Abstract
Krüppel-like factor 6 (Klf6) belongs to a family of zinc finger transcription factors known to play a role in development and tumor suppression. Although Klf6 is highly mutated in prostate cancer, its function in prostate development is unknown. We have generated a prostate-specific Klf6-deficient mouse model and report here a novel role for Klf6 in the regulation of prostate branching morphogenesis. Importantly, our study reveals a novel relationship between Klf6 and the Shh pathway. Klf6-deficiency leads to elevated levels of hedgehog pathway components (Shh, Ptc, and Gli) and loss of their localized expression, which in turn causes impaired lateral branching.
Klf6 belongs to the family of Krüppel-like zinc finger transcription factors that regulate cell proliferation and differentiation (1). All members of the Klf gene family contain a highly conserved zinc finger DNA binding domain at their C terminus and an activation domain at its N terminus, distinct to each Klf gene and accounting for their wide-ranging biological capabilities (2–4). Similar to other members of the Klf family (5, 6), Klf6 is reported to act as a tumor suppressor (7–10). Klf6 has been reported to be mutated in a large percentage of human prostate tumors (7), but its function during normal development has not been elucidated.
Prostate development, specifically epithelial branching morphogenesis, is a well studied process. Outgrowth and branching of the prostate epithelial buds into the enveloping mesenchyme occur during the first 3 weeks postnatally (11). The fully developed prostate in an adult mouse is composed of three different paired lobes, referred as the anterior, ventral, and dorsal-lateral lobes. In addition, the number of main ducts and complexity of ductal branching vary among the three lobes. A number of growth factors/pathways have been implicated in regulating prostatic epithelial proliferation and differentiation, including hedgehog (Hh),4 bone morphogenic proteins (BMPs), fibroblastic growth factors (FGFs), and Notch and Wnt pathways (12–19). For example, work done in our laboratory and others showed that sonic hedgehog (Shh) produced in the prostatic epithelium is a negative regulator of prostatic branching morphogenesis, mediated indirectly by periepithelial mesenchymal cells (12–15). In addition, while mesenchymal FGF10 stimulates prostatic epithelial growth (16), BMP4 is a mesenchymal factor that inhibits prostatic ductal budding and branching morphogenesis (17).
All Klf knockouts generated thus far are embryonic lethal (20–22), including the Klf6 knock-out mice (23). Because prostate development begins in late gestation and continues postnatally, we overcame embryonic lethality to study the role of Klf6 in prostate development and tumorigenesis by generating a prostate-specific deletion of the Klf6 gene using a Cre-lox recombination approach. We report here a novel role for Klf6 in the regulation of Shh-mediated epithelial branching morphogenesis in the anterior prostate.
MATERIALS AND METHODS
Generation, Genotyping, and Reverse Transcription-PCR of Klf6 Prostate-specific Mutant Mice
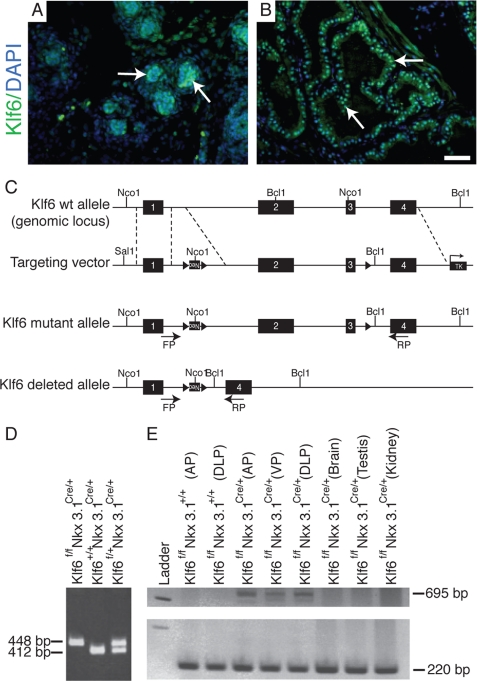
We generated prostate-specific deletion of Klf6 exons 2 and 3 by crossing Klf6f/f mice (C57BL/6, 129Sv) with Nkx3.1Cre/+ (C57BL/6, 129Sv) knock-in mice, which expressed Cre recombinase from the Nkx3.1 locus.5 The Nkx3.1Cre/+ knock-in mice used in the crosses were phenotypically identical to the published Nkx3.1 mutant mice (25) and, thus, exhibited normal prostate development. PIN lesions were detected in Nkx3.1Cre/+ mice only after they have been aged beyond 1 year.6 Data presented in this report were mostly analyzed at P5, and the latest time point presented is 1 year of age, where loss of one Nkx3.1 allele did not exert any abnormal effects on prostate development. Genotyping of Klf6 mutants was done by Southern analysis for the F0 and F1 generations (data not shown), and subsequent generations were genotyped by PCR analysis using the forward primer, 5′-GTC TCT TGA CAC CTT GAC TAT CTC TCC-3′, and the reverse primer, 5′-CTA CAG GAT TCG TCC CTC TGC-3′, from genomic DNA obtained from tail biopsies. We prepared RNA from TRIzol (Invitrogen) extractions from various prostate lobes, brain, testes, and kidney tissues. The RNA was then used for reverse transcription-PCR to confirm Klf6 deletion through detection of a 695-bp PCR product using forward primer 5′-GAA TAC TCT TGG AGT GCT AGG and reverse primer 5′-CTG CTC CTT CAG AGG TGC. RNA loading control was performed by employing a housekeeping gene called T-cell receptor delta chain (TCRD). TCRD was employed to ensure that equal RNA was loaded into each lane and was detected using forward primer 5′-CAA ATG TTG CTT GTC TGG TG and reverse primer 5′-GTC AGT CGA GTG CAC AGT TT. In addition, the Ptc-lacZ reporter mice, also known as Ptch1D11 (26), were obtained from Cruris Inc. They were intercrossed with Nkx3.1Cre;Klf6fl/fl to produce the experimental cohorts. Mice were genotyped by PCR.
Prostate Microdissection, ex Vivo Three-dimensional Ultrasound, and ex Vivo Micro-CT
Prostates were harvested and treated with 1% collagenase (Sigma) for 15 min at 37 °C. Individual ducts were teased apart using forceps (11). A Leica Z6 APO microscope was used to acquire images of the microdissected prostates. Tissues were fixed in 4% paraformaldehyde. High resolution ultrasound imaging was employed to evaluate the interior of anterior prostate. The fixed tissue samples were placed in a bath of phosphate-buffered saline to provide acoustic coupling for ex vivo three-dimensional ultrasound micro-imaging. Ultrasound imaging was performed with a high resolution dedicated small animal ultrasound imaging system (Vevo 770, VisualSonics, Toronto, Canada). Images were acquired with a single-element (mechanical sweep) ultrasound transducer operating at a center frequency of 55 MHz and a 4.5 mm focal length, resulting in an axial resolution of 30 μm and a lateral resolution of 50 μm. Three-dimensional acquisition was performed by the linear translation of the transducer that was controlled by a motorized drive mechanism, where images were acquired every 50 μm to provide a three-dimensional field-of-view of 7 × 7 × 10 mm. Images were processed with image analysis software provide with the Vevo 770 imaging.
Micro-CT was used to provide a quantitative analysis of the number of ductal tips and branch points. Microdissected tissues were soaked in 5% Isovue®/phosphate-buffered saline contrast agent for 2 h, blotted onto paper, and then suspended in soybean oil, which provides homogeneous and low x-ray absorption background for ex vivo micro-CT imaging. Samples were then imaged with the Scanco μCT40 (Bassendorf, Switzerland) micro-CT system at an isotropic voxel size of 12 μm. Images were acquired with the x-ray tube operating at an energy level of 45 kV and a current of 177 microamperes with an integration time of 300 ms. Acquired images were then analyzed using image analysis software from AnalyzeDirect (Lenexa, KS).
Histologic, Immunofluorescent Staining, and in Situ Hybridization Methods
We harvested prostate tissues from various genotypes and stages of development. Tissues for hematoxylin and eosin (H&E) histological staining purposes were formalin-fixed and paraffin embedded. Tissues were then sectioned and stained with H&E. Tissues for immunofluorescent staining were fixed in 4% paraformaldehyde/phosphate-buffered saline prior to Declere antigen retrieval (Cell Marque) using a pressure cooker for 10 min and then blocked in 10% donkey serum/phosphate-buffered saline. Primary antibodies used include rabbit anti-Klf6 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Ki67 (LabVision), rabbit anti-β-galactosidase (1:10,000, MP Biomedical, without antigen retrieval) (19), rabbit anti-androgen receptor (Upstate), rabbit anti-smooth muscle actin (LabVision), rabbit anti-desmin (EuroDiagnostica), rabbit anti-ck14 (Covance), mouse anti-ck8 (Novus), rabbit anti-phospho-Smad1/5/8 (1:500, Cell Signaling), and mouse anti-E-cadherin (BD Pharmingen). Secondary antibodies used to detect respective primary antibodies were anti-rabbit Alexa 488 (Invitrogen) and anti-mouse Alexa 594 (Invitrogen). Respective IgG controls were used on serial sections placed on each slide. Sections were mounted using Fluoromount containing 4′,6-diamidino-2-phenylindole stain (Dako).
Prostate tissues for non-radioactive in situ hybridization were harvested and fixed in 4% paraformaldehyde prior to embedding in an OCT compound and freezing on dry ice. Pretreatment of frozen tissue sections included proteinase K digestion (40 μg/ml, 6 min, and 25 °C) and acetylation (0.1 m triethanolamine, pH 8.0, 0.25% acetic anhydride, 10 min, 25 °C). Sections were hybridized with Riboprobes against Shh, Ptc, and Gli1 at 60 °C overnight, washed sequentially with 2× SSC, 1× SSC, 0.5× SSC, and blocked with 1× Digoxygenin-Block (Roche Applied Science) for 1 h before hybridization with alkaline phosphatase conjugated anti-Dig antibody (Roche Applied Science) for 1 h at room temperature.
Radioactive in situ hybridization was performed as previously described (27). PCR primers were designed to amplify a 415-bp fragment of mouse BMP4 spanning from nucleotides 948 to 1362 of mouse BMP4 reference sequence NM_007554.2 (upper, 5′-CAGGGCTTCCACCGTATAAAC-3′; lower, 5′-AATGGCGACGGCAGTT-3′). Primers included extensions encoding 27-nucletide T7 or T3 RNA polymerase initiation sites to allow in vitro transcription of sense or antisense probes, respectively, from the amplified products.
Formalin-fixed, paraffin-embedded sections 5-μm thick were deparaffinized, deproteinated in 10 μg/ml Proteinase K for 30 min at 37 °C, and further processed for in situ hybridization as previously described (Jubb et al. (27)). [33P]UTP-labeled sense and antisense probes were hybridized to the sections at 55 °C overnight. Unhybridized probe was removed by incubation in 20 μg/ml RNase A for 30 min at 37 °C, followed by a high stringency wash at 55 °C in 0.1 × SSC for 2 h and dehydration through graded ethanols. The slides were dipped in NTB nuclear track emulsion (Eastman Kodak), exposed in sealed plastic slide boxes containing desiccant for 4 weeks at 4 °C, developed, and counterstained with H&E. Images were analyzed in Metamorph (version 7.0r4, MDS Analytical, Sunnyvale, CA). Briefly, regions of interest (epithelial ductal units) were created manually to restrict analysis to the lumen. A threshold value was applied for each set of images (∼95% of maximum pixel value) to identify positively stained areas. The total area of each region and the positive stained area for each region were outputted directly to Excel (Microsoft, Redmond, WA). A value of 10.3 pixels per silver grain was used to calculate the total number of silver grains associated with the positively stained area.
RESULTS
Generation of Mice with Prostate-specific Deletion of Klf6
To understand the developmental role of Klf6 in the prostate, we first defined the spatial and temporal expression pattern of Klf6. Anti-Klf6 immunofluorescent staining revealed that Klf6 expression was highly expressed in the nuclei of prostate epithelium as early as postnatal day 1 (P1) (data not shown), P4 (arrow, Fig. 1A), and mature prostate (arrow, Fig. 1B). The specificity of the anti-Klf6 antibody was confirmed by the lack of immunofluorescent staining in the nuclei of cells in which the Klf6 were deleted (see below, Fig. 2, N and P).
FIGURE 1.
Klf6 expression and generation of mutant mice with prostate-specific Klf6 deletion. A and B, immunofluorescent staining of P4 and adult prostate, respectively, with anti-Klf6 revealed that nuclear Klf6 was expressed exclusively in the epithelial cells at both stages of development (arrow, A and B). Stromal staining was nonspecific and present in the anti-rabbit IgG control. C, a schematic drawing of the Klf6 wildtype (wt) genomic locus, targeting vector, Klf6 floxed mutant allele (Klf6f/f) and Klf6 deleted allele upon Nkx3.1-driven Cre recombinase-mediated deletion. The targeting vector was generated by inserting two loxP sites flanking a neo cassette between exons 1 and 2 and a loxP site just upstream of exon 3. Cre recombination would result in the excision of exons 2 and 3. D, genotyping of Klf6 mutant mice by PCR analysis. PCR of genomic DNA from wild-type mice would yield a 412-bp band, and the Klf6f/f mice would yield a 448-bp PCR product due to the presence of a loxP site. E, reverse transcription-PCR analysis of mRNA obtained from anterior (AP), dorsolateral (DLP), and ventral (VP) prostate of Klf6f/f mice with and without Cre-recombinase and from the brain, testis, and kidney of Klf6f/fNkx3.1Cre/+ tissues. Deletion of exons 2 and 3 from Klf6 would yield a 695-bp PCR product with the forward and reverse primer pair and equal loading was observed in all lanes through detection of a 220-bp PCR product of the TCRD gene. Bar, 50 μm for A and B.
FIGURE 2.
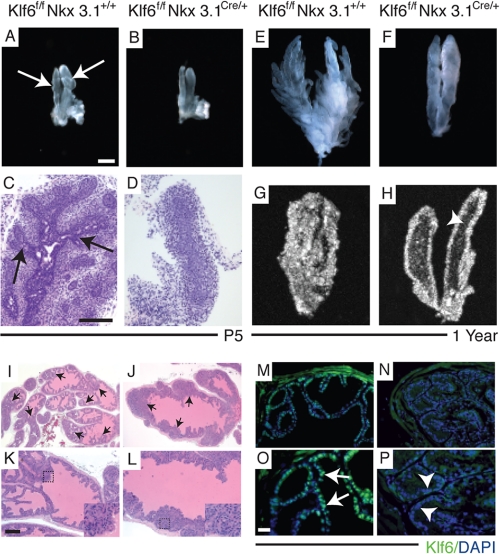
Abnormality in ductal branching in the Klf6-deficient prostate epithelium. Morphological ductal aberrations in Klf6-deficient anterior prostates were first observed at P5 and retained in adult mice. Light microscopy images (A, B, E, and F) and H&E staining (C and D) of Klff/fNkx3.1+/+ (A, C, and E) and Klff/fNkx3.1Cre/+ (B, D, and F) from P5 and 1-year-old anterior prostates that were microdissected prior to imaging. Only one side of the symmetrical anterior lobe was imaged, and three mice were imaged from each stage. Branching was visible in both microdissected (arrow, A) and H&E-stained (arrow, C) P5 sections from Klff/fNkx3.1+/+ but not in Klff/fNkx3.1Cre/+ (B and D). Ex vivo ultrasound imaging allowed visualization of the interior of anterior prostate of Klff/fNkx3.1+/+ (G) that appears dense and white due to extensive epithelial infolding in the ducts, and the single layer of while epithelial lumen that outlines the main ducts of Klff/fNkx3.1Cre/+ mice (H). Arrowhead (in H) points to the significantly enhanced luminal space present within the main duct of Klff/fNkx3.1Cre/+ anterior prostate. H&E staining of prostates obtained from 6-month-old (I and K) Klff/fNkx3.1+/+ and (J and L) Klff/fNkx3.1Cre/+ mice. Note that the widest region at the base of Klff/fNkx3.1+/+ prostate (K) fits into the field of view, whereas the narrowest region of Klff/fNkx3.1Cre/+ prostate (L) does not fit into the field of view. Epithelial infolding that spans the lumen was more extensive in Klff/fNkx3.1+/+ prostates (arrow, I) than Klff/fNkx3.1Cre/+ prostates (arrow, J). Insets represent higher magnification of the regions marked by the dotted square in K and L, respectively, that revealed nuclei with normal characteristics. Immunofluorescent staining of Klf6 expression in prostate tissues obtained from 8-week-old Klff/fNkx3.1+/+ (M and O) and Klff/fNkx3.1Cre/+ (N and P) mice. Klf6, which is a transcription factor, was highly localized to the nuclei of Klff/fNkx3.1+/+ prostate epithelium (arrows, O), and depletion of Klf6 in the Klff/fNkx3.1Cre/+ prostate was confirmed by the absence of green signal in the nuclei (arrowhead, P). I, J, M, and N, low magnification and high magnification (K, L, O, and P) of respective images. Bar in A, 500 μm for A and B, 1000 μm for E and F, and 750 μm for G and H. Bar in C, 50 μm for C and D. Bar in K, 200 μm for I and J and 100 μm for K–N. Bar in O, 50 μm for O and P.
To generate a prostate-specific deletion of exons 2 and 3 that encompass all three zinc fingers, which inactivates Klf6 transcription factor activity (Fig. 1C), we crossed Klf6 conditional mutant mice containing loxP sites flanking exons 2 and 3 (Klf6f/f) with Nkx3.1Cre/+ knock-in mice in which the Nkx3.1 promoter directs expression of Cre-recombinase to the prostate epithelium.5 To demonstrate prostate-specific deletion of exons 2 and 3 from Klf6, we performed reverse transcription-PCR analysis on mRNA from various genotypes and tissues. A 695-bp PCR product, amplified when exons 2 and 3 were deleted, could only be detected in Klf6f/fNkx3.1 Cre/+ prostate but not in brain, testes, and kidney or Klf6f/fNkx3.1+/+ prostate. The highest level of Cre-mediated deletion of Klf6 was detected in the anterior prostate (Fig. 1E).
Branching and Canalization Defects in Klf6-deficient Prostate
Progression of prostate branching morphogenesis varies from lobe to lobe. Main ducts are present in all lobes at birth, but lateral branching in the ventral prostates begins at birth (P1), the anterior prostate at P5, and the dorsolateral prostate by P10 (11, 28). Gross examination of all three lobes of Klf6f/fNkx3.1Cre/+ prostates revealed a prominent branching phenotype primarily in the anterior prostate, which was consistent with the highest level of Klf6 depletion. The branching defect in the Klf6f/fNkx3.1Cre/+ anterior prostates was evident as early as postnatal day 5 (P5) (Fig. 2, B and D). Impaired branching was 100% penetrant in the Klf6f/fNkx3.1Cre/+ mice, whereas all Klf6f/fNkx3.1+/+, Klf6f/+Nkx3.1Cre/+, and Klf6+/+Nkx3.1Cre/+ prostates developed normally. To rule out a possibility of a delay in secondary branching, we examined Klf6f/fNkx3.1Cre/+ prostates from 6-month-old (data not shown) and 1-year-old (Fig. 2F) mice and found that those mature prostates were still without secondary branches as first noted at P5. This was in contrast to the extensive secondary and tertiary branching observed in 1-year-old Klf6f/fNkx3.1+/+ mice (Fig. 2E). Distinguishing branching features in the dorsal and ventral prostates were not as apparent between Klf6f/fNkx3.1+/+ and Klf6f/fNkx3.1Cre/+ except for larger luminal spaces in the ducts (supplemental Fig. S1, A–D). This milder phenotype could be due to less efficient Klf6 deletion in those lobes, because the greatest level of Klf6 deletion was primarily in the anterior lobe, the remainder of our analyses focused therefore on detailed characterization of the anterior prostate.
Canalization of the anterior prostate begins around P2 and progresses in a proximal to distal manner with elaboration of epithelial infolding or tufting within the luminal space (28, 29). A novel ex vivo application of three-dimensional, high resolution ultrasound micro-imaging technology was employed to visualize the luminal space of intricately microdissected prostates. Extensive epithelial infolding in the smaller luminal space of 1-year-old Klf6f/fNkx3.1+/+ anterior prostates resulted in a dense, white, hyperechoic regions within the ultrasound image (Fig. 2G), which were also visible by H&E staining (Fig. 2, I and K). The absence of secondary branching from the two main ducts was evident in the Klf6f/fNkx3.1Cre/+ anterior prostates (Fig. 2H). Ex vivo ultrasound imaging revealed a dark, hypoechoic luminal space in the main ducts, suggesting that canalization was unaffected (arrowhead, Fig. 2H). The epithelial infolding in the Klf6f/fNkx3.1Cre/+ anterior prostate was significantly reduced resulting in a defined layer of epithelial cells outlining the main ducts (Fig. 2H). H&E-stained sections also revealed that the epithelial infolding rarely spanned across the lumen, and the eosinophilic lumen suggests a functional epithelium, capable of secretion (Fig. 2, J and L). Further histopathological analysis of the H&E-stained Klf6f/fNkx3.1Cre/+ sections did not reveal any signs of hyperproliferation, nuclear anomalies, or mitotic figures (insets, Fig. 2, J and L). Given the normal appearance of the luminal epithelium, we performed anti-Klf6 immunofluorescent staining to confirm that Klf6 protein was indeed depleted in the prostate epithelium. In Klf6f/fNkx3.1+/+ mice, nuclear Klf6 expression was detected in the prostate epithelium (arrow, Fig. 2O), whereas positive nuclear staining was absent from Klf6f/fNkx3.1Cre/+ epithelium (arrowhead, Fig. 2P).
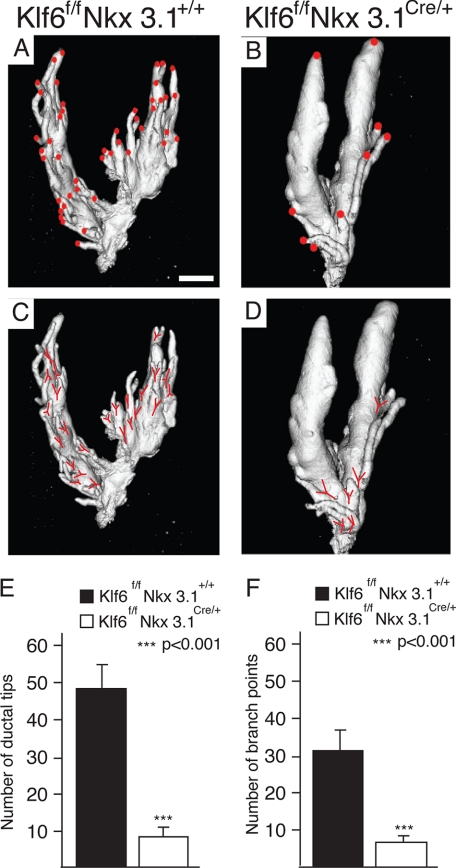
To quantify the number of ductal tips and branch points we used ex vivo micro-CT to image the anterior prostate. It was necessary to microdissect away connective tissues encapsulating the prostate to reveal adequately separated ducts ideal for micro-CT imaging. We found significantly (p < 0.001) more ductal tips (red dot) and branch points (red Y) in the anterior prostate of Klf6f/fNkx3.1+/+ mice (Fig. 3, A, C, and E) than Klf6f/fNkx3.1Cre/+ mice (Fig. 3, B, D, and F). It should be pointed out that, although Nkx3.1 null mice show a defect in prostatic branching morphogenesis (25, 30), the ductal epithelium in Nkx3.1+/− mice is essentially indistinguishable from wild type at 2 months of age and only shows much milder phenotype than that in Nkx3.1−/− mice at an age of 10–12 months or older (Fig. 4E of (30)). The branching defect due to conditional Klf6 deletion reported in the present study is far more severe than that found even in Nkx3.1 homozygous knockouts (Figs. 2F and 3 of the present study, as compared with Fig. 4 (I and J) of Ref. 25). These findings together argue strongly that Klf6 inactivation makes important contribution to the defect in prostate branching morphogenesis.
FIGURE 3.
Impaired lateral branching in Klf6-deficient anterior prostates. Microdissected prostate from Klff/fNkx3.1+/+ (A and C) and Klff/fNkx3.1Cre/+ (B and D) mice were imaged using ex vivo micro-CT and three-dimensional images were reconstructed. Ductal tips (A and B) and branch points (C and D) were marked on the three-dimensional images, counted (n = 4 from each genotype) and plotted in bar graphs, respectively (E and F). The number of ductal tips and branch points sprouting out of the main ducts are significantly more in (A, C, and E) Klff/fNkx3.1+/+ than (B, D, and F) Klff/fNkx3.1Cre/+ prostates. Statistical significance was calculated using Student's t test, and the bars represent mean ± S.E. Bar, 1 mm for A–D.
FIGURE 4.
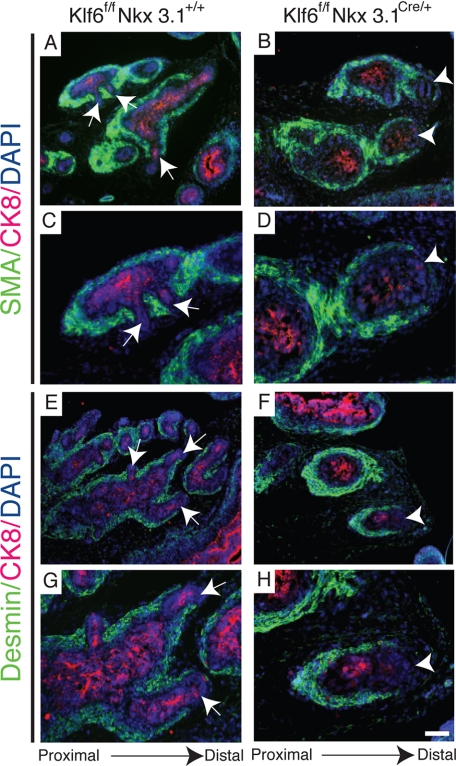
Smooth muscle formed along the sides of Klf6-deficient main ducts of the anterior prostate without intermittent smooth muscle deficient areas to permit lateral branching. Immunofluorescent staining of P5 Klff/fNkx3.1+/+ anterior prostates showed similar expression of early smooth muscle marker, SMA and late marker (A and C), desmin lining the ducts (E and G) but absent in areas where new lateral branches were formed (arrow). In Klff/fNkx3.1Cre/+ anterior prostates, SMA (B and D) and desmin (F and H) were expressed continuously along the entire length of the main ducts except for the distal tip (arrowhead). Low magnification (A, B, E, and F) and high magnification (C, D, G, and H) of respective images. Bar, 100 μm for A, B, E, and F, and 50 μm for C, D, G, and H.
Characterization of Gene Expression in Abnormal Prostate Resulting from Loss of Klf6
Murine prostate development begins in late gestation, in a process involving reciprocal interactions between the prostate epithelium and mesenchyme (31, 32). During development, the mesenchyme condenses around the ducts in a proximal to distal fashion to encase the ducts with smooth muscle actin (SMA), the earliest smooth muscle marker expressed, and desmin, a later marker (33). To determine the state of smooth muscle differentiation, we examined the expression of SMA and desmin at P5, which corresponds to the initiation of secondary branching in the anterior prostate. We found SMA (Fig. 4, A and C) and desmin (Fig. 4, E and G) expressed proximally but not at the distal tips (arrow) of newly branched lateral ducts of Klf6f/fNkx3.1+/+ prostates. In contrast, SMA (Fig. 4, B and D) and desmin (Fig. 4, F and H) were expressed along the entire length of each main duct of Klf6f/fNkx3.1Cre/+, but not at the distal tip (arrowhead). Interestingly, this is consistent with previous findings where the relative thickness of the smooth muscle layer is inversely proportional to epithelial ductal growth (34). This suggests that, although there was diminished smooth muscle expression around the emerging lateral ducts of Klf6f/fNkx3.1+/+ prostates, lateral epithelial branching in Klf6f/fNkx3.1Cre/+ was prevented as a result of a continuous layer of smooth muscle along the main ducts. On the other hand, it is also possible that the continuous layer of smooth muscle formed around the main ducts as a result of the absence of branching. To further understand this branching defect, we examined expression of androgen receptor by immunofluorescent staining, because several investigators had demonstrated that postnatal prostate branching morphogenesis and differentiation of the stroma are highly regulated by androgen (32, 33, 35). Androgen receptor was expressed continuously around the primary and secondary ducts of Klf6f/fNkx3.1+/+ (supplemental Fig. S2, A and C) and the main ducts of Klf6f/fNkx3.1Cre/+ prostates (supplemental Fig. S2, B and D). There were no apparent differences in proliferation by anti-Ki67 staining (supplemental Fig. S3, A and B), cell death by anti-active caspase-3 (data not shown) and epithelial differentiation by anti-CK8, anti-CK14 and anti-E-cadherin staining (supplemental Figs. S2 (E–H) and S3 (C and D)). Therefore, the lack of lateral branching could not be attributed to androgen receptor expression, proliferation, cell death, or epithelial differentiation in the mutant mice.
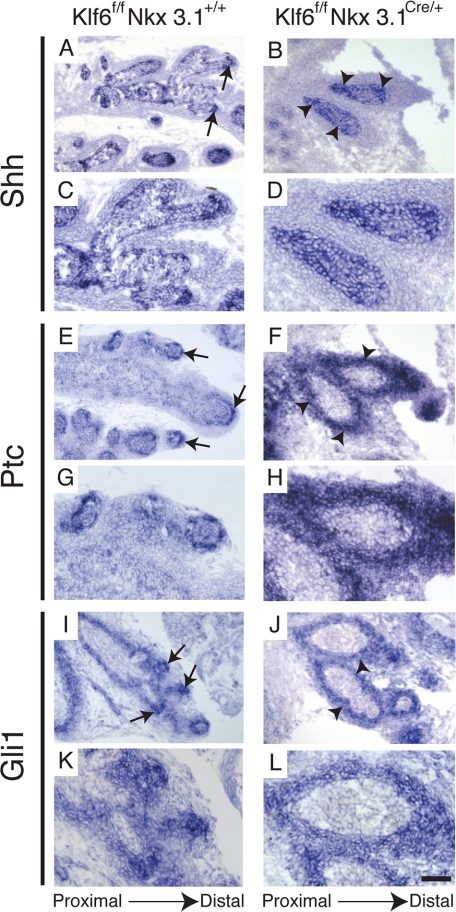
Factors in the epithelium and mesenchyme essential for proper ductal branching include Nkx3.1, Hh, BMP, FGF, Notch, and Wnt pathway components (12–19, 25, 36). Our group and others have previously shown that the activated Shh pathway could inhibit prostatic ductal branching (12–15, 37). The lack of branching in the Klf6f/fNkx3.1Cre/+ prostates prompted us to examine mRNA expression of Shh pathway components by in situ hybridization. In Klf6f/fNkx3.1+/+ prostates, highly localized epithelial Shh expression was detected in the distal tips, which was consistent with a previous reported study (37) (arrow, Fig. 5, A and C). However, Shh expression was dispersed throughout the entire length of the Klf6f/fNkx3.1Cre/+ main ducts (Fig. 5, B and D). Similarly, intense and dispersed expression of Ptc and Gli1, two well validated Hh target genes (38, 39), were detected throughout the stroma surrounding the main ducts of Klf6f/fNkx3.1Cre/+ (arrowhead, Fig. 5, F and J). This was in sharp contrast to the highly localized expression of Ptc and Gli1 observed in the stroma adjacent to Shh expression in the ductal tips of Klf6f/fNkx3.1+/+ that was previously shown in wild-type prostates (37) (arrow, Fig. 5, E, G, I, and K).
FIGURE 5.
Enhanced expression of Shh pathway components in Klf6-deficient prostate. In situ hybridization analysis of P7 Klff/fNkx3.1+/+ prostates revealed expression of Shh in the epithelium at the tips of lateral branches (arrow, A). Shh expression was elevated and dispersed throughout the main ductal epithelium of Klff/fNkx3.1Cre/+ prostates (arrowhead, B). Localized Ptc (arrow, E and G) and Gli1 (arrow, I and K) expression in Klff/fNkx3.1+/+ prostates were detected in the stromal region immediately adjacent to the Shh expressing epithelium at the ductal tips. However, in the Klff/fNkx3.1Cre/+ prostates, Ptc (arrowhead, F and H) and Gli1 (arrowhead, J and L) expression were highly elevated and localized expression to the stroma at the ductal tips was lost. Instead, Ptc and Gli expression were dispersed throughout the perimeter of the main ducts (arrowhead, F, H, J, and L). Low magnification (A, B, E, F, I, and G) and high magnification (C, D, G, H, K, and L) of respective images. Bars: 100 μm for A, B, E, F, I, and G, and 50 μm for C, D, G, H, and K.
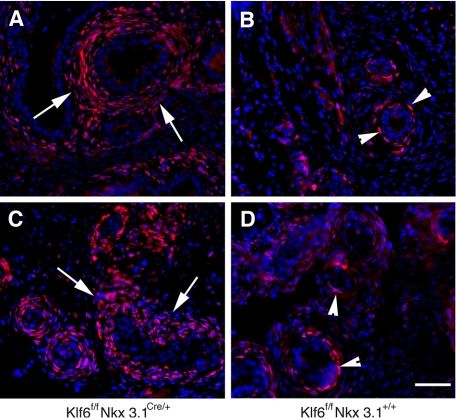
To confirm the change in expression pattern of Ptc, we crossed Klf6f/+Nkx3.1Cre/+ with Ptc-lacZ reporter mice (26) in which lacZ is driven under the Pct promoter, to obtain Klf6f/fNkx3.1Cre/+;Ptc-lacZ mice. This Ptc-lacZ reporter line is a well characterized and a sensitive indicator of endogenous Ptc expression or Hh signaling activity (26). Consistent with the in situ hybridization results, we found that there was increased β-galactosidase staining in the periepithelial stoma of Klf6f/fNkx3.1Cre/+;Ptc-lacZ mice compared with the staining observed in Klf6f/fNkx3.1+/+;Ptc-lacZ mice (Fig. 6). There were multiple cell layers of β-galactosidase-positive cells surrounding the epithelium in the Klf6f/fNkx3.1Cre/+;Ptc-lacZ mice (Fig. 6, A and C). In sharp contrast, only one to two layers of β-galactosidase-positive cells were seen in the Klf6f/fNkx3.1+/+;Ptc-lacZ control littermates (Fig. 6, B and D). Together these findings indicate that Klf6 expression leads to broader expression of Hh signaling around the developing prostate ducts.
FIGURE 6.
β-Galactosidase expression in Klf6f/fNkx3.1Cre/+;Ptc-lacZ reporter mouse prostates. Immunostaining of prostate tissue sections prepared from P7 Klf6f/fNkx3.1Cre/+;Ptc-lacZ (A and C) and Klf6f/fNkx3.1+/+;Ptc-lacZ (B and D) mice with anti-β-galactosidase antibody. The sections were counterstaining with 4′,6-diamidino-2-phenylindole (blue). Note that β-galactosidase-expressing cells are located in the mesenchymal cells surrounding the epithelium. Although there were multiple cell layers of β-galactosidase positive cells in the Klf6f/fNkx3.1Cre/+;Ptc-lacZ mice (A and C), only one to two layers of β-galactosidase-positive cells were seen in the Klf6f/fNkx3.1+/+;Ptc-lacZ control littermates. Slides were viewed using a Zeiss Axiophot epifluorescence microscope. Images were captured with Compix imaging systems using a cooled RGB charge-coupled device camera and analyzed using Adobe Photoshop CS. Bar, 50 μm.
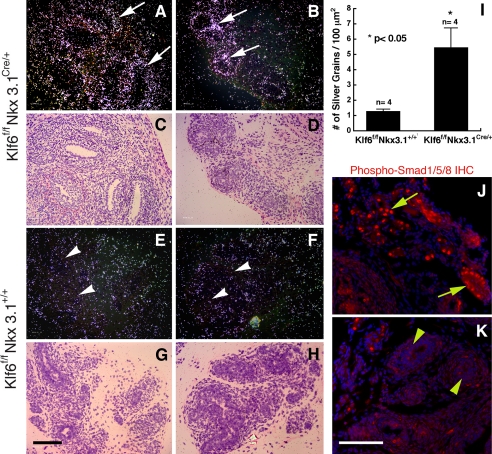
In an attempt to further understand how epithelial branching morphogenesis is inhibited in the Klf6f/fNkx3.1Cre/+ mice, we also performed radioactive in situ hybridization using a probe specific for BMP4, which was previously shown to be expressed in the prostatic stroma and can inhibit prostatic epithelial ductal budding and branching morphogenesis (17). As shown in Fig. 7, which contains tangential sections of P7 prostates, overall expression of BMP4 in the stroma surrounding the epithelium was enhanced in Klf6f/fNkx3.1Cre/+ mice (Fig. 7, A–D) compared with the expression levels observe in sections taken from Klf6f/fNkx3.1+/+ mice (Fig. 7, E–H). Upon quantification of the silver grains from the in situ hybridization, there was more than 4-fold BMP4 expression in the Klf6f/fNkx3.1Cre/+ mice as compared with the Klf6f/fNkx3.1+/+ mice (p < 0.05, Fig. 7I). Next, we investigated the status of signaling downstream of BMP4 by examining the phosphorylation status of Smad1/5/8. Immunofluorescence staining using anti-phospho-Smad1/5/8 antibody revealed enhanced nuclear staining in the prostate sections from Klf6f/fNkx3.1Cre/+ mice, whereas there were only a few positively stained nuclei in the Klf6f/fNkx3.1+/+ sections (Fig. 7, J and K). These findings together suggest that up-regulation of Hh signaling induced by the deletion of the Klf6 gene may inhibit prostate epithelial branching via up-regulation of BMP4.
FIGURE 7.
Expression of BMP4 is up-regulated in Klf6f/fNkx3.1Cre/+ mice. A and B, BMP4 expression detected by radioactive in situ hybridization in P7 Klf6f/fNkx3.1Cre/+ prostates. C and D, H&E staining from the same field of A and B. E and F, BMP4 expression in P7 Klf6f/fNkx3.1+/+ prostates. G and H, H&E staining from the same field of E and F. Note that the BMP4 signal is stronger and broader in the mutants compared with the control mice (arrows in A and B versus arrowheads in E and F). I, MetaMorph analysis of numbers of silver grains per μm2 indicates a significant increase in the prostate of Klf6f/fNkx3.1Cre/+ mice as compared with that of Klf6f/fNkx3.1+/+ mice. Sense control probes revealed no hybridization signal above background (not shown). J and K, phospho-Smad1/5/8 immunostaining of P7 prostates from Klf6f/fNkx3.1Cre/+ and Klf6f/fNkx3.1+/+ mice. Note that the staining is stronger, and the number of phospho-Smad1/5/8-positive nuclei is higher in the Klf6f/fNkx3.1Cre/+ prostate (yellow arrows in J) than the Klf6f/fNkx3.1+/+ prostate (yellow arrowheads in K). Bar in G, 100 μm for A–H; bar in K, 100 μm for J and K.
DISCUSSION
The Klf gene family is known to be crucial for development in vertebrates (4). They are expressed in a large number of tissues (40) and play an important role in the control of hematopoietic cell differentiation (4), erythroid cell maturation (21, 41), T-cell activation (42), blood vessel stability (20), and skin permeability (22). Using the prostate as a model, we describe here for the first time a role for Klf6 in prostate branching morphogenesis. Consistent with this notion, Klf14, another Klf family member, has been shown to be critical for alveolarization in the developing lung. Deletion of Klf14 leads to thinner lung alveolar walls and poor outgrowth of secondary septa, which results in defective blood-air exchange and respiratory failures in newborn mice (43). In the kidney, Klf6 is known to be expressed in the Wolffian duct, uteric bud, collecting ducts, and mesangium (44), an expression profile that would also be consistent with a role in branching morphogenesis.
Because systemic loss of Klf6 is embryonic lethal (23), we generated mice deficient for Klf6 specifically in the prostate. The resulting lateral branching defect in Klf6-deficient prostate was confirmed by light microscopy, H&E staining, and micro-CT imaging. Using a novel ex vivo ultrasound imaging modality, we were able to demonstrate that Klf6 plays a role in modulating the epithelial infolding and tufting, which results in decreased surface area of functional epithelium. The most prominent branching defects are seen in the anterior prostate, also known as a coagulating gland, which correlates with the highest level of Klf6 depletion (Fig. 1E). Despite the reduction in epithelial infolding and tufting whether or not prostatic function has been impaired in these mice remains to be determined.
The augmented branching defect in the anterior prostate is noteworthy, because the branching patterns in the anterior prostate are distinct from the dorsolateral prostate and ventral prostate. In addition, the embryologic origin of the anterior prostate is also different from the other prostate lobes (45, 46). Although branching patterns of the ventral prostate and dorsolateral prostates are believed to arise from the urogenital sinus epithelium invaginating into urogenital sinus mesenchyme, the anterior prostate is thought to be derived from urogenital sinus epithelium at the most dorsal aspect of the urogenital sinus invaginating into the mesenchyme of the seminal vesicle, which itself is derived from the mesodermal Wolffian duct. Hence, the resulting branched structure in the anterior prostate has unique patterns distinct from the other prostate lobes and occurs as a result of lateral branch events and not as a result of bifurcations at the elongating tips.
Consistent with previous reports from our group and others on the role of Shh signaling in prostate branching (12–15, 37), both Ptc-lacZ reporter mice crossed to Klf6f/fNkx3.1Cre/+ mice and in situ hybridization data in the present study indicate that impaired lateral branching in the Klf6-deficient prostate correlate with an up-regulation and loss of spatial localization of Shh, Ptc, and Gli1. Further support for the involvement of focal Hh signaling in prostate epithelial branching comes from a study by Bushman and co-workers (15), showing a close association between prostate ductal bud formation and localized expression of Shh at the growing tips of elongating prostate ducts. In addition, Shh regulates prostate branching morphogenesis in concert with FGF10 and BMP4 (37). Shh secreted from the epithelial ductal tips can down-regulate FGF10 that stimulates epithelial cell growth (16) and/or up-regulate BMP4 that inhibits epithelial cell growth (17) in the mesenchyme, which results in epithelial ductal branching (37). In the present study, we show that loss of Klf6 leads to dispersed, rather than focal, Shh expression throughout the epithelial ducts and corresponding dispersed Ptc and Gli1 expression in the adjacent mesenchyme (Fig. 5). This broader domain of Hh pathway activity leads to an up-regulation of BMP4 expression, resulting in reduced ductal branching. As proposed by Pu and coworkers, the interplay and localized expression pattern of Shh and BMP4 are key to conveying critical branching information (37).
Klf6 is reported to be a tumor suppressor that is inactivated in a large subset of prostate cancer (47). However, our detailed histological characterization of Klf6f/fNkx3.1+/+ mice as old as 2 years failed to reveal any prostate tumors.7 The lack of tumor phenotype in the Klf6 mutant mice could be due to compensations/redundancy with other Klf family members. It is also possible that loss of Klf6 might not be sufficient to initiate prostate tumorigenesis in the mouse and that loss/mutation of additional tumor suppressors might be required.
Supplementary Material
Acknowledgments
We thank Leisa Johnson, Mallika Singh, Hua Tian, and Zhenyu Gu for critical discussions. We also thank Margaret Fuentes, Christine Olsson, and Luz Orellana for assistance with the animal colony, Allison Bruce with Graphics, Jeffrey Eastham-Anderson for Metamorph analysis of BMP4 in situ hybridization signal, and Howard Stern for his assistance with some histopathological review.
This work was supported, in part, by National Institutes of Health Grant CA115985 (to M. M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Y. P. Hu, S. M. Price, Z. Chen, W. Banach-Petrosky, C. Abate-Shen, and M. M. Shen, unpublished observations.
M. Shen, unpublished data.
C. C. Leow, B. Wang, J. Ross, S. M. Chan, J. Zha, R. A. D. Carano, G. Frantz, M. M. Shen, F. J. de Sauvage, and W.-Q. Gao, unpublished observations.
- Hh
- hedgehog
- BMP
- bone morphogenic protein
- FGF
- fibroblastic growth factor
- Shh
- sonic hedgehog
- Micro-CT
- micro-computed tomography
- SMA
- smooth muscle actin
- H&E
- hematoxylin and eosin
- TCRD
- T-cell receptor delta chain.
REFERENCES
- 1.Bieker J. J. (2001) J. Biol. Chem. 276, 34355–34358 [DOI] [PubMed] [Google Scholar]
- 2.Huber T. L., Perkins A. C., Deconinck A. E., Chan F. Y., Mead P. E., Zon L. I. (2001) Curr. Biol. 11, 1456–1461 [DOI] [PubMed] [Google Scholar]
- 3.Laub F., Aldabe R., Friedrich V., Jr., Ohnishi S., Yoshida T., Ramirez F. (2001) Dev. Biol. 233, 305–318 [DOI] [PubMed] [Google Scholar]
- 4.Oates A. C., Pratt S. J., Vail B., Yan Y. I., Ho R. K., Johnson S. L., Postlethwait J. H., Zon L. I. (2001) Blood 98, 1792–1801 [DOI] [PubMed] [Google Scholar]
- 5.Zhao W., Hisamuddin I. M., Nandan M. O., Babbin B. A., Lamb N. E., Yang V. W. (2004) Oncogene 23, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C., Bhalala H. V., Qiao H., Dong J. T. (2002) Oncogene 21, 6567–6572 [DOI] [PubMed] [Google Scholar]
- 7.Narla G., Heath K. E., Reeves H. L., Li D., Giono L. E., Kimmelman A. C., Glucksman M. J., Narla J., Eng F. J., Chan A. M., Ferrari A. C., Martignetti J. A., Friedman S. L. (2001) Science 294, 2563–2566 [DOI] [PubMed] [Google Scholar]
- 8.Reeves H. L., Narla G., Ogunbiyi O., Haq A. I., Katz A., Benzeno S., Hod E., Harpaz N., Goldberg S., Tal-Kremer S., Eng F. J., Arthur M. J., Martignetti J. A., Friedman S. L. (2004) Gastroenterology 126, 1090–1103 [DOI] [PubMed] [Google Scholar]
- 9.Kimmelman A. C., Qiao R. F., Narla G., Banno A., Lau N., Bos P. D., Nuñez Rodriguez N., Liang B. C., Guha A., Martignetti J. A., Friedman S. L., Chan A. M. (2004) Oncogene 23, 5077–5083 [DOI] [PubMed] [Google Scholar]
- 10.Ito G., Uchiyama M., Kondo M., Mori S., Usami N., Maeda O., Kawabe T., Hasegawa Y., Shimokata K., Sekido Y. (2004) Cancer Res. 64, 3838–3843 [DOI] [PubMed] [Google Scholar]
- 11.Sugimura Y., Cunha G. R., Donjacour A. A. (1986) Biol. Reprod. 34, 961–971 [DOI] [PubMed] [Google Scholar]
- 12.Freestone S. H., Marker P., Grace O. C., Tomlinson D. C., Cunha G. R., Harnden P., Thomson A. A. (2003) Dev. Biol. 264, 352–362 [DOI] [PubMed] [Google Scholar]
- 13.Wang B. E., Shou J., Ross S., Koeppen H., De Sauvage F. J., Gao W. Q. (2003) J. Biol. Chem. 278, 18506–18513 [DOI] [PubMed] [Google Scholar]
- 14.Berman D. M., Desai N., Wang X., Karhadkar S. S., Reynon M., Abate-Shen C., Beachy P. A., Shen M. M. (2004) Dev. Biol. 267, 387–398 [DOI] [PubMed] [Google Scholar]
- 15.Lamm M. L., Catbagan W. S., Laciak R. J., Barnett D. H., Hebner C. M., Gaffield W., Walterhouse D., Iannaccone P., Bushman W. (2002) Dev. Biol. 249, 349–366 [DOI] [PubMed] [Google Scholar]
- 16.Thomson A. A., Cunha G. R. (1999) Development 126, 3693–3701 [DOI] [PubMed] [Google Scholar]
- 17.Lamm M. L., Podlasek C. A., Barnett D. H., Lee J., Clemens J. Q., Hebner C. M., Bushman W. (2001) Dev. Biol. 232, 301–314 [DOI] [PubMed] [Google Scholar]
- 18.Wang X. D., Leow C. C., Zha J., Tang Z., Modrusan Z., Radtke F., Aguet M., de Sauvage F. J., Gao W. Q. (2006) Dev. Biol. 290, 66–80 [DOI] [PubMed] [Google Scholar]
- 19.Wang B. E., Wang X. D., Ernst J. A., Polakis P., Gao W. Q. (2008) PLoS ONE 3, e2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo C. T., Veselits M. L., Barton K. P., Lu M. M., Clendenin C., Leiden J. M. (1997) Genes Dev. 11, 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuez B., Michalovich D., Bygrave A., Ploemacher R., Grosveld F. (1995) Nature 375, 316–318 [DOI] [PubMed] [Google Scholar]
- 22.Segre J. A., Bauer C., Fuchs E. (1999) Nat. Genet. 22, 356–360 [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto N., Kubo A., Liu H., Akita K., Laub F., Ramirez F., Keller G., Friedman S. L. (2006) Blood 107, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deleted in proof
- 25.Bhatia-Gaur R., Donjacour A. A., Sciavolino P. J., Kim M., Desai N., Young P., Norton C. R., Gridley T., Cardiff R. D., Cunha G. R., Abate-Shen C., Shen M. M. (1999) Genes Dev. 13, 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oro A. E., Higgins K. (2003) Dev. Biol. 255, 238–248 [DOI] [PubMed] [Google Scholar]
- 27.Jubb A. M., Pham T. Q., Frantz G. D., Peale F. V., Jr., Hillan K. J. (2006) Methods Mol Biol. 326, 255–264 [DOI] [PubMed] [Google Scholar]
- 28.Lung B., Cunha G. R. (1981) Anat. Rec. 199, 73–88 [DOI] [PubMed] [Google Scholar]
- 29.Shappell S. B., Thomas G. V., Roberts R. L., Herbert R., Ittmann M. M., Rubin M. A., Humphrey P. A., Sundberg J. P., Rozengurt N., Barrios R., Ward J. M., Cardiff R. D. (2004) Cancer Res. 64, 2270–2305 [DOI] [PubMed] [Google Scholar]
- 30.Schneider A., Brand T., Zweigerdt R., Arnold H. (2000) Mech. Dev. 95, 163–174 [DOI] [PubMed] [Google Scholar]
- 31.Cunha G. R., Donjacour A. (1987) Prog. Clin. Biol. Res. 239, 273–282 [PubMed] [Google Scholar]
- 32.Cunha G. R. (1994) Cancer 74, 1030–1044 [DOI] [PubMed] [Google Scholar]
- 33.Hayward S. W., Baskin L. S., Haughney P. C., Foster B. A., Cunha A. R., Dahiya R., Prins G. S., Cunha G. R. (1996) Acta Anat. (Basel) 155, 94–103 [DOI] [PubMed] [Google Scholar]
- 34.Nemeth J. A., Lee C. (1996) Prostate 28, 124–128 [DOI] [PubMed] [Google Scholar]
- 35.Donjacour A. A., Cunha G. R. (1993) Endocrinology 132, 2342–2350 [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M., Komuro I., Inagaki H., Jenkins N. A., Copeland N. G., Izumo S. (2000) Dev. Dyn. 219, 248–260 [DOI] [PubMed] [Google Scholar]
- 37.Pu Y., Huang L., Prins G. S. (2004) Dev. Biol. 273, 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramalho-Santos M., Melton D. A., McMahon A. P. (2000) Development 127, 2763–2772 [DOI] [PubMed] [Google Scholar]
- 39.Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T., Gumucio D. L. (2005) Development 132, 279–289 [DOI] [PubMed] [Google Scholar]
- 40.Turner J., Crossley M. (1999) Trends Biochem. Sci. 24, 236–240 [DOI] [PubMed] [Google Scholar]
- 41.Perkins A. C., Gaensler K. M., Orkin S. H. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12267–12271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo C. T., Veselits M. L., Leiden J. M. (1997) Science 277, 1986–1990 [DOI] [PubMed] [Google Scholar]
- 43.Hertveldt V., Louryan S., van Reeth T., Drèze P., van Vooren P., Szpirer J., Szpirer C. (2008) Dev. Dyn. 237, 883–892 [DOI] [PubMed] [Google Scholar]
- 44.Fischer E. A., Verpont M. C., Garrett-Sinha L. A., Ronco P. M., Rossert J. A. (2001) J. Am Soc. Nephrol. 12, 726–735 [DOI] [PubMed] [Google Scholar]
- 45.Cunha G. R., Lung B. (1979) in Accesory Glands of Male Reproductive Tract, Ann Arbor Science, Ann Arbor, MI, Vol. 6, pp. 1–28 [Google Scholar]
- 46.Cunha G. R., Alarid E. T., Turner T., Donjacour A. A., Boutin E. L., Foster B. A. (1992) J. Androl. 13, 465–475 [PubMed] [Google Scholar]
- 47.Narla G., Friedman S. L., Martignetti J. A. (2003) Am. J. Pathol. 162, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.