Abstract
In the 3′-untranslated region, the destabilizing adenine-uridine (AU)-rich elements (AREs) control the expression of several transcripts through interactions with ARE-binding proteins (AUBPs) and RNA degradation machinery. Although the fundamental role for AUBPs and associated factors in eliciting ARE-dependent degradation of cognate mRNAs has been recently highlighted, the molecular mechanisms underlying the specific regulation of individual mRNA turnover have not yet been fully elucidated. Here we focused on the post-transcriptional regulation of bcl-2 mRNA in human cell lines under different conditions and genetic backgrounds. In the context of an AUBPs silencing approach, HuR knockdown reduced the expression of endogenous bcl-2, whereas unexpectedly, a bcl-2 ARE-reporter transcript increased significantly, suggesting that HuR expression has opposite effects on endogenous and ectopic bcl-2 ARE. Moreover, evidence was provided for the essential, specific and dose-dependent role of the Bcl-2 protein in regulating the decay kinetics of its own mRNA, as ascertained by a luciferase reporter system. Altogether, the data support a model whereby the Bcl-2 protein is the major determinant of its own ARE-dependent transcript half-life in living cells and its effect overcomes the activity of ARE-binding proteins.
Eukaryotic cells coordinately regulate their functions to ensure appropriate responses to stimuli. Cell signaling involves a complex network of pathways tightly coupled to transcriptional and translational events. Between these two processes, modulation of the mRNA decay provides a crucial regulatory step (1). Most importantly, regulation of mRNA is a fundamental means of regulating both the level and the timing of gene expression along with the metabolic state of the cells, differentiation and stress stimuli (2). Furthermore, dysregulation of mRNA stability has been associated with human diseases including cancer, and inflammatory and neurodegenerative conditions (3, 4). The clinical relevance of post-transcriptional gene regulation by mRNA stability has also been shown (5).
mRNA decay is a highly regulated process, established through interactions between mRNA structures and corresponding subsets of binding proteins or noncoding small RNAs (6, 7). A variety of molecular determinants of mRNA stability have been described and, to a certain extent, characterized (8). Most cis-acting mRNA stability determinants are located in the 3′-untranslated region (3′-UTR).4 Among these, the AU-rich elements (AREs) are responsible for rapid decay of the mRNA and are recognized by a multimeric protein complex containing HuR and other RNA-binding proteins (9, 10). Recent studies have also indicated that microRNAs, hybridizing preferentially to the 3′-UTR, may interact with and affect the fate of ARE-containing mRNA (11, 12). These decay pathways are controlled by the interplay of function-specific as well as gene-specific RNA-protein determinants (13). Consistent with this hypothesis are previous biochemical studies showing that ARE-containing mRNAs can be differentially and coordinately regulated in response to particular extracellular stimuli (14).
Given the critical function of the Bcl-2 protein in mediating the processes of cell death and survival, in this study we focused on its cellular post-transcriptional regulation. Alterations in the level of Bcl-2 expression affect cell proliferation and cell death and are determinants in the pathogenesis and progression of several diseases such as cancer, neurodegenerative disorders, or autoimmune diseases among others (15–17). Despite extensive efforts, the molecular mechanisms leading to altered expression of bcl-2 in such different human pathologies have not been elucidated. Nevertheless, many attempts have been made to develop biological or pharmacological means of controlling bcl-2 expression and some promising results have been obtained (18–20). Therapeutic intervention targeting bcl-2 would undoubtedly benefit from a deeper understanding of the mechanisms ruling its regulation.
Here the post-transcriptional regulation of bcl-2 mRNA in multiple cellular systems and under different conditions has been explored. In detail, the small interfering RNA (siRNA) technology was exploited to alter the cellular repertoire of the mRNA degradation machinery to ascribe a role for proteins potentially involved in bcl-2 mRNA post-transcriptional regulation. In addition a luciferase reporter system was engineered to address questions regarding gene-specific and function-specific mechanisms of mRNA turnover regulation. Our findings point to a specific role of Bcl-2 in the ARE-dependent degradation of its own messenger in different cellular systems and conditions and, interestingly, they hint at a hierarchy of different regulatory mechanisms in modulating bcl-2 mRNA half-life.
EXPERIMENTAL PROCEDURES
Plasmid Construction
A DNA fragment containing the ARE from bcl-2 3′-UTR (bARE) was cloned downstream from the gene in the pGL4.71P vector to obtain the pGL4.71P-bARE plasmid as described previously (21). The primer pair 5′-CGTCTAGAACTTTTTTATGCTTACCATC-3′ and 5′-CGTCTAGACAATAGAAAAAAATCAACTT-3′ was used to amplify a 260-base pair segment containing the ARE sequence from the human c-myc 3′-UTR fragment (mARE). The latter was cloned into the pGL4.71P plasmid above to produce the pGL4.71P-mARE plasmid. pcDNA3-Bcl2 plasmid was constructed exciding the 930-base pair fragment from pB4 (22) containing the bcl-2 open reading frame and cloning it into the EcoRI restriction site of the pcDNA3 vector (Invitrogen). pGL4.71P-cARE plasmid containing the C2 region of the CDK5R1 3′-UTR sequence has been previously described (23).
For the FLAG-Bcl-2 construct, the bcl-2 open reading frame (NCBI_M14745) was cloned between the XhoI and NotI restriction sites of the pCI-NEO vector (Promega) carrying a FLAG epitope (DYKDDDDK) with the following primers pair: 5′-AAAACTCGAGATGGCGCACGCTG-3′ and 5′-AAAAGCGGCCGCTCACTTGTGGCCCAG-3′. Correct orientation of inserts was verified by sequencing.
Cell Cultures and Chemicals
Viable human embryonic kidney (HEK) 293, osteosarcoma U2OS, and neuroblastoma SK-N-BE cells were grown in complete Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum (FCS, HyClone Laboratories, Logan, UT), 2 mm glutamine, 50 IU/ml penicillin, and 50 mg/ml streptomycin (Sigma) in a humidified atmosphere of 5% CO2 at 37 °C. Burkitt lymphoma Daudi cells and follicular lymphoma Karpas 422 (K422) cells were maintained in RPMI 1640 (Sigma) supplemented with glutamine, antibiotics, and FCS added under standard conditions. The transcription blocker 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) was purchased from Sigma.
siRNA Transfections
3 × 105 cells were plated and cultured overnight. Thereafter gene-specific siRNAs were transfected in the presence of serum-free medium (Opti-MEM, Invitrogen) using Lipofectamine 2000 (Invitrogen) as a carrier, according to the manufacturer's instructions. Cells were re-transfected after 72 h with half-doses of siRNAs as indicated. siRNA targeting Bcl-2 (SMARTpool reagent) was purchased from Dharmacon, Inc. (Lafayette, CO), siRNAs targeting AUF1, TTP, HuR, KSRP, and TIA-1 were purchased from Qiagen (Hilden, Germany). The sequences of the siRNAs used in silencing experiments were as follows: siAUF1, AAGATTGACGCCAGTAAGAAC (24); siTTP, CGCTGCCACTTCATCCACAAC (11); siHuR, AAGAGGCAATTACCAGTTTCA (25); siKSRP, AGATCAACCGGAGAGCAAGA (26); and siTIA1, CTGGGCTAACAGAACAACTAA (27).
Cell Transfection
In transient transfection experiments, 3 × 104/well SK-N-BE and 1 × 104/well HEK293 cells were co-transfected in triplicate in 96-well plates with equal amounts (200 ng) of the pGL4.71P constructs and pGL3P control plasmid. 24 h post-transfection cells were lysed and luciferase activity was measured as described in the following section. To establish stably transfected HEK293 clones, 80% confluent cells were co-transfected with 10 μg of each pGL4.71P plasmid and 10 μg of pcDNA3 vector carrying the Geneticin (G418) resistance gene. Cells were selected for 3 weeks in the presence of 0.8 mg/ml G418 (Invitrogen). Resistant cells gave rise to a set of pGL4.71P-bARE and pGL4.71P-mARE clones. The expression of the Renilla hRlucP gene was analyzed in each clone by luciferase assays as described afterward. C7, B4, and D21 clones, stably transfected with pGL4.71P, pGL4.71P-bARE, and pGL4.71P-mARE, respectively, were selected for further experiments due to similar Renilla luciferase activity. Lipofectamine 2000 (Invitrogen) was used as transfection reagent in all the aforementioned experiments according to the manufacturer's instructions. Daudi cells stably expressing Bcl-2 protein were obtained by electroporating pcDNA3-Bcl2 plasmid in a Bio-Rad Gene Pulser at a setting of 220 V/960 microfarads. After electroporation, cells were incubated for 5 min on ice, then placed in 10 ml of RPMI supplemented with 20% FCS. Transfected Daudi cells were supplemented with 1 mg/ml G418 to select stably transfected clones.
Measurement of Luciferase Activity
After co-transfection of the two luciferase plasmids, both firefly and Renilla luciferase activities were measured sequentially from a single aliquot of the cell lysate using the Dual-Glo Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Relative Renilla luciferase light output was normalized to firefly luciferase output as well as to the protein concentration. Data were expressed as mean ± S.E. Statistical significance was calculated by using a Student's t test.
RNA Isolation, cDNA Synthesis, and Real Time Quantitative PCR
Total cellular RNA was extracted at the indicated times using the NucleoSpin RNA II columns (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. The RNA was then treated with RNase-free DNase (Invitrogen) and analyzed spectroscopically and by gel electrophoresis for purity and integrity, respectively. For cDNA synthesis, 0.1 μg/μl total RNA, DNase treated, was reverse-transcribed with the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) using standard conditions set by the manufacturer in a total volume of 50 μl. cDNAs levels of Renilla and firefly luciferases, bcl-2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were determined using Real Time PCR, through TaqMan technology (Applied Biosystems), using specific primers and probes. Reporters and bcl-2 amplification data were normalized versus the expression of the housekeeping gene GAPDH. Each reaction was performed in triplicate for a better statistical reliability of results. The PCR were carried out in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). The amplification plot and cycle threshold data were elaborated with ABI Prism 7000 SDS software (version 1.1).
mRNA Decay Assays
In mRNA decay assays cells were treated with DRB (20 μg/ml final concentration) 24 h post-transfection to suppress the transcription of the luciferase reporter gene. Cells were harvested at 0, 3, 5, and 7 h after treatment with DRB and RNA isolated for quantitative reverse transcriptase-PCR analysis.
Flow Cytometric Analysis
2 × 106 electroporated Daudi cells were harvested and washed with 1× phosphate-buffered saline. Cells were resuspended in a 3% paraformaldehyde solution and incubated on ice for 1 h. Fixed cells were pelleted and washed in pre-cooled 1× phosphate-buffered saline, then resuspended in 500 μl of RPMI medium containing 20% FCS and 0.5% Tween (Sigma). Cells were pelleted, resuspended in 50 μl of FCS, and incubated 15 min at room temperature. Permeabilized cells were centrifuged and the pellet was resuspended RPMI medium containing 20% FCS, 0.5% Tween, and 50 μl of fluorescein-conjugated 1:50 diluted anti-human Bcl-2 monoclonal antibody (Dako Italia, Milan, Italy). The cells were incubated at room temperature for 1 h in the dark, then pelleted and washed four times in pre-cooled 1× phosphate-buffered saline at 4 °C. Washed cells were resuspended in 350 μl of 1× phosphate-buffered saline before flow cytometric analysis.
Western Blot and Co-immunoprecipitation (IP) Assays
The Western blot assays were performed under standard conditions as previously described (28). Protein concentrations were measured using the Micro BCA Protein Assay (Thermo Fisher Scientific, Rockford, IL). Images of Western blots were acquired and analyzed using the Bio-Rad Versadoc Imaging System. Protein band densities were quantified using the Quantity One software provided with the station. For IP assays, protein A/G UltraLink Resin (Thermo Fisher Scientific) was precoated with 1.6 μg of IgG1, HuR, or Bcl-2 mouse monoclonal antibodies (Santa Cruz Biotechnology), washed with lysis buffer (20 mm Tris-HCl, pH 7.2, 50 mm KCl, 10 mm MgCl2, 0.5% Nonidet P-40, 1 mm dithiothreitol, protease, and RNase inhibitors), and incubated with 0.5 ml of cell lysates (16 h, 4 °C). After washes with modified lysis buffer (20 mm Tris-HCl, pH 7.2, 300 mm KCl, 10 mm MgCl2, 0.5% Nonidet P-40), samples were denatured, fractionated by SDS-PAGE, and analyzed by Western blot. For FLAG IP assays, 0.5 ml of U2OS cell lysate (10 mm Hepes, pH 7.6, 100 mm KCl, 5 mm MgCl2, 2.5 mm EDTA, 2 mm dithiothreitol, protease, and RNase inhibitors) were incubated for 2 h at 4 °C with protein A/G UltraLink Resin (Thermo Fisher Scientific), precoated with 2.0 μg of FLAG M2 mouse monoclonal antibody (Sigma). Samples were then washed and processed as described above.
RESULTS
ARE-dependent Destabilizing Activity on Reporter Transcript
Luciferase gene containing plasmids were generated and employed in both transient and stable transfection experiments in different human cell lines to validate the suitability of this reporter system to study the ARE-dependent destabilizing activity. Among reporters used in mammalian cells, the Renilla reniformis hRlucP luciferase gene incorporates a protein degradation sequence conferring high instability to the encoded reporter protein (29) and thus suitable for studying the rate of reporter response to cellular stimuli.
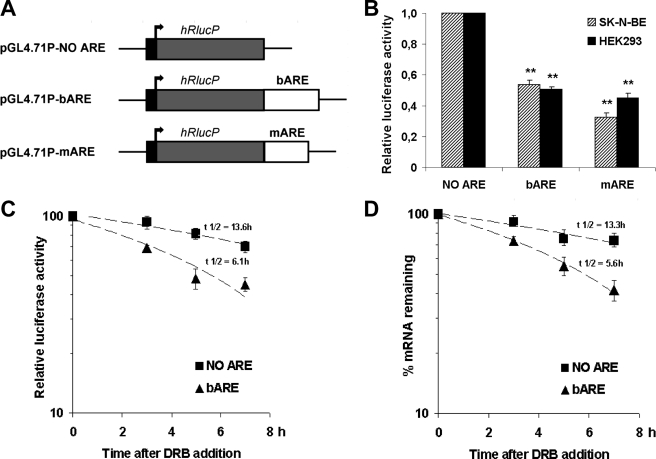
A DNA fragment containing the ARE bcl-2 (bARE) was cloned downstream of the stop codon of Renilla luciferase gene in the pGL4.71P vector to obtain the pGL4.71P-bARE plasmid. A second ARE fragment from c-myc 3′-UTR was inserted in the same position in the pGL4.71P vector to obtain the pGL4.71P-mARE plasmid, used as a control. Both the Renilla luciferase empty vector (pGL4.71P-NO ARE) and the ARE-containing constructs (Fig. 1A) were transiently co-transfected with the pGL3P firefly luciferase reporter plasmid into human neuroblastoma SK-N-BE and HEK293 cells. The Renilla luciferase activity was normalized to firefly luciferase activity, as shown in Table 1 and supplemental Table S1. The insertion of the ARE fragments downstream of the reporter gene led to a decrease in the Renilla luciferase activity in both transiently transfected human cell lines: bARE was able to reduce the reporter activity to about 50% and mARE to 32 and 45% in SK-N-BE and HEK293 cells, respectively (Fig. 1B). The destabilizing activity of ARE sequences was shown in HEK293 transiently transfected cells after the addition of the transcriptional blocker DRB. The relative luciferase activity normalized over protein concentration is shown in Fig. 1C. RlucP mRNA half-life was halved in the presence of bARE (Fig. 1D) following the same kinetics as the reporter protein activity, thus assessing the role of bARE on mRNA decay rates and validating the reporter system to study ARE-containing mRNAs.
FIGURE 1.
Validation of the reporter system. A, schematic representation of Renilla luciferase plasmids generated to investigate the effect of ARE sequences on the expression of the reporter gene. B, SK-N-BE and HEK293 human cell lines were transiently transfected with equal amounts of pGL3P firefly luciferase vector and pGL4.71P Renilla luciferase plasmids. Renilla luciferase normalized to firefly luciferase activity is shown with the normalized activity of the pGL4.71P-NO ARE plasmid being taken as 1. Data are the mean of four independent experiments in triplicate ± S.E. (error bars). **, p < 0.01 compared with the corresponding pGL4.71P-NO ARE value. C, time course of luciferase activity in HEK293 cells after DRB addition 24 h post-transfection. Renilla luciferase was normalized to firefly luciferase activity and to protein content. Data are the mean of three independent experiments ± S.E. (error bars). D, time course of hRlucP mRNA decay in HEK293 cells after DRB addition 24 h post-transfection. Total RNA was analyzed for Renilla luciferase transcript expression by real time quantitative PCR and normalized to firefly luciferase mRNA levels at each time point after DRB addition. Data are the mean of three independent experiments ± S.E. (error bars).
TABLE 1.
Ratio of luciferase activities in transiently transfected human cell lines
SK-N-BE and HEK293 cells were plated and after 24 h co-transfected with firefly and Renilla luciferase reporter plasmids as described under “Experimental Procedures.” 24 h post-transfection cells were lysed and luciferase activities measured in a counter plate reader for luminometry. Renilla luciferase activity values were normalized to firefly luciferase activity (i.e. Renilla luciferase light units/firefly luciferase light units) and the data from four independent experiments are shown as the ratio mean ± S.E.
| Renilla luciferase constructs |
Renilla/firefly luciferase activity |
|
|---|---|---|
| SK-N-BE cells | HEK293 cells | |
| pGL4.71P | 1.71 ± 0.17 | 1.70 ± 0.36 |
| pGL4.71P-bARE | 0.88 ± 0.06 | 0.85 ± 0.18 |
| pGL4.71P-mARE | 0.52 ± 0.05 | 0.75 ± 0.15 |
Role of AUBPs in Regulating bARE Reporter Transcript Stability
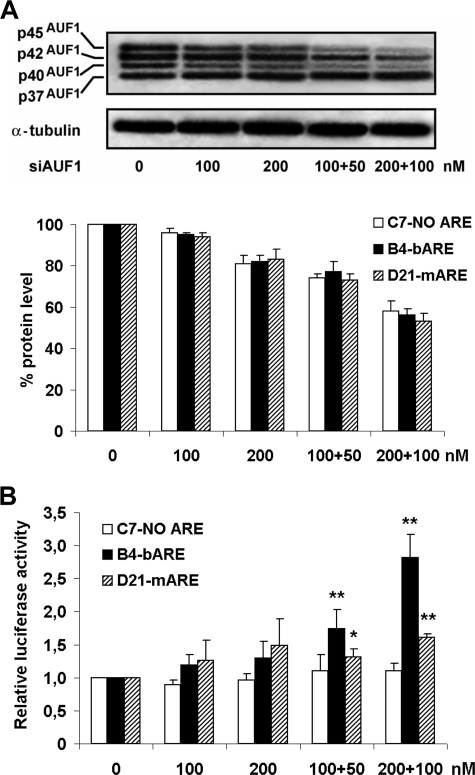
Three HEK293 clones, namely C7-NO ARE, B4-bARE, and D21-mARE, stably transfected with pGL4.71P-NO ARE, pGL4.71P-bARE, and pGL4.71P-mARE, respectively, expressed similar levels of reporter gene, as verified by real time reverse transcriptase-PCR and luciferase activity assays (not shown). In these stably transfected HEK clones a subset of either destabilizing or stabilizing AUBPs were silenced by siRNAs.
AUF1, expressed in four isoforms arising from differential splicing (30), was silenced with different siRNA doses by targeting the exon 1 region (siAUF1), present in all AUF1 isoforms. Albeit, the silencing effect was stronger on p45AUF1 and p40AUF1 isoforms, possibly less abundant in this cell line, as evaluated by Western blot (Fig. 2A); siAUF1 treatment significantly increased the Renilla luciferase activity (Fig. 2B) in B4-bARE and, to a lesser extent in D21-mARE cells.
FIGURE 2.
Increased luciferase activity by AUF1 silencing. A, HEK293 stably transfected cells lipofected with siAUF1 for 72 h and half-doses for a further 72 h were Western blot analyzed to test the levels of AUF1 isoforms. Upper panel, the gel refers to B4-bARE cells, similar findings were obtained from all siAUF1-treated clones in three independent experiments. Lower panel, densitometric quantifications of AUF1 Western blot bands, normalized to housekeeping loading control protein, are shown as percentage of unsilenced cells. The data shown are the mean of three independent experiments ± S.E. (error bars). B, Renilla luciferase activity was measured and data normalized to protein content. Values are expressed as relative luciferase activity versus cells treated with lipofectamine only. Data are the mean of three independent experiments ± S.E. (error bars). *, p < 0.05; **, p < 0.01 compared with vehicle-treated cells.
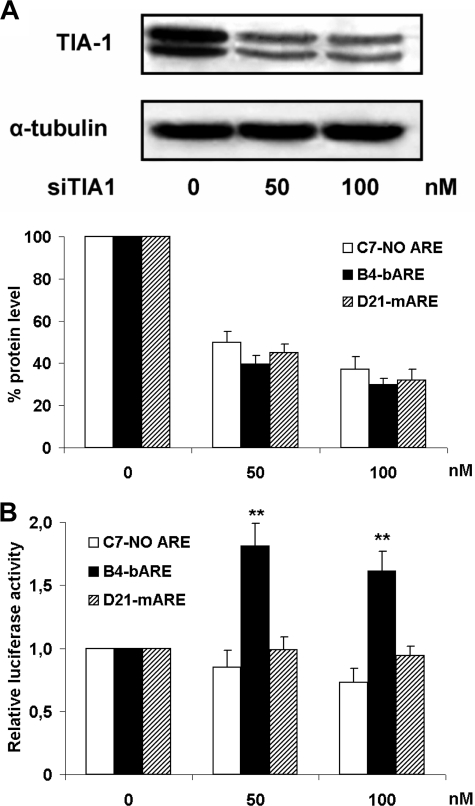
Furthermore, we considered the role of TIA-1, a translational silencer that also turns mRNA liable to the decay machinery (31). A reduction in TIA-1 protein level (Fig. 3A) with a siRNA (siTIA1), could increase the Renilla luciferase activity in B4-bARE cells from 60 to 80% compared with vehicle-treated cells (Fig. 3B). In contrast, TTP and KSRP were silenced without significant changes to luciferase activity, although the targeted protein levels decreased using siRNA treatment (data not shown).
FIGURE 3.
Increased luciferase activity by TIA-1 silencing. A, HEK293 stably transfected cells lipofected with siTIA1 for 72 h were lysed and cell extracts analyzed by Western blot to test the level of TIA-1 silencing. Upper panel, the gel is representative of B4-bARE cells, similar findings were obtained from all siTIA1-treated clones in three independent experiments. Lower panel, densitometric quantifications of TIA-1 Western blot bands, normalized to housekeeping loading control protein, are shown as percentage of unsilenced cells. The data shown are the mean of three independent experiments ± S.E. (error bars). B, Renilla luciferase activity was measured. Data normalized as in Fig. 2B are the mean of three independent experiments ± S.E. (error bars). **, p < 0.01 compared with vehicle-treated cells.
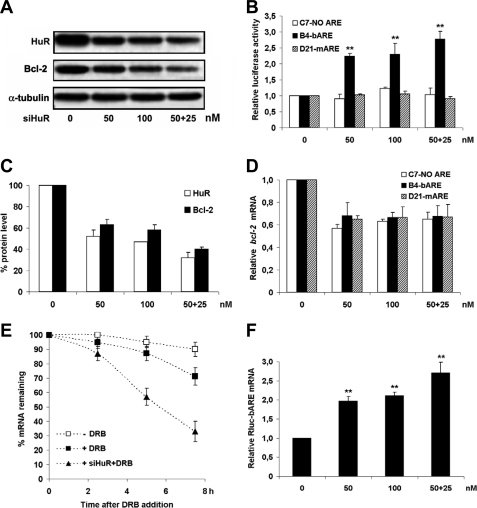
Silencing of HuR (siHuR) brought about quite unexpected findings. In contrast to the stabilizing effect of HuR reported in the literature (32), its down-regulation led to a 2-fold increase in the Renilla luciferase activity in B4-bARE cells (Fig. 4B). A double siRNA treatment with 50 + 25 nm doses showed a further increase in luciferase activity.
FIGURE 4.
Specific increase of bARE reporter activity by silencing HuR. A, stably transfected HEK293 clones treated with siHuR for 72 h were lysed and Renilla luciferase activity was measured. Cells were given a second half-dose of siHuR and lysed after further 72 h. Western blot analysis of HuR and Bcl-2 shown here refers to B4-bARE cells. Similar results were achieved in all siHuR-treated clones, in four independent experiments. B, Renilla luciferase activity was normalized as described in the legend to Fig. 2B. Data are the mean of four independent experiments ± S.E. (error bars). C, densitometric quantification of Bcl-2 and HuR Western blot bands, normalized to housekeeping loading control proteins, are shown as percentage of unsilenced cells. The data shown are the mean of four independent experiments ± S.E. (error bars). D, total RNA was analyzed for bcl-2 mRNA expression by real time quantitative PCR in cells treated with siHuR as in A. Data normalized versus GAPDH are expressed as relative bcl-2 mRNA level versus vehicle-treated cells. Data are the mean of three independent experiments ± S.E. (error bars). E, rate of bcl-2 mRNA degradation in B4-bARE cells treated with siHuR and further with DRB for transcription inhibition was determined as in D. Data are the mean of three independent experiments ± S.E. (error bars). F, total RNA was analyzed for Renilla luciferase mRNA expression by real time quantitative PCR in B4-bARE cells treated with siHuR as in A. Data normalized versus GAPDH are expressed as relative Renilla luciferase mRNA level versus vehicle-treated cells. Data are the mean of three independent experiments ± S.E. (error bars). **, p < 0.01 compared with vehicle-treated cells.
Based on the well established HuR stabilizing activity on ARE-containing mRNA (32), a decrease in reporter levels in both B4-bARE and D21-mARE clones was expected. The unforeseen findings inspired a detailed analysis of the mechanism responsible for increased reporter activity in B4-bARE siHuR-treated cells. As previously shown (33, 34), and confirmed here (Fig. 4, A and C), HuR silencing caused a decrease in the expression of a number of genes, including bcl-2. This reduction was also confirmed by bcl-2 mRNA levels in all tested cell clones (Fig. 4D). To evaluate whether mRNA quantitative changes depended on altered mRNA stability, a transcriptional blocker (DRB) was used in siHuR experiments. Real time reverse transcriptase-PCR assays on RNA extracted from treated cells clearly indicate that siHuR is able to destabilize endogenous bcl-2 mRNA as shown in Fig. 4E. Conversely, when Rluc-bARE transcript levels were measured in siHuR B4-bARE-treated cells, an increase comparable with the extent of the reporter activity increment was found (Fig. 4F).
Data presented herein and by others (35) show that HuR silencing can determine decreased expression of the mRNA and the relevant protein levels of a number of genes. The hypothesis that increased luciferase activity restricted to the bARE reporter may arise from the dominant effect of low Bcl-2 expression on the degradation rate of the bARE mRNA has been addressed.
Bcl-2 Role in Regulating bARE Reporter Transcript
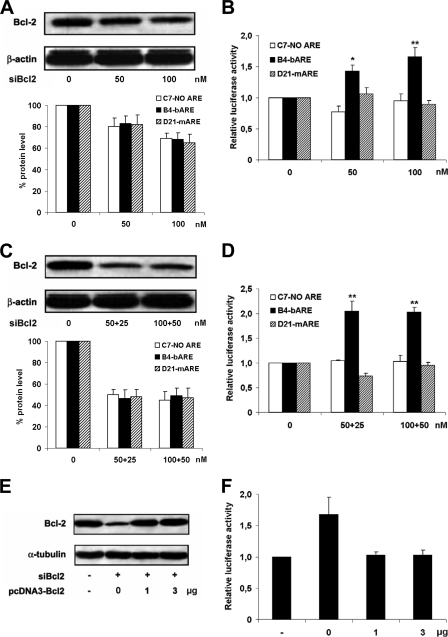
siRNAs directed against bcl-2 mRNA (siBcl2) were transfected at final concentrations of 50 and 100 nm into cell clones. Analysis of the Bcl-2 protein by Western blot showed effective and specific siBcl2 silencing (Fig. 5A). Lowering the amount of Bcl-2 significantly increased the bARE reporter activity (Fig. 5B), attesting the direct role of the Bcl-2 protein in triggering the degradation of bARE mRNA. The specificity of the effect of siBcl2 on the bARE reporter was assayed by lipofecting cells with a second siRNA dose after the first siBcl2 treatment. Half-doses of siRNA, added 72 h after the first siBcl2 lipofection, further down-regulated endogenous bcl-2 mRNA and protein in all cell clones as assayed by real time reverse transcriptase-PCR (data not shown) and Western blot, respectively (Fig. 5C). The stronger silencing of Bcl-2 caused a higher increase of luciferase activity restricted to B4-bARE reporter cells, as the level of reporter gene expression in C7-NO ARE and D21-mARE clones was unchanged compared with vehicle-treated cells (Fig. 5, B and D).
FIGURE 5.
Increased bARE reporter activity by Bcl-2 silencing restored by Bcl-2 rescuing. A, stably transfected HEK293 clones were treated with siBcl2 for 72 h and the expression of Bcl-2 was analyzed by Western blot. Upper panel, the gel shown here refers to B4-bARE-treated cells, similar findings were obtained with all siBcl2-treated clones in three independent experiments. Lower panel, densitometric quantifications of Bcl-2 Western blot bands, normalized to housekeeping loading control protein, are shown as percentage of unsilenced cells. The data shown are the mean of three independent experiments ± S.E. (error bars). B, Renilla luciferase activity was measured and data analyzed as described in the legend to Fig. 2B. Data are the mean of three independent experiments ± S.E. (error bars). C, HEK293 clones treated twice with siBcl2 were lysed after 144 h and Bcl-2 analyzed by Western blot as in A. Upper panel, the gel shown here refers to B4-bARE-treated cells, similar findings were obtained with all siBcl2-treated clones in four independent experiments. Lower panel, densitometric quantifications of Bcl-2 Western blot bands, normalized to housekeeping loading control protein, are shown as percentage of unsilenced cells. The data shown are the mean of four independent experiments ± S.E. (error bars). D, Renilla luciferase activity was measured and data normalized as above. Data are the mean of four independent experiments ± S.E. (error bars). E, B4-bARE cells were treated with 80 nm siBcl2 for 24 h and then transiently transfected with pcDNA3-Bcl2 plasmid for 48 h before sample collection. Bcl-2 levels were assayed by Western blot. F, Renilla luciferase activity was analyzed as above. Data are the mean of three independent experiments ± S.E. (error bars). *, p < 0.05; **, p < 0.01 compared with vehicle-treated cells.
To further establish the molecular mechanism underlying bcl-2 post-transcriptional regulation by its own gene product, Bcl-2 expression was rescued in B4-bARE cells upon siBcl2 treatment. Bcl-2 was expressed ectopically by transiently transfecting cells with pcDNA3-Bcl2, a plasmid encoding for a wild type Bcl-2 protein, but lacking the bcl-2 3′-UTR, and thus refractory to the bARE-dependent regulation (Fig. 5E). As shown in Fig. 5F, in Bcl2-silenced cells the restored expression of Bcl-2 was able to down-regulate Renilla luciferase activity to the levels of unsilenced cells.
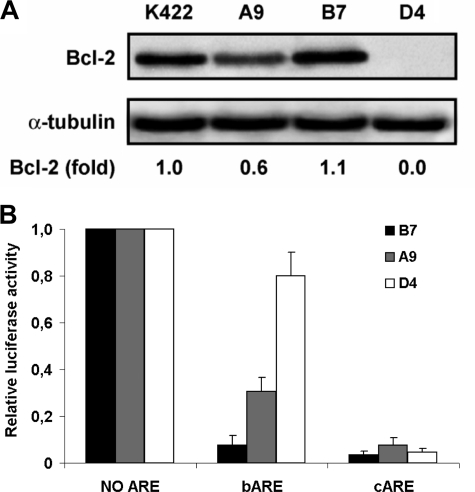
The dose-dependent role of Bcl-2 in regulating bARE-containing mRNA half-life was studied in another engineered cellular system. Cell clones expressing, in basal conditions, different levels of Bcl-2 protein were generated. We then correlated Bcl-2 levels to reporter activity of Renilla luciferase in transiently transfected cells. In particular, Daudi Burkitt lymphoma cells, not expressing Bcl-2 protein (36), were electroporated with pcDNA3-Bcl2 plasmid and clones stably expressing different levels of Bcl-2 protein were created after G418 selection. The Bcl-2 protein expression level was screened by flow cytometric analysis of immunostained cells (data not shown) and Western blot analysis of individual clones (Fig. 6A). One Daudi clone expressing high levels of Bcl-2 protein (B7), a second one expressing low levels of Bcl-2 protein (A9), and a mock cell clone (D4) were transiently transfected with Renilla luciferase plasmids as well as pGL3P control vector. Luciferase activity was measured to evaluate the correlation between different levels of Bcl-2 expression and the activity of the reporter transcript. In this study, the ARE containing C2 region of the CDK5R1 3′-UTR sequence (23) was used as a control (pGL4.71P-cARE), because the high level of c-myc expression in Daudi cells might interfere with the metabolism of the reporter transcript containing the c-myc 3′-UTR. Daudi clones were electroporated with reporter plasmids and luciferase activity was measured (Fig. 6B). An inverse correlation between the bARE-reporter activity and the cellular amount of Bcl-2 protein was found, thus demonstrating the dose-dependent role of Bcl-2 in destabilizing bARE-containing mRNA in living cells. No significant luciferase change according to Bcl-2 levels was obtained in c-ARE reporter cells.
FIGURE 6.
Inhibition of luciferase activity in Daudi clones expressing different Bcl-2 levels. A, Bcl-2 protein levels were determined by Western blot in Daudi cells stably transfected with pcDNA3-Bcl2. Fold of Bcl-2 expression in A9, B7, and D4 clones compared with follicular lymphoma K422 cells was obtained by densitometric analysis. B, Daudi clones electroporated with Renilla luciferase plasmids and pGL3P control vector were lysed after 48 h for luciferase activity determinations. Data were normalized as described in the legend to Fig. 1B. Data are the mean of three independent experiments ± S.E. (error bars).
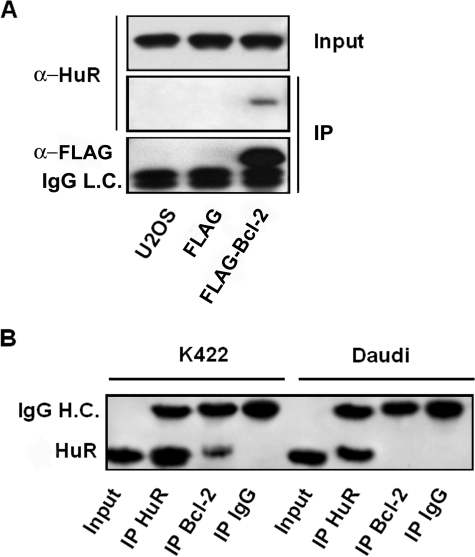
To get more insight into the mechanistic understanding of bcl-2 regulation, we then tested whether HuR associates with Bcl-2 by co-immunoprecipitation. U2OS cells were transiently transfected with a FLAG-Bcl2 expression plasmid, cell extracts were immunoprecipitated with anti-FLAG incubated protein A/G resin and analyzed by Western blotting. The expression plasmid encoding only the FLAG epitope was also transfected as a control to determine the background binding activities. As shown in Fig. 7A, HuR co-immunoprecipitated with FLAG-Bcl2. To better assess the HuR/Bcl-2 interaction, we next conducted IP assays on endogenous proteins in K422 cells. Wild type Daudi cells were used in IP assays as negative controls. Cell extracts prepared from both cell lines were immunoprecipitated with anti-HuR and anti-Bcl2-coated A/G resin. Mouse IgG-bound resin was used as negative control. Samples were separated by electrophoresis and probed for HuR. IP using anti-Bcl2 antibody revealed that HuR co-immunoprecipitated with endogenous Bcl2, as shown in Fig. 7B. No interaction was found in samples from Daudi cells used as a negative control not expressing Bcl-2 protein.
FIGURE 7.
HuR co-immunoprecipitation with Bcl-2 protein. A, U2OS cells were transfected with FLAG or with FLAG-Bcl2 expressing plasmid. 48 h later, cell lysates from transfected and untransfected cells were immunoprecipitated with anti-FLAG-incubated A/G resin. HuR immunoprecipitates were assessed by Western blot using anti-HuR antibody (middle panel). As a control, the amounts of HuR were verified in total cell lysates (upper panel) and the expression of FLAG-Bcl2 was confirmed using an anti-FLAG antibody (lower panel). The migration of the immunoglobulin light chain (IgG L.C.) is indicated at 25 kDa. B, cell lysates from K422 and Daudi cells were immunoprecipitated with the relevant primary antibody as indicated. Immunoprecipitates were subjected to Western blot for HuR. A sample of each cell lysate, labeled as Input, was also analyzed. The migration of the immunoglobulin heavy chain (IgG H.C.) and HuR are indicated at 50 and 36 kDa, respectively.
According to the findings, Bcl-2 is essential for the bARE-dependent degradation of the bcl-2 mRNA. Moreover, the rate of the messenger decay is inversely correlated to the protein amount, thus providing a potential mechanism for gene-specific regulation of mRNA decay. Bcl-2 may dynamically associate to HuR to wield this regulatory role.
DISCUSSION
mRNA decay processes play a key role in the global control of gene expression and in regulating cellular response to exogenous signals (1, 2). Recent findings have been remarkable in identifying the molecular components, elucidating the “mechanics” and spotting the cellular sites of mRNA turnover (8, 37, 38). According to the “RNA Regulon” theory multiple functionally related mRNAs are coordinately regulated by trans-acting factors (39). However, despite these tremendous achievements, the mechanisms by which cells coordinate the expression of specific genes in response to stimuli by post-transcriptional regulation remain unclear in many cases.
One enduring question is how individual and functionally related ARE-containing mRNAs can be differentially regulated in response to a variety of stimuli. An intriguing hypothesis has been proposed (13, 40) for the mechanism underlying the gene-specific regulation of ARE-containing mRNA turnover. On the basis of biochemical studies, Bcl-2 protein triggers the ARE-dependent degradation of its own transcript and regulates the rate of decay in a dose-response manner. The molecular mechanism by which cellular Bcl-2 regulates the rapid degradation of its own mRNA remains poorly defined. Here evidence is provided on the role of the Bcl-2 protein in leading the degradative enzymatic machinery to its cognate mRNA in cells under different genetic and growth conditions. This is of particular importance because of the critical function of Bcl-2 in a number of (pato-)physiological processes (41, 42). Most relevant are the mechanisms underlying its altered level of expression in a variety of human diseases and its tuning the response to different stimuli and chemotherapy treatments.
The object of this study is therefore to gain deeper comprehension of the post-transcriptional regulation of the bcl-2 transcript. Among the broad spectrum of determinants of bcl-2 mRNA half-life, we investigated the role of AUBPs required for the ARE-dependent mRNA degradation (9) with respect to bARE. MicroRNAs have also been implicated in the decay process of ARE-containing messengers (12) although their potential role has not been considered in this study.
The silencing of AUF1 and TIA-1 led to an increase in the bARE reporter activity. Most likely, down-regulation of the decay determinants stabilized the ARE-containing target. In particular, siRNA targeting AUF1 was able to down-regulate predominantly p40AUF1/p45AUF1 isoforms and this in turn determined an increase in luciferase activity of the bARE reporter and, to a lesser extent, of the mARE clone. These results, in keeping with others (24), provide additional support to the theory that the relative levels of individual isoforms, rather than the absolute amount of AUF1, determine mRNA stability of ARE-containing transcripts, consistent with the different ARE-binding functions of the isoforms.
It was not possible to ascribe a role for TTP and KSRP in regulation of bARE mRNA as their silencing had no effect on the reporter activity. Supplementary investigation is needed to explain these unexpected results because we have formerly shown by UV cross-linking and RNA immunoprecipitation assays that both AUBPs are able to bind to the bARE transcript (21).
Unanticipated findings were observed when HuR was silenced. This AUBP is known as a mRNA stabilizer (32) thus a decrease of ARE-containing reporter following HuR silencing was foreseen. Quite unexpectedly, HuR silencing caused specific up-regulation of the bARE reporter. Indeed, the mARE transcript decay rate was not modified by siHuR, as observed by others (43). We speculate that opposite actions of different AUBPs modulating mARE turnover may produce this net null result, but deeper investigation is needed.
The increased luciferase activity in bARE reporter cells prompted us to envisage a potential interplay between gene-specific and function-specific mechanisms of mRNA turnover regulation. In particular, we hypothesized that the HuR silencing effect on endogenous bcl-2 mRNA might, in turn, influence the bARE-specific reporter mRNA turnover. Indeed, the silencing of HuR caused a significant reduction of the levels of Bcl-2 protein and mRNA due to decreased mRNA stability. Altogether these results proved the stabilizing activity of HuR on endogenous bcl-2 mRNA but also revealed the opposite effects of HuR silencing on ectopic and endogenous bcl-2 ARE. As a consequence of HuR silencing, the Bcl-2 down-regulation can mediate a stabilizing effect on the bARE reporter mRNA that increases its luminescence. Most importantly, the unpredicted increase of bARE reporter activity following HuR silencing hints at a possible picture where the gene-specific effect triggered by endogenous Bcl-2 down-regulation is dominant on the function-specific outcome of HuR silencing.
We address this hypothesis and provide evidence of the specificity and the efficacy of the Bcl-2 protein in regulating the decay rate of its own mRNA in genetically defined cell lines and clones. Both transiently and stably transfected cells expressing the 3′-modified reporter gene showed the Bcl-2 specific regulation in different cell lines. The siRNA silencing of Bcl-2 expression in either transiently or stably transfected clones showed the increase of the bARE-reporter luminescence in a dose-response fashion. The Bcl-2 destabilizing function on its own mRNA observed in siBcl2-treated stably transfected clones was directly proved by restoring Bcl-2 expression in Bcl-2-silenced cells and by ectopically expressing the bcl-2 gene in (bcl-2−/−) Daudi cells.
Finally, HuR and Bcl-2 physically interact with each other, as ascertained by co-immunoprecipitation experiments, with Bcl-2 counteracting HuR activity on bARE, as indicated by the unanticipated findings that siHuR led to an increase of Rluc-bARE transcript levels due to reduced levels of Bcl-2. Altogether these data suggest a mechanism whereby Bcl-2 might displace HuR from its targets localized in the bARE sequence.
In human cellular systems with diverse genetic backgrounds and differentiation conditions, the Bcl-2 protein was determinant to regulate the rate of bARE turnover, as evaluated by the reporter activity. A negative feedback mechanism finely modulating Bcl-2 levels by a bARE-specific regulation is substantiated by the data presented herein and suggest that negative feedback might be a foremost general principle in mammalian regulation of mRNA turnover (44).
The results achieved might have implications for other ARE-containing RNAs, thus shedding light on the molecular mechanism by which gene-specific regulation of mRNA decay occurs. This is a crucial issue in the investigation of determinants of mRNA stability and gene expression regulation overall.
In conclusion, we can envisage a hierarchic network of different AUBPs and associated factors affecting bARE RNA stability involving Bcl-2 protein itself as the major determinant of its own transcript half-life in living cells. This mechanism would provide an autoregulatory loop able to rapidly vary the Bcl-2 level in response to different stimuli. Interestingly, the expression of many RNA-binding proteins has been recently suggested to be post-transcriptionally self-regulated (45–47), and tightly interdependent, suggesting a circuitry of self- and cross-regulatory interactions (48). Altogether these data reveal a hierarchy in the mechanism of post-transcriptional regulation of gene expression, at least for ARE-containing mRNAs, with the gene-specific elements prevailing on the function-specific factors.
Supplementary Material
Acknowledgments
We thank Roberto Gherzi, Paola Briata, and Pavel Ivanov (FLAG-Bcl-2 construct) for critical reading and discussion of the manuscript.
The work was supported by grants from Ministero dell'Istruzione, dell'Università e della Ricerca, Istituto Superiore Sanità (Rome, Italy), and the Italian Association for Cancer Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- 3′-UTR
- 3′-untranslated region
- ARE
- adenine-uridine-rich elements
- AUBP
- ARE-binding protein
- bARE
- bcl-2 ARE
- mARE
- c-myc ARE
- cARE
- CDK5R1 ARE
- HEK293
- human embryonic kidney 293
- K422
- Karpas 422
- DRB
- 5,6-dichloro-1-β-d-ribobenzimidazole
- siRNA
- small interfering RNA
- siBcl2
- siRNAs targeting bcl-2 mRNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siAUF1
- siRNA targeting AUF1 mRNA
- siTIA1
- siRNA targeting TIA-1 mRNA
- siHuR
- siRNA targeting HuR mRNA
- IP
- immunoprecipitation
- α-HuR
- anti-HuR mouse monoclonal antibody
- FCS
- fetal calf serum.
REFERENCES
- 1.Shim J., Karin M. (2002) Mol. Cells 14, 323–331 [PubMed] [Google Scholar]
- 2.Wilusz C. J., Wilusz J. (2004) Trends Genet. 20, 491–497 [DOI] [PubMed] [Google Scholar]
- 3.Audic Y., Hartley R. S. (2004) Biol. Cell 96, 479–498 [DOI] [PubMed] [Google Scholar]
- 4.Hollams E. M., Giles K. M., Thomson A. M., Leedman P. J. (2002) Neurochem. Res. 27, 957–980 [DOI] [PubMed] [Google Scholar]
- 5.Eberhardt W., Doller A., Akool el-S., Pfeilschifter J. (2007) Pharmacol. Ther. 114, 56–73 [DOI] [PubMed] [Google Scholar]
- 6.Newbury S. F. (2006) Biochem. Soc. Trans. 34, 30–34 [DOI] [PubMed] [Google Scholar]
- 7.Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garneau N. L., Wilusz J., Wilusz C. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 113–126 [DOI] [PubMed] [Google Scholar]
- 9.Barreau C., Paillard L., Osborne H. B. (2005) Nucleic Acids Res. 33, 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin W. J., Duffy A., Chen C. Y. (2007) J. Biol. Chem. 282, 19958–19968 [DOI] [PubMed] [Google Scholar]
- 11.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S. C., Gram H., Han J. (2005) Cell 120, 623–634 [DOI] [PubMed] [Google Scholar]
- 12.von Roretz C., Gallouzi I. E. (2008) J. Cell Biol. 181, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bevilacqua A., Ceriani M. C., Capaccioli S., Nicolin A. (2003) J. Cell. Physiol. 195, 356–372 [DOI] [PubMed] [Google Scholar]
- 14.Pan Y. X., Chen H., Kilberg M. S. (2005) J. Biol. Chem. 280, 34609–34616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akhtar R. S., Ness J. M., Roth K. A. (2004) Biochim. Biophys. Acta 1644, 189–203 [DOI] [PubMed] [Google Scholar]
- 16.Hughes P., Bouillet P., Strasser A. (2006) Curr. Dir. Autoimmun. 9, 74–94 [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V., Joos S., Zörnig M. (2004) Biochim. Biophys. Acta 1644, 229–249 [DOI] [PubMed] [Google Scholar]
- 18.Papucci L., Witort E., Bevilacqua A. M., Donnini M., Lulli M., Borchi E., Khabar K. S., Tempestini A., Lapucci A., Schiavone N., Nicolin A., Capaccioli S. (2008) Mol. Pharmacol. 73, 498–508 [DOI] [PubMed] [Google Scholar]
- 19.Wesarg E., Hoffarth S., Wiewrodt R., Kröll M., Biesterfeld S., Huber C., Schuler M. (2007) Int. J. Cancer 121, 2387–2394 [DOI] [PubMed] [Google Scholar]
- 20.Zeitlin B. D., Zeitlin I. J., Nör J. E. (2008) J. Clin. Oncol. 26, 4180–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bevilacqua A., Ghisolfi L., Franzi S., Maresca G., Gherzi R., Capaccioli S., Nicolin A., Canti G. (2007) Mol. Pharmacol. 71, 531–538 [DOI] [PubMed] [Google Scholar]
- 22.Reed J. C., Cuddy M., Slabiak T., Croce C. M., Nowell P. C. (1988) Nature 336, 259–261 [DOI] [PubMed] [Google Scholar]
- 23.Moncini S., Bevilacqua A., Venturin M., Fallini C., Ratti A., Nicolin A., Riva P. (2007) BMC Mol. Biol. 8, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. (2004) Nucleic Acids Res. 32, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M. (2006) Mol. Cell. Biol. 26, 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gherzi R., Lee K. Y., Briata P., Wegmüller D., Moroni C., Karin M., Chen C. Y. (2004) Mol. Cell 14, 571–583 [DOI] [PubMed] [Google Scholar]
- 27.López de Silanes I., Galbán S., Martindale J. L., Yang X., Mazan-Mamczarz K., Indig F. E., Falco G., Zhan M., Gorospe M. (2005) Mol. Cell. Biol. 25, 9520–9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghisolfi L., Papucci L., Bevilacqua A., Canti G., Tataranni G., Lapucci A., Schiavone N., Capaccioli S., Nicolin A. (2005) Mol. Pharmacol. 68, 816–821 [DOI] [PubMed] [Google Scholar]
- 29.Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., Kain S. R. (1998) J. Biol. Chem. 273, 34970–34975 [DOI] [PubMed] [Google Scholar]
- 30.Wagner B. J., DeMaria C. T., Sun Y., Wilson G. M., Brewer G. (1998) Genomics 48, 195–202 [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki S., Stoecklin G., Kedersha N., Simarro M., Anderson P. (2007) J. Biol. Chem. 282, 30070–30077 [DOI] [PubMed] [Google Scholar]
- 32.Brennan C. M., Steitz J. A. (2001) Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdelmohsen K., Lal A., Kim H. H., Gorospe M. (2007) Cell Cycle 6, 1288–1292 [DOI] [PubMed] [Google Scholar]
- 34.Cherradi N., Lejczak C., Desroches-Castan A., Feige J. J. (2006) Mol. Endocrinol. 20, 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazan-Mamczarz K., Hagner P. R., Corl S., Srikantan S., Wood W. H., Becker K. G., Gorospe M., Keene J. D., Levenson A. S., Gartenhaus R. B. (2008) Oncogene 27, 6151–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposti M. D., Hatzinisiriou I., McLennan H., Ralph S. (1999) J. Biol. Chem. 274, 29831–29837 [DOI] [PubMed] [Google Scholar]
- 37.Lin-Chao S., Chiou N. T., Schuster G. (2007) J. Biomed. Sci. 14, 523–532 [DOI] [PubMed] [Google Scholar]
- 38.Anderson P., Kedersha N. (2008) Trends Biochem. Sci. 33, 141–150 [DOI] [PubMed] [Google Scholar]
- 39.Keene J. D. (2007) Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 40.Bevilacqua A., Ceriani M. C., Canti G., Asnaghi L., Gherzi R., Brewer G., Papucci L., Schiavone N., Capaccioli S., Nicolin A. (2003) J. Biol. Chem. 278, 23451–23459 [DOI] [PubMed] [Google Scholar]
- 41.Levine B., Sinha S., Kroemer G. (2008) Autophagy 4, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 43.Nagaoka K., Tanaka T., Imakawa K., Sakai S. (2007) Exp. Cell Res. 313, 2937–2945 [DOI] [PubMed] [Google Scholar]
- 44.Legewie S., Herzel H., Westerhoff H. V., Blüthgen N. (2008) Mol. Syst. Biol. 4, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks S. A., Connolly J. E., Rigby W. F. (2004) J. Immunol. 172, 7263–7271 [DOI] [PubMed] [Google Scholar]
- 46.Tchen C. R., Brook M., Saklatvala J., Clark A. R. (2004) J. Biol. Chem. 279, 32393–32400 [DOI] [PubMed] [Google Scholar]
- 47.Lin N. Y., Lin C. T., Chen Y. L., Chang C. J. (2007) FEBS J. 274, 867–878 [DOI] [PubMed] [Google Scholar]
- 48.Pullmann R., Jr., Kim H. H., Abdelmohsen K., Lal A., Martindale J. L., Yang X., Gorospe M. (2007) Mol. Cell. Biol. 27, 6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.