Abstract
Flagella of the bacteria Helicobacter pylori and Campylobacter jejuni are important virulence determinants, whose proper assembly and function are dependent upon glycosylation at multiple positions by sialic acid-like sugars, such as 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid (pseudaminic acid (Pse)). The fourth enzymatic step in the pseudaminic acid pathway, the hydrolysis of UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose to generate 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose, is performed by the nucleotide sugar hydrolase PseG. To better understand the molecular basis of the PseG catalytic reaction, we have determined the crystal structures of C. jejuni PseG in apo-form and as a complex with its UDP product at 1.8 and 1.85 Å resolution, respectively. In addition, molecular modeling was utilized to provide insight into the structure of the PseG-substrate complex. This modeling identifies a His17-coordinated water molecule as the putative nucleophile and suggests the UDP-sugar substrate adopts a twist-boat conformation upon binding to PseG, enhancing the exposure of the anomeric bond cleaved and favoring inversion at C-1. Furthermore, based on these structures a series of amino acid substitution derivatives were constructed, altering residues within the active site, and each was kinetically characterized to examine its contribution to PseG catalysis. In conjunction with structural comparisons, the almost complete inactivation of the PseG H17F and H17L derivatives suggests that His17 functions as an active site base, thereby activating the nucleophilic water molecule for attack of the anomeric C–O bond of the UDP-sugar. As the PseG structure reveals similarity to those of glycosyltransferase family-28 members, in particular that of Escherichia coli MurG, these findings may also be of relevance for the mechanistic understanding of this important enzyme family.
The gastrointestinal pathogens Campylobacter jejuni and Helicobacter pylori have been shown to modify their flagellins with the sialic acid-like sugar 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid or pseudaminic acid (Pse),3 via O-linkage at up to 19 sites per flagellin monomer (1, 2). Not only is this sialic acid-like modification necessary for flagellar assembly and motility (1, 2), it has also been shown to be important for C. jejuni virulence (3). In addition to its role in autoagglutination of bacterial cells, Pse and related derivatives may also influence pathogenesis through bacterial adhesion, invasion, and immune evasion (4, 5), since sialic acids in humans have been shown to mediate a myriad of cell-cell and cell-molecule interactions (6). As flagellin glycosylation in these organisms is required for host colonization and ultimately virulence (3, 7, 8), these novel sugar biosynthetic pathways provide an excellent platform for therapeutic development.
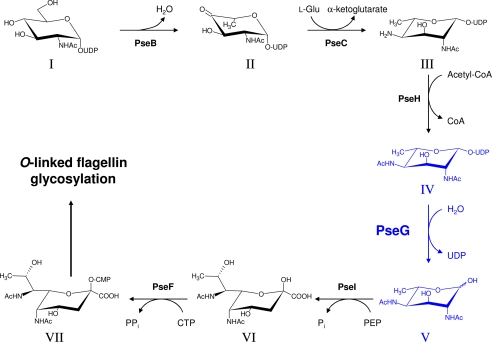
The reliance of H. pylori pathogenicity on Pse biosynthesis, in combination with the prevalence of H. pylori resistance to existing antibiotic treatments (9), prompted and led to the complete elucidation of the CMP-pseudaminic acid (CMP-Pse) biosynthetic pathway in both C. jejuni and H. pylori (10–15). The CMP-Pse biosynthetic pathway (Fig. 1) is similar to that of CMP-sialic acid, involving condensation of an N-acetylhexosamine intermediate with the three-carbon pyruvate molecule forming a nine-carbon sialic acid-like nonulosonate, although in contrast the CMP-Pse pathway consists of several more steps between the initial building block UDP-GlcNAc and the condensation reaction. PseG, a UDP-sugar hydrolase, produces the final 6-deoxy-N-acetylhexosamine intermediate in the CMP-Pse pathway by removing the nucleotide moiety from UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose or UDP-6-deoxy-AltdiNAc (Fig. 1). This sort of single enzymatic function is rare in nature, with the only other similar example being a GDP-mannose/GDP-glucose hydrolase (16), which belongs to the metal-dependent Nudix family of enzymes. In an elegant study, Liu and Tanner (11) demonstrated that PseG catalyzes nucleotide removal by a metal-independent C–O bond cleavage mechanism resulting in inversion of stereochemistry at C-1 of the product 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose or 6-deoxy-AltdiNAc, similar to the catalytic properties of some GT-B glycosyltransferases.
FIGURE 1.
Role of PseG within the CMP-pseudaminic acid biosynthetic pathway of C. jejuni and H. pylori. The biosynthetic step involving PseG is highlighted in blue. The enzymes and biosynthetic intermediates of the CMP-pseudaminic acid pathway are, in the following order, PseB (Cj1293/HP0840), NADP-dependent dehydratase/epimerase; PseC (Cj1294/HP0366), pyridoxal phosphate-dependent aminotransferase; PseH (Cj1313/HP0327), N-acetyltransferase; PseG (Cj1312/HP0326B), NDP-sugar hydrolase; PseI (Cj1317/HP0178), pseudaminic acid synthase; PseF (Cj1311/HP0326A), CMP-pseudaminic acid synthetase; and I, UDP-GlcNAc; II, UDP-2-acetamido-2,6-dideoxy-β-l-arabino-hexos-4-ulose; III, UDP-4-amino-4,6-dideoxy-β-l-AltNAc; IV, UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose; V, 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose; VI, pseudaminic acid; and VII, CMP-pseudaminic acid. Here, PEP refers to phosphoenolpyruvate. Pyranose rings are shown as their predominant chair conformation in solution as determined from nuclear Overhauser effects and JH,H coupling constants (13).
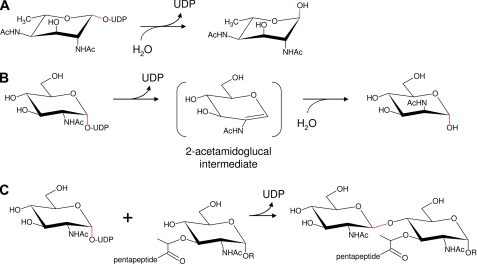
Together, glycosyltransferases and glycoside hydrolases compose the majority of enzymes in both eukaryotes and prokaryotes that manipulate glycosidic bonds. Glycosyltransferases of the Leloir classification use sugar-nucleotide derivatives as glycosyl donors resulting in transfer to acceptors such as a monosaccharide, oligosaccharide, or polysaccharide. It is therefore plausible that a “glycosyltransferase fold” in PseG has evolved to efficiently utilize water as an acceptor, instead of another carbohydrate, consequently behaving as a hydrolase (11). Based on structure, most glycosyltransferases fall into two groups, GT-A and GT-B, that exhibit different folds, respectively (17). For both families, depending on the particular enzyme, the outcome may result in either inversion or retention of stereochemistry for the donor anomeric carbon (see Fig. 2). In addition, GT-B family enzymes are metal-independent, lacking an important DXD motif present in most GT-A members. Based on the novelty of PseG and its role in H. pylori pathogenicity, we sought a greater structural and mechanistic understanding of this important enzyme.
FIGURE 2.
Functional comparison of enzymes belonging to the GT-B superfamily. A, UDP-sugar hydrolase PseG catalyzes the removal of UDP from UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-Alt or UDP-6-deoxy-AltdiNAc. B, UDP-GlcNAc hydrolyzing 2-epimerase NeuC catalyzes the removal of UDP and the formation of ManNAc from UDP-GlcNAc. C, GlcNAc transferase MurG catalyzes the formation of undecaprenyl-phosphoryl-muramyl-pentapeptide-GlcNAc via formation of a glycosidic linkage between UDP-GlcNAc and undecaprenyl-phosphoryl-muramyl-pentapeptide. R represents the phosphoryl-undecaprenyl moiety, with the pentapeptide having the specific sequence l-Ala-d-γGlu-l-Lys-d-Ala-d-Ala. Both A and C activities result in an initial inversion of stereochemistry at C-1 for the donor substrate. In contrast, the activity for B results in an initial retention of C-1 stereochemistry. Enzymatically altered anomeric bonds are indicated in red.
Here we report the crystal structure of PseG alone at 1.8 Å resolution and in complex with UDP, a product of the reaction, at 1.85 Å resolution. Although very few homologs have been identified based on sequence similarity alone, PseG bears the closest structural similarity to MurG, a GT-B family member (18). In addition, computational docking and molecular dynamics simulations were performed to gain insight into the binding mode of the PseG substrate UDP-6-deoxy-AltdiNAc. Based on the crystallographic and modeled structures, several potential active site residues were selected for mutagenesis and kinetic analyses to further characterize the PseG active site. The relevance of these findings to the structurally related MurG family of enzymes is discussed.
EXPERIMENTAL PROCEDURES
Construction of PseG H17F, H17L, H17N, Y78F, and N255A Substitution Derivatives
Site-directed mutagenesis was performed using the QuikChange II mutagenesis kit (Stratagene) according to the manufacturer's instructions with pNRC17.1 (13) as template and the mutagenic oligonucleotides listed in supplemental Table S1. The newly constructed plasmids were sequenced as described previously (13).
C. jejuni PseG Purification
Protein production was performed using the expression strain BL21[DE3] (Novagen, Madison, WI) containing pseG-His6 from C. jejuni 11168 cloned in-frame in pET30a (Novagen) yielding the plasmid pNRC17.1 (13) or mutant derivatives. The original pNRC17.1 plasmid encodes for a PseG E155K substitution (PseGHis6), in addition to those described here. This PseG E155K derivative or PseGHis6 was found to have activity similar to that of wild-type enzyme. For CD spectroscopic and kinetic analyses, PseGHis6 and substitution derivatives were purified as described previously (13), except that 1 mm MgCl2 was absent from lysis and dialysis buffers (see supplemental Figs. S1 and S2 and supplemental Table S2). Protein concentration was measured spectrophotometrically using A2800.1% values (PseGHis6, PseGHis6-H17F, PseGHis6-H17L, PseGHis6-H17N, and PseGHis6-N255A 0.826; PseGHis6-Y78F 0.780).
For crystallization, PseGHis6 was expressed in 1 liter of TB medium containing 30 μg ml−1 kanamycin. The culture was first grown for 2 h at 37 °C, followed by induction with 100 μm isopropyl β-d-thiogalactopyranoside for an overnight period at room temperature. The same induction protocol was followed to express the selenomethionine (SeMet)-containing protein in the Escherichia coli metA− auxotroph DL41(DE3) in LeMaster medium (19) containing 25 mg liter−1 l-SeMet. Cells were harvested by centrifugation (4000 × g, 4 °C, 30 min) and resuspended in lysis buffer containing 50 mm Tris-Cl, pH 7.5, 400 mm NaCl, 1% (v/v) Triton X-100, 5% (v/v) glycerol, 1 mm DTT, 20 mm imidazole, and a mixture of protease inhibitors (10 μm leupeptin, 0.5 mm benzamidine, and 0.1 mm PefablocTM). Cells were disrupted by sonication for a total of 2 min with alternating cycles of 15 s on and 15 s off. The lysate was clarified by ultracentrifugation (100,000 × g, 4 °C, 45 min), and the protein supernatant was incubated with 1 ml of pre-equilibrated nickel-nitrilotriacetic acid-agarose (Qiagen, Mississauga, Ontario, Canada) for 1 h with gentle shaking. Nickel-nitrilotriacetic acid beads were washed first with buffer (50 mm Tris-Cl, pH 7.5, 1 m NaCl, 5% (v/v) glycerol, 1 mm DTT, 20 mm imidazole, pH 7.5) followed by a similar buffer containing 0.4 m NaCl and 40 mm imidazole to remove nonspecifically bound proteins. Elution was performed in the same buffer containing 250 mm imidazole, pH 7.5. Purity of the eluted protein was assessed by SDS-PAGE. A total of 9 mg of pure native protein and 5 mg of SeMet-labeled protein was obtained using this procedure. The protein buffer was changed to 20 mm Tris-Cl, pH 7.5, 0.2 m NaCl, 5% (v/v) glycerol, 5 mm DTT, 1 mm MgCl2 prior to concentration by ultrafiltration to a final concentration of 9 mg ml−1 as determined by the method of Bradford (20).
Enzymatic Synthesis and Purification of PseG Substrate
Large scale enzymatic synthesis of UDP-6-deoxy-AltdiNAc was accomplished using a 45-ml reaction containing 1 mm UDP-GlcNAc, 1 mm pyridoxal-5′-phosphate, 10 mm l-glutamate, 1.5 mm acetyl-CoA, and ∼10 mg each of H. pylori PseBHis6, His6PseC, and His6PseH, similar to methods described previously (13). After passage through an Amicon Ultra-15 filter (10,000 molecular weight cutoff), the sugar preparation was lyophilized and desalted/purified using a Superdex Peptide 10/300 GL column (Amersham Biosciences) in 25 mm ammonium bicarbonate, pH 7.9. For kinetic analysis, the sample was subjected to a final polishing step using a Mono Q 4.6/100 PE column (Amersham Biosciences) with a 25–500 mm ammonium bicarbonate gradient, pH 7.9, over 20 column volumes. Purified fractions were pooled and lyophilized. Finally, quantification was determined using the molar extinction coefficient of UDP (ϵ260 = 10,000).
CD Spectral Analysis of PseG and Substitution Derivatives
CD spectra were recorded on a Jasco J-600 spectropolarimeter (Utrecht, Netherlands) with a 0.05-cm quartz cuvette at ambient temperature (20–22 °C). The instrument was calibrated with ammonium (+)-10-camphorsulfonate. Wavelengths from 190 to 260 nm were measured with a 0.2-nm step resolution, a 2.0-nm bandwidth, and 20-nm/min scan speed. All CD experiments were performed with protein samples in a buffer consisting of 25 mm sodium phosphate, pH 7.3, 25 mm NaCl. The final concentrations of the proteins were 0.098–0.104 mg/ml, calculated from the absorption at 280 nm. Four scans were collected and averaged, and the data were smoothed using the Jasco software. Data are expressed per peptide bond as mean residue ellipticity [θ]×10−3 (degrees cm2 dmol−1).
Kinetic Measurements
For routine analysis of enzyme activity, PseGHis6 or respective substitution derivatives were incubated at 37 °C in 25 mm sodium phosphate buffer, pH 7.3, 25 mm NaCl, with UDP-6-deoxy-AltdiNAc, and reactions were analyzed by capillary electrophoresis as described earlier (14). For kinetic analysis, substrate conversion was measured using a continuous coupled assay for UDP formation by monitoring NADH oxidation (21). Specifically, enzyme reactions (150 μl each) were performed in a 96-well microplate at 37 °C with 25 mm sodium phosphate, pH 7.3, 25 mm NaCl, 5 mm MgCl2, 2 mm phosphoenolpyruvate, 0.2 mm NADH, 12 units each of lactate dehydrogenase and pyruvate kinase, and variable UDP-6-deoxy-AltdiNAc concentrations (0.03–2 mm). Reactions were initiated by the addition of PseGHis6 derivatives (1.5–90 nm final concentration) as required. Substrate conversion was extrapolated from A340 versus [NADH] standard curves using a microplate reader (Bio-Tek Instruments Inc., Winooski, VT), where initial [NADH] was determined using the molar extinction coefficient for NADH (ϵ340 = 6220) and a quartz cuvette. Kinetic constants were calculated using initial velocities and Eadie-Hofstee plots with the program GraphPad Prism 3.
Characterization of PseGHis6
Dynamic light scattering was performed using a DynaPro plate reader (Wyatt Technologies, Santa Barbara, CA) using purified PseG at a concentration of 14 mg ml−1 in 20 mm Tris-Cl, pH 7.5, 0.2 m NaCl, 5% (v/v) glycerol, 5 mm DTT, 1 mm MgCl2. The mass of native and SeMet-substituted PseGHis6 was determined by electrospray ionization mass spectrometry using an Agilent 1100 Series LC-MSD mass spectrometer (Agilent Technologies, Mississauga, Ontario, Canada) and analyzed using Agilent Chemstation version A.09.01 software.
Crystallization
Initial crystallization screening was performed in 96-well sitting drop plates using crystallization screens from Hampton Research (Aliso Viejo, CA) with a Hydra II crystallization robot (Thermo Fisher Scientific, Hudson, NH). Well diffracting crystals were obtained from two reservoir conditions (a and b), following optimization from initial screens, using the hanging drop vapor diffusion method at 20 °C as follows: condition a, 0.1 m BisTris, pH 6.5, 0.2 m ammonium sulfate, 23% (v/v) PEG 3350; condition b, 0.1 m BisTris, pH 6.5, 50 mm CaCl2, 25% (v/v) PEG monomethyl ether 550. Crystals of SeMet-substituted protein were obtained under the same conditions but by increasing the precipitant concentration by 2–3% (v/v). PseGHis6 crystals obtained by co-crystallization in the presence of 1 mm UDP-6-deoxy-AltdiNAc, 5 mm UDP-glucosamine, or 5 mm UDP-N-acetylglucosamine were obtained under the same conditions. Crystals obtained with UDP-N-acetylglucosamine showed electron density only for UDP and were used to determine the co-crystal structure. For cryoprotection prior to x-ray diffraction data collection, crystals were transferred to a solution containing reservoir solution supplemented with 15% glycerol (v/v) for crystals from condition a and with reservoir solution alone for crystals from condition b, picked up with a nylon loop and flash-cooled directly in the N2 cold stream at 100 K (Oxford Cryosystems, Oxford, UK). Apo- and UDP-bound PseGHis6 crystals belong to space group P41 with the apo-form having unit cell dimensions of a = b = 93.6, c = 42.7 and Z = 4, and a Matthews coefficient of 2.9 Da Å−1 (22), whereas UDP-bound PseGHis6 crystals have a unit cell of a = b = 94.4, c = 43.6 and Z = 4.
Structure Determination and Refinement
X-ray diffraction data were collected at beamlines X12B and X29, National Synchrotron Light Source, Brookhaven National Laboratory, using a Quantum-4 CCD detector (X12B) or Quantum-315 detector (X29) (Area Detector Systems Corp., Poway, CA). For phase determination, data collected at the selenium anomalous peak were used in the selenium single-wavelength anomalous dispersion method (19). All three selenium sites present in the asymmetric unit were identified using SHELXD (23) with cross-correlation values of 42.3 and 25.1 for all and weak reflections, respectively. Initial phases were calculated using SHELXE (23), and the resulting maps were used directly for automated model building with ARP/wARP (24). The resulting model was 70% complete with remaining residues fit manually using the program COOT (25). The final model of PseGHis6 was refined using Refmac5 (26) to a final Rwork and Rfree of 0.193 and 0.237, respectively, for all reflections to 1.8 Å. No σ-cutoff was used in the refinement. The apo-PseGHis6 structure was used as the starting model for molecular replacement, phase calculation, and refinement with data sets obtained for PseGHis6 co-crystallized with ligands. In all cases, the resulting electron density maps clearly indicated the presence of bound UDP, which was modeled using the program COOT (25). The PseGHis6-UDP complex was refined to 1.85 Å resolution with a final Rwork and Rfree of 0.177 and 0.211, respectively. Final refinement statistics for both models are given in Table 1. Coordinates for apo- and UDP-bound PseGHis6 have been deposited in the RCSB Protein Data Bank (27) with accession codes 3HBM and 3HBN, respectively.
TABLE 1.
X-ray crystallographic data
| Data collection | ||
|---|---|---|
| Data set | Apo | UDP complex |
| Space group | P41 | P41 |
| Unit cell | ||
| a | 93.65 Å | 94.44 Å |
| b | 93.65 Å | 94.44 Å |
| c | 42.70 Å | 43.57 Å |
| α = β = γ | 90.0 | 90.0 |
| Za | 4 | 4 |
| Resolution | 1.8 Å | 1.85 Å |
| Wavelength | 0.9791 Å | 0.98 Å |
| Observed hkl | 253,712 | 197,191 |
| Unique hkl | 34,208 | 32,759 |
| Redundancy | 7.4 | 6.0 |
| Completeness | 98.8 | 99.1 |
| Rsymb | 0.073 | 0.056 |
| I/σ(I) | 22.6 | 17 |
| Refinement | ||
| Resolution | 33.1 to 1.80 Å | 39.56 to 1.85 Å |
| Rwork (no. of hkl)c | 0.193 (32,485) | 0.177 (31,109) |
| Rfree (no. of hkl) | 0.237 (1718) | 0.211 (1631) |
| Average B-factors | ||
| Protein | 23.4 | 21.4 |
| Solvent | 34.4 | 32.6 |
| SO43− | 47.4 | |
| Glycerol | 39.4 | |
| UDP | 29.0 | |
| Cl− | 45.1 | |
| Ramachandran plot | ||
| Favored | 99.6% | 99.6% |
| Allowed | 0.4% | 0.4% |
| Disallowed | 0% | 0% |
| r.m.s. bond | 0.011 Å | 0.010 Å |
| r.m.s. angles | 1.238° | 1.255° |
| PDB code | 3HBM | 3HBN |
a Number of molecules in the unit cell is given.
b Rsym = (Σ|Iobs − Iavg|)/ΣIavg.
c Rwork = (Σ|Fobs − Fcalc|)/ΣFobs.
Molecular Modeling of Free and Bound Substrate
To facilitate molecular modeling of PseG-bound substrate, several energetically favorable free-substrate conformations (OGly-methylated) were calculated. All “free” structures were first optimized using the AM1 Hamiltonian as implemented in Hyperchem 6.0, starting from idealized ring structures, including several boat structures. Three different ring structures 4C1, 1C4, and 5S1 were found. Each structure then had the hydroxyl rotamers optimized by using the conformational search routine available in Hyperchem. These three structures were then optimized using the Amsterdam Density Functional (ADF-2006) DFT QM program. Optimized structures were checked by frequency calculations (analytical second derivatives) for true convergence. Structures that exhibited negative frequencies were re-optimized using the coefficients of the negative modes to adjust the Cartesian coordinates and the optimization, with the frequency calculations repeated until all modes were positive. All structures were optimized as internal coordinates using the triple ζ plus basis set with no frozen core. Keywords used for the DFT calculations were LDA, SCF, VWN, METAGGA, and HFEXCHANGE (see ADF manual). Full solvation was considered parameterized to water. For further free conformer (4C1, 1C4, and 5S1) details, see supplemental Fig. S3.
Substrate docking was performed on the UDP-bound PseGHis6 crystal structure, after removal of UDP, C-terminal fusion tag extension Leu-Glu-His6, and water molecules. The protonation state of PseG was assigned with H++ (28). The UDP-6-deoxy-AltdiNAc substrate was built in Sybyl 6.9 (Tripos, Inc., St. Louis, MO), starting from the PseG-bound conformation of UDP to which the 6-deoxy-AltdiNAc sugar moiety was appropriately connected. Three substrate conformations were separately built, corresponding to the 1C4 chair, 4C1 chair, and 5S1 twist-boat sugar conformations calculated for the free OGly-methylated sugar as described earlier. Flexible docking of the sugar part of the substrate was carried out by Monte Carlo minimization (MCM) conformational sampling (29) applied to protein-ligand binding (30). The crystallographic binding mode of the UDP product was adopted for the UDP part of the substrate. Three independent docking runs of 10,000 MCM cycles were carried out, initiated from the expected 1C4, 4C1, and 5S1 conformations for the 6-deoxy-AltdiNAc sugar moiety. Conformational sampling included all acyclic and cyclic rotatable bonds starting from the P-α atom and covered the pyrophosphate and 6-deoxy-AltdiNAc moieties. In each MCM cycle, at least two dihedral angles were simultaneously and randomly perturbed. The O-5–C-1 endocyclic sugar bond was used for pyranose ring opening and reforming during sampling. The minimized energy of the complex formed the basis of accepting or rejecting the resulting conformation according to the Metropolis probability criterion at 300 K. Energy minimization was carried out with the AMBER force field (31, 32) and a distance-dependent (4R) dielectric constant. Ligand partial charges were calculated with Molcharge (OpenEye, Inc., Santa Fe, NM) based on the AM1-BCC method (33) and the 1C4-sugar conformation of the substrate. The protein region allowed to move during the energy minimization step included 36 residues around the putative sugar-binding pocket (Ser10–His17, Arg20, Asp76–Gly79, Asp101–Glu102, Val114–Asn115, Met164–Lys170, Ser232–Leu236, Asn238, Tyr252–Gln256, Arg259). The uridine portion of the substrate was held fixed during MCM.
Separate hybrid quantum mechanics (QM)/molecular mechanics (MM) molecular dynamics (MD) simulations were carried out for PseG complexes with the substrate in 1C4 or 5S1 conformations. These simulations were initiated from PseG-bound substrate geometries generated by MCM flexible docking runs as described above but without internal sampling of the sugar ring to prevent the 1C4 to 5S1 conversion. Before starting QM/MM MD, 3 ns of classical MD simulations were performed to equilibrate the protein and allow the substrate to accommodate in the binding cavity, using the AMBER force field (31, 32) in AMBER 9 (34). AM1-BCC partial charges (33) were assigned to the substrate. Each complex was solvated in a truncated octahedron TIP3P water box (35), and electroneutrality was achieved by adding Na+ counterions. Applying harmonic restraints with force constants of 10 kcal mol−1 Å−2 to all solute atoms, the system was energy-minimized first, followed by heating from 100 to 300 K over 25 ps in the canonical ensemble (constant number of particles, volume, and temperature, NVT) and by equilibrating to adjust the solvent density under 1 atm pressure over 25 ps in the isothermal-isobaric ensemble (constant number of particles, pressure, and temperature, NPT) simulation. The harmonic restraints were then gradually reduced to 0 with four rounds of 25-ps simulations. After an additional 25-ps simulation, a 3-ns production NPT run was obtained with snapshots collected every 1 ps, using a 2-fs time step and 9-Å nonbonded cutoff. The Particle Mesh Ewald method (36) was used to treat long range electrostatic interactions, and bond lengths involving bonds to hydrogen atoms were constrained by SHAKE (37). The QM/MM MD simulation started from the last snapshot of the classical MD simulation and was carried out in AMBER 10 (38). The entire substrate molecule was included in the QM region described with the PM3 Hamiltonian (39), whereas the solvated protein was treated at the molecular mechanics level using the AMBER force field. The SHAKE option was lifted, and the MM correction for peptide linkage was applied for the QM region. The QM/MM simulations were run for 120 ps with a time step of 1 fs and snapshots collected every 50 fs.
Standard analyses of MD trajectories were carried out with PTRAJ in AMBER 10. Water O and H occupancy plots and solute (PseG-substrate complex) average structures were generated from 120- and 90-ps QM/MM MD trajectories for the 5S1 and 1C4 conformations, respectively. Each average structure was first fitted with a water molecule that was added into the water density identified at the putative catalytic position, and then energy-minimized in vacuum with the hybrid PM3-AMBER potential for 5000 steps, including the catalytic water along with the substrate in the QM region. PseG-substrate binding affinities were estimated with SIETRAJ (40) by calculating the average solvated interaction energy (SIE) (41). Relative protein strain energies were estimated by calculating the average solvated conformational energy of substrate-bound PseG. The SIE and solvated conformational energy were averaged over 160 snapshots at 0.5-ps intervals from the last 80 ps of QM/MM MD trajectories (see supplemental material for further details on SIE and solvated conformational energy calculations).
RESULTS AND DISCUSSION
Sequence Relationships
Surprisingly, there is very low sequence similarity among PseG hydrolases from different bacterial species. Based on a PSI-BLAST analysis (42), C. jejuni PseG (274 residues) shares only ∼24–28% sequence identity with other putative bacterial PseG proteins. In fact, from this analysis, only weak sequence similarity is observed between C. jejuni and H. pylori PseG proteins, yet their identical enzymatic activity has been confirmed in vitro (13). The other possible (∼24–28% identity) PseG proteins identified include FlmD from Aeromonas punctata (43) and Aeromonas hydrophilia (44) that play a role in flagellar assembly and, additionally, for the former, in O-antigen biosynthesis. These pseG or flmD sequences are found in gene clusters highly similar to pseudaminic acid biosynthetic genes (13), and in fact, flagellin from A. punctata was shown to be glycosylated with pseudaminic acid (45). As well, the rkpO genes from Sinorhizobium meliloti Rm41 (46) and Rhizobium sp. NGR234 (47), associated with the synthesis of K-antigen capsular polysaccharides, are required for pseudaminic acid biosynthesis in these organisms, suggesting that these proteins also are PseG family members exhibiting low sequence similarity with their C. jejuni counterpart. Similar sequences have also been identified in the O-antigen biosynthetic loci of Pseudomonas aeruginosa serotype O7 and O9, ORF11 and ORF9, respectively (48). As these O-antigen biosynthetic loci also possess pseudaminic acid-like biosynthetic genes, and the fact that this sugar is known to decorate the lipopolysaccharide and pili of P. aeruginosa (49, 50), we propose that these too are PseG members. Importantly, when performing a multiple sequence alignment using MUSCLE (51) with the PseG sequences described, a minimum consensus sequence (DX5GXGHX2R) for this family of proteins was identified (supplemental Fig. S4).
PseG Structure
PseG was purified as a C-terminally His6-tagged fusion protein and crystallized in both apo-form as well as bound to UDP. Electrospray ionization mass spectrometry analysis of purified PseGHis6 gave a mass of 32,376 Da (calculated mass 32,383 Da), whereas the selenium-substituted PseGHis6 sample gave a mass of 32,516 Da (calculated mass 32524 Da), consistent with the expected incorporation of three SeMet residues. Although the dynamic light scattering experiment showed the protein was polydisperse in solution, this did not adversely affect our efforts to obtain well diffracting crystals of this protein.
The structures of PseGHis6 in apo- and UDP-bound forms have been determined by selenium single-wavelength anomalous dispersion and refined with good geometry to Rwork = 0.193, Rfree = 0.237 at 1.8 Å resolution, and Rwork = 0.177, Rfree = 0.211 at 1.85 Å resolution, respectively (Table 1). The apo-PseGHis6 structure contains three bound sulfate molecules, whereas the complex with UDP, in addition to the ligand, includes two molecules of glycerol (Fig. 3). The structure consists of the entire sequence, with the exception of Phe129, which was not modeled in the apo-PseGHis6 structure because of poor density. Both models contain a C-terminal extension consisting of vector sequence from the C-terminal His6 fusion tag, 275Leu-Glu-His6282, which is ordered in the crystal structures. This C-terminal extension forms both direct and water-mediated H-bonds with the PseG molecule itself, as well as with a surface-exposed cluster of negatively charged residues (Asp30, Asp44, Glu52, Glu67, and Glu68) from a symmetry-related PseG molecule. Based on the crystal structures and analysis by size exclusion chromatography (result not shown), we surmise that the enzyme is a monomer.
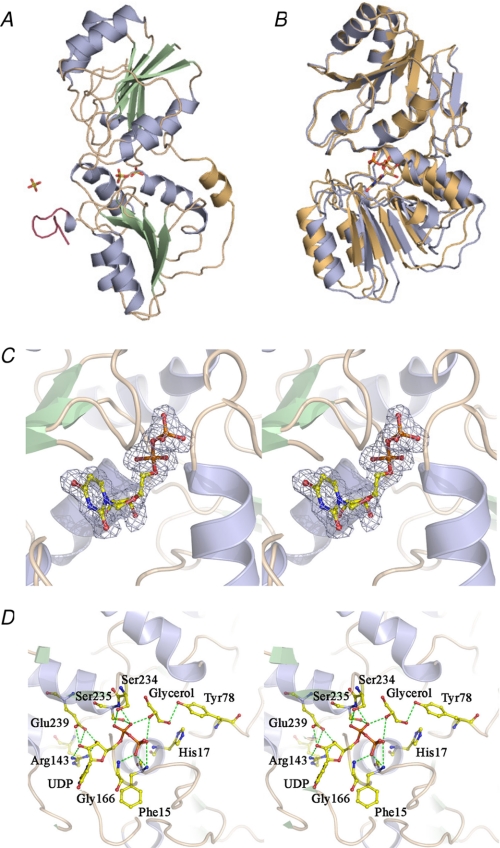
FIGURE 3.
Structures of PseGHis6. A, apo-PseGHis6, colored by secondary structure (α-helix, pale blue; β-strands, pale green; and loop regions, wheat), shows the N-terminal domain (top) and C-terminal domain (bottom) connected by the linking α-helix (pale orange). The His6 tag sequence at the C terminus of the protein is colored raspberry. Sulfate molecules are shown in stick representation and colored by a CPK scheme. B, superposition of the apo- (pale blue) and UDP-bound (pale orange) PseGHis6 structures based on 142 common C-α atoms from the N-terminal domain. C, stereo Fo − Fc (omit) electron density for UDP bound to PseGHis6 contoured at a level of 2.5σ. D, H-bonding interactions between PseGHis6 and UDP, as well as with the bound glycerol molecule in the active site.
The overall structure of PseGHis6 is composed of distinct N- and C-terminal domains, each having a classical α/β-fold (Fig. 3A). The N-terminal domain (residues 1–142) harbors a central seven-stranded parallel β-sheet (↑3-↑2-↑1-↑4-↑5-↑6-↑7) flanked on either side by three α-helices, whereas the C-terminal domain (residues 153–282) has a six-stranded parallel β-sheet (↑3-↑2-↑1-↑4-↑5-↑6) surrounded by five α-helices. A short helix (residues 143–152) links the N- and C-terminal domains. Most of the inter-domain interactions are water-mediated, with one of the sulfates (301) acting as a bridge through H-bonding to the NPhe-15 from the N-terminal domain and the NGly-166 and OG1Thr-167 of the C-terminal domain. The inter-domain interactions are contributed by residues situated in the loops between β1/α1 and at the C-terminal ends of β4-β7 of the N-terminal domain and the β1/α1 and β2/α2 loops as well as helices α4 and α5 of the C-terminal domain. The domain interface consists of both polar and non-polar residues; however, only one direct hydrogen bond is observed between ND2Asn-238 and OVal-114. The two domains are structurally related and can be superposed with an r.m.s.d. of 1.6 Å for 126 C-α pairs. The very low sequence identity and similarity (9 and 26 residues, respectively) from the structure-based alignment of the two domains are suggestive of an ancient gene duplication followed by divergence in sequence.
UDP-binding Site
Attempts to obtain a PseGHis6 co-crystal structure with the sugar product alone were unsuccessful. However, co-crystallization experiments with UDP-GlcNAc, UDP-GlcN, or the natural substrate UDP-6-deoxy-AltdiNAc yielded a PseGHis6 complex with bound UDP. Activity measurements in solution of PseGHis6 with UDP-GlcNAc did not yield any significant activity, suggesting that the bound UDP observed in our structures is the result of either low UDP contamination of UDP-sugar preparations or spontaneous hydrolysis of the UDP-sugars either in the reservoir or upon storage (52).
Superposition of the apo- and UDP-bound forms of PseGHis6 shows small but readily detectable domain movements, resulting in partial closure of the two domains upon UDP binding. This domain movement is clearly revealed upon superposition of the two structures, with an overall r.m.s.d. for all C-α atoms of the apo- and UDP-bound forms of 0.90 Å, with the r.m.s.d. decreasing to 0.39 Å when only the 142 residues of their respective N-terminal domains are used in the superposition (Fig. 3B). The UDP- or product-binding site, seen as a tunnel, is situated at the domain interface and stretches across the entire interface (supplemental Fig. S5).
In the PseGHis6-UDP complex (Fig. 3C), UDP binds within a cleft between the two domains, interacting with β2 and the β4-α4 loop of the C-terminal domain and the β1-α1 loop of the N-terminal domain. The conserved, consensus sequence motif identified based on alignment of PseG-related sequences, DX5GXGHX2R, lines one side of the PseG active site cleft and makes contacts with the β-phosphate of UDP. UDP is held mainly through interactions with residues of the C-terminal domain, with fewer contributions from the N-terminal domain. The 2′- and 3′-hydroxyl groups of the ribose are positioned by the side chains of Arg143 and Glu239, along with water-mediated interactions involving the 3′-hydroxyl. The phosphate groups exhibit H-bonds with the main-chain nitrogen atoms of Phe15, Gly16, Gly166, Ser235, and Leu236 along with OHSer-234 (Fig. 3D). The uracil base is positioned within a hydrophobic pocket formed by the side chains of Phe15, Cys163, Ala189, Ile219, Leu236, and the main-chain atoms of Gly165, Gly166, and Thr190. Water-mediated H-bonds are found between N-3, O-2, and O-4 of uracil with the His216 main chain and through additional waters to other residues, including Arg143, Glu145, Ile219, and Glu217.
Molecular Modeling of the PseG-Substrate Complex
Adjacent to the β-phosphate group of UDP, a cavity is observed at the N- and C-terminal domain interface suggestive of a putative binding site for the substrate sugar moiety, 6-deoxy-AltdiNAc. This cavity contains a bound molecule of glycerol in the PseGHis6-UDP complex (Fig. 3D and supplemental Fig. S5B). The glycerol is involved in an intricate network of hydrogen bonding interactions, including direct interactions with the α- and β-phosphates of UDP, direct contacts with Gly16, His17, Tyr78, Ser234, Ser235, and the Arg20–Asp101 salt bridge, along with indirect hydrogen-bond interactions via several buried water molecules. The candidate sugar-binding pocket is also lined by other residues, including Ile13, Gly14, Thr167, Ile169, Ser232, Tyr252, Val253, Asn255, and Gln256. The general location of the PseGHis6-bound glycerol is similar to that of the GlcNAc moiety of UDP-GlcNAc bound to MurG (18).
As efforts to co-crystallize the PseGHis6-substrate complex using either wild-type or mutant enzymes were unsuccessful, we employed protein-ligand docking and molecular dynamics simulations to arrive at a plausible model of PseG-substrate binding. Flexible docking of the sugar part of the UDP-6-deoxy-AltdiNAc substrate was first carried out by MCM conformational sampling (29, 30) of acyclic and cyclic rotatable bonds in the sugar and pyrophosphate parts of the substrate, while maintaining the crystallographically observed binding mode of the uridine part. The three feasible pyranose ring conformations of the 6-deoxy-AltdiNAc sugar moiety, the 1C4 chair, 4C1 chair, and 5S1 twist-boat (supplemental Fig. S3), were incorporated into UDP-6-deoxy-AltdiNAc substrate models and independently subjected to MCM docking to PseG. All three MCM docking simulations converged toward the same lowest energy structure of the complex, corresponding to the twist-boat sugar ring conformation. Compelling evidence for the propensity of the 6-deoxy-AltdiNAc sugar, in the free state, to acquire a twist-boat conformation is the finding that the β-l-altropyranose sugar ring is flexible and found to adopt a twist-boat conformation (53). In addition, as stated above, ab initio structure calculations of OGly-methylated 6-deoxy-AltdiNAc (supplemental Fig. S3) show the 5S1 twist-boat as a low energy conformation along with the 1C4 and 4C1 chairs.
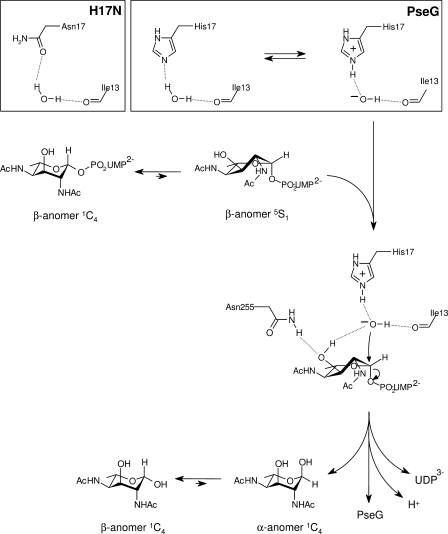
The predicted PseG-substrate binding mode was further investigated by hybrid QM/MM unconstrained molecular dynamics simulations in explicit solvent. Separate simulations were carried out for the substrate in the 1C4 and 5S1 sugar ring conformations observed experimentally in the free state (53). Analysis of the QM/MM MD trajectories indicate that these 1C4 and 5S1 sugar ring conformations are stable in the solvated PseG-binding site, i.e. they did not interconvert or transition to other conformations during the course of the simulations (supplemental Fig. S6A). The most common feature of the PseG-substrate binding modes obtained from these simulations is that the substrate sugar C-1 atom approaches the His17 side chain (supplemental Fig. S6B) with which it interacts via a structured water molecule in both the 1C4 and 5S1 conformations (Fig. 4). This water molecule that appears structured during both QM/MM MD simulations is also present in the PseGHis6-UDP crystal structure (Wat110), and it is anchored via H-bond interactions by the His17NE atom and Ile13 main-chain oxygen atom. Hence, these models predict that a water molecule at this location is the nucleophile that attacks the substrate at the sugar C-1 atom, and that His17 serves a key catalytic role in activating this water molecule.
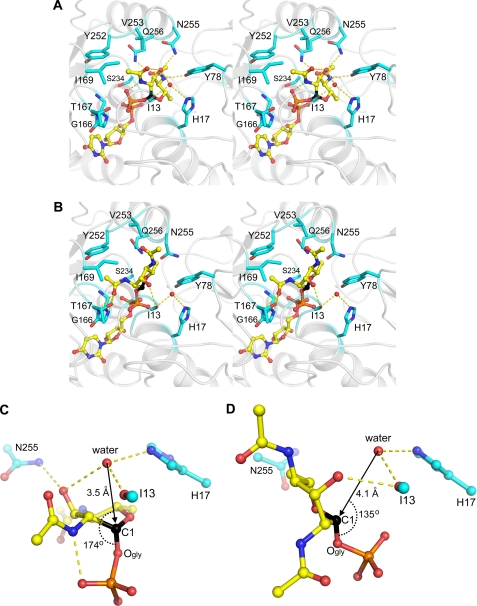
FIGURE 4.
Minimized average structures of the PseG-UDP-6-deoxy-AltdiNAc complexes obtained from QM/MM molecular dynamics simulations (see under “Experimental Procedures” for details). Stereo views of the substrate-binding site show PseG residues (C atoms in cyan) in contact with the sugar moiety of the substrate (C atoms in yellow, anomeric C-1 atom in black) from models corresponding to the 5S1 twist-boat conformation (A) and 1C4 chair conformation of the sugar ring (B). For Ile13, only the main-chain carbonyl group is rendered. Hydrogen bonds are shown as dashed lines, and the putative water nucleophile is shown as a red sphere. Hydrogen atoms are omitted for clarity. Geometric details in the ground state show the principal putative catalytic residues and the position of the putative catalytic water molecule relative to the anomeric scissile bond C-1–OGly of the substrate, from models corresponding to the 5S1 twist-boat conformation (C) and the 1C4 chair conformation of the sugar ring (D). Rendering scheme for C and D is as in A and B.
A closer examination of the QM/MM MD models appears to favor the twist-boat conformation of the sugar as representing the Michaelis complex. The putative catalytic water molecule is more favorably positioned for an Sn2 attack with inversion of configuration at the sugar C-1 atom in the case of the 5S1 conformation than in the 1C4 conformation (Fig. 4, C and D). In the 5S1 conformation, the water molecule is closer to the C-1 atom (3.5 versus 4.1 Å for 1C4) and more in-line with the scissile anomeric C-1–OGly bond (174° versus 135° for 1C4). Such geometrical differences in the ground state would incur a lower energy barrier to the transition state of the hydrolytic reaction in the case of the twist-boat conformation. The QM/MM calculations also indicate a more polarized anomeric C-1–OGly bond in the case of bound 5S1 conformation by 12% (0.11 e), i.e. the C-1 atom is more positively charged and the OGly atom is more negatively charged relative to the 1C4 conformation (supplemental Fig. S6, C and D). Thus, the twist-boat conformation may resemble electronically the transition state of the reaction. The QM/MM MD simulations also indicate that accommodation of the 5S1 conformation is accompanied by smaller perturbations and increased rigidity of the PseG-binding site in comparison with the 1C4 conformation (supplemental Fig. S6, E and F). The 5S1 substrate conformation is also favored by a more negative solvated interaction energy (40, 41) with PseG (by 1.7 kcal/mol) and a lower solvated conformational energy of the complexed PseG protein (by 11 kcal/mol) (supplemental Table S3). In conclusion, although we cannot rule out the possibility of a PseG-substrate-binding mode involving a 1C4 chair conformation, all of our computational data support the likelihood of it involving a twist-boat conformation of the 6-deoxy-AltdiNAc sugar moiety.
Direct intermolecular interactions between PseG and the modeled 5S1 twist-boat conformation of the 6-deoxy-AltdiNAc sugar are detailed in Fig. 4A. The 3-hydroxyl and 4-N-acetyl substituents of the sugar are predicted to form hydrogen bonds with the Tyr78, Asn255, and Gln256 residues of PseG. Substantial packing is achieved between the 4-N-acetyl and C-6-methyl substituents buried in a deep pocket that is partially solvated and partially filled by the glycerol molecule co-crystallized with the PseGHis6-UDP complex (supplemental Fig. S5). This PseGHis6-bound glycerol molecule mimics the interactions established by the 4-N-acetyl and C-6-methyl substituents of the modeled substrate. Direct contacts in this pocket also implicate PseG residues Gly16, His17, Ser234, and the Arg20–Asp101 salt bridge. The methyl group of the more solvent-exposed 2-N-acetyl moiety is predicted to form non-polar contacts in a surface cradle formed by Ile169, Tyr252, and Val253. The 2-N-acetyl substituent also establishes intramolecular hydrogen bonds with the 1-phosphate and 3-hydroxyl substituents. The latter group is also found in proximity (3.4 Å) of the putative catalytic water molecule. In the case of the 1C4 conformation, predicted substrate interactions with the enzyme appear less intimate and less complementary (Fig. 4B), as reflected quantitatively by a less favorable SIE value relative to the 5S1 conformation (supplemental Table S3). The sugar moiety does not fill the pocket occupied by the trapped glycerol molecule in the PseGHis6-UDP complex, which remains filled by several water molecules that connect to a water channel sandwiched between substrate and enzyme (supplemental Fig. S7). In contrast with the bound 5S1 conformation, the 4-N-acetyl of the bound 1C4 conformation is not engaged in direct H-bond contacts with the protein and, together with the C-6-methyl, is partially solvent-exposed. The 2-N-acetyl substituent is buried instead, with the carbonyl oxygen forming H-bonds with the Thr167 hydroxyl and the backbone amides of Gly165 and Gly166, and with the methyl group sandwiched between Gly165 and Tyr252. The 3-hydroxyl substituent forms a hydrogen bond with the main-chain carbonyl of Ile13.
Kinetic Characterization of PseG Mutants
For kinetic analysis, the PseG hydrolase reaction was monitored using a continuous coupled assay for UDP release (21) in a 96-well microplate format. The kinetic constants obtained for PseGHis6 were similar to those reported by Liu and Tanner (11) yielding an apparent kcat of 25 ± 0.73 s−1 and apparent Km of 0.25 ± 0.014 mm (Table 2). Three active site residues were targeted for mutagenesis, based on the PseG-substrate model, to examine their contribution to catalysis. All substitution derivatives tested exhibited Michaelis-Menten kinetics (supplemental Fig. S8), with varying levels of activity, and did not appear to induce gross protein misfolding or degradation as assessed by CD spectroscopy and SDS-PAGE analyses, respectively (supplemental Figs. S1 and S2 and supplemental Table S2). Substitution of His17 to Phe or Leu had the most dramatic effect on turnover, resulting in near nonmeasurable activity, confirming the catalytic importance of this residue (Table 2). In contrast, an H17N substitution retained ∼10% turnover, suggestive of possible functional complementation by the Asn side chain (see Fig. 5). Another important residue highlighted by the model was Asn255, and evidently, an N255A substitution harbored only ∼1% turnover relative to wild type. The last substitution, Y78F, was the least affected kinetically, which may indicate a possible role for the phenyl ring that is conserved here (see below). Finally, PseGHis6 is specific for UDP-6-deoxy-AltdiNAc, in that no activity was observed with UDP-4-amino-4,6-dideoxy-β-l-AltNAc or UDP-2,4-diacetamido-2,4,6-trideoxy-α-d-glucopyranose (or UDP-2,4-diacetamido-bacillosamine). This may be explained by the molecular modeling of the PseG-substrate complex, where the 4-N-acetyl and 6-methyl substituents of UDP-6-deoxy-AltdiNAc in the twist-boat conformation are buried within a deep cavity resulting in substantial packing and intermolecular interactions (Fig. 4 and supplemental Figs. S5 and S6). As such, the positively charged 4N amino group of the former and the C-6 epimer stereochemistry of the latter may preclude a proper induced fit within the active site. We note that the residues altered in this study (His17, Tyr78, and Asn255) are invariant residues within the PseG hydrolase family (supplemental Fig. S4).
TABLE 2.
Apparent kinetic values for PseGHis6 and respective substitution derivatives, calculated using Eadie-Hofstee plots
ND means not determined.
| PseG derivative | Km | kcat |
|---|---|---|
| mm | s−1 | |
| Wild type | 0.25 ± 0.014 | 25 ± 0.73 |
| Y78F | 0.88 ± 0.062 | 8.3 ± 0.42 |
| H17N | 1.3 ± 0.085 | 2.8 ± 0.13 |
| N255A | 0.44 ± 0.043 | 0.27 ± 0.014 |
| H17F | ND | ∼0.005 |
| H17L | ND | ∼0.005 |
FIGURE 5.
Proposed catalytic mechanism for PseG based on modeling of the UDP-sugar complex and activity analysis of substitution derivatives. Here, PseG employs a single displacement mechanism involving C–O bond cleavage via direct attack of the anomeric carbon by a hydroxide nucleophile. Specifically, the residue His17 performs a base-catalyzed attack of water by abstracting a proton. The activated water is anchored with its other proton hydrogen-bonded to the main-chain carbonyl of Ile13, and it is further stabilized by the partially positively charged edge of the aromatic ring of Tyr78 (not shown). Several PseG residues participate in direct intermolecular interactions with the sugar moiety and stabilize the Michaelis complex, including Asn255 that appears as a determinant in selecting the twist-boat conformation already present in the free substrate. This twist-boat conformation of the sugar appears primed for catalysis, as it exposes the C-1 atom and favors attack by the His17-activated water, resulting in inversion of stereochemistry at C-1 and cleavage of the C-1–OGly bond. The liberated sugar product is expected to attain an equilibrium distribution of α and β anomers that interconvert nonenzymatically in solution. The inset shows a proposed scheme for His17 acting as a general base, abstracting a proton from water, or via the H17N substitution H-bonding with the proposed catalytic water molecule.
Implications for the Mechanism of Catalysis
Mechanistic studies of PseG, employing labeling using H218O, are most consistent with a concerted mechanism in which a water molecule attacks C-1 of the UDP-6-deoxy-AltdiNAc sugar, followed by cleavage of the C–O anomeric bond (11). This conclusion is consistent with both (a) incorporation of the 18O label in the sugar product and not the UDP leaving group and (b) the inversion of configuration of the C-1 hydroxyl. This mechanism is distinct from that used by the UDP-N-acetylglucosamine 2-epimerase NeuC (SiaA) from Neisseria meningitidis, which proceeds via formation of the putative intermediate, 2-acetamidoglucal (Fig. 2B) (54). Here an anti-elimination of UDP from the substrate is performed, with the carboxylate moieties of Glu122 and Asp131 important in catalysis, possibly through activation of a water molecule necessary to perform nucleophilic attack at C-1 of the 2-acetamidoglucal intermediate.
From a mechanistic point of view, the reaction catalyzed by PseG is similar to that catalyzed by E. coli GDP-mannose mannosylhydrolase, although, unlike PseG, GDP-mannose mannosylhydrolase is a member of the Nudix family and utilizes a divalent cation in catalysis (16, 55, 56). Both PseG and GDP-mannose mannosylhydrolase, however, participate in C-O bond cleavage with inversion of configuration of C-1 of the sugar. His124 of GDP-mannose mannosylhydrolase has been identified as a base responsible for activating a water molecule, with coordination of the Mg2+ ion to the water contributing to activation (57).
Our hybrid QM/MM molecular dynamics simulations of PseG complexes with the substrate in the 5S1 twist-boat and 1C4 chair conformations point to a water molecule (Wat110 in the PseGHis6-UDP crystal structure) that is hydrogen-bonded to His17 and is also suitably positioned for nucleophilic attack to the anomeric C-1 atom of UDP-6-deoxy-AltdiNAc, particularly in the case of the twist-boat conformation. This water molecule would contact the C-1 atom in-line with the C-1–OGly anomeric bond and is consistent with an Sn2 reaction leading to inversion of configuration at C-1. The putative catalytic water molecule is also hydrogen-bonded to the main-chain carbonyl of Ile13 and further stabilized by the edge of the Tyr78 aromatic ring (Fig. 4, A and B). This may explain the higher turnover observed for the Y78F substitution derivative, with both residues containing an aromatic ring (Table 2).
The catalytic mechanism inferred from our crystallographic, modeling, and kinetic data is shown in Fig. 5. The His17 residue would polarize a water molecule at the described location adjacent to the sugar C-1 atom of the substrate. This polarization would involve an equilibrium between the ionization states OH−-His17+ and water-His17. The hydroxide anion, tethered to His17+ and Ile13, would then attack the sugar C-1 atom of the substrate. In the case of the H17N substitution, the carbonyl oxygen atom of Asn17 would provide a weaker water polarization, as reflected by a 10-fold drop in the measured kcat value (Table 2). A substrate-binding mode with the sugar ring in a twist-boat conformation appears to favor the nucleophilic attack in-line with the C-1–OGly anomeric bond for inversion of configuration at C-1. In contrast, the 1C4 chair conformation displays a less optimal ground state geometry for nucleophilic attack, given the constraints imposed by the binding site geometry and also possibly relating to a reduced access to the C-1 atom. The predicted twist-boat conformation of the sugar ring would be facilitated mainly through interactions with Asn255, which is tethered to the 3-OH substituent of the sugar, in turn poised for hydrogen bonding to the catalytic water molecule, consistent with the mutagenesis data (Table 2). It is therefore possible that the enzyme takes advantage of the relatively stable twist-boat sugar ring conformation of 6-deoxy-AltdiNAc to facilitate the attack. Recent crystallographic and theoretical studies have shown substrate binding in twist-boat conformations in several systems, notably retaining and inverting β-glycosidases (see Refs. 58, 59 and references therein). This nucleophilic attack promotes the Sn2-like displacement of UDP to generate the C-1-inverted α anomer of the 6-deoxy-AltdiNAc sugar. It is expected that the α to β conversion of 6-deoxy-AltdiNAc would take place nonenzymatically in solution (11).
PseG-MurG Comparative Structural Analysis and Insight into the Mechanism of MurG Glycosyltransferases
Within SCOP (60), PseG would be classified within the UDP-glucosyltransferase/glycogen phosphorylase fold (61), characterized by two dissimilar α/β domains that share similar central, parallel β-sheets. This fold currently includes nine families of carbohydrate-active enzymes, including several glycosyltransferases (18, 62–67), sugar phosphorylases (68, 69), and a UDP-sugar epimerase (70). Although there is a lack of significant sequence similarity between the two, C. jejuni PseG and MurG from E. coli (PDB codes 1F0K and 1NLM (18, 71)) bear striking structural resemblance. MurG is a membrane-associated UDP-GlcNAc:lipid I glycosyltransferase involved in peptidoglycan biosynthesis and is currently the most structurally similar protein to PseG. Relative to C. jejuni PseG, E. coli MurG contains some additional insertions around the central β-sheets, as well as a C-terminal α-helical extension (supplemental Fig. S4). The best structural alignment between PseG-UDP and molecule A of MurG bound to UDP-GlcNAc (PDB code 1NLM (18)), carried out using Swiss PDB Viewer (72), gives an r.m.s.d. of ∼1.6 Å for 146 common C-α atoms, the vast majority of which reside in the N-terminal domain (supplemental Fig. S9A). According to this structural alignment, the putative catalytic base His17 of PseG is structurally conserved in MurG (His19), whereas there is a clear translation of the bound UDP moiety between PseG and MurG. An alternative fit of the two enzymes carried out on the bound UDP moiety and including select C-αs from the C-terminal β-sheet results in a displacement of the N-terminal domains together with a misalignment of the conserved putative catalytic histidine (supplemental Fig. S9).
One characteristic observed in all GT-B transferases is substrate-induced domain movement. The shift observed in PseGHis6 upon binding of UDP relative to the apo-structure is smaller (∼1.4 Å) in comparison with MurG (∼3.4 Å). It is possible that a sulfate ion situated near the domain interface in the apo-PseGHis6 structure, which coincides with the β-phosphate position of UDP in the PseGHis6-UDP complex, serves to hold the two domains together via interactions with loop regions of both the domains. However, when comparing the ligand-bound structures of PseG and MurG, it is evident that the two domains are arranged into a more open conformation in the case of the substrate-bound MurG structure relative to the UDP-bound (or the substrate-bound) PseG structure (supplemental Fig. S9). This may be a consequence of the differing size of acceptor nucleophiles utilized by these two enzymes.
Despite different inter-domain openings leading to a translation of the bound UDP moiety relative to the aligned N-terminal domains, a combined structure-based alignment between PseG and MurG followed by alignment of related sequences showed conservation between several invariant residues across known PseG homologs and residues near the substrate-binding site of E. coli MurG (supplemental Fig. S4). These residues in PseG(MurG) include Gly16(Gly18), His17(His19), and Gln256(Gln289) in the putative sugar-binding site, Glu239(Glu269) involved in anchoring the ribose moiety, and Gly165(Gly191) involved in anchoring the β-phosphate. The E269D mutation in MurG has a significant impact on substrate binding with little impact on turnover, indicating that binding of the ribose moiety is critical for MurG function (18, 73). In addition, although there is structural conservation of side chains between the PseG and E. coli MurG residues Asn255(Gln288) (supplemental Fig. S4), this conservation is not observed for the MurG family in general (71). This may be expected as our study suggests Asn255 is involved in stabilizing the catalytic twist-boat conformation of the PseG substrate, a substrate conformation that is not adopted in the MurG reaction (18). Moreover, the Tyr78 residue altered in this study is not found to be conserved in E. coli MurG. However, Asn128 of MurG appears to occupy a similar space in structural comparisons where, like Tyr78 of PseG, it interacts with the 4-substituent of the donor sugar. Coincidentally, an N128A MurG substitution was found to greatly affect turnover (18). This agrees with our mechanistic model, whereby these residues may additionally facilitate the coordination of the acceptor nucleophile. As such, their differences may not only be due to different donors but perhaps more importantly to the unique acceptors, respectively.
The kinetic mechanism of MurG has been investigated, and it is found to follow a compulsory Bi-Bi mechanism, with binding of the donor UDP-sugar prior to the lipid-linked N-acetylmuramic acid (lipid I) (74), although the identity of the catalytic base that activates the C-4 hydroxyl moiety of the acceptor substrate remains unknown (Fig. 2C). The significant impact of the H19A substitution on MurG activity, which lowers the kcat by more than 3 orders of magnitude (18), prompted recent speculations that it may represent the catalytic base of MurG (75). However, His19 is located remotely from the donor sugar, 9.3 Å across the inter-domain cleft to the anomeric bond. Our mutagenesis and structural data, supporting a catalytic role for His17 as a general base in the PseG inverting glycoside hydrolase, strengthen the putative catalytic role of the structurally conserved His19 in the MurG inverting glycosyltransferase.
Supplementary Material
Acknowledgments
We thank Alexei Soares and Howard Robinson for assistance in synchrotron x-ray data collection. We also thank Annie Aubry, Anna Burianova, and Pernilla Berin for technical assistance in the preparation of site-directed mutants; Eunice Ajamian for SEC analysis; and Drs. Jean-Robert Brisson, Martin Tanner, and Warren Wakarchuk for helpful discussions. Data for this study were measured at beamlines X12B and X29 of the National Synchrotron Light Source, Brookhaven National Laboratory. Financial support for the National Synchrotron Light Source, Brookhaven National Laboratory, comes principally from the Office of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and the National Institutes of Health, NCRR.
This work was supported by the National Research Council Canada (to S. M. L., E. O. P., and M. C.) and Canadian Institutes of Health Research Operating Grant GSP-48370 (to M. C.). This is publication number NRC49599 from the National Research Council Canada.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, Figs. S1–S9, and additional references.
The atomic coordinates and structure factors (codes 3HBM and 3HBN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Pse
- pseudaminic acid, 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-nonulosonic acid
- 6-deoxy-AltdiNAc
- 2,4-diacetamido-2,4,6-trideoxy-l-altropyranose
- DTT
- dithiothreitol
- GT
- glycosyltransferase
- MCM
- Monte Carlo minimization
- MD
- molecular dynamics
- r.m.s.d.
- root mean square deviation
- sialic acid
- 5-acetamido-3,5-dideoxy-d-glycero-d-galacto-nonulosonic acid
- SIE
- solvated interaction energy
- UDP-6-deoxy-AltdiNAc
- UDP-2,4-diacetamido-2,4,6-trideoxy-β-l-altropyranose
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SeMet
- selenomethionine
- QM
- quantum mechanics
- MM
- molecular mechanics
- PDB
- Protein Data Bank.
REFERENCES
- 1.Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 2.Schirm M., Soo E. C., Aubry A. J., Austin J., Thibault P., Logan S. M. (2003) Mol. Microbiol. 48, 1579–1592 [DOI] [PubMed] [Google Scholar]
- 3.Guerry P., Ewing C. P., Schirm M., Lorenzo M., Kelly J., Pattarini D., Majam G., Thibault P., Logan S. (2006) Mol. Microbiol. 60, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knirel Y. A., Shashkov A. S., Tsvetkov Y. E., Jansson P. E., Zãhringer U. (2003) Adv. Carbohydr. Chem. Biochem. 58, 371–417 [DOI] [PubMed] [Google Scholar]
- 5.Hsu K. L., Pilobello K. T., Mahal L. K. (2006) Nat. Chem. Biol. 2, 153–157 [DOI] [PubMed] [Google Scholar]
- 6.Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black R. E., Levine M. M., Clements M. L., Hughes T. P., Blaser M. J. (1988) J. Infect. Dis. 157, 472–479 [DOI] [PubMed] [Google Scholar]
- 8.Josenhans C., Vossebein L., Friedrich S., Suerbaum S. (2002) FEMS Microbiol. Lett. 210, 165–172 [DOI] [PubMed] [Google Scholar]
- 9.Megraud F. (2007) Gut 56, 1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou W. K., Dick S., Wakarchuk W. W., Tanner M. E. (2005) J. Biol. Chem. 280, 35922–35928 [DOI] [PubMed] [Google Scholar]
- 11.Liu F., Tanner M. E. (2006) J. Biol. Chem. 281, 20902–20909 [DOI] [PubMed] [Google Scholar]
- 12.Schoenhofen I. C., Lunin V. V., Julien J. P., Li Y., Ajamian E., Matte A., Cygler M., Brisson J. R., Aubry A., Logan S. M., Bhatia S., Wakarchuk W. W., Young N. M. (2006) J. Biol. Chem. 281, 8907–8916 [DOI] [PubMed] [Google Scholar]
- 13.Schoenhofen I. C., McNally D. J., Brisson J. R., Logan S. M. (2006) Glycobiology 16, 8C–14C [DOI] [PubMed] [Google Scholar]
- 14.Schoenhofen I. C., McNally D. J., Vinogradov E., Whitfield D., Young N. M., Dick S., Wakarchuk W. W., Brisson J. R., Logan S. M. (2006) J. Biol. Chem. 281, 723–732 [DOI] [PubMed] [Google Scholar]
- 15.Ishiyama N., Creuzenet C., Miller W. L., Demendi M., Anderson E. M., Harauz G., Lam J. S., Berghuis A. M. (2006) J. Biol. Chem. 281, 24489–24495 [DOI] [PubMed] [Google Scholar]
- 16.Legler P. M., Massiah M. A., Bessman M. J., Mildvan A. S. (2000) Biochemistry 39, 8603–8608 [DOI] [PubMed] [Google Scholar]
- 17.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 18.Hu Y., Chen L., Ha S., Gross B., Falcone B., Walker D., Mokhtarzadeh M., Walker S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) EMBO. J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 21.Gosselin S., Alhussaini M., Streiff M. B., Takabayashi K., Palcic M. M. (1994) Anal. Biochem. 220, 92–97 [DOI] [PubMed] [Google Scholar]
- 22.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 23.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 24.Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 27.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon J. C., Myers J. B., Folta T., Shoja V., Heath L. S., Onufriev A. (2005) Nucleic Acids Res. 33, W368–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Scheraga H. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6611–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangarajan E. S., Ruane K. M., Sulea T., Watson D. C., Proteau A., Leclerc S., Cygler M., Matte A., Young N. M. (2008) Biochemistry 47, 1827–1836 [DOI] [PubMed] [Google Scholar]
- 31.Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. (1995) J. Am. Chem. Soc. 117, 5179–5197 [Google Scholar]
- 32.Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. (2004) J. Comput. Chem. 25, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 33.Jakalian A., Bush B. L., Jack D. B., Bayly C. I. (2000) J. Comput. Chem. 21, 132–146 [Google Scholar]
- 34.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 36.Darden T., York D., Pedersen L. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 37.Ryckaert J. P., Ciccotti G., Berendsen H. J. C. (1977) J. Comput. Phys. 23, 327–341 [Google Scholar]
- 38.Walker R. C., Crowley M. F., Case D. A. (2008) J. Comput. Chem. 29, 1019–1031 [DOI] [PubMed] [Google Scholar]
- 39.Stewart J. J. P. (1989) J. Comput. Chem. 10, 209–220 [Google Scholar]
- 40.Cui Q., Sulea T., Schrag J. D., Munger C., Hung M. N., Naïm M., Cygler M., Purisima E. O. (2008) J. Mol. Biol. 379, 787–802 [DOI] [PubMed] [Google Scholar]
- 41.Naïm M., Bhat S., Rankin K. N., Dennis S., Chowdhury S. F., Siddiqi I., Drabik P., Sulea T., Bayly C. I., Jakalian A., Purisima E. O. (2007) J. Chem. Inf. Model 47, 122–133 [DOI] [PubMed] [Google Scholar]
- 42.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gryllos I., Shaw J. G., Gavín R., Merino S., Tomás J. M. (2001) Infect. Immun. 69, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canals R., Vilches S., Wilhelms M., Shaw J. G., Merino S., Tomás J. M. (2007) Microbiology 153, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 45.Schirm M., Schoenhofen I. C., Logan S. M., Waldron K. C., Thibault P. (2005) Anal. Chem. 77, 7774–7782 [DOI] [PubMed] [Google Scholar]
- 46.Kiss E., Kereszt A., Barta F., Stephens S., Reuhs B. L., Kondorosi A., Putnoky P. (2001) Mol. Plant-Microbe Interact. 14, 1395–1403 [DOI] [PubMed] [Google Scholar]
- 47.Le Quéré A. J., Deakin W. J., Schmeisser C., Carlson R. W., Streit W. R., Broughton W. J., Forsberg L. S. (2006) J. Biol. Chem. 281, 28981–28992 [DOI] [PubMed] [Google Scholar]
- 48.Raymond C. K., Sims E. H., Kas A., Spencer D. H., Kutyavin T. V., Ivey R. G., Zhou Y., Kaul R., Clendenning J. B., Olson M. V. (2002) J. Bacteriol. 184, 3614–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knirel Y. A., Vinogradov E. V., L'vov V. L., Kocharova N. A., Shashkov A. S., Dmitriev B. A., Kochetkov N. K. (1984) Carbohydr. Res. 133, C5–C8 [DOI] [PubMed] [Google Scholar]
- 50.Castric P., Cassels F. J., Carlson R. W. (2001) J. Biol. Chem. 276, 26479–26485 [DOI] [PubMed] [Google Scholar]
- 51.Edgar R. C. (2004) BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vetting M. W., Frantom P. A., Blanchard J. S. (2008) J. Biol. Chem. 283, 15834–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally D. J., Schoenhofen I. C., Mulrooney E. F., Whitfield D. M., Vinogradov E., Lam J. S., Logan S. M., Brisson J. R. (2006) ChemBioChem. 7, 1865–1868 [DOI] [PubMed] [Google Scholar]
- 54.Murkin A. S., Chou W. K., Wakarchuk W. W., Tanner M. E. (2004) Biochemistry 43, 14290–14298 [DOI] [PubMed] [Google Scholar]
- 55.Legler P. M., Massiah M. A., Mildvan A. S. (2002) Biochemistry 41, 10834–10848 [DOI] [PubMed] [Google Scholar]
- 56.Legler P. M., Lee H. C., Peisach J., Mildvan A. S. (2002) Biochemistry 41, 4655–4668 [DOI] [PubMed] [Google Scholar]
- 57.Gabelli S. B., Azurmendi H. F., Bianchet M. A., Amzel L. M., Mildvan A. S. (2006) Biochemistry 45, 11290–11303 [DOI] [PubMed] [Google Scholar]
- 58.Davies G. J., Ducros V. M., Varrot A., Zechel D. L. (2003) Biochem. Soc. Trans. 31, 523–527 [DOI] [PubMed] [Google Scholar]
- 59.Biarnés X., Nieto J., Planas A., Rovira C. (2006) J. Biol. Chem. 281, 1432–1441 [DOI] [PubMed] [Google Scholar]
- 60.Andreeva A., Howorth D., Chandonia J. M., Brenner S. E., Hubbard T. J., Chothia C., Murzin A. G. (2008) Nucleic Acids Res. 36, D419–D425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrabl J. O., Grishin N. V. (2001) J. Mol. Biol. 314, 365–374 [DOI] [PubMed] [Google Scholar]
- 62.Larivière L., Gueguen-Chaignon V., Moréra S. (2003) J. Mol. Biol. 330, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 63.Mulichak A. M., Losey H. C., Walsh C. T., Garavito R. M. (2001) Structure 9, 547–557 [DOI] [PubMed] [Google Scholar]
- 64.Gibson R. P., Turkenburg J. P., Charnock S. J., Lloyd R., Davies G. J. (2002) Chem. Biol. 9, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 65.Buschiazzo A., Ugalde J. E., Guerin M. E., Shepard W., Ugalde R. A., Alzari P. M. (2004) EMBO J. 23, 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horcajada C., Guinovart J. J., Fita I., Ferrer J. C. (2006) J. Biol. Chem. 281, 2923–2931 [DOI] [PubMed] [Google Scholar]
- 67.Ni L., Chokhawala H. A., Cao H., Henning R., Ng L., Huang S., Yu H., Chen X., Fisher A. J. (2007) Biochemistry 46, 6288–6298 [DOI] [PubMed] [Google Scholar]
- 68.Lin K., Rath V. L., Dai S. C., Fletterick R. J., Hwang P. K. (1996) Science 273, 1539–1542 [DOI] [PubMed] [Google Scholar]
- 69.Geremia S., Campagnolo M., Schinzel R., Johnson L. N. (2002) J. Mol. Biol. 322, 413–423 [DOI] [PubMed] [Google Scholar]
- 70.Campbell R. E., Mosimann S. C., Tanner M. E., Strynadka N. C. (2000) Biochemistry 39, 14993–15001 [DOI] [PubMed] [Google Scholar]
- 71.Ha S., Walker D., Shi Y., Walker S. (2000) Protein Sci. 9, 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 73.Crouvoisier M., Auger G., Blanot D., Mengin-Lecreulx D. (2007) Biochimie 89, 1498–1508 [DOI] [PubMed] [Google Scholar]
- 74.Chen L., Men H., Ha S., Ye X. Y., Brunner L., Hu Y., Walker S. (2002) Biochemistry 41, 6824–6833 [DOI] [PubMed] [Google Scholar]
- 75.Bouhss A., Trunkfield A. E., Bugg T. D., Mengin-Lecreulx D. (2008) FEMS Microbiol. Rev. 32, 208–233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.