Abstract
Earlier studies have reported a role for lipooligosaccharides (LOSs) in sliding motility, biofilm formation, and infection of host macrophages in Mycobacterium marinum. Although a LOS biosynthetic gene cluster has recently been identified in this species, many structural features of the different LOSs (LOS-I–IV) are still unknown. This clearly hampers assessing the contribution of each LOS in mycobacterial virulence as well as structure-function-based studies of these important cell wall-associated glycolipids. In this study, we have identified an M. marinum isolate, M. marinum 7 (Mma7), which failed to produce LOS-IV but instead accumulated large amounts of LOS-III. Local genomic comparison of the LOS biosynthetic cluster established the presence of a highly disorganized region in Mma7 compared with the standard M strain, characterized by multiple genetic lesions that are likely to be responsible for the defect in LOS-IV production in Mma7. Our results indicate that the glycosyltransferase LosA alone is not sufficient to ensure LOS-IV biosynthesis. The availability of different M. marinum strains allowed us to determine the precise structure of individual LOSs through the combination of mass spectrometric and NMR techniques. In particular, we established the presence of two related 4-C-branched monosaccharides within LOS-II to IV sequences, of which one was never identified before. In addition, we provided evidence that LOSs are capable of inhibiting the secretion of tumor necrosis factor-α in lipopolysaccharide-stimulated human macrophages. This unexpected finding suggests that these cell wall-associated glycolipids represent key effectors capable of interfering with the establishment of a pro-inflammatory response.
A key feature of all members of the genus Mycobacterium is a cell wall of unique and complex structure, which plays an important role in antibiotic resistance and pathogenesis of mycobacteria by modulating the host immune system and phagocytic cell functions (1). The mycobacterial cell wall includes essentially two types of lipids, the mycolic acids, which are very long chain fatty acids covalently bound to the arabinogalactan polysaccharide attached to a peptidoglycan backbone (2), and a vast array of extractable lipids/glycolipids (3). The latter include the ubiquitous trehalose dimycolate (TDM)3 and phosphatidyl mannosides (PIM) (4) as well as a vast array of species-specific lipids such as phenol glycolipids (5), phthiocerol dimycocerosates (5), sulfolipids (4), glycopeptidolipids, and lipooligosaccharides (LOSs).
LOSs were found and described in Mycobacterium kansasii (6–8), Mycobacterium gastri (8, 9), Mycobacterium szulgai (10), Mycobacterium malmoense (11), Mycobacterium gordonae (12), Mycobacterium butyricum (13), Mycobacterium mucogenicum (14), the Canetti variant of Mycobacterium tuberculosis (15) and, more recently in Mycobacterium marinum (Mma) (16). However, they remain among the less studied mycobacterial glycolipids at a biosynthetic, structural, and functional point of view. To date, only three genes have been experimentally demonstrated to be involved in the late steps of LOS biosynthesis in M. marinum (16, 17), and one gene encodes a polyketide synthase responsible for the synthesis of the polymethyl-branched fatty acid in the Mycobacterium smegmatis LOS (18).
LOSs represent highly antigenic glycoconjugates exposed to the cell surface and useful target molecules for serotyping in a given mycobacterial species. Their precise role in mycobacteria virulence as well as in the colony morphology remains unclear (19, 20). Early studies demonstrated that rough variants of M. kansasii, devoid of all LOSs, induce chronic systemic infections in mice, whereas smooth variants containing LOSs are rapidly cleared from the organs of infected animals (19, 21). It was therefore proposed that LOSs may act as avirulence factors by masking other cell wall-associated virulence factors. Accordingly, LOSs are absent in most clinical isolates of M. tuberculosis as well as in the laboratory strain H37Rv. A recent genetically based comparison of the LOS biosynthetic cluster in M. marinum and M. tuberculosis revealed that only about one-third of the genes are conserved between the two species, with the genetic locus of M. tuberculosis H37Rv containing fewer genes (17). Although recent studies suggested a possible role of LOSs in sliding motility, biofilm formation, and infection of macrophages by M. marinum (17), the precise contribution of LOSs to M. marinum pathogenesis or virulence is seriously hampered by the restricted number of isogenic strains deficient in their production and the lack of precise structural data of LOS variants. LOSs from different mycobacterial species exhibit considerable variations in the glycan core. A previous work identified the presence of four major LOS variants in M. marinum, designated LOS-I to LOS-IV (16). Through partial characterization, the structure of LOS-I was previously established as 3-O-Me-Rhap-(1–3)-Glcp-(1–3)-Glcp-(1–4)-Glcp-(1–1)-Glcp. Although all LOSs were shown to contain this common oligosaccharidic core substituted by an additional Xylp unit, LOS-II, -III, and -IV are further substituted by other unidentified monosaccharides, designated X and YZ, which leave their exact sequence largely unknown (16).
In this study, we report the identification of a natural mutant of M. marinum, devoid of LOS-IV production, which allowed the production of large amounts of LOS-III and the determination of the fine structure of all LOSs. In addition, the availability of all LOS variants with defined structures has opened the possibility to undertake structure-function relationship studies. These molecules were therefore used in in vitro assays to uncover their potent biological roles.
EXPERIMENTAL PROCEDURES
M. marinum Strains and Growth Culture Conditions
M. marinum isolates, originating from infected fish or humans, used in this study were described previously (22). The reference strain M, a human isolate (23), was also included in this work. Bacteria were grown at 30 °C on plates containing Middlebrook 7H10 supplemented with oleic acid/albumin/dextrose/catalase enrichment or in Sauton's broth medium.
DNA Manipulation
Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were purchased from New England Biolabs. M. marinum genomic DNA was prepared as described previously (24). Probes were labeled with digoxigenin (DIG-dUTP-5′) using the PCR DIG probe synthesis kit (Roche Applied Science), and Southern blot hybridization was performed as described earlier (24). Detection was performed using the DIG luminescence detection kit (Roche Applied Science) according to the manufacturer's recommendations.
Cloning of Rv1500 (losA) from M. tuberculosis
PCR amplification of Rv1500, the gene homologous to losA in M. tuberculosis, was performed using H37Rv genomic DNA along with the upstream primer 5′-TGC GCC TCT CAA TCG TAA CGA CTA T-3′ and downstream primer 5′-AAA GGA ATT CTG CAG ATG TAA GAT GG-3′, which contains an EcoRI site (underlined). The 1360-bp product was then digested with EcoRI and cloned into the MscI/EcoRI-restricted pMV261, a mycobacterial shuttle vector containing the hsp60 promoter (25), thus giving rise to pMV261-Rv1500. Mma7 was transformed with this plasmid or pMV261, and polar lipids were extracted.
Lipid Extraction and Analysis
Polar and apolar lipids were extracted according to established procedures (16). Polar lipid extracts were examined by two-dimensional TLC on plates of Silica Gel 60 (Merck), using chloroform/methanol/water (60:30:6, v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v) in the second direction. Glycolipids were visualized by spraying plates with orcinol/sulfuric acid reagent (0.2%, w/v, orcinol in H2SO4/water, 1:4, v/v) followed by charring. Lipids were also visualized by spraying with 5% ethanolic molybdophosphoric acid and charring. Phosphate-containing lipids were stained and visualized using the Dittmer and Lester reagent.
Purification of Glycolipids
To purify LOSs, the crude polar extract was dissolved in chloroform/methanol (2:1, v/v) and applied in a DEAE-cellulose column (2 × 30 cm) for purification. The column was eluted with chloroform/methanol (500 ml; 2:1, v/v) and increasing concentrations of ammonium acetate (5–500 mm) in chloroform/methanol (2:1, v/v). The purification process was monitored by one-dimensional TLC using chloroform/acetic acid/methanol/water (40:25:3:6, v/v). Glycolipids were visualized by spraying plates with orcinol/sulfuric acid reagent followed by charring. LOS-I, -II, and -III were co-eluted from the DEAE column using chloroform/methanol (2:1, v/v), whereas LOS-IV was eluted from DEAE in the presence of 40–60 mm ammonium acetate in chloroform/methanol (2:1, v/v). DEAE fractions containing LOS-I to LOS-III were concentrated and applied on a silicate gel column for a second purification step. The column was eluted with chloroform (500 ml) and chloroform/methanol with increasing volumes of methanol (1–80%). LOS-I was eluted in a fraction containing 15% methanol, and LOS-II and LOS-III were co-eluted in the presence of 20% methanol. LOS-I to LOS-IV were further purified into individual species by preparative TLC on 20 × 20-cm high performance TLC plates of Silica Gel 60 (Merck) and run in chloroform/acetic acid/methanol/water (40:25:3:6, v/v). Glycolipids and lipids were revealed by iodine vapors. Spots corresponding to the glycolipids were scraped from the plates, and the glycolipids were extracted using chloroform/methanol (2:1, v/v) and finally purified on Sep-pack C18 cartridge (Waters, Milford, CT).
Purification of TDM was realized from extracted apolar glycolipids. Briefly, apolar glycolipids were dissolved in chloroform and applied on Florisil® column chromatography (Acros Organics). The column was eluted with chloroform (500 ml) and chloroform/methanol with increasing volumes of methanol (1–50%). The purification process was monitored by one-dimensional high performance TLC using chloroform/methanol (90:10, v/v). TDM was visualized by spraying plates with orcinol/sulfuric acid reagent followed by heating the plates at 120 °C. TDM was eluted in fraction containing 10% of methanol. TDM-containing fractions were pooled and applied on a second Florisil® column chromatography eluted with chloroform (500 ml) and chloroform/methanol with increasing volumes of methanol (1–20%). Purity of TDM was checked by MALDI-MS and NMR.
Lipooligosaccharide De-O-acylation
LOSs were de-O-acylated in chloroform/methanol (2:1, v/v) containing 0.1 mm NaOH at 37 °C for 2 h. Solvents were evaporated, and the resulting oligosaccharides were dissolved in water and further purified on a carbograph column (Alltech carbograph SPE column).
Permethylation and Linkage Analysis
Permethylation (or perdeuteromethylation) was performed according to the procedure of Ciucanu and Kerek (26). Briefly, compounds were incubated overnight in a suspension of 200 mg/ml NaOH in dry DMSO (300 ml) and iodomethane (200 ml) (or iododeuteromethane). The methylated (or deuteromethylated) products were extracted in chloroform and washed with water. The reagents were evaporated, and the sample was dissolved in methanol prior to mass spectrometry analysis. Permethylated samples were hydrolyzed with 4 m trifluoroacetic acid for 2 h at 100 °C and reduced with 10 mg/ml sodium NaBH4 in 0.5 m aqueous ammonium hydroxide solution at room temperature for 4 h, and the reaction was terminated by adding a few drops of acetic acid. Samples were dried and acetylated with acetic anhydride at 100 °C for 4 h. The reagent was removed under nitrogen stream and sample dissolved in chloroform. The chloroform phase was then washed several times with water before gas chromatography coupled to mass spectrometry (GC-MS) analysis using a Carlo Erba GC 8000 gas chromatograph equipped with a 25-m × 0.32-mm CP-Sil5 CB low bleed/MS capillary column, 0.25-μm film phase (Chrompack). Temperature of the Ross injector was 280 °C, and samples were analyzed using a temperature program starting at 100 °C for 1 min, followed by a temperature ramp of 5 °C min−1 to 300 °C. Electron ionization-mass spectra were obtained using a Finnigan Automass II mass spectrometer.
Matrix-assisted Laser Desorption Ionization-Mass Spectrometry (MALDI-MS) Analyses
The molecular masses of polysaccharides were measured by MALDI-TOF on a Voyager Elite reflectron mass spectrometer (PerSeptive Biosystems, Framingham, MA), equipped with a 337 nm UV laser. Native and permethylated samples were prepared by mixing directly on the target 1 ml of the methanol (native and permethylated)-diluted oligosaccharide solution and 1 ml of 2,5-dihydroxybenzoic acid matrix solution (10 mg/ml dissolved in acetonitrile/H2O).
Electrospray Ionization-MS Analysis
Nano-electrospray mass spectrometric analyses were performed using a QSTAR Pulsar quadrupole time-of-flight mass spectrometer (AB/MDS Sciex, Toronto, Canada) equipped with a nanoelectrospray ion source (Protana, Odense, Denmark). Samples were dissolved at a concentration of 10 pmol/μl in methanol/water (50:50, v/v), 0.1% formic acid and sprayed from gold-coated “medium length” borosilicate capillaries (Protana, Odense, Denmark). A potential of 800 V was applied to the capillary tip, and the declustering potential and the focusing potential were set at 120 and 180 V, respectively. For the recording of conventional mass spectra, TOF data were acquired by accumulation of 10 multiple channel acquisition scans over the m/z range 400–2000. In the collision-induced dissociation (MS/MS) analyses, multiple charged ions were fragmented using nitrogen as collision gas (5.3 × 10−3 torr), and the collision energy varied between 40 and 80 eV to obtain maximal fragmentation. The collision-induced dissociation spectra were recorded on the orthogonal TOF analyzer over a range of m/z 100–1400. Signal detection was performed with a multichannel plate detector and time to digital conversion. Resolution was measured as the full width at half-maximum and was 7000 in the measured mass ranges. External calibration was performed prior to each measure using a 1 pmol/μl solution of glufibrinopeptide in methanol/water (50:50, v/v).
NMR Analysis
NMR experiments were performed on a Bruker NMR spectrometer 9.4 teslas (where 1H resonated at 400.33 MHz and 13C at 100.66 MHz) and on a Bruker NMR spectrometer 14.1 teslas (where 1H resonated at 600.13 MHz and 13C at 150.91 MHz) equipped with a cryo-probe head. Prior to NMR spectroscopic analyses, LOS-I–IV were repeatedly exchanged in CD3OD (99.97% purity, Euriso-top®, CEA Saclay, France) with intermediate drying and then dissolved in CDCl3/CD3OD (2:1, v/v) (Euriso-top®). 1H and 13C chemical shifts are given in parts/million downfield from internal tetramethylsilane at 0 ppm. OS-II and OS-IV were repeatedly exchanged in 2H2O (99.97% 2H, Euriso-top®) with intermediate freeze-drying and finally dissolved in 2H2O (Euriso-top®) in Shigemi® tubes (Allison Park). Chemical shifts (parts/million) were calibrated taking methyl groups from internal acetone at δ1H 2.225 and 13C at 31.55 ppm. All experiments were recorded at 300 K without sample spinning. The Bruker pulse programs were used and optimized (pulse lengths and delays) for each one- or two-dimensional experiments.
Cell Culture Conditions and Quantification of TNF-α Secretion by ELISA
RPMI 1640 medium, l-glutamine were purchased from Invitrogen, and fetal calf serum (FCS) was from Dominique Dutscher (Brumath, France). The 1,25-dihydroxyvitamin D3 was provided by Calbiochem. The human TNF-α protein detection kit was from ABCys. By LPS from Salmonella typhimurium was purchased from Alexis Corp. Lipomannan (LM) was purified from MmaM using procedures reported previously (27, 28). The model bacterial lipopeptide Pam3Cys-SKKKK (Pam3CSK4) was purchased from EMC microcollections GmbH (Germany).
The human promonocytic leukemia THP-1 cell line was grown in RPMI 1640 medium supplemented with 10% FCS, 2 mm l-glutamine, and 2 × 10−5 m β-mercaptoethanol in a 5% CO2 air-humidified atmosphere at 37 °C. THP-1 cells were differentiated by treatment for 72 h with 50 nm 1,25-dihydroxyvitamin D3 (29). Viability of cells was over 96% as determined by trypan blue dye exclusion. To investigate the effect of LOSs on TNF-α secretion, differentiated THP-1 cells were seeded in 96-well plastic culture plates at a density of 2 × 105 cells/well, in RPMI 1640 medium supplemented with 5% FCS and l-glutamine. Purified LOSs as well as TDM were resuspended in ethanol and hexane, respectively, and sonicated for 3 min. These glycolipids were coated on plates at the indicated concentrations, which were subsequently dried at 37 °C to ensure complete solvent evaporation. Control wells were layered with solvent without glycolipids. Additionally, purified LM (used as positive control) was dissolved in apyrogen water and sonicated before addition to the cells. After 6 h of incubation, culture supernatants of triplicates were then collected and centrifuged prior to quantitation of cytokines by ELISA, according to the manufacturers' instructions. Cytokines concentrations present in the supernatants were obtained in comparison with a standard curve generated with recombinant human TNF-α.
To investigate the TNF-α-inhibitory properties of LOS-II and LOS-IV on stimulated macrophages, THP-1 cells were pretreated with LOSs for 20 h prior to the addition of either LPS (20 ng/ml) or Pam3CSK4 (20 ng/ml) and incubated for a further 6 h. Supernatants were processed for cytokine quantification as mentioned above. Viability of the cells was checked by flow cytometry (FACSCalibur flow cytometer) by using propidium iodide.
Statistical significance was analyzed with a Student's t test for unpaired data. Values of p < 0.05 were considered to be significant.
RESULTS
Identification of an M. marinum Isolate Defective in LOS-IV Production
LOSs are highly antigenic glycoconjugates exposed at the cell surface of several mycobacterial species, but their precise role in virulence and colony morphology is still a matter of debate (19, 20). Nevertheless, recent studies on M. marinum mutants carrying transposon insertions in the LOS biosynthetic cluster indicated that LOSs participate in sliding motility, biofilm formation and infection of host macrophages (17). Deciphering the biological properties of LOSs requires fine structural determination and to date, only a partial structure of M. marinum LOSs is available (16). Structure-function relationship studies are also hampered by the lack of detailed structural information, which can be attributed to the failure of obtaining high yields of the purified LOS variants, and particularly of the more polar ones (16). We reasoned that screening a set of M. marinum isolates from various origins would allow us to select “natural” mutants affected in their quantitative and/or qualitative LOS content, representing interesting sources for subsequent LOS production and purification. Therefore, we included in our study unrelated M. marinum strains isolated either from humans with fish tank granuloma (Mma20, Mma21, and Mma98) or from different poikilothermic species (Mma7, Mma11), which, when injected into adult zebrafish, developed an acute or a chronic disease (22).
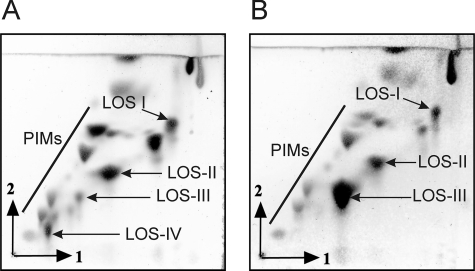
Polar lipids were extracted from all M. marinum strains and compared with the glycolipid content of the reference M strain (MmaM) (23). Glycolipid profiles were recorded on two-dimensional TLC stained by orcinol/sulfuric acid or ethanolic molybdophosphoric acid. Lipid phosphates were stained and visualized using the Dittmer and Lester reagent. The polar glycolipid pattern from MmaM (Fig. 1A) was similar to the one recorded previously for the M. marinum ATCC 927 strain (16), characterized by the presence of PIMs and different LOS-type species (LOS-I, -II, -III, and -IV). Comparison of polar glycolipids established that, among all six strains, five exhibited a very similar profile (data not shown). However, one isolate, Mma7, displayed a profoundly altered polar lipid pattern, characterized by the absence of a compound exhibiting low chromatographic mobility in both directions and the concomitant accumulation of a higher mobility compound (Fig. 1B). Based on the literature and the standard molecule chromatographic behaviors, these two compounds were tentatively assigned as members of the LOS family. Mma7 appeared to accumulate large amounts of LOS-III but failed to produce LOS-IV (Fig. 1B).
FIGURE 1.
Two-dimensional TLC patterns of M. marinum polar glycolipids. Following extraction from MmaM (A) or Mma7 (B), polar glycolipids were separated on two-dimensional TLC using the solvent system chloroform/methanol/water (60:30:6, v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v) in the second direction. Glycolipids were detected with orcinol/sulfuric acid stain.
Comparative Analyses of LOS-I, -II, -III in MmaM and Mma7
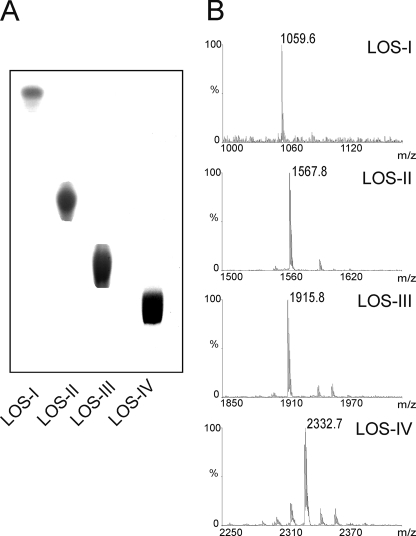
To confirm LOS-III accumulation in Mma7, LOSs were extracted and purified individually from both the MmaM and Mma7 strains and their structures compared. The final step of purification was performed by one-dimensional preparative TLC that allowed us to generate four compounds tentatively assigned to LOS-I to -IV and that appeared essentially pure by TLC analysis (Fig. 2A). Individual LOSs were then permethylated and analyzed by MALDI-MS (Fig. 2B). Permethylated LOS-I, LOS-II, and LOS-III from Mma7 gave prominent signals at m/z 1059.6 [M + Na]+, 1567.8 [M + Na]+, and 1915.8 [M + Na]+, respectively, in agreement with m/z values previously established (16). Identical values were obtained for permethylated LOS-I to LOS-III purified from MmaM (data not shown), which indicates that both strains synthesize LOSs with identical structures. In particular, these results demonstrate that Mma7 accumulates LOS-III in large quantities. Accordingly, LOS-IV could only be purified from MmaM, supporting the fact that this compound was not produced in Mma7. Consistently with a previous report (16), MALDI-MS analysis of permethylated LOS-IV gave a signal at m/z 2332.7 [M + Na]+ (Fig. 2B).
FIGURE 2.
TLC and mass spectrometric analyses of LOSs from M. marinum 7 strains. Intact purified LOSs fractions were separated by one-dimensional TLC with chloroform/methanol/water solvent (60:30:6, v/v) (A) and then analyzed by MALDI-MS as permethylated derivatives (B).
Local Genome Comparison Reveals Multiple Lesions with the LOS Biosynthetic Cluster in Mma7
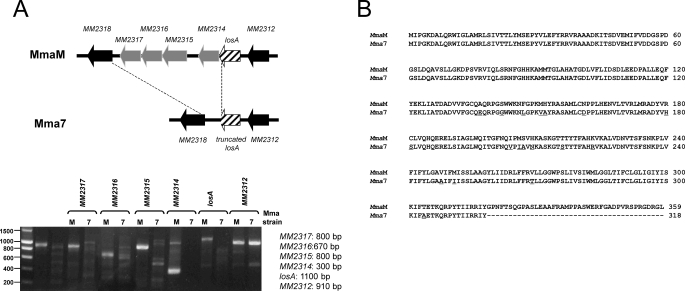
The altered LOS profile in Mma7 (Fig. 1B) is reminiscent of the one observed by Alexander et al. (30) in an M. marinum mutant carrying a transposon in a gene homologous to Rv1500, originally assigned to participate in PIM7 synthesis. This gene was subsequently renamed losA after it was established that it participates in LOS-IV rather than PIM7 synthesis based on both biochemical studies (16) and genetic studies in which inactivation of Rv1500 did not alter expression of the PIM pattern in M. tuberculosis (31). This prompted us to investigate whether the presence of mutations within losA in Mma7 could explain the observed LOS alteration. Therefore, losA was amplified by PCR, and as shown in Fig. 3A, a band of the expected size was amplified using MmaM, but not Mma7, genomic DNA. Southern blotting using losA as a probe generated a positive signal in Mma7, although with a different pattern compared with MmaM, suggesting the possibility of mutations within this gene (data not shown). Indeed, sequence analysis clearly indicated the presence of multiple lesions in the losA gene of Mma7, including substitutions and an important deletion and the 3′-end of the gene. The derived amino acid sequence of LosA shows that it carries 18 amino acid substitutions and a truncation of the last 41 residues (Fig. 3B). To test whether these mutations, presumably responsible for a nonfunctional protein, would account for the lack of LOS-IV production, Mma7 was transformed with a functional copy of LosA, as described under “Experimental Procedures.” However, no complementation of LOS-IV production was observed, because the LOS profile of this strain remained identical to the one of Mma7 (data not shown). This raised the possibility of additional mutations (presumably in other LOS biosynthetic genes) preventing LOS-III to LOS-IV conversion. We thus undertook the local comparison of the genes surrounding losA in Mma7 and compared it with MmaM. Amplification of the four genes MM2314 to MM2317 adjacent to losA failed to produce a signal of the expected size in Mma7 as compared with MmaM (Fig. 3A). Primers were therefore designed to amplify short gene segments, which were then sequenced and subsequently used as probes for a comparative genomic walk. Using this approach, major changes in the organization of the LOS cluster were identified, culminating with an important deletion spanning from the 3′-end of losA to MM2318 (Fig. 3A). In addition, sequences of the genes lying downstream of this deleted region (starting from MM2318) contained, like losA, multiple mutations (data not shown). Taken collectively, our genetic studies clearly indicated that the LOS biosynthetic cluster organization/sequence is severely impeded in Mma7, very likely explaining the failure to produce LOS-IV. Our data also highlighted the fact that losA, although required (16), is not sufficient to ensure LOS-IV biosynthesis.
FIGURE 3.
Genomic analysis of the losA region in MmaM and Mma7. A, illustration of the LOS biosynthetic cluster surrounding losA in MmaM and Mma7 (upper panel). Agarose gel showing the different amplicons (MM2312 to MM2317) obtained using either MmaM or Mma7 genomic DNA. Expected sizes of the PCR products in MmaM are indicated on the right (lower panel). B, comparison of LosA amino acid sequences in MmaM and Mma7. Point mutations found in Mma7 are underscored.
However, although our results clearly established that Mma7 is deficient in LOS-IV biosynthesis and suggested that structures of the remaining LOS-I to LOS-III in Mma7 are identical to those present in the reference M strain, the fine structure of LOSs remains largely unknown. In particular, no data are currently available on the nature and substitution patterns of terminal monosaccharide X of LOS-II and -III and monosaccharide YZ in LOS-IV (16). Therefore, an exhaustive analysis of LOS-II that contains a single copy of the X monosaccharide was undertaken to identify X. We also analyzed the substitution pattern of the X-X terminal disaccharide in LOS-III and partially characterized the YZ monosaccharide in LOS-IV. Structures of LOS-II and -IV were derived from the MmaM strain. However, because LOS-III is a minor component in MmaM, we took advantage of Mma7 as a source of LOS-III, as this molecule strongly accumulates in this strain.
Analysis of LOS-II Sugar Moiety
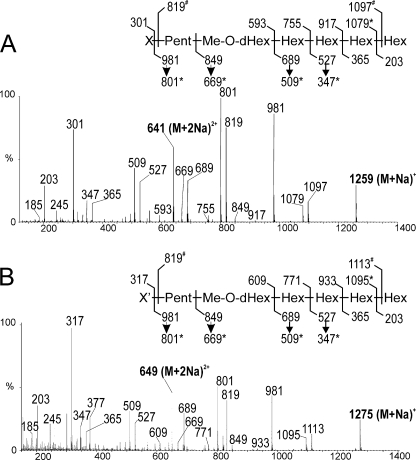
Initially, the oligosaccharide moiety of LOS-II was analyzed from both intact and deacylated LOS-II. However, given the higher quality of analyses performed on the deacylated molecule, only these results will be presented in detail. LOS-II was deacylated by alkaline hydrolysis to generate an oligosaccharide (OS-II). Analysis of native OS-II by MALDI-MS generated a major signal at m/z 1259.6 [M + Na]+ (supplemental Fig. S1), consistent with a monosaccharide composition of X1Pent1deHex1Hex4Me1, where X is an unidentified monosaccharide with an average molecular mass of 296 mass units in native form and Pent is pentose. Composition was confirmed by ES-MS/MS analysis of the corresponding doubly charged ion at m/z 641 [M + 2Na]2+ (Fig. 4A). In particular, [M + Na]+ B/Y couple fragmentation ions, according to standard nomenclature (49), at m/z 301/981 confirmed the molecular mass of the unknown X residue present in the nonreducing position of OS-II. Furthermore, it established the position of a methyl group on the deHex residue as well as the sequence of OS-II as X-Pent-MedeHex-Hex4, in agreement with the literature (16). Then MALDI-MS analysis of the permethylated derivative of OS-II generated a signal at m/z 1567.8 [M + Na]+ (supplemental Fig. S1A). It is noteworthy that permethylated derivatives of OS-II and LOS-II exhibited similar m/z values, indicating that permethylation of LOS-II in alkaline solution completely removed fatty acids. Furthermore, comparison of native and permethylated OS-II m/z values at m/z 1259.6 and 1567.8 respectively, as well as permethylated and per-deuteromethylated OS-II values at m/z 1567.8 and 1634.2, respectively, unambiguously indicated the presence of 22 potentially methylated functional groups in OS-II (supplemental Fig. S1A). This permitted us to calculate that the free monosaccharide X in the permethylated form has an average molecular mass of 394, and to propose that X possesses seven functional groups in free form and six when linked to LOS-II.
FIGURE 4.
Mass spectrometry fragmentation patterns of native OS-II and OS-II′. ES-MS/MS analyses of doubly charged ion at m/z 641 corresponding to OS-II (A) and doubly charged ion at m/z 649 corresponding to OS-II′ (B) are shown.
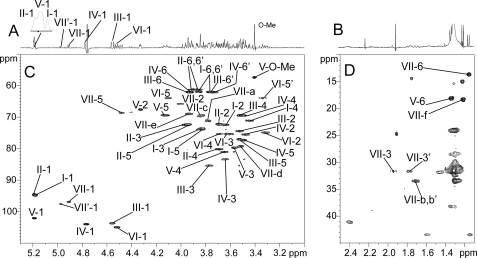
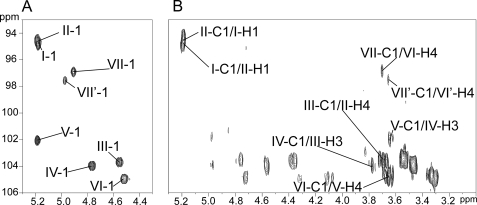
The nature of monosaccharides and linkage patterns of OS-II were further established by 1H homonuclear and 1H and 13C heteronuclear NMR experiments. Entire spin systems of all constituting monosaccharides were established by two-dimensional 1H-1H TOCSY (supplemental Fig. S2) and 1H-13C HSQC NMR (Fig. 5) experiments. Glycosyl residues were identified on the basis of spin systems, chemical shift values, and their relevant vicinal coupling constants patterns. 1H and 13C parameters of the individual monosaccharides are summarized in Table 1. NMR parameters of intact LOS-II sugar moiety were very similar to OS-II and mainly differ from the trehalose unit, considering it bears the lipid moiety (data not shown). The anomeric region of the 1H-13C HSQC NMR spectrum of OS-II permitted the identification of eight signals associated with distinct monosaccharides labeled I to VII′ (Fig. 6A). According to their 1H/13C chemical shifts and 3J1,2 coupling constant values, the I, II, V, VII, and VII′-H1 signals were all associated with monosaccharides in α-anomeric configurations (I-H1 δ 5.18 (3J1,2 = 3.5 Hz); II-H1 δ 5.19 (3J1,2 = 3.6 Hz); V-H1 δ 5.18 (3J1,2 = 1.0 Hz); VII-H1 δ 4.91 (3J1,2 = 4.0 Hz); and VII′-H1 δ 4.97 (3J1,2 = 4.0 Hz)), whereas III, IV, and VI-H1 were associated with monosaccharides in β-anomeric configurations (III-H1 δ 4.55 (3J1,2 = 7.7 Hz), IV-H1 δ 4.76 (3J1,2 = 7.3 Hz), and VI-H1 δ 4.52 (3J1,2 = 8.0 Hz)) (Table 1). Spin systems associated with I- and II-H1/C1 signals at 5.18/94.7 and 5.19/94.5 ppm typified α-glucopyranosyl units (Table 1). Upfield resonance of carbons I-C1 and II-C1 around 94.6 ppm (Fig. 6A) as well as clear 3JH-C inter-correlations I-C1/II-H1 and II-C1/I-H1 on the HMBC spectrum (Fig. 6B) demonstrated that both α-Glc residues are part of a trehalose unit. Large 3J1,2, 3J2,3, and 3J3,4 values (Table 1) for spin systems associated with the β-anomeric signals III-1 (δ 4.55/103.6) and IV-1 (δ 4.76/104.0) typified III and IV as β-glucopyranosyl units. Identification of a spin system (3J1,2, 1.0 Hz; 3J2,3, 4.6 Hz; 3J3,4; 9.5 Hz; 3J4,5, 9.6 Hz) from V-H1 representative of a manno configuration and of a methyl group at δ 1.34 indicative of a 6-deoxyhexopyranosyl unit permitted the identification of V as an α-rhamnopyranosyl unit (Table 1). From the HMBC experiment, the 3JH-C vicinal connectivity between the V-H3 at 3.56 ppm and O-methyl carbon at 57.3 ppm established that residue V is an α-3-O-Me-Rhap unit (data not shown). Then, the anomeric signal resonance at 4.52/104.9 ppm of residue VI was attributed to a β-xylopyranosyl unit because of H5ax and H5eq assignment and large vicinal coupling constant values (3J1,2 = 8.0, 3J2,3 = 8.9, 3J3,4 = 9.6, and 3J4,5ax = 11.7) (Table 1). The remaining anomeric signals VII-1 and VII′-1 at 4.91/96.9 and 4.97/97.5 ppm were then assigned to two yet unidentified monosaccharides. Further analyses (see below) permitted us to assign the VII signal to the unidentified monosaccharide X that was observed by mass spectrometry analysis in a terminal nonreducing position of LOS-II (Fig. 4A). Similarly, VII′ signal was associated with X′, a close relative of X present in a terminal nonreducing position of a minor equivalent of LOS-II, LOS-II′. Differential relative integrations of H1 signals (I + II + V = 3.10; VII′ = 0.35; VII = 0.75; IV = 0.98; II = 1.00; VI = 1.02) established that X and X′ were present in less than a single residue per glycolipid, whereas all others were present as a single copy. Furthermore, the sum of VII and VII′-H1 signal integrations (VII + VII′ = 1.1) strongly suggests that X and X′ are part of two similar molecules called LOS-II (substituted by X) and LOS-II′ (substituted by X′) that only differ by the nature of their respective nonreducing terminal monosaccharide. Accordingly, subsequent NMR and MS analyses permitted us to establish that the LOS-II fraction consisted in fact of the mixture of two glycolipids, LOS-II and LOS-II′, that were individualized based on their differential fragmentation patterns and NMR parameters. We subsequently initiated the structural determination of the unknown monosaccharide X, and we demonstrated the existence of the minor but structurally related glycolipid LOS-II′ and established its exact glycosidic sequence.
FIGURE 5.
1H-13C NMR analysis of OS-II fraction. Details of the anomer and bulk regions of the 1H NMR spectrum (A) and the 1H-13C HSQC NMR spectrum of OS-II (C) are shown. Details of the methyl and methylene resonances containing regions of the 1H NMR spectrum (B) and of 1H-13C HSQC NMR spectrum of OS-II (D) are shown.
TABLE 1.
1H and 13C chemical shifts (in ppm) and coupling constant values (in Hz) of OS-II monosaccharides (measured at 300 K in D2O using acetone as internal reference)
Values in parentheses are coupling constants; values in bold correspond to substituted carbons. ND indicates not determined.
| Sugar units | H1/C1 (3JH1-H2) | H2/C2 (3JH2-H3) | H3/C3 (3JH3-H4) | H4/C4 (3JH4-H5) | H5/C5 | H6,6′/C6 | Others |
|---|---|---|---|---|---|---|---|
| (I) α-Glcp | 5.18/94.7 (3.5) | 3.63/72.3 (10.0) | 3.84/73.9 (9.4) | 3.44/71.1 (9.4) | 3.82/73.7 | 3.85, 3.76/61.9 | |
| (II) α-Glcp 1→1 | 5.19/94.5 (3.6) | 3.69/72.1 (10.0) | 3.96/72.3 (9.4) | 3.69/80.1 (8.9) | 3.95/72.3 | 3.91, 3.85/61.2 | |
| (III) β-Glcp 1→4 | 4.55/103.6 (7.7) | 3.52/74.5 (8.7) | 3.77/85.3 (9.3) | 3.53/69.4 ND | 3.52/76.9 | 3.93, 3.76/62.0 | |
| (IV) β-Glcp 1→3 | 4.76/104.0 (7.3) | 3.47/75.5 (8.4) | 3.64/83.3 (9.4) | 3.48/69.4 ND | 3.49/77.2 | 3.92, 3.73/62.0 | |
| (V) 3-O-Me-α-Rhap 1→3 | 5.18/102.0 (1.0) | 4.32/67.5 (4.6) | 3.56/79.6 (9.5) | 3.64/80.9 (9.6) | 4.13/69.4 | 1.34/18.0 | 3.40/57.3 (O-Me) |
| (VI) β-Xylp 1→4 | 4.52/104.9 (8.0) | 3.32/74.9 (8.9) | 3.62/75.2 (9.6) | 3.68/75.2 (11.7) | 3.33, 4.10/63.9 | ||
| 4-C-branched sugars | |||||||
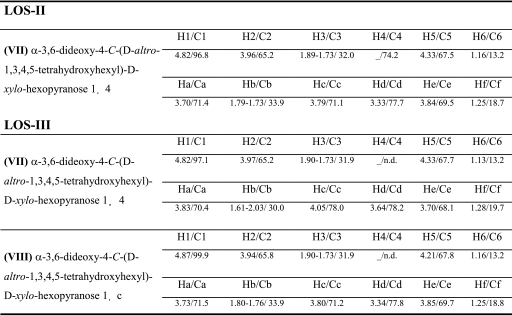
| (VII) α-3,6-Dideoxy-4-C-(d- altro-1,3,4,5-tetrahydroxyhexyl)- d-xylo-hexopyranose 1→4 | H1/C1 (3JH1-H2) | H2/C2 (3JH2-H3) | H3,3′/C3(3JH3-H3′) | H4/C4 | H5/C5 | H6/C6 | |
| 4.91/96.9 (4) | 4.00/65.7 (4/9) | 1.94, 1.77/31.7 (11) | −/75.8 | 4.47/68.6 | 1.15/13.6 | ||
| Ha/Ca | Hb,b′/Cb | Hc/Cc | Hd/Cd | He/Ce | Hf/Cf | ||
| 3.78/71.1 | 1.73, 1.68/33.5 | 3.84/69.4 | 3.52/79.2 | 3.93/68.8 | 1.21/18.2 | ||
| (VII′) α-6-dideoxy-4-C-(d- altro-1,3,4,5-tetrahydroxyhexyl)- galacto-hexopyranose 1→4 | H1/C1 (3JH1-H2) | H2/C2 (3JH2-H3) | H3/C3 | H4/C4 | H5/C5 | H6/C6 | |
| 4.97/97.5 (3.5) | 3.78/69.6 (10.1) | 3.84/72.5 | −/77.7 | 4.40/68.4 | 1.22/14.9 | ||
| Ha/Ca | Hb,b′/Cb | Hc/Cc | Hd/Cd | He/Ce | Hf/Cf | ||
| 4.12/69.4 | 1.82, 1.75/34.8 | 3.83/69.4 | 3.50/79.2 | 3.93/68.8 | 1.21/18.2 | ||
FIGURE 6.
Sequence analysis of OS-II by NMR. Expanded areas of two-dimensional 1H-13C HSQC spectrum (A) and 1H-13C HMBC spectrum of OS-II fraction (B) are shown.
The glycosidic sequence of OS-II was established from the HMBC experiment by observing scalar coupling correlations (3JH-C) between 1H and 13C implicated in glycosidic linkages (Fig. 6B). Trehalose linkage pattern was first confirmed by crossed correlations I-C1/II-H1 and II-C1/I-H1. Then substitutions of II-VI residues in C3 or C4 positions were established by a continuous series of III-C1/II-H4, IV-C1/III-H3, V-C1/IV-H3, VI-C1/V-H4, and VII-C1/VI-H4 3JH-C correlations. Positions of glycosidic linkages were supported by specific downfield 13C resonance shifts by a value of 4–10 ppm on carbon involved in glycosidic linkages (Table 1). Altogether, in accordance with ES-MS/MS fragmentation pattern of native OS-II (Fig. 4A), NMR data clearly indicated that OS-II presents the following linear monosaccharide sequence: X-(1→4)-β-Xylp-(1→4)-α-3-O-Me-Rhap-(1→3)-β- Glcp-(1→3)-β-Glcp-(1→4)-α-Glcp-(1→1)-α-Glcp.
Further determination of the nature of monosaccharide X was exclusively based on NMR experiments. The entire spin system of the ring protons was deduced from the COSY and TOCSY spectra of OS-II (supplemental Figs. S2 and S3). The observation of VII-H2 signal with an octuplet shape at δ 4.00 and the identification of two shielded VII-H3 signals (VII-H3 at δ 1.94 and VII-H3′ at δ 1.77) clearly typified the VII unit as a 3-deoxysugar. The presence of a methylene group in C3 position was further confirmed by the observation of two 1JH-C correlations from C3 carbons (δ 31.7) with its corresponding protons at δ 1.94 and at δ 1.77, respectively (Fig. 5D). Triplet and quadruplet shapes of H3 and H3′, induced by vicinal coupling constants 3J3,2 = 9 Hz, 3J3′,2 = 4 Hz and geminal coupling constant 2J3,3′ = 11 Hz, allowed us to assign H3 as axial and H3′ as equatorial protons (Table 1). Analysis of relayed COSY showed that H3 and H3′ were only coupled to H2, indicating that C4 is a quaternary carbon (data not shown). The VII-C4 resonance was determined on the HMBC spectrum by observing the internal 2JH-C correlations between VII-C4 and VII-H3′, VII-H5 and 3JH-C correlation between VII-C4 and VII-H6 signal (supplemental Fig. S4). Similarly, C5 resonance was established because of intra-residue 3JH-C connectivities of VII-C5 with H1 and H3′. Finally, VII-H6 was identified at 1.15 ppm on COSY spectrum through a correlation with VII-H5 (supplemental Fig. S3B). Parameters of H6/C6 at 1.15/13.6 established from an HSQC experiment (Fig. 5D and Table 1) typified a methyl group. The ring configuration was defined as xylo based on data from a nuclear Overhauser effect spectroscopy experiment (data not shown) and vicinal coupling constants pattern previously defined. Indeed, NOE effect from VII-H3 to VII-H5 indicated the axial orientation of VII-H5 (data not shown). Furthermore, Ha-VII NOE contacts with VII-H6 and VII-H5 suggesting a spatial proximity of this proton and an equatorial orientation of the side chain (data not shown). Thus, NMR data permitted us to identify the X monosaccharide as a 3,6-dideoxyhexopyranose in xylo configuration further substituted by a side chain in the C4 position.
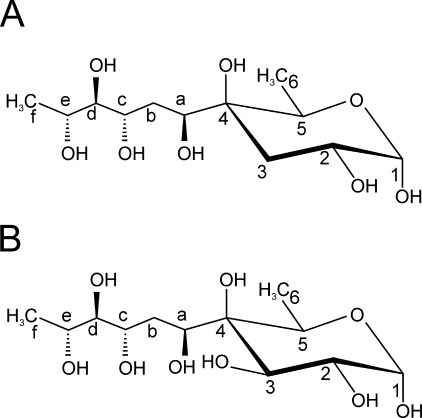
Accordingly, linkage of side chain to the quaternary ring carbon was directly observed on HMBC spectrum because of clear 3JH-C and 2JH-C connectivities of C3 and C4 with an extra ring proton Ha at 3.78 ppm (supplemental Fig. S4). This interpretation is further supported by strong NOE effects from Ha to H5 and H6 on rotating frame Overhauser enhancement spectrum (data not shown). The proton resonance VII-Ha was used as starting point to define the spin system of the side chain. From COSY 90 spectrum analysis, VII-Ha was correlated with two methylene protons VII-Hb at δ 1.73 and VII-Hb′ at δ 1.68 (3JHa,Hb = 3.8 Hz and 3JHa,Hb′ = 9.8 Hz) (supplemental Fig. S3B). Then Hb protons permitted us to successively identify Hc, Hd, and He protons at δ 3.84, 3.52, and 3.93, respectively, all associated with CHOD groups. Finally, He correlates with a final Hf proton at 1.21 ppm associated with a methyl group (supplemental Fig. S3B). Resonance chemical shifts of associated carbons, defined by the 1H-13C HSQC experiment, Cb and Cf at 33.5 and 18.2 ppm confirmed previous assignments as –CH2– and –CH3 groups, respectively (Fig. 5D and Table 1). Similarly, Ca, Cc, Cd, and Ce chemical shifts values between 68.8 and 79.2 ppm are in agreement with carbons of CHOD groups. Altogether, NMR data identified the side chain of X monosaccharide branched in the C4 position as a 1,3,4,5-tetrahydroxyhexyl chain. A complete set of NOE contacts between protons of the tetrahydroxyhexyl chain (data not shown) strongly suggested that the configuration of the side chain is altro, which finally established the structure of previously unknown monosaccharide X as a 3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose.
The nature of the monosaccharide X was further confirmed by analyzing its fragmentation pattern in MS. LOS-II was permethylated, hydrolyzed, reduced by BD4Na, and per-acetylated. Derived monosaccharides were analyzed by GC-MS in electronic impact mode. Chromatographic mobility and fragmentation patterns of individual partially methylated alditol acetate derivatives showed the presence of terminal glucose (rt = 19.56 min), 4-substituted glucose (rt = 22.07 min), 3-substituted glucose (rt = 21.51 min), 3,4-substituted rhamnose identified as 4-substituted 3-O-methyl-rhamnose (rt = 19.04 min), and 4-substituted xylose (rt = 18.58 min) (data not shown), in accordance with NMR and ES-MS/MS analyses. An additional monosaccharide derivative was observed at rt = 30.54 min that exhibits a fragmentation pattern totally compatible with a terminal 3,6-dideoxy-4-C-(1,3,4,5-tetrahydroxyhexyl)-hexopyranose derivative (supplemental Fig. S5A). In particular, the presence of a partially hydroxylated six-carbon side chain was evidenced by the fragment ion couple 262/205 between C4 and Ca positions. Also, positions of hydroxyl groups were evidenced because of prominent cleavage ions between O-methylated Cc and Cd and between O-methylated Cd and Ce. Finally, structure information determined by GC-MS techniques is in total agreement with mass spectrometry and NMR data, which confirmed the previous assignment of the X monosaccharide.
A polysaccharide chain composed of a 4-C-branched monosaccharide that presents a structure identical to X residue was previously reported in Pseudomonas caryophylli (32). Conformational studies permitted its identification as a 3,6-dideoxy- 4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose that was named caryophyllose (33). The fact that X presented strictly identical NMR parameters than α-caryophyllose (α-Car) (34) confirmed the configuration of the side chain as altro configuration, as we established by nuclear Overhauser effect spectroscopy experiment and demonstrated its α-anomery (Fig. 7A).
FIGURE 7.
Structures of α-3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose (A) and α-6-deoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-galactopyranose (B).
Structural Heterogeneity of the LOS-II Sugar Moiety
As stated above, preliminary observation of NMR data indicated that the so-called LOS-II fraction consisted in fact of a mixture of two closely related molecules, LOS-II and LOS-II′, differing by the nature of their terminal nonreducing monosaccharide residue. LOS-II is capped by the residue VII (VII-H1/C1 at 4.91/96.9 ppm), identified as a α-caryophyllose, whereas LOS-II′ is characterized by the VII′ residue (VII′-H1/C1 at 4.97/97.5 ppm). Spin system of VII′ residue was totally resolved by 1H-1H COSY, TOCSY, 1H-13C HSQC, and HMBC experiments on OS-II fraction (Table 1). It exhibited a general pattern similar to VII, demonstrating it is also a 4-C-branched hexopyranose (Table 1). In particular on COSY spectrum, correlation of VII′-H5 with methyl protons at δ 1.22 clearly established it is a α-6-deoxyhexose (supplemental Fig. S3B). Furthermore, absence of the H4 signal permitted us to detect the presence of a quaternary carbon on the C4 position, which chemical shift was established at 77.7 ppm because of 2JH-C correlation between VII′-H5 and VII′-C4 on 1H-13C HMBC spectrum. As for residue VII, the Ca/Ha linked to C4 was identified from 1H-13C HMBC spectrum by 2JH-C scalar coupling connectivity between VII′-Ha and VII′-C4 (supplemental Fig. S4) and from rotating frame Overhauser enhancement spectrum because of a NOE effect between VII′-H5 and VII′-Ha (data not shown). Then the spin system of side chain was identified on relayed COSY experiment starting from the proton VII′-Ha, which demonstrated that it has the same structure than the one substituting VII (Table 1). Considering the closeness of 1H and 13C NMR parameters from the chains associated with VII′, with the exception of VII′-Ha, with the one associated with VII residues, it is most likely that it also exhibits an altro configuration. However, despite these similarities, VII′ differs from the VII residue by the presence of a hydroxyl group on the C3 position, as demonstrated by the large downfield shift of VII′-H3 from 1.94 to 3.84 ppm and upfield shift of VII′-H2 from 4.00 to 3.78 ppm (Table 1 and supplemental Fig. S2). It is also obvious from the loss of vicinal coupling constants between H2 and H3eq and the geminal constant between two H3. Moderate downfield shift of VII′-Ha (δ 4.12) compared with VII-Ha (δ 3.78) may also result from the closeness of the modified C3 position. On the TOCSY analysis, the 3J2,3 value of 10.1 Hz indicated that H3 is in axial position. Altogether, NMR data identified the X′ residue as a novel 4-C-branched sugar corresponding to a α-6-deoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-galactopyranose (Fig. 7B).
A closer examination of the 1H-13C HMBC spectrum revealed a clear correlation between VII′-C1 and a 1H signal at δ 3.64, tentatively attributed to the H4 of a β-Xylp residue labeled VI′ (Fig. 6B). This residue exhibits identical parameters to the VI residue, only differing by the minute upfield shift of its H4 from 3.68 to 3.64 ppm. This indicated that 3-C-hydroxylation of α-caryophyllose induces a slight modification of the VI-H4 parameter. Altogether, NMR data established the presence of two highly related molecules in the LOS-II fraction as follows: LOS-II terminally substituted by a α-3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose and LOS-II′ substituted by a α-6-deoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-galactopyranose. Both molecules were directly observed from the native and permethylated OS-II fraction by MALDI-MS. Indeed, MALDI-MS of the native OS-II fraction shows two major signals at m/z 1259 and 1275 attributed to [M + Na]+ adducts of OS-II and OS-II′ (supplemental Fig. S1A). To rule out the possibility that the signal at m/z 1275 may originate from the [M + K]+ adduct of OS-II, we also analyzed permethylated and per-deuteromethylated derivatives of the OS-II fraction. After derivatization, OS-II and OS-II′ could still be differentiated according to the m/z values of their respective derivatives as [M + Na]+ adducts (supplemental Fig. S1A). Indeed, the 30-mass unit increment between permethylated oligosaccharides and the 33-mass unit increment between per-deuteromethylated oligosaccharides established that OS-II and OS-II′ differed by the presence of a single hydroxyl group. MS-MS fragmentation patterns of [M + 2Na]2+ adducts of native OS-II and OS-II′ confirmed that the supplementary hydroxyl group was located on the X′ monosaccharide at the nonreducing end of OS-II′, in perfect accordance with the NMR data (Fig. 4B).
Analysis of LOS-III
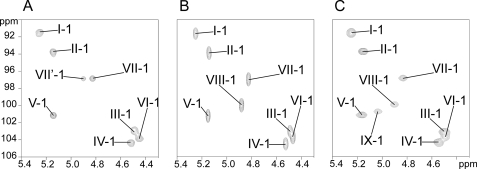
MALDI-MS analysis of the permethylated derivative of LOS-III already established that it differed from LOS-II by an additional caryophyllose (Δ348 mass units) (Fig. 2B). Comparative NMR analyses demonstrated that LOS-II and LOS-III exhibited essentially identical NMR parameters (data not shown), and as a consequence both share a very similar glycan sequence. Indeed, comparison of LOS-II and LOS-III 1H-13C HSQC anomeric region spectra showed that all I- to VII-C1/H1 signals had identical NMR parameters, and that LOS-III differed exclusively from LOS-II by the presence of an additional VIII-H1/C1 signal at δ 4.87/99.9, tentatively assigned as the anomer of a second α-caryophyllose residue (Fig. 8, A and B). As shown in Table 2, the spin system of VII from LOS-II and LOS-III essentially differ for their respective tetrahydroxyhexyl chains. In particular, a large downfield shift of LOS-III VII-Cc compared with LOS-II VII-Cc from 71.1 to 78.0 ppm, as well as a moderate downfield shift of LOS-III VII-Hc (Δδ +0.26), strongly suggested that LOS-III VII is substituted in the Cc position by VIII. Accordingly, neighboring proton and carbon parameters are also moderately affected (Table 2). The LOS-III VIII spin system then confirmed its assignment as an additional 3,6-dideoxy-4-C-(1,3,4,5-tetrahydroxyhexyl)-hexopyranose residue. Furthermore, perfect matching between the 1H/13C chemical shifts of LOS-II VII and LOS-III VIII residues established that VIII is a α-3,6-dideoxy- 4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose in a terminal nonreducing position. Moderate downfield shifts of LOS-VIII H1/C1 parameters (Δδ +0.05/3.1) compared with LOS-II VII can easily be rationalized by the different nature of the substituted monosaccharide, β-Xyl for LOS-II and α-Car for LOS-III. The linkage pattern of the α-caryophylloses in LOS-III was further confirmed by methylation analysis in GC-MS. Two relevant peaks at rt = 30.55 and 32.23 min were associated with caryophyllose based on their electron ionization-MS fragmentations patterns. Peaks at rt = 30.55 min presented an identical fragmentation pattern than in LOS-II and was identified as terminal caryophyllose (data not shown). Peak at rt = 32.23 min showed the presence of an additional O-acetyl group at position c indicating that this position is involved in interglycosidic linkage (supplemental Fig. S5B). Thus, the OS-III monosaccharide sequence was identified as α-Car-(1→c)-α-Car-(1→4)-β-Xylp-(1→4)-α- 3-O-Me-Rhap-(1→3)-β-Glcp-(1→3)-β-Glcp-(1→4)-α-Glcp- (1→1)-α-Glcp.
FIGURE 8.
Comparison of anomeric signals from LOS-II, -II, and -IV. Anomeric area of the two-dimensional 1H-13C HSQC spectra from LOS-II (A), LOS-III (B), and LOS-IV (C) is shown.
TABLE 2.
1H and 13C chemical shifts (in parts/million) of 4-C-branched monosaccharides from LOS-II and LOS-III (measured at 300 K in D2O using acetone as internal reference)
ND indicates not determined.
Analysis of LOS-IV
Contrary to the other LOSs, LOS-IV was retained on a DEAE column during chloroform/methanol elution (2:1, v/v) and required ammonium acetate salt to be eluted, suggesting the presence of an acidic function, very likely to be localized on the terminal YZ unidentified sugar residue. To delineate its oligosaccharide sequence, the glycan moiety was released by de-O-acetylation to generate OS-IV. Native OS-IV analyzed by MALDI-MS produced a major signal at m/z 1867.5 [M + Na]+ (supplemental Fig. S1B), suggesting that it exhibits an apparent m/z value of 1844 mass units. Surprisingly, ES-MS analysis permitted us to calculate for the same molecule an apparent m/z value of 1889 through the observation of a doubly charged signal at m/z 967.47 [M + 2Na]2+ (data not shown). Discrepancy of 45 m.u. between the two analyses suggested that native OS-IV undergoes partial degradation upon laser desorption, probably from the YZ residue. This hypothesis was confirmed by deducing OS-IV native molecular mass from MALDI-MS analyses of permethylated and per-deuteromethylated OS-IV derivatives (supplemental Fig. S1). Comparison of [M + Na]+ signals at m/z 2332 and 2422 established that OS-IV possesses 30 methylatable functional groups of which 4 would be localized on the terminal YZ residue, allowing us to deduce that the molecular mass of native OS-IV is indeed 1889 mass units. Altogether, mass spectrometry analyses permitted us to estimate the molecular mass of the free YZ residue to 393 Da, which is way above the molecular mass of all known monosaccharides, including caryophyllose (296 Da).
Comparison of LOS-III and LOS-IV 1H-13C HSQC spectra showed a single additional anomeric signal at 4.94/101.5 ppm assigned to the monosaccharide residue IX (Fig. 8, B and C). This unique supplementary anomeric signal demonstrated that the unknown YZ consists of a single monosaccharide unit despite its high molecular mass (Fig. 8C). Complete spin systems of constituting monosaccharides were established from OS-IV in identical condition than for OS-II (supplemental Table S1). Comparison of NMR parameters of OS-IV with OS-II demonstrated that OS-IV exhibited an oligosaccharidic moiety similar to other LOSs with a α-Car-(1→c)-α-Car-(1→4)-β-Xylp-(1→4)-α-3-O-Me-Rhap-(1→3)-β-Glcp-(1→3)- β-Glcp-(1→3)-α-Glcp-(1→1)-α-Glcp sequence further substituted by an additional monosaccharide residue labeled IX in Cc position of terminal α-Car residue (VIII residue). Position of substitution was assigned because of large downfield shift of OS-IV VIII-Cc to 79.1 ppm, compared with unsubstituted OS-II VII-Cc at 69.4 ppm (Table 1 and supplemental Table S1). The substitution pattern of Car residues was then confirmed by methylation analysis in GC-MS (data not shown) that showed a unique partially methylated and acetylated derivative of Car residue with a methyl group in the Cc position. NMR parameters of IX residue ring protons and 3J connectivities established through 1H/13C HSQC and 1H/1H COSY experiments permitted us to assign it as a 4,6-dideoxy-galactopyranose in α-anomery (supplemental Table S1). Distinctive chemical shift of IX-H4/C4 at 4.28/56.3 strongly suggests that IX-C4 is linked to a nitrogen atom (supplemental Table S1). Furthermore, observation of a 3JH,C correlation on 1H/13C HMBC spectrum (data not shown) between IX-H4 and a carbon at 170.2 ppm clearly typified the presence of a –CO-NH– group substituting IX in the C4 position. Altogether, the data established that LOS-IV is capped by α-4,6-dideoxy-Galp substituted in the C4 position by an aglycone group through an amide linkage.
Role of LOSs in Modulating the Macrophage TNF-α Response
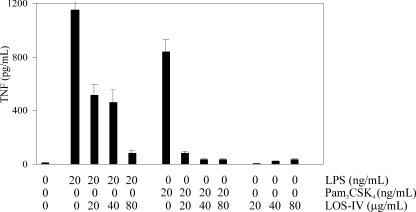
The mycobacterial cell envelope is important in directing host-pathogen interactions (3, 4). Several cell envelope-associated lipids, glycolipids, or lipoglycans have recently been significantly studied because they are considered to be important virulence factors and because of their immunomodulatory properties (1). Paradoxically, little attention has been paid regarding the potential immunomodulatory functions of LOSs. We therefore took advantage of the availability of purified LOS-I to LOS-IV with defined structural variations to examine their propensity to modulate the TNF-α response in the human monocytic cell line THP-1. First, the capacity of the different LOS subtypes to trigger TNF-α secretion was investigated, but none of these glycolipids were able to induce a TNF-α response (supplemental Fig. S6), in contrast to LM and TDM, both known as potent proinflammatory-inducing factors (35–37). We next examined whether increasing concentrations of LOS-IV may inhibit TNF-α secretion in macrophages stimulated with either LPS or Pam3CSK4 (20 ng/ml), known to activate macrophage through TLR4- and TLR2-dependent signaling pathways, respectively. As presented in Fig. 9, LOS-IV strongly inhibited the release of TNF-α when the glycolipid was added 20 h before LPS or Pam3CSK4 stimulation. LPS-induced release of TNF-α was inhibited by LOS-IV in a dose-dependent manner by 55–93%. The release of TNF-α induced by Pam3CSK4 was inhibited by 90% from 20 μg/ml of LOS-IV. Taken together, these results suggest that LOSs represent potent glycolipids interfering with the macrophage TNF-α response.
FIGURE 9.
Inhibitory effect of LOS-IV on TNF-α secretion in differentiated THP-1 cells. Cells were preincubated for 20 h with increasing concentrations of LOS-IV isolated from MmaM and then stimulated with LPS or Pam3CSK4 (20 ng/ml). Culture supernatants were collected after 6 h of induction and assayed by ELISA for TNF-α secretion. The results presented are from one representative experiment of three independent experiments with similar results. Comparable results were obtained using different batches of THP-1 cells and different LOS-IV preparations. Data are expressed as means ± S.D. of triplicate plates. Statistical analyses were performed by using Student's t test to compare LOS-IV-pretreated cells with nontreated cells. The p values were <0.0023 and <0.0003 in comparison with LPS- or Pam3CSK4-stimulated cells, respectively.
DISCUSSION
M. marinum causes a systemic tuberculosis-like disease in a large number of poikilothermic animals and is often used as a model to study mycobacterial pathogenesis (38–41). M. marinum isolates can be divided into two distinct types based on genetic diversity and virulence, developing either an acute or a chronic disease in fish (22). In this study, we analyzed the apolar and by polar lipid profiles in various strains inducing either an acute or a chronic disease. The major difference was found in Mma7, a strain originally isolated from the butterfly fish (22), which exhibited an altered LOS profile, characterized by a defect of LOS-IV synthesis and a concomitant accumulation of LOS-III. Interestingly, although most zebrafish inoculated with Mma7 or Mma11 (isolated from butterfly fish and sea bass, respectively) survived 56 days postinfection, those inoculated with Mma7 showed no signs of infection for 8 weeks, whereas those infected with Mma11 showed skin lesions (ulceration) after 45 days (22). Because Mma11 produces a standard LOS profile, similarly to the reference M strain (data not shown), our results suggest that the differences between strains inducing chronic and acute diseases are unlikely to be caused by differences observed in the LOS profiles. These results tend to indicate that the marked differences in pathogenicity of the M. marinum strains are not because of direct activities of LOSs. Ren et al. (17) demonstrated that M. marinum strains carrying transposon in MM2309 and MM2332 are defective in LOS-II and accumulate an intermediate between LOS-I and -II respectively, and are affected in sliding motility, biofilm formation, and infection of host macrophages. However, whether LOSs participate directly to the pathogenicity of M. marinum requires to be investigated in more detail by derivating isogenic strains that are completely deficient in LOS synthesis.
We subsequently established that the failure of Mma7 to synthesize LOS-IV was associated with the presence of multiples genetic lesions present in the LOS biosynthetic gene cluster (17). First, LosA, which was reported as a key glycosyltransferase involved in transferring sugars to LOS-III to form LOS-IV (16), contains several point mutations and is lacking its C terminus in Mma7. Second, a comparative genomic analysis indicated that this LOS cluster contains important mutations, including a 3.8-kb deletion downstream of the truncated losA gene. Thus, it can be inferred that the LOS biosynthetic cluster organization/sequence is severely compromised in Mma7 and is very likely responsible for the loss of LOS-IV production. In addition, the failure to restore LOS-IV biosynthesis in Mma7 following introduction of a functional losA gene indicates that losA, although necessary (16), is not sufficient to ensure LOS-IV biosynthesis. Our partial characterization of the YZ substituent in LOS-IV indicates that it consists of a single high molecular mass N-acylamino-monosaccharide substituted by a very large side chain, rather than a disaccharide as proposed earlier (16). Interestingly, another acylamino sugar presented a much simpler side chain has already been identified in M. kansasii as a 4,6-dideoxy-2-O-methyl-3-C-methyl-4-(2′-methoxypropionamido)hexopyranose (42). Therefore, the structural complexity of this sugar unit probably results from the action of an important number of enzymes. Thus, it appears conceivable that, in addition to LosA, multiple enzymes are operating to ensure both the synthesis and transfer of YZ to LOS-III, and that the deletion and/or mutations in these genes in Mma7 compromises LOS-IV production in this strain. Work is currently in progress to establish the exact structure of the remaining YZ monosaccharide.
One surprising finding of this study is the characterization of a polysaccharide chain composed of a 4-C-branched monosaccharide structurally identical to the one described in P. caryophylli (32) identified as a 3,6-dideoxy-4-C-(d-altro-1,3,4,5-tetrahydroxyhexyl)-d-xylo-hexopyranose and subsequently designated caryophyllose (33). Caryophyllose is part of a family of unusual 4-C-branched monosaccharides of which members have been identified from various Gram-negative bacteria, including Yersinia species (43, 44) and Burkholderia brasiliensis (45). All 4-C-branched monosaccharides described so far are surprisingly based on a 3,6-dideoxyhexose core, which strongly suggests a conserved origin for this family of molecules. To our knowledge, α-caryophyllose has only been described so far in P. caryophylli. A related monosaccharide differing in the nature of the side chain has been previously identified at the nonreducing position of an LOS of M. gastri (9). The presence of this very rare and unusual monosaccharide in two totally unrelated bacteria suggests the presence of a common biosynthetic pathway that ultimately leads to α-caryophyllose biosynthesis. Furthermore, we identified a novel monosaccharide that derives from the caryophyllose but differs by the presence of an additional hydroxyl group in C3 position. To our knowledge, hydroxylated caryophyllose has never been observed before. This study describes a novel family of 4-C-branched 6-deoxyhexoses. In addition, this study reveals an additional level of complexity within the LOS structures of M. marinum. Indeed, its presence first permitted us to define another LOS subtype, designated LOS-II′, in which the terminal α-caryophyllose is replaced by its 3-C-hydroxylated equivalent (OH-Car) that co-exists with LOS-II. Differential integration of the NMR signals revealed that within the LOS-II mixture, LOS-II′ represents a quantitatively minor compound compared with LOS-II. Although it was initially identified at the terminal nonreductive position of LOS-II′, OH-Car was also observed within the glycan sequences of both LOS-III and LOS-IV, defining again multiple subtypes of LOS-III and LOS-IV. Indeed, both NMR and mass spectrometry analyses (data not shown) provided conclusive evidence that LOS-III and -IV fractions were in fact constituted by mixtures of subtypes in which OH-Car could replace Car residues in both positions, thus presumably generating four isoforms for each molecule. However, the OH-Car residue containing LOS-III and -IV isoforms appeared as minor molecules (<10%).
Our study also unraveled a role of LOS in the inhibition of the TNF-α response in LPS-stimulated human macrophages. This unexpected finding is somehow reminiscent to the observation that phenolic glycolipids (PGL), produced by a subset of M. tuberculosis isolates of the Beijing family, are responsible for the “hyperlethality” phenotype of these strains in mice (46). Indeed, loss of PGL was found to correlate with an increase in the release of the pro-inflammatory cytokines in vitro, including TNF-α. Moreover, the addition of purified PGL to macrophages inhibited the secretion of TNF-α in a dose-dependent manner but did not induce the release of this cytokine (46). This anti-inflammatory effect has recently been confirmed using PGL purified from M. marinum (47). Our data suggest that, in addition to PGL, LOS can also confer the ability to inhibit the release of key inflammatory effector molecules by the innate immune system of the host. Whether this inhibition manifests itself in the hyperlethal phenotype of virulence M. marinum strains in infected fish remains, however, to be further demonstrated. Recent work demonstrated that loss of TNF signaling was responsible for an increase of M. marinum-infected zebrafish mortality and M. marinum intracellular growth and an acceleration in granuloma formation, followed by necrotic death of overladen macrophages and granuloma breakdown (48). It is therefore tempting to speculate that LOSs, by inhibiting TNF signaling, directly influence these physiopathological processes.
Our results point out an important immunomodulatory role of LOS, which may also be of high biological significance in the context of infection with other pathogenic species. In particular, LOSs are found in the Canetti strain, a smooth variant of M. tuberculosis (15). Further work is now required to address whether LOSs participate and contribute in the Canetti strain to pathogenesis or virulence by inhibiting the pro-inflammatory response in the Canetti strain.
Supplementary Material
Acknowledgments
We are grateful to Marlène Mortuaire for technical advice in cell culture and to Wilbert Bitter for kindly providing the M. marinum strains.
This work was supported by Grant ANR-05-MIIM-025 from the French National Research Agency (to Y. G. and L. K.), the Ministère de l'Enseignement Supérieur (to Y. R.), Université des Sciences et Technologies de Lille, and Institut Fédératif de Recherche Grant (IFR 147).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- TDM
- trehalose dimycolate
- COSY
- correlation spectroscopy
- FCS
- fetal calf serum
- GC-MS
- gas chromatography-mass spectrometry
- HMBC
- heteronuclear multiple-bound correlation
- LM
- lipomannan
- LOS
- lipooligosaccharide
- MALDI-MS
- matrix-assisted laser desorption ionization-time of flight-mass spectrometry
- Mma
- Mycobacterium marinum
- PIM
- phosphatidylinositol mannoside
- TNF
- tumor necrosis factor
- TOCSY
- total correlation spectroscopy
- NOE
- nuclear Overhauser effect
- ELISA
- enzyme-linked immunosorbent assay
- DIG
- digoxigenin
- HSQC
- heteronuclear single quantum coherence
- α-Car
- α-caryophyllose
- PGL
- phenolic glycolipid
- LPS
- lipopolysaccharide
- ES-MS
- electrospray-mass spectrometry
- rt
- retention time.
REFERENCES
- 1.Karakousis P. C., Bishai W. R., Dorman S. E. (2004) Cell. Microbiol. 6, 105–116 [DOI] [PubMed] [Google Scholar]
- 2.Kremer L., Baulard A. R., Besra G. S. (2000) in Molecular Genetics of Mycobacteria ( Hatfull G. F., Jacobs W. R., Jr. eds) pp. 173–190, American Society for Microbiology, Washington, DC [Google Scholar]
- 3.Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 4.Daffé M., Draper P. (1998) Adv. Microb. Physiol. 39, 131–203 [DOI] [PubMed] [Google Scholar]
- 5.Minnikin D. E., Kremer L., Dover L. G., Besra G. S. (2002) Chem. Biol. 9, 545–553 [DOI] [PubMed] [Google Scholar]
- 6.Hunter S. W., Murphy R. C., Clay K., Goren M. B., Brennan P. J. (1983) J. Biol. Chem. 258, 10481–10487 [PubMed] [Google Scholar]
- 7.Hunter S. W., Jardine I., Yanagihara D. L., Brennan P. J. (1985) Biochemistry 24, 2798–2805 [DOI] [PubMed] [Google Scholar]
- 8.Gilleron M., Puzo G. (1995) Glycoconj. J. 12, 298–308 [DOI] [PubMed] [Google Scholar]
- 9.Gilleron M., Vercauteren J., Puzo G. (1993) J. Biol. Chem. 268, 3168–3179 [PubMed] [Google Scholar]
- 10.Hunter S. W., Barr V. L., McNeil M., Jardine I., Brennan P. J. (1988) Biochemistry 27, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 11.McNeil M., Tsang A. Y., McClatchy J. K., Stewart C., Jardine I., Brennan P. J. (1987) J. Bacteriol. 169, 3312–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besra G. S., McNeil M. R., Khoo K. H., Dell A., Morris H. R., Brennan P. J. (1993) Biochemistry 32, 12705–12714 [DOI] [PubMed] [Google Scholar]
- 13.Khoo K. H., Suzuki R., Morris H. R., Dell A., Brennan P. J., Besra G. S. (1995) Carbohydr. Res. 276, 449–455 [DOI] [PubMed] [Google Scholar]
- 14.Muñoz M., Raynaud C., Lanéelle M. A., Julián E., López Marín L. M., Silve G., Ausina V., Daffé M., Luquin M. (1998) Microbiology 144, 137–148 [DOI] [PubMed] [Google Scholar]
- 15.Daffe M., McNeil M., Brennan P. J. (1991) Biochemistry 30, 378–388 [DOI] [PubMed] [Google Scholar]
- 16.Burguière A., Hitchen P. G., Dover L. G., Kremer L., Ridell M., Alexander D. C., Liu J., Morris H. R., Minnikin D. E., Dell A., Besra G. S. (2005) J. Biol. Chem. 280, 42124–42133 [DOI] [PubMed] [Google Scholar]
- 17.Ren H., Dover L. G., Islam S. T., Alexander D. C., Chen J. M., Besra G. S., Liu J. (2007) Mol. Microbiol. 63, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 18.Etienne G., Malaga W., Laval F., Lemassu A., Guilhot C., Daffé M. (2009) J. Bacteriol. 191, 2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belisle J. T., Brennan P. J. (1989) J. Bacteriol. 171, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemassu A., Lévy-Frébault V. V., Lanéelle M. A., Daffé M. (1992) J. Gen. Microbiol. 138, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 21.Collins F. M., Cunningham D. S. (1981) Infect. Immun. 32, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Sar A. M., Abdallah A. M., Sparrius M., Reinders E., Vandenbroucke-Grauls C. M., Bitter W. (2004) Infect. Immun. 72, 6306–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., Jagels K., Lord A., Moule S., Mungall K., Norbertczak H., Quail M. A., Rabbinowitsch E., Walker D., White B., Whitehead S., Small P. L., Brosch R., Ramakrishnan L., Fischbach M. A., Parkhill J., Cole S. T. (2008) Genome Res. 18, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer L., Gurcha S. S., Bifani P., Hitchen P. G., Baulard A., Morris H. R., Dell A., Brennan P. J., Besra G. S. (2002) Biochem. J. 363, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F. (1991) Nature 351, 456–460 [DOI] [PubMed] [Google Scholar]
- 26.Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 27.Guerardel Y., Maes E., Elass E., Leroy Y., Timmerman P., Besra G. S., Locht C., Strecker G., Kremer L. (2002) J. Biol. Chem. 277, 30635–30648 [DOI] [PubMed] [Google Scholar]
- 28.Guérardel Y., Maes E., Briken V., Chirat F., Leroy Y., Locht C., Strecker G., Kremer L. (2003) J. Biol. Chem. 278, 36637–36651 [DOI] [PubMed] [Google Scholar]
- 29.Vey E., Zhang J. H., Dayer J. M. (1992) J. Immunol. 149, 2040–2046 [PubMed] [Google Scholar]
- 30.Alexander D. C., Jones J. R., Tan T., Chen J. M., Liu J. (2004) J. Biol. Chem. 279, 18824–18833 [DOI] [PubMed] [Google Scholar]
- 31.Malm S., Walter K., Engel R., Maass S., Pfau S., Hübner G., Lindner B., Holst O., Ehlers S., Bange F. C. (2008) Int. J. Med. Microbiol. 298, 645–655 [DOI] [PubMed] [Google Scholar]
- 32.Adinolfi M., Corsaro M. M., De Castro C., Lanzetta R., Parrilli M., Evidente A., Lavermicocca P. (1995) Carbohydr. Res. 267, 307–311 [Google Scholar]
- 33.Adinolfi M., Corsaro M. M., De Castro C., Evidente A., Lanzetta R., Mangoni L., Parrilli M. (1995) Carbohydr. Res. 274, 223–232 [Google Scholar]
- 34.Molinaro A., De Castro C., Lanzetta R., Manzo E., Parrilli M. (2001) J. Am. Chem. Soc. 123, 12605–12610 [DOI] [PubMed] [Google Scholar]
- 35.Vignal C., Guérardel Y., Kremer L., Masson M., Legrand D., Mazurier J., Elass E. (2003) J. Immunol. 171, 2014–2023 [DOI] [PubMed] [Google Scholar]
- 36.Briken V., Porcelli S. A., Besra G. S., Kremer L. (2004) Mol. Microbiol. 53, 391–403 [DOI] [PubMed] [Google Scholar]
- 37.Ryll R., Kumazawa Y., Yano I. (2001) Microbiol. Immunol. 45, 801–811 [DOI] [PubMed] [Google Scholar]
- 38.Clay H., Davis J. M., Beery D., Huttenlocher A., Lyons S. E., Ramakrishnan L. (2007) Cell Host Microbe 2, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosma C. L., Humbert O., Ramakrishnan L. (2004) Nat. Immunol. 5, 828–835 [DOI] [PubMed] [Google Scholar]
- 40.Davis J. M., Clay H., Lewis J. L., Ghori N., Herbomel P., Ramakrishnan L. (2002) Immunity 17, 693–702 [DOI] [PubMed] [Google Scholar]
- 41.Appelmelk B. J., den Dunnen J., Driessen N. N., Ummels R., Pak M., Nigou J., Larrouy-Maumus G., Gurcha S. S., Movahedzadeh F., Geurtsen J., Brown E. J., Eysink, Smeets M. M., Besra G. S., Willemsen P. T., Lowary T. L., van Kooyk Y., Maaskant J. J., Stoker N. G., van der Ley P., Puzo G., Vandenbroucke-Grauls C. M., Wieland C. W., van der Poll T., Geijtenbeek T. B., van der Sar A. M., Bitter W. (2008) Cell. Microbiol. 10, 930–944 [DOI] [PubMed] [Google Scholar]
- 42.Hunter S. W., Fujiwara T., Murphy R. C., Brennan P. J. (1984) J. Biol. Chem. 259, 9729–9734 [PubMed] [Google Scholar]
- 43.Gorshkova R. P., Zubkov V. A., Isakov V. V., Ovodov Y. S. (1984) Carbohydr. Res. 126, 308–312 [DOI] [PubMed] [Google Scholar]
- 44.Gorshkova R. P., Zubkov V. A., Isakov V. V., Ovodov IuS. (1987) Bioorg. Khim. 13, 1146–1147 [PubMed] [Google Scholar]
- 45.Mattos K. A., Todeschini A. R., Heise N., Jones C., Previato J. O., Mendonça-Previato L. (2005) Glycobiology 15, 313–321 [DOI] [PubMed] [Google Scholar]
- 46.Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., Kaplan G., Barry C. E., 3rd (2004) Nature 431, 84–87 [DOI] [PubMed] [Google Scholar]
- 47.Robinson N., Kolter T., Wolke M., Rybniker J., Hartmann P., Plum G. (2008) Traffic 9, 1936–1947 [DOI] [PubMed] [Google Scholar]
- 48.Clay H., Volkman H. E., Ramakrishnan L. (2008) Immunity 29, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domon B., Costello C. E. (1988) Biochemistry 27, 1534–1543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.