehyde was added directly to the cultured cells to a final concentration of 1% for 10 min at 37 °C. The cross-linking was stopped by adding glycine to a final concentration of 0.125 m. After 5 min at 37 °C, cells were lysed directly in the plates by resuspension in cell lysis buffer for 5 min. Then the samples were pelleted, resuspended in nuclei lysis buffer, and sonicated to chromatins with an average size of 500 bp using a BioRuptor (Diagenode, Liege, Belgium). After preclearing with a 50% slurry of protein A-G beads preincubated with salmon sperm DNA and bovine serum albumin for 4 h at 4 °C, the chromatins were incubated with the anti-HIC1 antibodies, normal rabbit IgG, or with no antibodies overnight. The antibody-bound chromatin was then pooled down for 30 min with protein A-G beads, washed extensively, and eluted two times with 250 μl of elution buffer. After addition of 20 μl of 5 m NaCl, the cross-linking was reversed by overnight incubation at 65 °C. The immunoprecipitated DNAs as well as whole cell extract DNAs (input) were purified by treatment with RNase A and then proteinase K followed by purification on Nucleobond Extract II (Macherey-Nagel). The purified DNAs were used for PCR analyses using the relevant primers for SIRT1, CXCR7, and GAPDH (see supplemental Table S3).
For ChIP upon ChIP experiments, we started with 8-fold more cells than in the single ChIP experiment. After the first round of immunoprecipitation, the beads were pooled by centrifugation in TE buffer and incubated in 100 μl of elution buffer for 10 min at 65 °C. After centrifugation, the supernatant was diluted in 900 μl of IP buffer and incubated with the second antibody as in a single ChIP experiment.
RESULTS
HIC1 Expression in U2OS Cells Using an Adenoviral Vector Results in a Proliferation Arrest
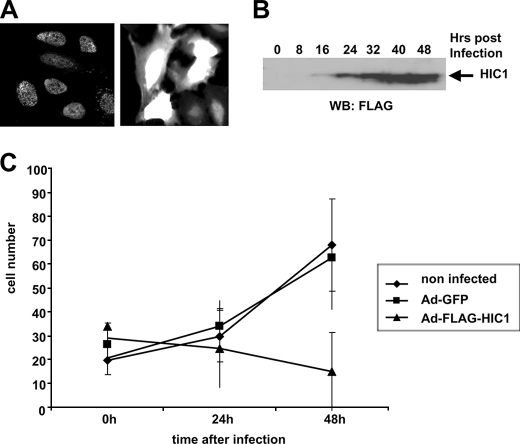
U2OS cells were infected with Ad-FLAG-HIC1 or Ad-GFP at a multiplicity of infection of 100 that was determined experimentally to allow infection of 90–100% of the cells as shown by immunofluorescence analyses at 24 h after infection. HIC1 proteins display a nuclear punctated localization typical of BTB/POZ proteins (Fig. 1 A).
FIGURE 1.
HIC1 overexpression in U2OS osteosarcoma cells induces a proliferation arrest. A, expression of HIC1 and GFP in infected U2OS cells. U2OS cells were infected with Ad-FLAG-HIC1 (left panel) or Ad-GFP (right panel), and 24 h later the expression of HIC1 and GFP was analyzed. The HIC1 protein is detected by immunofluorescence with the anti-FLAG M2 monoclonal antibody in a punctate nuclear pattern. B, expression of HIC1 proteins in Ad-FLAG-HIC1-infected U2OS cells. Equal amounts of total protein extracts were subjected to Western blotting (WB) using the anti-FLAG monoclonal antibody. C, overexpression of HIC1 through adenoviral infection induces a growth arrest in U2OS cells. U2OS osteosarcoma cells were seeded in 60-mm plates and infected with Ad-FLAG-HIC1 or Ad-GFP at a multiplicity of infection of 100 to allow infection of at least 90% of the cells. At the indicated times, cell counts were made on five microscopic fields, and each point is representative of the average number of noninfected control cells, Ad-FLAG-HIC1, and Ad-GFP U2OS-infected cells.
Examination of the kinetics of infection over 48 h demonstrated that Ad-FLAG-HIC1 proteins can be detected by Western blotting as early as 16 h post-infection (Fig. 1B). Ectopic overexpression of HIC1 resulted in a proliferation arrest starting around 16 h, coinciding with the detectability of the FLAG-HIC1 protein (Fig. 1C). U2OS cells infected with the control Ad-GFP virus did not undergo proliferation arrest.
Microarray Analyses Identify HIC1-activated and HIC1-repressed Genes
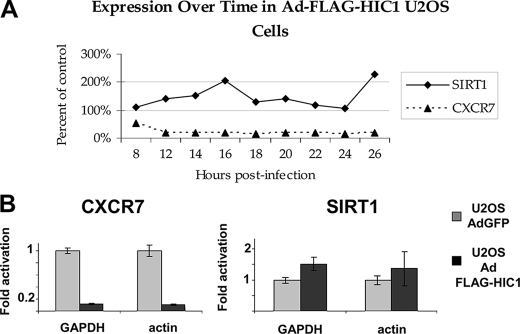
To obtain a profile of HIC1-regulated genes, we prepared total RNAs from U2OS cells infected at a multiplicity of infection of 100 with Ad-FLAG-HIC1 or Ad-GFP as control and extracted every 2 h starting at 8 until 26 h post-infection. Multiple infection time points provided independent sets of experiments that allowed the identification of early regulated genes that are more likely to be direct target genes. These RNAs were then used to interrogate the Affymetrix Human Genome U133A chip containing 14,500 transcripts. Direct comparisons were performed between Ad-FLAG-HIC1 and Ad-GFP-infected cells for each time of infection. These analyses revealed that 81 genes were down-regulated more than 3-fold, whereas 23 were up-regulated more than 3-fold after 16 and 24 h of infection, respectively (Tables 1 and 2). Upon closer analysis of the differentially expressed genes, it became apparent that the repressed genes were observed early post-infection, whereas the activated genes were predominantly found later in the infection kinetics, suggesting that they are not direct HIC1 target genes in keeping with the known function of HIC1 as a transcriptional repressor. Strikingly, SIRT1, a direct HIC1 target gene in WI38 cells (10), was not repressed in the Ad-FLAG-HIC1-infected U2OS cells; rather it was slightly activated (Fig. 2A). These microarray data were confirmed by qRT-PCR using RNAs from the 16-h time point (Fig. 2B).
TABLE 1.
Genes repressed more than 3-fold in Ad-FLAG-HIC1-infected U2OS cells at 16 h of infection
| Repressed gene name | Relative expression at 16 h | Affymetrix probe set no. | Chromosomal location |
|---|---|---|---|
| % | |||
| SNAP 43, SNAPC1 | 9 | 205443_at | 14q22 |
| TRIM16, TRIM16L | 17 | 204341_at | 17p11.2 |
| MAP2K3 | 18 | 215498_s_at | 17q11.2 |
| PLEC1 | 18 | 201373_at | 8q24 |
| RDC1, CXCR7 | 20 | 212977_at | 2 |
| TNS3 | 20 | 217853_at | 7p12.3 |
| LIF | 20 | 205266_at | 22q12.2 |
| WDR6 | 21 | 217734_s_at | 15q21 |
| AMIGO2 | 21 | 222108_at | 7 |
| MAP2K5 | 21 | 211371_at | 15q22.2-q22.31 |
| GRSF1 | 22 | 215030_at | 4q13.3 |
| FADS1 | 23 | 208963_x_at | 11q12.2-q13.1 |
| CYP24A1 | 23 | 206504_at | 20q13 |
| ADORA2B | 23 | 205891_at | 17p12-p11.2 |
| KIAA1026 | 25 | 213478_at | 1p36.13 |
| RIN2 | 26 | 209684_at | 20p11.22 |
| LRP8 | 26 | 205282_at | 1p34 |
| MCAM | 27 | 209086_x_at | 11q23.3 |
| ADRB2 | 27 | 206170_at | 5q31-q32 |
| TMEM16B | 28 | 220111_s_at | 12 |
| CCND1 | 29 | 208712_at | 11q13 |
| AHNAK2 | 29 | 212992_at | 14 |
| KCNJ6 | 30 | 214126_at | 21q22.13-q22.2 |
| LRP8 | 30 | 208433_s_at | 1p34 |
| CA12 | 30 | 203963_at | 15q22 |
| DRAL, SLIM3, FHL2 | 31 | 202949_s_at | 2q12-q14 |
| INHBA | 31 | 210511_s_at | 7p15-p13 |
| NOV | 31 | 214321_at | 8q24.1 |
| SFXN3 | 32 | 220974_x_at | 10q24.2 |
| SNAP-25 | 32 | 202508_s_at | 20p12-p11.2 |
| EPHA2 | 32 | 203499_at | 1p36 |
| SOX9 | 33 | 202935_s_at | 17q24.3-q25.1 |
| CXCL14 | 33 | 218002_s_at | 5q31 |
TABLE 2.
Genes activated more than 3-fold in Ad-FLAG-HIC1-infected U2OS cells at 24 h of infection
| Up-regulated gene name | Relative expression at 24 h | Affymetrix probe set no. | Chromosomal location |
|---|---|---|---|
| % | |||
| CA2 | 974 | 209301_at | 8q22 |
| MMP12 | 744 | 204580_at | 11q22.3 |
| ID4 | 644 | 209291_at | 6p22-p21 |
| SMPDL3A | 464 | 213624_at | 6q22.31 |
| LPPR4 | 460 | 213496_at | 1p21.2 |
| CCNE2 | 448 | 205034_at | 8q22.1 |
| C14orf45 | 447 | 220173_at | 14q24.3 |
| ACTA2 | 442 | 200974_at | 10q23.3 |
| TNFAIP8 | 413 | 210260_s_at | 5q23.1 |
| CDO1 | 407 | 204154_at | 5q22-q23 |
| SLC7A11 | 393 | 207528_s_at | 4q28-q32 |
| CYP7B1 | 386 | 207386_at | 8q21.3 |
| TMEM62 | 384 | 218776_s_at | 15q15.2 |
| KIAA0895 | 371 | 213424_at | 7p14.2 |
| PLA2G4A | 356 | 210145_at | 1q25 |
| FST | 355 | 204948_s_at | 5q11.2 |
| ABHD3 | 333 | 213017_at | 18q11.2 |
| SPP1 | 333 | 209875_s_at | 4q21-q25 |
| GPR161 | 328 | 214104_at | 1q24.2 |
| LGALS8 | 325 | 210732_s_at | 1q42-q43 |
| TMEM47 | 316 | 209656_s_at | Xp11.4 |
| SRGN | 310 | 201859_at | 10q22.1 |
| ITPR1 | 307 | 203710_at | 3p26-p25 |
FIGURE 2.
CXCR7 is down-regulated in Ad-FLAG-HIC1-infected U2OS cells, whereas SIRT1 is activated. A, expression levels of CXCR7 and of SIRT1. Total RNAs from U2OS cells (HIC1 null) infected with Ad-FLAG-HIC1 and Ad-GFP were prepared at the indicated times after infection (from 8 to 26 h), and Affymetrix HG U133A chips were used to measure the gene expression. Expression values were normalized to Ad-GFP-infected control cells at the same time points. The Percent of control corresponds to the ratio between the expression levels of CXCR7 and SIRT1 measured in Ad-GFP and in Ad-FLAG-HIC1-infected cells at each time point. B, confirmation of the microarray results for CXCR7 and SIRT1 by quantitative RT-PCR. qRT-PCR analyses were performed using total RNAs isolated from U2OS cells infected (time course point 16 h) with Ad-GFP (gray boxes) or with Ad-FLAG-HIC1 (black boxes) for CXCR7 (left column) and SIRT1 (right column). Values were normalized to GAPDH or actin as indicated.
Thus, SIRT1 is not a HIC1 target gene in the Ad-FLAG-HIC1-infected U2OS cells as has also been shown in an Ad-HIC1-infected human D245 medulloblastoma cell line (21). Nevertheless, our results are in close agreement with the well established function of HIC1 as a transcriptional repressor (5–7).
Down-regulation of CXCR7 in Ad-FLAG-HIC1-infected U2OS Cells
Among the genes most markedly repressed in HIC1-overexpressing U2OS cells, we decided to focus our efforts on the orphan G protein-coupled receptor RDC1 because of its relevance to cancer and the presence of HIC1 consensus binding sites in the regulatory region of the gene. RDC1 has been recently “deorphanized” as the scavenger chemokine receptor CXCR7 that binds chemokine CXCL11 and CXCL12, also known as SDF-1 (stromal-derived factor) (22, 23). CXCR7 is expressed in a variety of breast, lung, prostate, and others cancers (24) and promotes their growth in vivo (25). After infection by Ad-FLAG-HIC1, CXCR7 was specifically down-regulated 5–7-fold (Fig. 2A), as confirmed by Northern blot analyses (data not shown) and by qRT-PCR (Fig. 2B, left panel). Collectively, these data suggested that CXCR7 is a target gene of HIC1 in U2OS cells infected by Ad-FLAG-HIC1.
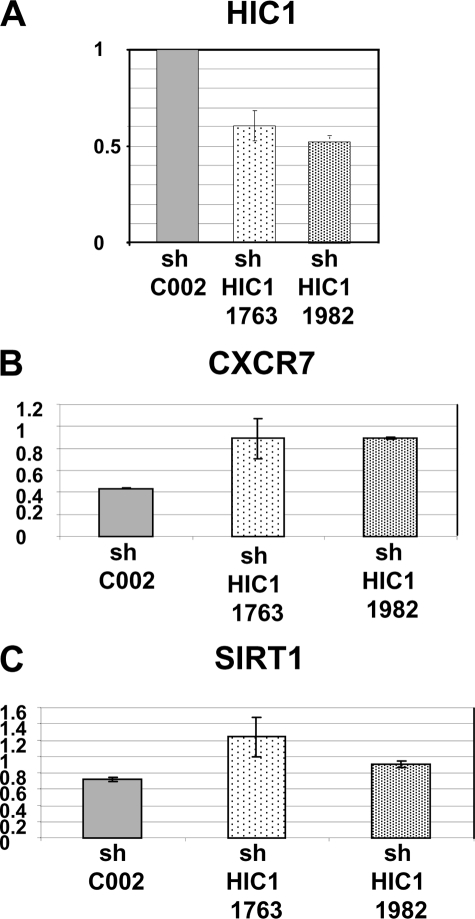
HIC1 mRNA Knockdown Increases CXCR7 and SIRT1 Expression in Normal WI38 Fibroblasts
All the previous results were obtained by ectopic HIC1 expression through infection with adenovirus of transformed cell lines not expressing endogenous HIC1. To validate CXCR7 as a HIC1 transcriptional target in a more physiological situation, we used RNA interference with a lentiviral shRNA against HIC1 to reduce endogenous HIC1 expression in normal human WI38 fibroblasts. These cells were chosen because they express HIC1 (supplemental Fig. S1) and have previously been used to validate SIRT1 as a direct HIC1 target gene (10). To that end, we generated WI38 cell lines expressing a nontargeting control shRNA (SHC002) and two cell lines expressing shRNAs targeting HIC1 (shHIC1 1763 and shHIC1 1982), as shown previously in HL60 acute myeloid leukemia cells (19). qRT-PCR analyses demonstrated an efficient, roughly 50%, knockdown of HIC1 in the WI38 shHIC1 1763 and 1982 cell lines as compared with the cells infected with the control shRNA (Fig. 3A). Similar analyses using the same RNA samples detected a 2-fold increase in CXCR7 expression (Fig. 3B) and a 1.5-fold up-regulation of SIRT1 (Fig. 3C). These results demonstrate that knockdown of endogenous HIC1 in normal human WI38 fibroblasts results in up-regulation of CXCR7 as well as of SIRT1, as shown previously (10).
FIGURE 3.
Inactivation of endogenous HIC1 in normal WI38 fibroblasts up-regulates CXCR7 and SIRT1 expression. A, WI38 cells were infected with the lentiviruses expressing a control shRNA (SHC002) and two shRNAs (SH1763 and SH1982) targeting HIC1 (Mission shRNAs; Sigma) as described previously (19). Total RNAs were extracted, and expression levels of HIC1 mRNA were assessed with real time quantitative PCR as described (9, 19). B and C, similarly, the expression levels of CXCR7 and SIRT1 were assessed using specific oligonucleotides (see supplemental Table S2). Values were normalized to actin.
Repression of CXCR7 Promoter by HIC1
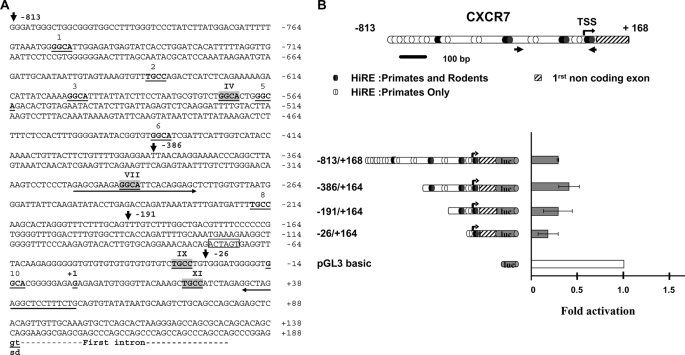
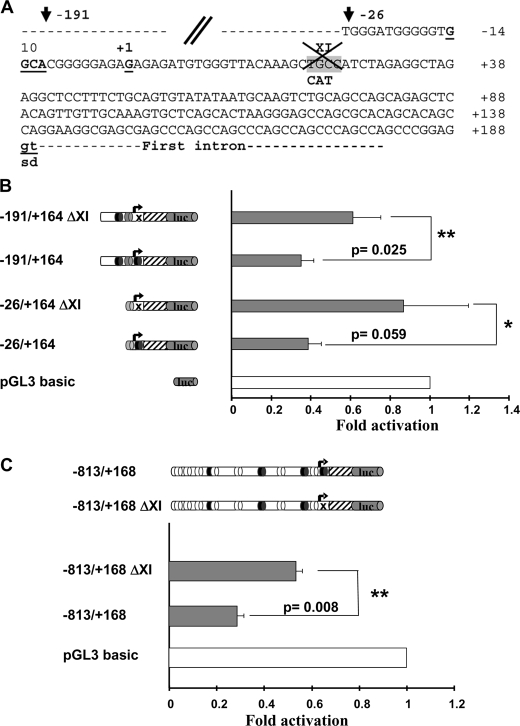
All the above described results demonstrate that CXCR7 is a HIC1 target gene. To determine whether CXCR7 is a direct target gene of HIC1, we first scanned its promoter region for the presence of consensus HIC1-responsive elements (HiRE, 5′-(C/G)NG(C/G)GGGCA(C/A)CC-3′) centered on a GGCA (reverse TGCC) core motif (5). These analyses identified 11 putative HiRE to which HIC1 could directly bind through its Krüppel-like C2H2 zinc fingers, some of which are conserved in primate (human and chimpanzee) and rodent (rat and mouse) genomes (Fig. 4A and supplemental Fig. S2).
FIGURE 4.
CXCR7 is down-regulated by HIC1. A, nucleotide sequence of the 5′ region of the human CXCR7 gene. The transcription start site as well as the first noncoding exon are described in GenBankTM under accession number NM_020311; gi 114155149. The conserved gt dinucleotides of the splice donor site (sd) are indicated in boldface letters and underlined. Upstream genomic sequences were extracted from the human genome sequence (NC_000002). Potential HiREs are indicated with the 4-bp core consensus 5′-GGCA-3′ (reverse complement strand TGCC) shown in boldface and underlined (5). These sites are numbered from 1 to 11. The sites indicated by roman numerals and highlighted in light gray boxes are those conserved in the primate (human and chimpanzee) and rodent (mice and rat) genomes (see also supplemental Fig. 3 for a CLUSTAL alignment). The 5′ boundaries of the various deletions in the CXCR7 promoter obtained by PCR are indicated by arrows with the numbering relative to the +1 position of the transcription initiation site. A unique SpeI site (position −75 to −70) used to obtain the −863/+168 ΔXI construct is boxed. Primers used to amplify the CXCR7 promoter fragment in the ChIP experiment presented in Figs. 6 and 7 are indicated by an arrow. B, from top to bottom, a schematic drawing of the 5′ promoter region and the first noncoding exon of CXCR7 is shown. Numbering is relative to the transcription start site (TSS) (bent arrow, position +1). The potential HiREs (5) are shown as black ovals for those found in the primate and rodent genomes or as white ovals for those found only in the primate genomes. The small arrows indicate the position of the primers used to amplify the relevant region of CXCR7 in the ChIP and ChIP upon ChIP experiments. The left panel shows a schematic drawing of various fragments of the human CXCR7 first noncoding exon 5′ sequences subcloned in the luciferase reporter plasmid pGL3 basic. Reporter constructs were transfected in triplicate into U2OS cells and assayed for luciferase activity. Luciferase (luc) activity was normalized for transfection efficiency using a co-transfected β-galactosidase reporter. Repression of transcription of each construct by HIC1 was calculated by first dividing luciferase activity in the absence of HIC1 by the activity in the presence of HIC1 (normalized for transfection efficiency using a co-transfected β-galactosidase reporter). The value obtained for each construct was then divided by the repressive effect elicited by HIC1 on the empty pGL3 basic vector to obtain the final fold of activation. Results, expressed relative to a value of 1.0 for cells transfected with the pGL3 empty vector, are expressed as the mean of three different experiments, and error bars represent standard deviations.
To directly assess the ability of HIC1 to repress transcription of CXCR7 through these sites, we cloned ∼0.8 kbp of genomic DNA upstream of the transcription start site and first noncoding exon of CXCR7, as defined in GenBankTM (NM_020311; gi 114155149), and performed luciferase promoter-reporter assays. To this end, the −813/+168 promoter region of CXCR7 was cloned in the pGL3 basic reporter vector, and a series of deletion constructs were made that gradually eliminated the putative HiREs (Fig. 4, A and B). These constructs were then transfected alone or with the pcDNA3 FLAG-HIC1 expression vector into U2OS cells, and promoter activities were thus measured in the absence or presence of HIC1. As shown in Fig. 4B, transient transfection of HIC1 significantly repressed the strong CXCR7 promoter activity in the −813/+168, −386/+164, and −191/+164 constructs. Interestingly enough, this repression is still elevated in the smaller construct −26/+164.
These results suggest that the regulatory region primarily involved in the HIC1-mediated repression of CXCR7 is located in the −26/+164 upstream region of the promoter. Notably, this region contains only one of the two phylogenetically conserved and adjacent HiREs (sites IX and XI) found in the same reverse orientation (5′-TGCC-3′) as in the SIRT1 promoter (10) (Fig. 4A and supplemental Fig. S2). The HiRE site IX, which is present in the −191/+164 construct, does not seem to be essential because its absence in the −26/+164 construct does not significantly impair the HIC1-mediated repression (Fig. 4B).
We thus introduced into this XI HiRE site a mutation (TGC into CAT) previously shown to abolish HIC1 binding (Fig. 5A) (5). The −191/+164 and −26/+164 constructs both display a high basal activity repressed by HIC1 expression (Fig. 4B). However, these two mutated ΔXI constructs are not significantly repressed in the presence of transfected HIC1 (Fig. 5B). Thus, these results demonstrate that this conserved site XI is essential for HIC1-mediated repression, whereas the nonconserved site 10 has little or no effect. In addition, the mutation of this XI site in the context of the longest promoter construct (−813/+168) results in a significant decrease of repression by HIC1 (Fig. 5C). In conclusion, the CXCR7 promoter is negatively regulated by HIC1 in transient transfection assays, strongly suggesting that CXCR7 is a direct HIC1 target gene.
FIGURE 5.
Phylogenetically conserved HiRE XI is essential for the transcriptional repression of CXCR7. A, nucleotide sequence of the 5′ region of the human CXCR7 gene showing the mutation introduced into the conserved HiRE XI site (TGC in the core motif replaced by CAT) to impede HIC1 binding (5). B, reporter constructs schematically shown in the left panel were transfected in triplicate into U2OS cells and assayed for luciferase activity. Luciferase (luc) activity is shown in the right panel (gray boxes). Repression of transcription of each construct by HIC1 was calculated exactly as detailed in legend to Fig. 4B. Results, expressed relative to a value of 1.0 for cells transfected with the pGL3 empty vector, are expressed as the mean of four different experiments, and error bars represent standard deviations. The p values are indicated (*, p values < 0.5; **, p values < 0.05). C, same experiments were performed with the −813/+168 wild-type and ΔXI constructs. The results are the mean of two independent transfections in triplicate with the p value indicated as in B.
CXCR7 and SIRT1 Are Direct HIC1 Target Genes in Two Different Human Cell Types Expressing Endogenous HIC1 Proteins
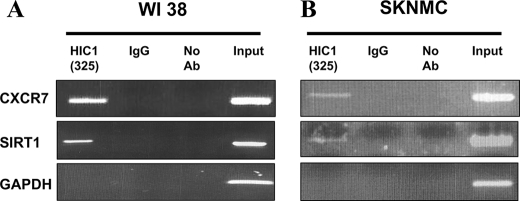
To confirm that CXCR7 is indeed a direct target gene of HIC1 in a more physiologically relevant system, we performed ChIP assays in two different human cell types that express endogenous HIC1 proteins as follows: normal human fibroblast WI38 cells (10) and Ewing/PNET SK-N-MC cells (supplemental Fig. S1). As a positive control for specificity, we used primers flanking the previously identified HiRE in the SIRT1 promoter in our ChIP-PCR analysis (10). As negative control, we used primers located in the GAPDH promoter. As shown in Fig. 6, we were able to specifically amplify the region encompassing the HiREs in the SIRT1 promoter from both WI38 and SK-N-MC chromatin immunoprecipitated by our polyclonal anti-HIC1 antibody (6) but not by normal rabbit IgG. Primers designed to amplify the 5′ region upstream of the GAPDH promoter did not yield a product, further demonstrating the specificity of this ChIP assay.
FIGURE 6.
ChIP analyses of HIC1 on CXCR7 and SIRT1 promoters. Normal human WI38 fibroblasts (A) or transformed human SK-N-MC Ewing/PNET cells (B) that both express endogenous HIC1 proteins (see supplemental Fig. 1) were cross-linked with 1% formaldehyde. Cross-linked chromatin immunoprecipitated (IP) with polyclonal anti-HIC1 antibody (Ab) (325), with rabbit IgG, or no antibody were used in PCR amplification with primers flanking the functional HiREs identified by our luciferase assays in the CXCR7 promoter. The region previously identified in the SIRT1 promoter in WI38 cells (10) was used as positive control, whereas PCR with the 5′ promoter of GAPDH was used as an internal nonbinding control (10). An H2O control corresponding to PCR without DNA yielded no amplified products (data not shown). Representative gels of one ChIP among three experiments are shown.
To test the promoter of CXCR7, we designed primers sets to amplify the region mediating the repressive effects of HIC1 on this promoter in transient transfection assays and containing adjacent HiREs (Fig. 4B, black arrows, and supplemental Table S3). These primers were used in PCR experiments using the same WI38 and SK-N-MC chromatin samples as above. CXCR7 was readily amplified from the HIC1-immunoprecipitated WI38 and SK-N-MC chromatins but not from chromatin immunoprecipitated by the control rabbit IgG, as shown previously for SIRT1 (Fig. 6, A and B). Taken together, these results demonstrate that endogenous HIC1 proteins are recruited onto the SIRT1 and CXCR7 promoters in vivo both in normal WI38 fibroblasts and in transformed SK-N-MC cells.
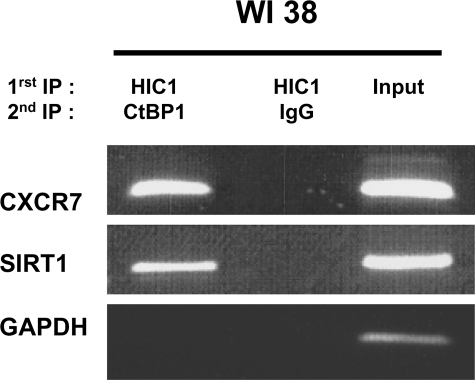
Co-occupancy of HIC1 and Its Co-repressor CtBP on the CXCR7 and SIRT1 Promoters
We have previously identified CtBP as a co-repressor of HIC1 (6, 11). We thus performed sequential chromatin immunoprecipitation assays (ChIP upon ChIP) to determine whether these two proteins are found co-localized on endogenous HIC1 target genes. In brief, WI38 chromatin was subjected to a first round of immunoprecipitation as described above. Then the bound material was eluted, and equal amounts were subjected to a second round of immunoprecipitation with anti-CtBP antibodies or normal rabbit IgG. A band corresponding to the relevant regions of the SIRT1 (10, 11) and CXCR7 promoters was detected in the HIC1/CtBP but not in the HIC1/IgG ChIP upon ChIP (Fig. 7). Primers designed to amplify the 5′ region upstream of the GAPDH promoter did not yield a product, further demonstrating the specificity of our assay.
FIGURE 7.
ChIP upon ChIP assays demonstrate that HIC1 and CtBP might form a stable complex on SIRT1 and CXCR7 promoters. Normal human WI38 fibroblasts were cross-linked with 1% formaldehyde. Cross-linked chromatin was sonicated and immunoprecipitated with polyclonal anti-HIC1 antibody (325) (1st IP HIC1). The bound material was eluted, divided in two, and subjected to a second round of immunoprecipitation with anti-CtBP antibodies (2nd IP CtBP1) or with normal rabbit IgG (2nd IP IgG). PCR amplifications were performed using primers flanking the functional HiREs previously identified in SIRT1 (10) and in CXCR7 (this study). PCR with the 5′ promoter of GAPDH was used as an internal nonbinding control as described in Fig. 6. Representative gels of one ChIP upon ChIP among two experiments are shown.
In conclusion, HIC1 and the co-repressor CtBP could form a complex at the SIRT1 and CXCR7 promoters in normal WI38 fibroblasts, likely resulting in their direct transcriptional repression.
DISCUSSION
Since its discovery in 1995 (2), a large body of information has been accumulated about HIC1, which is now recognized as a bona fide tumor suppressor gene (reviewed in Ref. 4). Hic1+/− heterozygous mice spontaneously develop an age- and gender-dependent spectrum of tumors (16), and the epigenetically silenced Hic1 gene cooperates with p53 to determine cancer prevalence, progression, and spectrum (26). In fact, HIC1 is implicated in a complex regulatory feedback loop with p53 and the class III deacetylase SIRT1 (10) through direct transcriptional regulation mechanisms (2, 8, 9) as well as post-translational modifications (12, 15, 27, 28). HIC1 encodes a sequence-specific transcriptional repressor belonging to the BTB/POZ and Krüppel C2H2 zinc finger subfamily (2). However, despite this wealth of information, only four genes directly regulated by HIC1 have been identified to date either by induction, SIRT1 (10), or by promoter analysis and mutagenesis such as FGF-BP1 and E2F1. The FGF-BP1 gene is repressed by HIC1 following transforming growth factor-β treatment (29). Recently, it has been shown that HIC1 represses the E2F1 promoter in serum-starved HSF8 human fibroblasts through recruitment of Brg1 (13) and that E2F1 activates the transcription of HIC1 (40), thus generating another regulatory loop. Finally, microarray analyses based on the Ptch1+/− Hic1+/− model of medulloblastoma identified ATOH1 (21). In addition to its role as a tumor suppressor gene, Hic1 is also essential for normal development as shown by the embryonic lethal phenotype of mice bearing homozygous Hic1 deletion (30). As the four known HIC1 target genes are unlikely to fully account for the effects of HIC1 deletion, it stands to reason that the majority of HIC1 target genes remains to be discovered.
This study is the second attempted genomic analysis of HIC1 target genes through microarray gene expression profiling (21). Expression changes at various time points were monitored after HIC1 re-expression in transformed U2OS cells that have lost endogenous HIC1 expression but are wild type for p53, retinoblastoma protein, and Brg1. This model is physiologically relevant because double-cis Hic1+/− p53+/− heterozygous mice develop osteosarcomas and can also be correlated with human osteosarcomas that frequently harbor hypermethylation of HIC1 in tumors with p53 mutations (26). We used RNAs extracted from 8 to 26 h post-infection allowing us to identify genes repressed at the earliest time points after infection, which are more likely to represent a “first wave” of direct target genes. At 16 h, Ad-FLAG-HIC1-infected U2OS cells overexpressed moderately the FLAG-HIC1 protein but were already growth-arrested (Fig. 1).
In our time course experiment, we identified a total of 81 genes whose expression was down-regulated at least 3-fold after 16 h of infection with Ad-FLAG-HIC1, whereas 23 genes were up-regulated at least 3-fold after 26 h (Tables 1 and 2). Strikingly, SIRT1, one of the four HIC1 target genes described so far, was not repressed but was rather activated (∼1.5-fold) in each Ad-FLAG-HIC1 sample (Fig. 2A) as confirmed by qRT-PCR (Fig. 2B). Although clearly a target gene in fibroblasts (10, 11, 13), SIRT1 may not be an universal HIC1 target (21). Indeed, a recent study has failed to detect any correlation between the SIRT1 mRNA levels and the HIC1 status in diffuse large B-cell lymphomas showing either wild-type HIC1 expression (8 tumors) or complete HIC1 inactivation (10 tumors) (31). In two animal models of medulloblastoma, SIRT1 mRNA levels from Ptch1+/− Hic1+/− tumors and Ptch1+/− tumors were similar to normal age-matched cerebellum. Moreover, in the D245 medulloblastoma cell line infected with Ad-HIC1, SIRT1 is slightly activated (1.3-fold), whereas ATOH1 is strongly repressed (18-fold) (21).
One would not expect the other previously described HIC1 target genes FGF-BP1, ATOH1, and E2F1 to be found on our list of potential target genes because FGF-BP1 is only repressed after transforming growth factor-β treatment of cell lines that can undergo smooth muscle differentiation (29) and ATOH1 encodes a neuronally specific transcription factor. E2F1 is a target of HIC1 in normal HSF8 fibroblasts (13). However, in transformed cell lines such as U2OS, the re-expression of HIC1 may not be sufficient to counteract the deregulation of E2F1 expression. This is reminiscent of the lack of repression of SIRT1 observed upon re-expression of HIC1 in medulloblastoma cell lines (21) or in U2OS (this study).
We have first focused our studies on RDC1, a recently deorphanized G-protein-coupled receptor strongly repressed in our Ad-FLAG-HIC1-infected U2OS cells (Fig. 2). CXCR7, formerly known as RDC1, has been shown to be a second receptor, in addition to CXCR4, for the chemokine SDF-1 (stromal cell-derived factor 1)/CXCL12 (22). Although CXCR7 binds CXCL12 even in cells lacking CXCR4, CXCR7 may not function as a classical chemokine receptor, as evidenced by the absence of ligand-induced CXCR7-mediated calcium mobilization or cell migration (23, 32). In fact, recent studies in zebrafish have demonstrated that CXCR7 does not act as a signaling receptor but instead participates in chemokine-guided cell migration by ligand sequestration (33). These results however did not totally rule out the possibility that CXCR7 could induce signal transduction as suggested by studies on tumor growth and survival.
The SDF-1/CXCR4 chemotactic pathway is a key player in the cross-talk between various tumor cells and their microenvironment. First, CXCR4 is essential for the metastatic spread of breast cancers to organs where CXCL12 is expressed (34). Second, stromal fibroblast-derived CXCL12 can stimulate survival and growth of neoplastic breast cells in a paracrine fashion and can promote tumor angiogenesis by recruiting circulating endothelial cells to the tumor microenvironment (endocrine effect) (35). Similarly, CXCR7 is by far the strongest cellular gene induced during transformation of dermal microvascular endothelial cells by Kaposi sarcoma-associated herpesvirus, and upon ectopic expression of CXCR7, NIH3T3 become tumorigenic in nude mice (36). CXCR7 is highly expressed in human primary breast and lung tumors, and it is expressed in both malignant cells and in the tumor-associated vasculature but not in normal blood vessels (25). Finally, inhibition of CXCR7 by various strategies such as small interfering RNA interference, specific high affinity small molecule antagonist, or intrakines severely reduces proliferation of carcinoma cells in vitro (37) as well as in vivo tumor growth in animal models (32). Taken together, these results demonstrate that CXCR7 has key functions in promoting tumor development and progression through specific pathways and mechanisms that still remain to be elucidated. In addition, quantitative histologic analyses confirmed that CXCR7 expression increases with increasing tumor grade of prostate tumors (24). Reciprocally, HIC1 is hypermethylated in prostate tumors, and hypermethylation of one HIC1 allele is even already observed in histologically normal prostate and in benign hyperplastic tissues (38, 39). Our results showing that CXCR7 is a direct target of HIC1 could tie together these two sets of data.
In conclusion, we have identified the scavenger chemokine receptor CXCR7 as an additional bona fide HIC1 target gene. Our results demonstrate that HIC1 is a direct repressor of the CXCR7 gene and suggest that in tumors with loss of HIC1 expression the resulting increase in CXCR7 expression could participate in tumor progression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant 5K12HD001399-05 from the NICHD (a Child Health Research career development award to B. R. R.). This work was also supported in part by funds from CNRS, Association for International Cancer Research (St. Andrews, United Kingdom), the Pasteur Institute and Association pour la Recherche Contre le Cancer 3586 and 3983 (to D. L.), Hope Street Kids Grants the Swiss Federation against Cancer/Oncosuisse Grant OCS-01823-02-2006, and the Werner and Hedy Berger-Janser Foundation of Cancer Research (to M. P. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S2.
- CtBP
- C-terminal binding protein
- GFP
- green fluorescent protein
- Ad-GFP
- adenovirus encoding green fluorescent protein
- qRT
- quantitative reverse transcription
- shRNA
- short hairpin RNA
- ChIP
- chromatin immunoprecipitation
- HiRE
- HIC1-responsive element
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Makos M., Nelkin B. D., Reiter R. E., Gnarra J. R., Brooks J., Isaacs W., Linehan M., Baylin S. B. (1993) Cancer Res. 53, 2719–2722 [PubMed] [Google Scholar]
- 2.Wales M. M., Biel M. A., el Deiry W., Nelkin B. D., Issa J. P., Cavenee W. K., Kuerbitz S. J., Baylin S. B. (1995) Nat. Med. 1, 570–577 [DOI] [PubMed] [Google Scholar]
- 3.Rood B. R., Zhang H., Weitman D. M., Cogen P. H. (2002) Cancer Res. 62, 3794–3797 [PubMed] [Google Scholar]
- 4.Fleuriel C., Touka M., Boulay G., Guérardel C., Rood B. R., Leprince D. (2009) Int. J. Biochem. Cell Biol. 41, 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinte S., Stankovic-Valentin N., Deltour S., Rood B. R., Guérardel C., Leprince D. (2004) J. Biol. Chem. 279, 38313–38324 [DOI] [PubMed] [Google Scholar]
- 6.Deltour S., Pinte S., Guerardel C., Wasylyk B., Leprince D. (2002) Mol. Cell. Biol. 22, 4890–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deltour S., Guerardel C., Leprince D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerardel C., Deltour S., Pinte S., Monte D., Begue A., Godwin A. K., Leprince D. (2001) J. Biol. Chem. 276, 3078–3089 [DOI] [PubMed] [Google Scholar]
- 9.Britschgi C., Rizzi M., Grob T. J., Tschan M. P., Hügli B., Reddy V. A., Andres A. C., Torbett B. E., Tobler A., Fey M. F. (2006) Oncogene 25, 2030–2039 [DOI] [PubMed] [Google Scholar]
- 10.Chen W. Y., Wang D. H., Yen R. C., Luo J., Gu W., Baylin S. B. (2005) Cell 123, 437–448 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q., Wang S. Y., Fleuriel C., Leprince D., Rocheleau J. V., Piston D. W., Goodman R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Stankovic-Valentin N., Deltour S., Seeler J., Pinte S., Vergoten G., Guérardel C., Dejean A., Leprince D. (2007) Mol. Cell. Biol. 27, 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B., Chambers K. J., Leprince D., Faller D. V., Wang S. (2009) Oncogene 28, 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 15.Kwon H. S., Ott M. (2008) Trends Biochem. Sci. 33, 517–525 [DOI] [PubMed] [Google Scholar]
- 16.Chen W. Y., Zeng X., Carter M. G., Morrell C. N., Chiu Yen R. W., Esteller M., Watkins D. N., Herman J. G., Mankowski J. L., Baylin S. B. (2003) Nat. Genet. 33, 197–202 [DOI] [PubMed] [Google Scholar]
- 17.Makos M., Nelkin B. D., Lerman M. I., Latif F., Zbar B., Baylin S. B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard D., Gosselin K., Monte D., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C. (2004) Cancer Res. 64, 472–481 [DOI] [PubMed] [Google Scholar]
- 19.Britschgi C., Jenal M., Rizzi M., Mueller B. U., Torbett B. E., Andres A. C., Tobler A., Fey M. F., Tschan M. P. (2008) Br. J. Haematol. 141, 179–187 [DOI] [PubMed] [Google Scholar]
- 20.Pinte S., Guérardel C., Deltour-Balerdi S., Godwin A. K., Leprince D. (2004) Oncogene 23, 4023–4031 [DOI] [PubMed] [Google Scholar]
- 21.Briggs K. J., Corcoran-Schwartz I. M., Zhang W., Harcke T., Devereux W. L., Baylin S. B., Eberhart C. G., Watkins D. N. (2008) Genes Dev. 22, 770–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balabanian K., Lagane B., Infantino S., Chow K. Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. (2005) J. Biol. Chem. 280, 35760–35766 [DOI] [PubMed] [Google Scholar]
- 23.Thelen M., Thelen S. (2008) J. Neuroimmunol. 198, 9–13 [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Shiozawa Y., Wang J., Wang Y., Jung Y., Pienta K. J., Mehra R., Loberg R., Taichman R. S. (2008) J. Biol. Chem. 283, 4283–4294 [DOI] [PubMed] [Google Scholar]
- 25.Miao Z., Luker K. E., Summers B. C., Berahovich R., Bhojani M. S., Rehemtulla A., Kleer C. G., Essner J. J., Nasevicius A., Luker G. D., Howard M. C., Schall T. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15735–15740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Cooper T. K., Zahnow C. A., Overholtzer M., Zhao Z., Ladanyi M., Karp J. E., Gokgoz N., Wunder J. S., Andrulis I. L., Levine A. J., Mankowski J. L., Baylin S. B. (2004) Cancer Cell 6, 387–398 [DOI] [PubMed] [Google Scholar]
- 27.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 28.Langley E., Pearson M., Faretta M., Bauer U. M., Frye R. A., Minucci S., Pelicci P. G., Kouzarides T. (2002) EMBO J. 21, 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briones V. R., Chen S., Riegel A. T., Lechleider R. J. (2006) Biochem. Biophys. Res. Commun. 345, 595–601 [DOI] [PubMed] [Google Scholar]
- 30.Carter M. G., Johns M. A., Zeng X., Zhou L., Zink M. C., Mankowski J. L., Donovan D. M., Baylin S. B. (2000) Hum. Mol. Genet. 9, 413–419 [DOI] [PubMed] [Google Scholar]
- 31.Stöcklein H., Smardova J., Macak J., Katzenberger T., Höller S., Wessendorf S., Hutter G., Dreyling M., Haralambieva E., Mäder U., Müller-Hermelink H. K., Rosenwald A., Ott G., Kalla J. (2008) Oncogene 27, 2613–2625 [DOI] [PubMed] [Google Scholar]
- 32.Burns J. M., Summers B. C., Wang Y., Melikian A., Berahovich R., Miao Z., Penfold M. E., Sunshine M. J., Littman D. R., Kuo C. J., Wei K., McMaster B. E., Wright K., Howard M. C., Schall T. J. (2006) J. Exp. Med. 203, 2201–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q., Raz E. (2008) Cell 132, 463–473 [DOI] [PubMed] [Google Scholar]
- 34.Smith M. C., Luker K. E., Garbow J. R., Prior J. L., Jackson E., Piwnica-Worms D., Luker G. D. (2004) Cancer Res. 64, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 35.Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L., Weinberg R. A. (2005) Cell 121, 335–348 [DOI] [PubMed] [Google Scholar]
- 36.Raggo C., Ruhl R., McAllister S., Koon H., Dezube B. J., Früh K., Moses A. V. (2005) Cancer Res. 65, 5084–5095 [DOI] [PubMed] [Google Scholar]
- 37.Meijer J., Ogink J., Roos E. (2008) Br. J. Cancer 99, 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Morton R. A., Jr., Watkins J. J., Bova G. S., Wales M. M., Baylin S. B., Isaacs W. B. (1996) J. Urol. 156, 512–516 [DOI] [PubMed] [Google Scholar]
- 39.Li L. C., Okino S. T., Dahiya R. (2004) Biochim. Biophys. Acta 1704, 87–102 [DOI] [PubMed] [Google Scholar]
- 40.Jenal M., Trinh E., Britschgi C., Britschgi A., Roh V., Vorburger S. A., Tobler A., Leprince D., Fey M. F., Helin K., Tschan M. P. (2009) Mol. Cancer Res., in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.