Abstract
Two lymphoid cell-specific proteins, RAG1 and RAG2 (RAG), initiate V(D)J recombination by assembling a synaptic complex with recombination signal sequences (RSSs) abutting two different antigen receptor gene coding segments, and then introducing a DNA double strand break at the end of each RSS. Despite the biological importance of this system, the structure of the synaptic complex, and the RAG protein stoichiometry and arrangement of DNA within the synaptosome, remains poorly understood. Here we applied atomic force microscopy to directly visualize and characterize RAG synaptic complexes. We report that the pre-cleavage RAG synaptic complex contains about twice the protein content as a RAG complex bound to a single RSS, with a calculated mass consistent with a pair of RAG heterotetramers. In the synaptic complex, the RSSs are predominantly oriented in a side-by-side configuration with no DNA strand crossover. The mass of the synaptic complex, and the conditions under which it is formed in vitro, favors an association model of assembly in which isolated RAG-RSS complexes undergo synapsis mediated by RAG protein-protein interactions. The replacement of Mg2+ cations with Ca2+ leads to a dramatic change in protein stoichiometry for all RAG-RSS complexes, suggesting that the cation composition profoundly influences the type of complex assembled.
To generate diverse surface antigen receptor molecules, developing lymphocytes undergo a series of site-specific DNA rearrangements to assemble functional antigen receptor genes from component gene segments (1). This DNA rearrangement process, known as V(D)J recombination, is initiated when two lymphoid cell-specific proteins, called RAG1 and RAG2, assemble a multiprotein synaptic complex with a pair of antigen receptor gene segments and subsequently introduce a DNA double strand break at the end of each gene segment (2). A recombination signal sequence (RSS)3 that abuts each participating gene segment serves as the binding site of the RAG proteins and directs the location of DNA cleavage. Each RSS contains conserved heptamer and nonamer sequences that are separated by either 12 or 23 bp of DNA of more varied sequence (12RSS and 23RSS, respectively); efficient V(D)J recombination generally only occurs between two RSSs in which the lengths of DNA separating the heptamer and nonamer differ (the 12/23 rule). The RAG proteins mediate DNA cleavage via a nick-hairpin mechanism, breaking the DNA between the RSS heptamer and the coding segment; these reaction products are subsequently processed and repaired by the non-homologous end-joining pathway (1, 3).
Previous studies suggest that RAG synaptic complexes are assembled through the stepwise binding of a 12RSS followed by the capture of a 23RSS (4–6). In vitro biochemical studies suggest synapsis is mediated by a RAG1/2 heterotetramer, but there remains disagreement over the stoichiometry of RAG1 in these complexes (7). In addition, fluorescence resonance energy transfer techniques have recently been applied to examine the orientation of DNA strands within the synaptic complex. The data obtained from these experiments led the authors to favor a model in which the RSSs adopt a bent and crossed configuration in the synaptic complex, although an alternative model in which synaptic complexes containing RSSs in parallel and antiparallel configurations assemble with similar frequency could not be formally excluded (8). For most in vitro biochemical studies, the synaptic complex has been assembled by incubating the RAG proteins with a pair of oligonucleotide substrates, one containing a 12RSS, and one containing a 23RSS. Whether the RAG proteins and the RSSs adopt the same configuration in synaptic complexes assembled with oligonucleotide substrates as those assembled with longer, more physiological substrates remains to be verified, but some studies suggest there are DNA length-dependent differences in RAG-mediated RSS binding and cleavage activity (9, 35).
To directly observe and analyze RAG-RSS synaptic complexes assembled on long DNA substrates that more closely model an RSS embedded in chromosomal DNA, we used atomic force microscopy (AFM), given its previously demonstrated success for visualizing synaptic complexes in other systems (10–14), and its ability to reveal structural details for synaptic complexes that correlate well with independently obtained crystallographic data (15). AFM has also been recently applied to study the bending of 12RSS substrates by RAG1 and RAG2 (16). We report here the first successful visualization of RAG-RSS synaptic complexes by AFM, and describe their characterization with respect to DNA arrangement and the protein stoichiometry within the complexes. These data provide new and important insight into how RAG-RSS synaptic complexes are assembled and organized.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Murine core RAG1 and RAG2, fused at the amino terminus to maltose-binding protein (MBP), were coexpressed in HEK293 cells and purified as described previously (17). Human high mobility group B1 protein (HMGB1) appended with an amino-terminal His6 tag, and lacking the carboxyl-terminal basic motif and acidic tail, was expressed in the Escherichia coli strain BL21(DE3)pLysS and purified by immobilized metal affinity chromatography and ion exchange chromatography as previously described (18).
RSS Substrates
The plasmid V(D)J recombination substrate pJH200 (19) was subjected to PCR using unlabeled primers, JH6876F (5′-CCGCCTCTCCCCGCGCGTTGG-3′) and JH7285R (5′-GACGACATGGCTCGATTGGCG-3′) to amplify a 409-bp 12RSS substrate, and unlabeled or radiolabeled primers JH7182F (5′-CCTCAGAACTCCATCTGGATT-3′) and JH292R (5′-CCATATCACCAGCTCACCGTC-3′) to amplify a 551-bp 23RSS substrate. Amplification reactions (100 μl) were assembled containing pJH200 template (20 ng), appropriate primer pairs (5 pmol each), and native Taq polymerase (2.5 units; Invitrogen) in PCR buffer (20 mm Tris-HCl, pH 8.4, 50 mm KCl, 200 μm each dNTP, 1.5 mm MgCl2). PCR were subjected to initial denaturation (95 °C, 5 min), 25 cycles of amplification (95 °C × 15 s, 50 °C × 30 s, and 72 °C × 20 s), and a final extension (72 °C × 5 min) using a PTC-100 Thermal Cycler (MJ Research, Waltham, MA). PCR products were fractionated on a 2% agarose gel and gel purified using the QIAQuick Gel Extraction kit (Qiagen Inc., Valencia, CA). Unlabeled DNA fragments of various lengths were similarly amplified by PCR from pJH299 or its derivative lacking the 23RSS (20) and purified using a QIAQuick PCR Cleanup kit.
Electrophoretic Mobility Shift Assay (EMSA)
Binding reactions to assemble RAG pre-synaptic or synaptic complexes on oligonucleotide substrates were assembled as previously described (17). RAG binding reactions containing PCR-generated fragments were similarly prepared.
Preparation of RAG-RSS Complexes for AFM Analysis
Protein-DNA complexes assembled with the RAG proteins and the 12RSS and 23RSS substrates were prepared in two different ways. In the first method, all components were added to the initial reaction. Binding reactions (10 μl) contained 400 ng of purified RAG proteins, 150 ng of HMGB1, 14 nm 23RSS fragment, and 1.3 nm 12RSS fragment in binding buffer (25 mm MOPS, pH 8.0, and 5 mm CaCl2 or MgCl2). The reactions were incubated at room temperature for 10 min. Protein-DNA complexes were fixed with 0.5% glutaraldehyde for 10 min, quenched with 2 m Tris, pH 7.4 (3.5 μl), and diluted 10–20 times for deposition onto APS-mica. These reactions contained an 11-fold excess of the 23RSS fragment compared with the 12RSS fragment. The protein:DNA ratio of each RAG protein to the 23RSS fragment is 12:1, a ratio found by mobility shift assays to yield optimum complex formation. In the second method, the RAG proteins were incubated with the 12RSS and 23RSS fragments separately in half-reactions. The samples were incubated at room temperature for 10 min, and then combined and incubated for another 10 min. These complexes were prepared for sample deposition as described above.
Atomic Force Microscopy, Sample Preparation, and Imaging
Freshly cleaved muscovite ruby mica was incubated in a mixture of 1-(3-aminopropyl)silatrane (APS) solution for 30 min to prepare APS-mica, as described previously for the preparation of various protein-DNA complexes (12–14, 21, 22). The sample droplets (5 μl) were deposited on APS-mica for 2 min, then washed with deionized water, and dried with argon. The mica was attached to a metal disc with double stick tape for imaging. Images were acquired in tapping mode in air using the Multimode SPM Nanoscope IV system (Veeco/Digital Instruments, Santa Barbara, CA). Silicon-etched tapping mode probes (TESP; Veeco/Digital Instruments, Inc.) were used. They had nominal spring constants of ∼42 N/m and a resonant frequency of ∼320 Hz. Image processing, the cross-section, and contour length measurements were performed using Femtoscan (Advanced Technologies Center, Moscow, Russia).
Data Analysis
The volume of the RAG proteins in RAG-RSS complexes was approximated as a segment of a sphere with a diameter measured at half-maximal height of the protein as described (23). To estimate the mass of the RAG proteins in the RAG-RSS complexes, a volume-to-mass conversion factor was derived from AFM studies of the SfiI tetramer bound to DNA (12). This factor, 1.92, was obtained by dividing the average volume calculated for the SfiI-DNA complexes analyzed by AFM (238 nm3) by the expected molecular mass of SfiI tetramer (124 kDa). Crystallographic analysis of the Sfi-DNA complex (15) has recently corroborated the data obtained using AFM (12). The lengths of 12RSS and 23RSS fragments were used for assigning the fragments in the RAG-RSS complexes. The fragment length was obtained by drawing a line over the middle of the DNA fragment. The position of the protein complex on the DNA fragment was obtained by dividing the distance between the end of the fragment and the middle of the protein molecule sitting on the fragment by the whole length of the fragment. The mean values and the mean ± S.E. shown in the histograms were obtained by using Origin 6.0 Software.
RESULTS
Assembly of RAG-RSS Complexes in Preparation for AFM
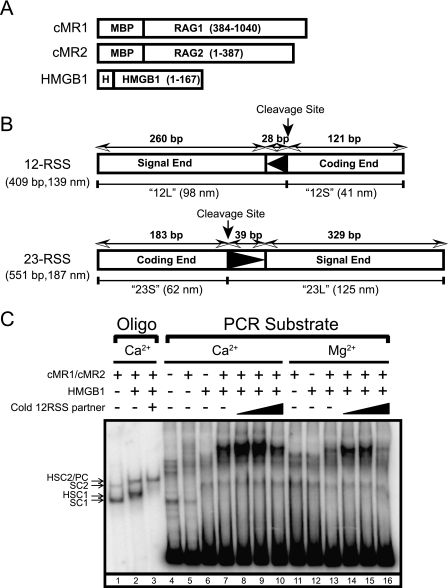
In preparation for our analysis of RAG-RSS complexes by AFM, we coexpressed wild-type truncated catalytically active “core” forms of RAG1 (residues 384–1040; full-length RAG1 contains 1040 amino acids) and RAG2 (residues 1–387; full-length RAG2 contains 527 amino acids) fused at the amino terminus with maltose-binding protein in HEK293 cells, and purified them by amylose affinity chromatography (Fig. 1A). We chose to use truncated RAG proteins for this study due to their relative ease of purification and better behavior in DNA binding assays compared with their full-length counterparts (24). The DNA substrates we prepared for AFM experiments are PCR-generated fragments containing a single 12RSS (409 bp) or 23RSS (551 bp) amplified from the plasmid V(D)J recombination substrate pJH200 (19) (Fig. 1B). These substrates were designed such that the DNA ends on either side of RSSs were all of different lengths, so that, based on our previous analysis of the SfiI-DNA synaptic complex (12), the ends can be distinguished by AFM in RAG-RSS synaptic complexes. Lengths of the two free RSS substrates imaged with AFM were 139 ± 1 and 191 ± 3 nm (data not shown) both of which are very close to expected values (139 and 187 nm, respectively).
FIGURE 1.
Proteins and DNA substrates used in this study. A, RAG1, RAG2, and HMGB1 fusion proteins are depicted (encoded residues in parentheses) and designated at the left. Full-length RAG1, RAG2, and HMGB1 contain 1040, 527, and 216 amino acids, respectively. MBP and polyhistidine (H) sequences are also indicated. B, PCR-generated 12RSS and 23RSS substrates are depicted and designated at the left. Lengths of the RSSs and the long and short arms are shown in bp and nm (12L, 12S, 23L, and 23S, respectively). C, EMSA of RAG-RSS complexes assembled on oligonucleotide and PCR-generated substrates in Ca2+ or Mg2+ in the absence or presence of HMGB1 and cold partner RSS.
In initial studies, we sought to determine whether the PCR-generated DNA substrates support assembly of RAG-RSS complexes detectable by EMSA under conditions suitable for the analysis of these RAG-RSS complexes using AFM. Toward this end, we prepared a radiolabeled PCR-generated 23RSS substrate and incubated the substrate with purified RAG proteins in binding reactions containing Ca2+ or Mg2+ in the absence or presence of purified HMGB1 and cold partner 12RSS. HMGB1 is an architectural DNA binding and bending factor that when added with partner RSS provide conditions known to promote formation of functional RAG synaptic complexes in vitro (25, 26). As controls, synaptic complexes were also assembled using oligonucleotide substrates in the presence of Ca2+. Binding reactions were then fractionated by EMSA to visualize discrete protein-DNA complexes (Fig. 1C). As expected from previous studies, when purified RAG proteins are incubated with an oligonucleotide 23RSS substrate alone in a binding reaction containing Ca2+, two protein-DNA complexes are observed, called stable complexes 1 and 2 (SC1 and SC2, respectively). Both the SC1 and SC2 complexes were earlier shown by our laboratory to contain a RAG1 dimer, but the RAG2 stoichiometry differed between the two complexes, with SC1 containing monomeric RAG2, and SC2 containing two RAG2 molecules (27). In previous results with short oligonucleotide substrates, the addition of HMGB1 to the binding reaction is found to supershift both SC1 and SC2, forming HSC1 and HSC2, respectively. Adding cold 12RSS partner supershifted HSC2 slightly to form a paired complex (PC), but reduced the abundance of HSC1. In contrast to results obtained with oligonucleotide substrates, the RAG proteins or HMGB1 alone do not form discrete protein-DNA complexes on the long PCR-generated 23RSS substrate in the absence of HMGB1 (Fig. 1C). However, when both the RAG and HMGB1 proteins were incubated together with the long 23RSS substrate, a single discrete protein-DNA complex was observed. The abundance of this complex was increased in the presence of low concentrations of cold partner RSS, but reduced when the concentration of partner RSS was increased, likely due to competitive inhibition of RAG binding. Moreover, both Ca2+ and Mg2+ support formation of this stable protein-DNA complex, although the abundance of the complex is slightly lower in Mg2+ than in Ca2+. Notably, the protein-DNA complex assembled with the long 23RSS substrate exhibits much slower mobility in the EMSA than the PC assembled using the oligonucleotide 23RSS substrate.
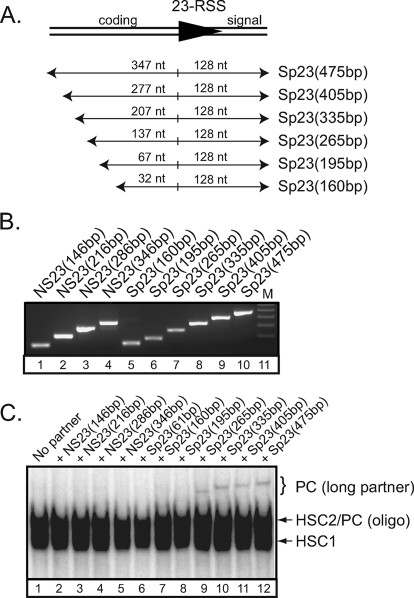
We examined the effect of the DNA length more carefully. We generated a panel of unlabeled PCR-generated substrates lacking or containing a 23RSS. In these substrates, the length of DNA flanking the nonamer is constant, whereas the coding flank is incrementally lengthened (Fig. 2A). The amplification and recovery of the nonspecific and specific DNA fragments was comparable (Fig. 2B). Next, we incubated the RAG proteins with a radiolabeled oligonucleotide 12RSS substrate in binding reactions containing Mg2+ and HMGB1 in the absence or presence of cold nonspecific or specific partner DNA of different lengths and separated the RAG-RSS complexes by EMSA (Fig. 2C). We find that in the presence of cold nonspecific partner DNA, only RAG-RSS complexes comigrating with those formed in the absence of partner RSS were detected (Fig. 2C, compare lane 1 to lanes 2–5). In the presence of a cold oligonucleotide 23RSS partner, which contains 16 bp of coding sequence, the mobility of the RAG-RSS complex was not shifted significantly (Fig. 2C, compare lanes 1 and 6), consistent with data shown in Fig. 1C. Similar results were obtained when the coding flank of the 23RSS partner is lengthened from 16 to 32 or 67 bp (Fig. 2C, compare lane 6 to lanes 7–8). Interestingly, however, a higher-order protein-DNA complex becomes apparent when the coding flank was lengthened to 137 bp and beyond (Fig. 2C, lane 9); the mobility of this complex becomes progressively slower as the length of the coding flank is increased (Fig. 2C, compare lanes 9–12). These data together argue that the higher-order RAG protein complex represents a synaptic complex, and that its formation depends on 23RSS partner length.
FIGURE 2.
DNA length-dependent formation of higher-order RAG-RSS complexes. A, diagram of DNA fragments containing a 23RSS are designated at the right, with lengths of the coding and signal ends indicated above each fragment. B, nonspecific and specific 23RSS DNA fragments (NS23 and Sp23) of different lengths were amplified by PCR, and purified DNA was visualized on an agarose gel stained with ethidium bromide. C, EMSA of RAG-RSS complexes assembled on a radiolabeled 12RSS in Mg2+ with HMGB1 in the absence or presence of cold nonspecific or specific 23RSS partner as indicated above the gel.
Imaging of Synaptic RAG-RSS Complex by AFM
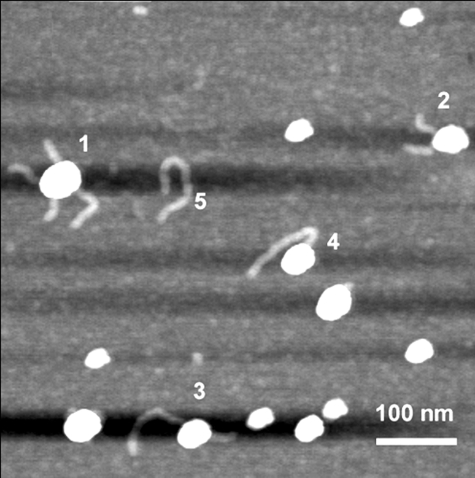
Because EMSA provides evidence that assembly of RAG synaptic complexes on long PCR-generated substrates is feasible, and that visualized complexes may be larger than their counterparts assembled on oligonucleotide substrates, we wished to determine whether RAG synaptic complexes could be directly visualized by AFM and, if so, whether analysis of these complexes could provide information on RAG protein stoichiometry. Toward this end, samples from binding reactions were deposited on mica surface functionalized with 3-aminopropylsilatrane (APS-mica) and imaged by AFM as described previously (e.g. Refs. 12–14, 16, 21, and 22, and references therein). Two notable changes to the binding reactions were made to facilitate visualization of RAG-RSS complexes by AFM. First, we found that full-length HMGB1 impaired RAG-RSS complex deposition,4 necessitating the use of a truncated form of HMGB1 lacking the carboxyl-terminal 48 residues that includes a basic motif and the acidic tail (Fig. 1A) (18). Second, RAG-RSS complexes were subjected to glutaraldehyde cross-linking before AFM because it was necessary for maintaining the integrity of the protein-DNA complex during sample dilution and deposition onto APS-mica. Glutaraldehyde has been used previously to stabilize RAG-RSS complexes for visualization by EMSA (28–31). In addition, glutaraldehyde has been successfully used in various EM studies and recently in AFM imaging of various nucleoprotein complexes including synaptic complexes of DNA with SfiI (12) and EcoRII (13). Importantly, studies of glutaraldehyde cross-linked SfiI-DNA complexes by AFM and uncross-linked SfiI-DNA complexes by crystallography suggest that AFM can provide information on the organization of protein-DNA complexes that is accurately reflected in complexes analyzed by crystallography (15). Evidence from several laboratories provide experimental support for a model of RAG synaptic complex formation in which the RAG proteins first bind to one RSS and then “capture” an appropriate RSS partner (the “capture model”) (4–6). In an alternative model of synapsis, the RAG proteins may first assemble partial (presynaptic) complexes on individual RSSs that subsequently associate with one another through RAG protein-protein interactions (the “association model”). To avoid biasing our experiments to favor one model of assembly over the other, we tried different methods of complex assembly. In the first method, all components were mixed simultaneously and the sample was prepared for AFM. Such an approach should not exhibit a bias for either model of assembly. In the second method, the RAG proteins were incubated with the 12RSS and 23RSS fragments separately in identical solutions before the reactions with each fragment were combined, an approach that likely favors the association model over the capture model of synaptic complex assembly (see “Experimental Procedures” for specifics). In both cases, samples were prepared at room temperature in the presence of Mg2+, a condition known to support 12/23-regulated synapsis and cleavage in vitro (26, 32). The samples prepared by both approaches were comparable in terms of the yield of different types of complexes visualized in AFM images. A representative example of a large scale image on which various complexes appear is shown in Fig. 3. Free protein and DNA molecules, as well as various protein-DNA complexes were observed. Typically, about 30% of the DNA was associated with protein in these images.
FIGURE 3.
A large scale AFM image of RAG complexes with DNA. Various complexes are indicated: 1, synaptic complex; 2, presynaptic 12RSS complex; 3, presynaptic 23RSS complex; 4, end-bound complex; and 5, naked DNA (23RSS).
Specificity and Estimated Mass of RAG Synaptic Complexes Assembled in Mg2+
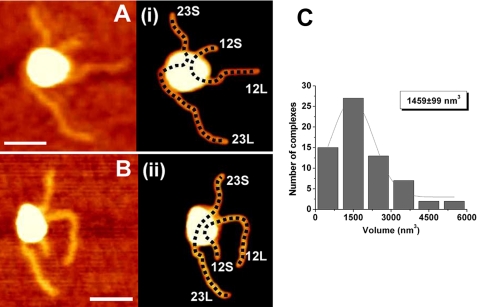
We first investigated the structure of RAG synaptic complexes containing both 12RSS and 23RSS. Synaptic complexes were selected from dozens of images, one of which is represented by Fig. 3. The yield of such complexes among other protein-DNA complexes was quite low (10%), and they were analyzed without bias to the shape of the complex. Two frames of selected rather typical images of synaptic complexes are shown in Fig. 4, A and B. Due to the large size of the protein complexes, the identification of individual DNA fragments within the complexes was not unequivocal. For each complex, length measurements for various combinations of DNA fragments were performed. Those fragment lengths that corresponded most closely to the fragment lengths of the original 12RSS and 23RSS substrates were identified and the DNA traces corresponding to these pairwise combinations are shown as dotted lines in plates i and ii of Fig. 4, A and B. It is clear even visually that other possible combinations provide pairwise values for DNA fragment sizes that differ considerably from the expected lengths for the 12RSS and 23RSS substrate, with the difference being most striking for the 12RSS substrate. This issue is illustrated by the data shown in supplemental Fig. S1, where the length measurements for different paths are shown. In these cases, the lengths of short and long fragments in other alignments are 87 and 228 nm (A) and 149 and 176 nm (B) versus the expected values of 139 and 187 nm. One interesting finding from this analysis is that the DNA paths deviate more greatly from predicted values in complexes drawn to adopt a crossover configuration than those drawn to adopt a side-by-side configuration, suggesting the latter configuration is preferred over the former. The image in Fig. 4A (plate i) also illustrates that the orientation of the two RSSs is in a parallel configuration (12L proximal to 23L). However, there are examples where the strands adopt an antiparallel orientation (12L proximal to 23S; Fig. 4B (plate ii)). The parallel orientation was found to be 2-fold more frequent than the antiparallel orientation. To investigate whether the RAG proteins are specifically bound to the RSS in synaptic complexes, the protein position was mapped on the DNA. Histograms of these measurements are shown under supplement data (supplemental Fig. S2, A and B). The relative positions calculated from these histograms were 0.36 ± 0.01 and 0.37 ± 0.01 nm for the 12RSS and 23RSS substrates, respectively. These values are very close to that predicted for the position of the RSS on both substrates (0.33 and 0.37 nm, respectively), suggesting that the analyzed complexes are indeed specific synaptic complexes.
FIGURE 4.
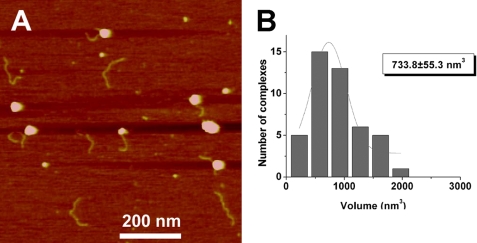
AFM images of RAG synaptic complexes. Representative AFM images are shown in panels A and B along with corresponding rendered images of the complexes (plates i and ii) included for clarity to illustrate the arm designations. The bar length is 40 nm. The DNA path is shown with a dotted line on the rendered image. The lengths measured for 12RSS and 23RSS in this configuration for panel i are 137 and 187 nm, respectively. The expected lengths of 12RSS and 23RSS substrates are 139 and 187 nm, respectively. A histogram of protein volumes calculated for the synaptic complexes is shown in C. The protein volume corresponding to the maximum of the Gaussian fit is shown in the inset.
We took advantage of AFM providing the height of the sample to determine the volume of protein found in RAG-RSS complexes. The values of protein volume in the synaptic complexes were plotted as a histogram (Fig. 4C). Gaussian approximation provides the value for the maximum at 1459 ± 99 nm3. We derived a protein volume-to-mass conversion factor based on AFM data obtained from analysis of tetrameric SfiI-DNA synaptic complexes (12). The calculated volume-to-mass ratio in the SfiI-DNA complex is about 1.92. Using this conversion factor, the experimentally observed mass for the RAG synaptic complex is ∼760 kDa. This value is much larger than the ∼408 kDa mass predicted for the RAG1/RAG2 heterotetramer of the MBP-RAG1 and RAG2 fusion proteins used in this study (assuming a 1:1 ratio of RAG1 (118 kDa/monomer) and RAG2 (86 kDa/monomer)), but is close to the 816-kDa mass predicted for a RAG1/RAG2 hetero-octamer (predicted volume of 1566 nm3). Based on these estimations, the RAG protein stoichiometry in synaptic complexes is close to the hetero-octamer.
Specificity and Estimated Mass of RAG Complexes Bound to a Single RSS in Mg2+ (Presynaptic Complex)
Because the RAG synaptic complexes visualized by AFM were larger than expected for the RAG heterotetramer, we wished to determine whether this feature extended to RAG complexes bound to a single RSS. Therefore, we investigated the structure of RAG complexes bound to individual 12RSS and 23RSS substrates (presynaptic complexes). A typical image of such complexes bound to an individual 12RSS is shown in Fig. 5A. Measurements of the DNA contour length in these complexes yields a Gaussian maximum of 135 ± 1 and 183 ± 3 nm for the 12RSS and 23RSS, respectively, values that are very close to the lengths of the free DNA fragments (see Fig. 1B). These data suggest that when the RAG proteins bind a single RSS internally, they do not change the DNA contour length (i.e. no DNA wrapping around the protein). We also measured the relative positions of the RAG proteins in complexes assembled on individual 12RSS and 23RSS substrates, obtaining values that are similar to data calculated above from synaptic complexes (0.35 ± 0.01 and 0.32 ± 0.03 for the 12RSS and 23RSS, respectively). Finally, protein volume measurements were calculated for a RAG complex bound to a 12RSS (Fig. 5B). Gaussian approximation provides the value for the maximum at 734 ± 55 nm3. Using the mass-to-volume conversion factor, we estimate that the mass of this complex is about 382 kDa, which is close to the ∼408 kDa mass predicted for a RAG1/2 hetero-tetramer (predicted volume of 783 nm3). Based on these estimations, the RAG protein stoichiometry in presynaptic complexes is close to the tetrameric form.
FIGURE 5.
AFM data of RAG presynaptic complexes. A, large scale AFM image of 12RSS-RAG presynaptic complexes. B, histogram of protein volumes measured for presynaptic complexes. The protein volume corresponding to the maximum of the Gaussian fit is shown in the inset.
Nonspecific RAG Binding
We next compared these data with results obtained from analysis of RAG complexes assembled on substrate DNA lacking an RSS. For this experiment, we used a 527-bp DNA fragment generated by PCR described previously (13). Two types of RAG-DNA complexes were visualized. In the first type of complex, RAG binding occurred anywhere in the “middle” of the fragment, with two DNA ends protruding from the protein complex (internal binding). Internally bound RAG complexes were uniformly distributed along the DNA (supplemental Fig. S3A), suggesting that RAG binding occurred nonspecifically. In the second type of complex, RAG binding occurred at the end of the DNA fragment (end binding). Among the complexes analyzed, 27% were internally bound complexes, whereas 73% were end-bound complexes. The average DNA fragment length with protein bound internally (173.5 ± 0.5 nm) was close to the expected length of 179 nm (supplemental Fig. S3B). Protein volume analysis was also performed with a nonspecific RAG-DNA complex using the DNA substrate as described above. For internally bound complexes, the mean protein volume for the nonspecific complex was 352 ± 21 nm3 (supplemental Fig. S4). This value is about half of the volume observed for RAG complexes specifically bound to a 12RSS substrate (Fig. 5B). Thus, the content of RAG complexes internally bound to specific and nonspecific substrates differs by ∼2-fold.
Analysis of Pre-synaptic and Synaptic Complexes Formed in the Presence of Ca2+
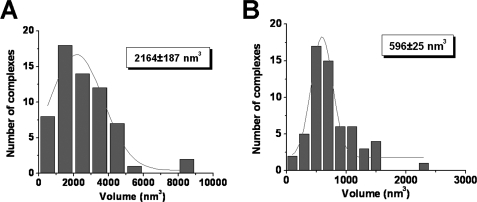
The data shown above were obtained in the presence of Mg2+ that is a critical cofactor for RAG-mediated RSS synapsis and cleavage. Although under the conditions used for these experiments (25 versus 37 °C) the kinetics of cleavage are very slow,5 we thought it would be instructive to characterize RAG complexes assembled under conditions where the enzymatic activity is suppressed. This is typically achieved by replacing Mg2+ with Ca2+ in the RAG in vitro cleavage reaction. RAG-RSS complexes were assembled using the same approach as for the Mg2+ experiments and large scale and zoomed images of various types of complexes were obtained (supplemental Fig. S5, A and B, respectively). We find that the abundance and distribution of RAG complexes with a single RSS or a pair of RSSs is similar when assembled in Ca2+ as compared with Mg2+. Moreover, mapping data (supplemental Fig. S6, A and B) suggest that the RAG proteins are bound specifically to the fragments at positions consistent with the location of the RSS (0.36 ± 0.01 and 0.32 ± 0.01 nm, respectively). However, volume measurements revealed unexpected results. We find that the distribution maximum for the RAG synaptic complex assembled in Ca2+ corresponds to 2164 ± 187 nm3 (Fig. 6A), a value that is 1.5 times larger than the value obtained for its counterpart assembled in the presence of Mg2+ (1459 ± 99 nm3). In contrast, the average value for pre-synaptic complexes assembled in Ca2+ is 596 ± 25 nm3 (Fig. 6B). This value is substantially less then that obtained for its counterpart assembled in the presence of Mg2+ (734 ± 55 nm3). Given the differences in the sizes of the protein-DNA complexes observed, we wondered whether the size of the free protein also exhibits metal ion dependence. To test this possibility, we measured the sizes of the unbound proteins prepared in the presence of Mg2+ or Ca2+, obtaining values of 510 ± 12 and 265 ± 17 nm3, respectively (supplemental Fig. S7, A and B). These data correspond to estimated masses of about 265 and 138 kDa, respectively, suggesting that the divalent cation type influences the stoichiometry of free protein under these experimental conditions. Note that this analysis was performed using the same set of images that had been used for the analyses of the protein-DNA complexes described above, obviating potential concerns regarding sample variability in different preparations. Taken together, these comparative studies suggest that the stoichiometry of free and bound RAG proteins depends on the type of divalent cation used in the binding reaction.
FIGURE 6.
Histogram of protein volumes measured for RAG synaptic and presynaptic complexes prepared in the presence of Ca2+ cations. Volume measurements were calculated for synaptic complexes (A) and presynaptic complexes (B). The distribution maxima for both histograms are shown in the insets.
DISCUSSION
The AFM data presented here have enabled us to directly analyze synaptic RAG-RSS complexes assembled under various conditions on long DNA substrates. A number of interesting and surprising conclusions emerged from the analysis of free RAG protein and RAG pre-synaptic and synaptic complexes identified in these AFM images. First, RAG complexes assembled on a single RSS are about half the size of RAG complexes bound to a pair of RSSs, an observation that has potentially important implications for understanding the composition and assembly of RAG synaptic complexes. Second, we detected only a few RAG complexes containing a pair of identical RSS substrates, suggesting that the 12/23 rule is operative under these conditions. Third, the location of the RAG proteins on the 12RSS and 23RSS substrates is consistent with specific binding to the RSS. Fourth, the length of the DNA fragments is not shortened when bound by the RAG proteins, suggesting that the DNA is not wrapped around the protein core. Fifth, RAG protein complexes bound specifically and nonspecifically to DNA are characterized by a distinct protein stoichiometry, suggesting that specific binding defines complex size. Finally, the size of the free and DNA-bound RAG complexes differs depending on what divalent metal cation is used in the binding reaction. This observation suggests that although the RAG proteins are capable of binding DNA in the presence of either cation, the biophysical properties of the complexes formed are not the same. In the sections below we elaborate on each of these issues.
Implications for the Stoichiometry of Synaptic and Presynaptic Complexes
Previous studies have shown that the RAG proteins can form two discrete protein-DNA complexes on an oligonucleotide substrate containing a single RSS: one containing a dimer of RAG1 and a monomer of RAG2, and another that contains two molecules of each protein (a RAG1/2 heterotetramer). Synaptic complexes are thought to either retain a RAG1/2 heterotetramer configuration (33) or contain one or more additional subunits of RAG1 (6, 31), as well as an unknown number of HMGB1 molecules. The MBP-RAG1, MBP-RAG2, and HMGB1 proteins used in these studies have predicted molecular masses of 118, 86, and 21.6 kDa, respectively. The estimated mass of RAG complexes assembled on a single RSS visualized by AFM is about 382 kDa, which is slightly less than expected for a RAG1/2 heterotetramer (408 kDa). The estimated molecular mass of synaptic complexes visualized by AFM is about 760 kDa. A model of a RAG synaptic complex containing a pair of RAG1/2 heterotetramers (1:1 ratio of RAG1 and RAG2) yields an expected molecular mass of 815 kDa, which is also slightly more but still reasonably close to the mass of synaptic complexes calculated by AFM, without accounting for any associated HMGB1 molecules. However, adding two HMGB1 molecules to the synaptic complex would not dramatically increase its mass, given the low molecular weight of the truncated form of HMGB1 used in this study. The small differences between expected and observed molecular masses of RAG complexes cannot be taken into serious consideration due to the relatively large distribution of protein volumes determined by analysis of AFM images. However, because AFM measures the volume (shape) of the proteins rather than their molecular masses, and the molecular mass estimates were obtained based on volume measurements in which a protein is modeled as a hemisphere, it is possible that differences between the anticipated and observed masses may reflect deviations from a hemispheric model that are intrinsic to the proteins themselves. Biochemical evidence in support of the latter scenario has been reported (34), and it is therefore reasonable to speculate that geometric characteristics of free and bound RAG proteins may differ.
Implications for Models of Synaptic Complex Assembly
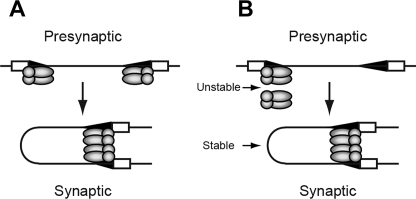
The AFM data show that a RAG complex binds an individual 12RSS template very specifically with an estimated mass close to that predicted for a RAG1/2 heterotetramer. The RAG synaptic complex is also characterized by sequence-specific binding of the RAG proteins, but the protein content of the synaptic complex is about twice that observed for presynaptic RAG complexes. These results are most consistent with a model of synaptic complex assembly in which two isolated RAG-RSS complexes form a synaptic complex mediated through RAG protein-protein interactions (Fig. 7A), as this model predicts an increase in protein content from a presynaptic to a synaptic complex. However, evidence from previous in vitro (5, 6) and in vivo (4) studies have been interpreted to favor a capture model of synaptic complex assembly in which the RAG protein content does not change between presynaptic and synaptic complexes. These apparently discordant results can be reconciled by considering the underlying limitations and assumptions of these studies. Previous in vitro studies showing that the complement of RAG proteins does not change between pre-synaptic and synaptic complexes were assembled using short, mostly oligonucleotide substrates rather than longer, more physiologically relevant DNA templates. However, a study by Huye et al. (35) suggests that there are DNA length-dependent differences in how the RAG proteins assemble synaptic complexes and cleave DNA in trans. It is therefore possible that although short oligonucleotide substrates can be bound by both RAG1 subunits in a RAG1/2 (heterotetramer) core protein complex, longer DNA fragments may be unable to do so, due to steric interference or electrostatic repulsion of the DNA flanking the RSS. In the latter case, two RAG core protein complexes may be required to bridge the two RSSs to assemble a functional synaptic complex. This possibility may partly explain conflicting results regarding the stoichiometry of RAG1 in the synaptic complex. EMSA-based studies using oligonucleotide substrates suggest the synaptic complex contains a RAG1 dimer, whereas kinetic studies of RAG-mediated cleavage of longer DNA fragments containing RSSs paired in cis using wild-type and catalytically inactive RAG1 heterodimers published by Landree et al. (31) suggest that synaptic complexes assembled on these substrates contains a pair of RAG1 dimers (6). In this model, the RAG1 subunits may play separable roles in mediating protein-protein and protein-DNA interactions and catalyzing strand cleavage and strand-transfer reactions, by analogy to the Mu transposasome (36), and must communicate the status of RSS occupancy of each core protein complex to enable synapsis and cleavage of RSSs regulated by the 12/23 rule.
FIGURE 7.
Models for RAG synaptic complex assembly. RAG1 (shaded ovals) and RAG2 (shaded circles) form a RAG core protein complex, shown here as a heterotetramer. In an association model of assembly (A), a core RAG protein complex binds at separate 12- and 23RSSs (small and large shaded triangles, respectively), and assembles a synaptic complex mediated by RAG protein-protein interactions. In an alternative model (B), association of an unbound RAG core complex with a complex bound to a 12RSS is unstable, but may transiently interact to capture a 23RSS partner, forming a stable synaptic complex. In the synaptic complex, the DNA strands do not cross over one another. RAG1 and RAG2 are shown bound to a single RSS as a heterotetramer based on previous studies using EMSA (28) and mass estimates obtained by AFM presented here. The synaptic complex is shown to contain a pair of RAG1/2 heterotetramers based on the approximate doubling of protein content between RAG complexes bound to a single RSS and those bound to a pair of RSSs. HMGB1 molecules, which are expected to be integrated into RAG-RSS complexes, are not shown for simplicity.
How does one consider this model in light of in vivo data published by Curry et al. (4)? In these experiments, nicks were detected at the 12RSS but not at the 23RSS in developing B cells, which is most easily explained by a model in which a RAG complex binds and nicks a 12RSS, and subsequently captures a 23RSS, nicking the 23RSS and cleaving both RSSs in rapid succession. To reconcile these results with our studies, we consider the possibility that association between RAG core protein complexes (i.e. two RAG1/2 heterotetramers) is weak unless both are bound to DNA. In this view, pre-synaptic complexes containing the protein complement found in synaptic complexes would be difficult to detect biochemically because the RAG protein core not bound to DNA would interact weakly with a DNA-bound RAG protein core. This association could be strengthened upon capture of an appropriate RSS partner, enabling the synaptic complex to be more readily visualized by AFM (Fig. 7B). Evidence for the transient formation of pre-synaptic complexes containing a RAG1/2 hetero-octamer is supported by the analysis of RAG complexes assembled in the presence of both 12RSS and 23RSS substrates. Under these conditions, both pre-synaptic complexes (molecules 2 and 3 in Fig. 3) and complete synaptic complexes (molecule 1) can be visualized, and the volume measurements for pre-synaptic complexes show a broad size distribution, ranging from tetrameric to octameric stoichiometry (data not shown). We hypothesize that complexes as large as octamers detected in these experiments likely originated from synaptic complexes that had lost one of the associated DNA fragments, as we did not observe RAG1/2 octamers when complexes were assembled only in the presence of the 12RSS substrate. On the other hand, interpreting the order of synaptic complex formation based on the differential detection of nicks at the 12RSS and 23RSSs may be complicated by the fact that RAG recruitment to antigen receptor loci may be facilitated in part by interactions between the RAG proteins and other cellular factors, such as methylated histones (37, 38) and transcription factors (39, 40), which could influence the efficiency of RAG binding to the 23RSS. Thus, in our view, the evidence favoring one model of RAG synaptic complex assembly over another is not yet definitive.
Implications for the Models of the Synaptic Complex Structure
AFM images provide insights into the arrangements of the DNA templates within synaptosomes. We have shown recently that length measurements of appropriately designed DNA templates allow one to predict the path of DNA templates within synaptic complexes (12), and that the predictions derived from analysis of SfiI-DNA synaptosomes detected by AFM were in perfect correlation with crystallographic data (15). Application of this approach was instrumental for understanding the arrangement of the DNA helices within RAG synaptic complexes. The length measurements illustrated on Fig. 4, A (plate i) and B (plate ii), showed that predicted RSS lengths in the protein-DNA complex could only be obtained if the two DNA strands were aligned side-by-side in a specific orientation. Statistical analysis of 63 clearly identified synaptic complexes showed that a parallel orientation of the helices is preferred over antiparallel orientation by ∼2-fold. This observation stands in contrast to results obtained from fluorescence resonance energy transfer experiments on RAG synaptic complexes assembled on oligonucleotide substrates (8), which supported a model in which the bound 12- and 23RSS substrates adopted a crossed configuration. This apparent discrepancy may be attributed to the potential for DNA length-dependent differences in the manner in which the RAG proteins bind short oligonucleotides and long DNA fragments as discussed above.
Another interesting feature that we would like to point out in the images of RAG synaptic complexes (Fig. 4, A and B (plates i and ii)) is that predicted paths of the DNA strands are not straight. We recently showed that DNA in RAG presynaptic complexes visualized by AFM is bent (16), and this is probably also true for DNA bound in RAG synaptic complexes as well. This is especially likely because HMGB1 used to facilitate RAG-mediated synaptic complex formation is known to possess intrinsic DNA bending activity (41) and also promote RAG-mediated DNA bending in the context of RAG-RSS complexes (42). DNA bending may facilitate subsequent compaction of the protein complex, which may partly explain why the complexes are smaller than expected based on the volume measurements.
The Structure of Synaptic and Pre-synaptic Complexes Depends on the Divalent Cation Type
The AFM data revealed an unexpectedly strong effect of the divalent cation type on the structure of the RAG presynaptic and synaptic complexes as well as free protein. It is generally accepted that replacing Mg2+ with Ca2+ cations primarily affects the enzymatic activity of the RAG proteins without affecting protein-DNA complex assembly. We show here that the stoichiometry of all types of complexes as well as the morphology of free proteins depends on the type of cation, although the sequence specificity of RAG binding to the RSS substrate remains high regardless of the cation type. Interestingly, the size of presynaptic complexes assembled in the presence Ca2+ cations is smaller than expected for a RAG1/2 tetramer (predicted mass of 310 versus 408 kDa), but reasonably close to that predicted for a RAG1 dimer and a RAG2 monomer (322 kDa) as determined previously in an SC1 RAG-RSS complex (43). The presynaptic complex observed in the presence of Mg2+ cations contains a protein mass more consistent with a RAG1/2 heterotetramer (382 versus 408 kDa). The size of synaptic complexes also vary according to the type of cation: in the presence of Ca2+ synaptic complexes appear to have greater mass than predicted for a RAG1/2 hetero-octamer. Such large complexes in the presence of Mg2+ cations are very rare events. Although protein-DNA interactions within the synaptic complex may also be influenced by the cation type, the large effect that cation type has on the size of free RAG proteins suggests that cations play a larger role in determining the nature of protein-protein assemblies. The cation type may also alter the conformational dynamics and shapes of the proteins, which, as discussed above, could potentially account for differences between the measured and anticipated masses of protein complexes analyzed by AFM.
In summary, we present here the first direct visualization of the RAG synaptic complex at the nanometer scale. These experimental findings suggest that the synaptic complex contains two core RAG complexes with a mass consistent with a pair of RAG1/2 heterotetramers; each core RAG complex binds a separate RSS, holding them in a side-by-side configuration through protein-protein interactions between the two core RAG complexes. These studies set the stage for future efforts to characterize the factors and forces that maintain the stability of the synaptic complex using different AFM modalities.
Supplementary Material
Acknowledgments
We thank Alex Lushnikov for help during the initial stages of this work. The AFM experiments were performed in the University of Nebraska Medical Center Nanoimaging core facility with the use of the AFM instruments purchased with the National Institutes of Health SIG Grant 1S10 SS023400.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI055599 (to P. C. S.) and R01 GM0062235 and S10 RR023400 (to Y. L. L.). This work was also supported by National Science Foundation Grant 061590 (to Y. L. L.), Research Facilities Improvement Program of the National Institutes of Health National Center for Research Resources Grant C06 RR17417-01 (to P. C. S.), and the Nebraska Research Initiative (to Y. L. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- RSS
- recombination signal sequence
- HMGB1
- high mobility group B1 protein
- EMSA
- electrophoretic mobility shift assay
- APS
- 1-(3-aminopropyl)silatrane
- MBP
- maltose-binding protein
- SC
- stable complex
- AFM
- atomic force microscopy
- MOPS
- 4-morpholinepropanesulfonic acid
- 12RSS
- 12-bp recombination signal sequence
- 23RSS
- 23-bp recombination signal sequence.
REFERENCES
- 1.Bassing C. H., Swat W., Alt F. W. (2002) Cell 109, ( suppl.) S45–S55 [DOI] [PubMed] [Google Scholar]
- 2.Hesslein D. G., Yang S. Y., Schatz D. G. (2006) Mol. Immunol. 43, 326–334 [DOI] [PubMed] [Google Scholar]
- 3.Lieber M. R., Lu H., Gu J., Schwarz K. (2008) Cell Res. 18, 125–133 [DOI] [PubMed] [Google Scholar]
- 4.Curry J. D., Geier J. K., Schlissel M. S. (2005) Nat. Immunol. 6, 1272–1279 [DOI] [PubMed] [Google Scholar]
- 5.Jones J. M., Gellert M. (2002) EMBO J. 21, 4162–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundy C. L., Patenge N., Matthews A. G., Oettinger M. A. (2002) Mol. Cell. Biol. 22, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson P. C. (2004) Immunol. Rev. 200, 90–114 [DOI] [PubMed] [Google Scholar]
- 8.Ciubotaru M., Kriatchko A. N., Swanson P. C., Bright F. V., Schatz D. G. (2007) Mol. Cell. Biol. 27, 4745–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Swanson P. C. (2009) Nucleic Acids Res. 37, 2211–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassell G., Moision R., Rabani E., Segall A. (1999) Nucleic Acids Res. 27, 1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang M., Cecconi C., Kim H., Bustamante C., Rio D. C. (2005) Genes Dev. 19, 1422–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lushnikov A. Y., Potaman V. N., Oussatcheva E. A., Sinden R. R., Lyubchenko Y. L. (2006) Biochemistry 45, 152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shlyakhtenko L. S., Gilmore J., Portillo A., Tamulaitis G., Siksnys V., Lyubchenko Y. L. (2007) Biochemistry 46, 11128–11136 [DOI] [PubMed] [Google Scholar]
- 14.Lyubchenko Y. L., Shlyakhtenko L. S. (2009) Methods 47, 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanamee E. S., Viadiu H., Kucera R., Dorner L., Picone S., Schildkraut I., Aggarwal A. K. (2005) EMBO J. 24, 4198–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlicek J. W., Lyubchenko Y. L., Chang Y. (2008) Biochemistry 47, 11204–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergeron S., Anderson D. K., Swanson P. C. (2006) Methods Enzymol. 408, 511–528 [DOI] [PubMed] [Google Scholar]
- 18.Bergeron S., Madathiparambil T., Swanson P. C. (2005) J. Biol. Chem. 280, 31314–31324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesse J. E., Lieber M. R., Gellert M., Mizuuchi K. (1987) Cell 49, 775–783 [DOI] [PubMed] [Google Scholar]
- 20.Kriatchko A. N., Anderson D. K., Swanson P. C. (2006) Mol. Cell. Biol. 26, 4712–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shlyakhtenko L. S., Gall A. A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y. L. (2003) Ultramicroscopy 97, 279–287 [DOI] [PubMed] [Google Scholar]
- 22.Lyubchenko Y. L. (2004) Cell Biochem. Biophys. 41, 75–98 [DOI] [PubMed] [Google Scholar]
- 23.Henderson R. M., Schneider S., Li Q., Hornby D., White S. J., Oberleithner H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8756–8760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson P. C., Volkmer D., Wang L. (2004) J. Biol. Chem. 279, 4034–4044 [DOI] [PubMed] [Google Scholar]
- 25.Hiom K., Gellert M. (1998) Mol. Cell 1, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 26.van Gent D. C., Hiom K., Paull T. T., Gellert M. (1997) EMBO J. 16, 2665–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanson P. C. (2002) Mol. Cell. Biol. 22, 7790–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiom K., Gellert M. (1997) Cell 88, 65–72 [DOI] [PubMed] [Google Scholar]
- 29.Akamatsu Y., Oettinger M. A. (1998) Mol. Cell. Biol. 18, 4670–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santagata S., Aidinis V., Spanopoulou E. (1998) J. Biol. Chem. 273, 16325–16331 [DOI] [PubMed] [Google Scholar]
- 31.Landree M. A., Kale S. B., Roth D. B. (2001) Mol. Cell. Biol. 21, 4256–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eastman Q. M., Leu T. M., Schatz D. G. (1996) Nature 380, 85–88 [DOI] [PubMed] [Google Scholar]
- 33.Swanson P. C. (2002) Mol. Cell. Biol. 22, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciubotaru M., Ptaszek L. M., Baker G. A., Baker S. N., Bright F. V., Schatz D. G. (2003) J. Biol. Chem. 278, 5584–5596 [DOI] [PubMed] [Google Scholar]
- 35.Huye L. E., Purugganan M. M., Jiang M. M., Roth D. B. (2002) Mol. Cell. Biol. 22, 3460–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams T. L., Jackson E. L., Carritte A., Baker T. A. (1999) Genes Dev. 13, 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews A. G., Kuo A. J., Ramón-Maiques S., Han S., Champagne K. S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D. N., Walter K. L., Utz P. J., Shi Y., Kutateladze T. G., Yang W., Gozani O., Oettinger M. A. (2007) Nature 450, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Subrahmanyam R., Chakraborty T., Sen R., Desiderio S. (2007) Immunity 27, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Espinoza C. R., Yu Z., Stephan R., He T., Williams G. S., Burrows P. D., Hagman J., Feeney A. J., Cooper M. D. (2006) Nat. Immunol. 7, 616–624 [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Xiao G., Zhang Y., Wen X., Gao X., Okada S., Liu X. (2008) Nat. Immunol. 9, 794–801 [DOI] [PubMed] [Google Scholar]
- 41.Thomas J. O., Travers A. A. (2001) Trends Biochem. Sci. 26, 167–174 [DOI] [PubMed] [Google Scholar]
- 42.Aidinis V., Bonaldi T., Beltrame M., Santagata S., Bianchi M. E., Spanopoulou E. (1999) Mol. Cell. Biol. 19, 6532–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson P. C., Desiderio S. (1999) Mol. Cell. Biol. 19, 3674–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.