Abstract
We have previously shown that the Na/K-ATPase binds and inhibits Src. Here, we report the molecular mechanism of Na/K-ATPase-mediated Src regulation and the generation of a novel peptide Src inhibitor that targets the Na/K-ATPase/Src receptor complex and antagonizes ouabain-induced protein kinase cascades. First, the Na/K-ATPase inhibits Src kinase through the N terminus of the nucleotide-binding domain of the α1 subunit. Second, detailed mapping leads to the identification of a 20-amino acid peptide (NaKtide) that inhibits Src (IC50 = 70 nm) in an ATP concentration-independent manner. Moreover, NaKtide does not directly affect the ERK and protein kinase C family of kinases. It inhibits Lyn with a much lower potency (IC50 = 2.5 μm). Third, highly positively charged leader peptide conjugates including HIV-Tat-NaKtide (pNaKtide) readily enter cultured cells. Finally, the following functional studies of pNaKtide demonstrate that this conjugate can specifically target the Na/K-ATPase-interacting pool of Src and act as a potent ouabain antagonist in cultured cells: 1) pNaKtide, unlike PP2, resides in the membranes. Consistently, it affects the basal Src activity much less than that of PP2. 2) pNaKtide is effective in disrupting the formation of the Na/K-ATPase/Src receptor complex in a dose-dependent manner. Consequently, it blocks ouabain-induced activation of Src, ERK, and hypertrophic growth in cardiac myocytes. 3) Unlike PP2, pNaKtide does not affect IGF-induced ERK activation in cardiac myocytes. Taken together, we suggest that pNaKtide may be used as a novel antagonist of ouabain for probing the physiological and pathological significance of the newly appreciated signaling function of Na/K-ATPase and cardiotonic steroids.
The Na/K-ATPase is expressed in most eukaryotic cells and is essential for maintaining the transmembrane ion gradient by pumping Na+ out of and K+ into cells (1). Structurally, the enzyme consists of two non-covalently linked α and β subunits. Similar to other P-ATPases, the Na/K-ATPase α subunit has 10 transmembrane domains with both the N and C termini located in the cytoplasm (2, 3). Based on the published crystal structures of Na/K-ATPase (4), the α subunit consists of several well-characterized domains. The actuator (A)2 domain consists of the N terminus and the second cytosolic domain (CD2) connected to transmembrane helices M2 and M3, and the highly conserved discontinuous phosphorylation (P) domain is close to the plasma membrane, while the nucleotide-binding (N) domain is relatively isolated (2). There is a significant amount of movement of both the A and N domains during the ion-pumping cycle as in the SR Ca2+-ATPase (4–6). It appears that the A domain rotates, while the N domain closes during the transport cycle. Interestingly, these domains have also been implicated in interacting with many protein partners, including inositol 1,4,5-trisphosphate receptors, phosphoinositide 3-kinase, phospholipase C-γ (PLC-γ), ankyrin, and cofilin (7–12).
Src, a member of the Src family non-receptor kinases, plays an important role in the signal transduction pathways of many extracellular stimuli such as cytokines, growth factors, and stress responses (13) and has been considered as a promising target for therapeutic intervention in certain cancers (14) and bone diseases (15). Several endogenous inhibitors of Src have been documented previously, including the C-terminal Src kinase, CSK-homologous kinase, Wiscott-Aldrich syndrome protein, RACK1, and caveolin (16–19).
Previously, we and others (20) have demonstrated that binding of cardiotonic steroids (CTS) such as ouabain to the Na/K-ATPase stimulates multiple protein kinase cascades. Moreover, the knock-out of Src prevents these cascades from being activated (10, 21, 22). More recently, we have observed that the Na/K-ATPase interacts directly with Src via at least two binding motifs. One of these interactions is between the CD2 of the α1 subunit and the Src SH2, and the other involves the third cytosolic domain (CD3) of the α1 subunit and the Src kinase domain. We propose that the formation of the Na/K-ATPase/Src complex serves as a receptor for ouabain to stimulate the aforementioned protein kinase cascades. Specifically, the CD3-Src kinase interaction maintains Src in an inactive form whereas the binding of ouabain to the Na/K-ATPase disrupts this interaction, resulting in the assembly and activation of different pathways including ERK cascades, PLC/PKC pathway and mitochondrial production of reactive oxygen species (23). Thus, the Na/K-ATPase functions as an endogenous negative Src regulator. This proposition is consistent with the fact that the basal Src activity is inversely correlated to the amount of Na/K-ATPase α1 subunit in both cultured cells (24) and in α1 heterozygous mouse tissues (25). Therefore, to better understand how the molecular interactions between the Na/K-ATPase and Src regulate Src activity, we have further mapped the Src-binding domains in the CD3 of α1. These studies led to the identification of a peptide Src inhibitor (pNaKtide) and the demonstration that pNaKtide can act as a novel ouabain antagonist capable of inhibiting ouabain-induced activation of protein kinase cascades and hypertrophic growth in cardiac myocytes.
EXPERIMENTAL PROCEDURES
Materials
PP2, a Src kinase inhibitor, and staurosporine, a nonspecific PKC inhibitor, were obtained from Calbiochem. The following antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): monoclonal anti-Src antibody (B12), polyclonal anti-ERK1/2 antibody, monoclonal anti-phosphor-ERK1/2 antibody, and goat anti-rabbit and goat anti-mouse secondary antibodies. The monoclonal anti-His antibody was from GE Healthcare (Buckinghamshire, England). The monoclonal anti-α1 antibody (α6F) was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA). Glutathione beads were from Amersham Biosciences (Uppsala, Sweden), and ProBond Purification System was from Invitrogen (Carlsbad, CA). Recombinant human Src, Lyn, and IGF-1 were obtained from Upstate Biotechnology (Lake Placid, NY). The CMV promoter-driven pEYFP-C1 was purchased from Clontech (Palo Alto, CA). The pGEX-4T-1 and pTrc-His A vectors were from GE Healthcare and Invitrogen, respectively. All the peptides were synthesized with the purity above 95%. Identity and purity were confirmed by high performance liquid chromatography mass spectroscopy.
Plasmid Constructs
The preparation of plasmid constructs expressing GST fusion proteins were prepared as previously described (23). GST-CD3 (amino acid residues 350–785), GST-ND (amino acid residues 379–594), GST-ND2 (amino acid residues 379–475), GST-ND2R (amino acid residues 476–594), GST-ND1 (amino acid residues 379–435), and GST-ND1R (amino acid residues 436–594) expression vectors were constructed based on the sequence of pig kidney Na/K-ATPase α1 subunit (see Fig. 1A). His-tagged c-Src constructs were generated by excising the corresponding c-Src cDNA from the GST-Src vector (26) and then inserting them into pTrc-His A vector. Src-ECFP expression vector for FRET analysis was constructed by cloning the full-length c-Src in-frame into pECFP-N1 vector. pEYFP-ND1, pEYFP-ND, and pEYFP-CD3 were made by directional subcloning of the corresponding cDNAs from the pEYFP-rat α1 vector. All constructs were verified by DNA sequencing.
FIGURE 1.
Identification of the N terminus of N domain as a Src-interacting motif from the Na/K-ATPase α1 subunit. A, schematic presentations of different GST fusion proteins. B and D, Coomassie Blue staining of purified GST-ND, GST-CD3, and GST-ND1, GST-ND1R, GST-ND2, GST-ND2R. C and E, binding of GST-ND and GST-ND1 to Src. Purified His-Src (200 ng) was incubated with 5 μg of GST fusion proteins in 0.5% Triton X-100 PBS for 30 min and followed by four washes with the same buffer. A representative Western blot from three independent experiments shows the pull-down products probed with anti-His antibody.
Cell Preparation, Culture, and Transient Transfections
Pig kidney proximal LLC-PK1 and mouse fibroblast SYF and SYF + Src cells were obtained from American Type Culture Collection (Manassas, VA). TCN23-19 cells were generated from LLC-PK1 cells as described (24). Cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin (100 units/ml)/streptomycin (100 μg/ml). LLC-PK1 and TCN23-19 cells were serum-starved for 24 h, whereas SYF and SYF + Src cells were cultured in the medium containing 0.5% fetal bovine serum for 24 h and used for the experiments. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Experiments were performed 24 h after transfection.
Primary cultures of neonatal rat cardiac myocytes were prepared as described previously with minor modifications (27). Myocytes were dispersed from ventricles of 1–2-day-old Sprague-Dawley rats by digestion with 0.04% collagenase II (Worthington) and 0.05% pancreatin (Sigma) at 37 °C. Noncardiomyocytes were eliminated by preplating for 1.5 h at 37 °C. Myocytes were plated at a density of 8 × 102 cells/mm2 in 100-mm Corning cell culture dishes in Dulbecco's modified Eagle's medium-M199 (4:1) containing 10% (v/v) fetal bovine serum (24 h, 37 °C) and then incubated in serum-free medium for 48 h before experiments were carried out. All research on rats was done according to procedures and guidelines approved by the Institutional Animal Care and Use Committee.
Preparation of Na/K-ATPase, GST-fused Proteins, and His-tagged Proteins
Na/K-ATPase was purified from pig kidney outer medulla using the Jorgensen method as previously described (27), and the preparations with specific activities between 1200 and 1400 μmol Pi/mg/h were used in this work. GST-fused proteins or His-tagged proteins were expressed in Escherichia coli BL21 (Invitrogen) and purified using glutathione beads or ProBond Purification System. Soluble GST-fused proteins were eluted from the glutathione beads with elution buffer (10 mm reduced glutathione, 0.1% Triton X-100, 50 mm Tris-HCl, pH 8.0) and then dialyzed in the buffer containing 0.1% Triton X-100, 50 mm Tris-HCl, pH 8.0, to remove remnant glutathione.
In Vitro GST Pull-down Assay
To map the interaction motifs in the CD3 of Na/K-ATPase, the GST pull-down assay was performed as previously described (23). Briefly, 5 μg of GST-fused proteins were conjugated on glutathione beads and incubated with 100 ng of purified His-Src in 500 μl of phosphate-buffered saline (PBS) in the presence of 0.5% Triton X-100 at room temperature for 30 min. The beads were washed with the same buffer four times. The bound His-Src was resolved on 10% SDS-PAGE and detected by Western blot with anti-His antibody.
Immunoprecipitation and Immunoblot Analysis
Following the indicated treatment, cells were washed with ice-cold PBS, and then lysed in ice-cold radioimmune precipitation assay buffer containing 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium orthovanadate, 1 mm NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 50 mm Tris-HCl, pH 7.4. Cell lysates were cleared by centrifugation at 16,000 × g for 15 min. For immunoprecipitation analysis, the supernatants were incubated with a polyclonal anti-α1 antibody. Immunoprecipitates were then separated by SDS-PAGE and transferred to an Optitran membrane and analyzed. The signal was detected using the enhanced chemiluminescence kit (Pierce) and quantified using a Bio-Rad GS-670 imaging densitometer as previously described (28). To measure the activation of ERK1/2 and Src, blots were first probed with anti-phosphor-ERK1/2 or anti-pY418 antibodies, and then the same membrane was stripped and reprobed with anti-ERK1/2 or -Src for loading control.
Kinase Activity Assay of Src, Lyn, and PKC
To determine whether the Na/K-ATPase constructs or peptides affect Src/Lyn kinase activity, the purified Src (4.5 units) or Lyn (20 ng) was incubated with different amount of the purified GST-fused Na/K-ATPase constructs or peptides in PBS for 30 min at 37 °C. Afterward, 2 mm ATP/Mg2+ was added. The reaction continued for 5 min at 37 °C and was stopped by addition of SDS sample buffer. Afterward, the Src pY418 and Lyn pY396 were measured by anti-pY418 antibody to indicate Src/Lyn activation (26).
Activity of PKC was measured by PepTag phosphorylation assay for non-radioactive detection of PKC (Promega) as described in the product instructions. Briefly, 40 ng of PKC were incubated for 30 min at 30 °C with the reaction mixture containing 5 μl of reaction buffer, 5 μl of PepTag C1 (0.4 μg/μl), 5 μl of PKC activator solution, 1 μl of peptide protection solution, and 10 μm staurosporine or peptides. The reaction mixture was then subjected to electrophoresis on a 0.8% agarose gel at 100 V for 20 min. After electrophoresis, negatively charged phosphorylated PepTagC1 peptide migrated toward the anode, while non-phosphorylated PepTagC1 peptide migrated toward the cathode. The percentage of the phosphorylated PepTagC1 was an indicator of the PKC activity.
Peptide-loading in Saponin-permeabilized LLC-PK1 Cells
To deliver peptides into LLC-PK1 cells, saponin-based permeabilization procedure was adopted as reported previously (29). When cells reached full confluence, media were removed and saved at 37 °C. Cells were then slowly brought down in temperature by two sequential 2-min incubations of PBS. Cells were incubated with the permeabilization buffer (20 mmol/liter HEPES, pH 7.4, 10 mmol/liter EGTA, 140 mmol/liter KCl, 50 μg/ml saponin, 5 mmol/liter NaN3, and 5 mmol/liter oxalic acid, dipotassium salt) containing the desired peptides for 10 min in an ice bath. Then cells were gently washed with chilled PBS and placed on ice for a 20-min recovery. Room temperature PBS was added for 2 min. This step was repeated with PBS at 37 °C. Original cell media were then added back to the cells for 30 min before used for experiments.
FRET Analysis by Acceptor Photobleaching in Fixed Cells
FRET analysis by acceptor photobleaching was performed as described previously (23). Briefly, Src-ECFP and EYFP-rat α1 plasmids were cotransfected into LLC-PK1 cells. After 24 h, cells grown on glass coverslips were exposed to peptides for 1 h. After washing with PBS solution, cells were fixed with ice-cold methanol for 15 min at −20 °C. The mounted coverslips were then used for FRET measurement with the Leica DMIRE2 confocal microscope (Wetzlar, Germany). The cells that expressed both Src-ECFP and EYFP-rat α1 were chosen to perform the FRET analysis. A membrane region of interest (ROI 1) containing the EYFP-α1 was photobleached by applying full power of 515 nm laser, and the emission of CFP excited by 456 nm laser was recorded before (Dpre) and after (Dpost) YFP photobleaching. Meanwhile, the emission of YFP was recorded before (Apre) and after (Apost) photobleaching. The FRET efficiency was then calculated by the Ep value, corrected ratio of (Dpost − Dpre)/Dpre in the photobleached region comparing it to the nonbleached region (ROI 2) in the same experiment.
Cellular Distribution Analysis of YFP-ND1 and NaKtides in Live Cells
LLC-PK1 cells were cultured on coverslips and then transiently transfected with pEYPF-ND1 plasmid. After 24 h, the cells were fixed for 15 min with ice-cold methanol, washed with PBS for three times and then blocked with Signal Enhancer (Invitrogen). The cells were then incubated with a mouse anti-Na/K-ATPase α1 monoclonal antibody (Upstate) in PBS containing 1% bovine serum albumin for 1 h at room temperature. After three washes with PBS, cells were exposed to AlexaFluor 555-conjugated anti-mouse secondary antibody for 1 h at room temperature, washed, and mounted onto slides. Image visualization was performed using a Leica DMIRE2 confocal microscope. To assess cellular distribution of NaKtides, the LLC-PK1 cells and cardiac myocytes were exposed to 1 μm FITC-tagged NaKtides, washed twice with PBS, and imaged by monitoring FITC fluorescence using a Leica DMIRE2 confocal microscope.
Measurement of Hypertrophic Growth in Cultured Cardiac Myocytes
Cultured neonatal cardiac myocytes were exposed to ouabain with or without preincubation with pNaKtide for 60 min. After 48 h, cells were trypsinized, and cell number and volume were measured with a Z2 Coulter Counter (Beckman Coulter, Fullerton, CA).
Analysis of Data
Data are given as the mean ± S.E. Statistical analysis was performed using the Student's t test, and significance was accepted at p < 0.05.
RESULTS
Identification of ND1 as a Src-interacting Domain from the Na/K-ATPase α1 Subunit
We have shown that the CD3 of the α1 subunit interacts with and inhibits Src (23). As depicted in Fig. 1A, the Na/K-ATPase CD3 consists of both P and N domains. The three-dimensional structure of Na/K-ATPase indicates that the N domain is exposed whereas the P domain is relatively close to the membrane (4). As Src is a cytosolic protein, it is therefore likely that the N domain interacts with the Src kinase domain. To test this hypothesis, we constructed GST-ND and tested whether it binds to purified His-Src. GST-CD3 was used as a positive control whereas purified GST served as a negative control. Pull-down analyses confirmed that the N domain of α1 subunit interacted with Src (Fig. 1, B and C). As depicted in Fig. 1A, the N domain contains over 200 amino acid residues. To further map the binding motif in the N domain, we constructed ND2 and ND2 remaining (ND2R) GST fusion proteins as illustrated in Fig. 1A and tested whether the interaction occurs outside the ATP-binding pocket (ND2R). The GST pull-down assay showed that Src primarily interacted with GST-ND2, while a minor trace of Src was detected in GST-ND2R beads (Fig. 1, D and E). Further structural analysis of the ND2 reveals that the N terminus of ND2 is highly exposed and does not contribute to ATP binding (30, 31). Moreover, the corresponding domain in SR Ca2+-ATPase is known to bind phospholamban (32). Therefore, we tested whether the N terminus of ND interacts with Src. As illustrated in Fig. 1A, we constructed two more GST fusion proteins (ND1 and ND1R), and assessed their binding to Src using GST pull-down assay. As depicted in Fig. 1E, we observed that the ND1, but not the ND1R, interacted with Src. Moreover, when the binding activities of GST-ND1 and GST-ND were compared in the same assay, we found that both pulled-down comparable amount of His-Src (data not shown).
ND1 as a Potent Src Inhibitor Can Target the Na/K-ATPase/Src Complex in Cultured Cells
To test whether ND1 inhibits Src as did the CD3, we incubated Src with 100 ng of soluble GST fusion proteins in the test tube and measured Src pY418 levels by Western blot. GST-CD3 was used in the experiment as a positive control, and GST-ND was also tested. As depicted in Fig. 2A, both GST-ND1 and GST-ND were as effective as the positive control in inhibiting Src activity. Moreover, the inhibitory effect of ND1 on Src was dose-dependent (IC50, 50 nm) (Fig. 2B). To further test whether ND1 could be used to inhibit Src in live cells, we measured Src pY418 level in LLC-PK1 cells transfected with different YFP expression vectors. We found that expression of YFP-ND1, like YFP-ND and YFP-CD3, reduced Src activity when compared with that of YFP-transfected cells (Fig. 2C).
FIGURE 2.
Regulation of Src by ND1. A, soluble GST fusion proteins (100 ng) were incubated with recombinant Src (4.5 units) for 15 min in PBS and assayed for Src activity as described under “Experimental Procedures.” B, dose-dependent inhibition of Src by GST-ND1. C and D, LLC-PK1 cells were transiently transfected with pEYFP, pEYFP-ND1, pEYFP-ND, or pEYFP-CD3 plasmid for 24 h. Representative Western blots (C) show the expression of YFP and YFP fusion proteins in the transfected cells and the effects of these proteins on pY418 Src and total Src in cell lysates. Bar graph (D) shows the combined data from 3 to 5 independent experiments. **, p < 0.01 compared with the control.
The above studies demonstrate the effectiveness of ND1 as a Src inhibitor in cultured cells. To further test whether ND1 can target the plasma membrane Na/K-ATPase/Src complex, we performed the following three sets of experiments. First, as depicted in Fig. 3A, we observed that YFP-ND1 was expressed as a soluble protein. However, we did detect a pool of YFP-ND1 co-localizing with Na/K-ATPase α1 in the plasma membrane. Second, to test whether this pool of YFP-ND1 has the potential to interact with Src, we co-transfected LLC-PK1 cells with YFP-ND1 and Src-CFP, and then performed FRET analysis using acceptor photobleaching protocols (23). The data show an energy transfer from Src-CFP to YFP-ND1, resulting in a significant FRET (13.4 ± 2.4%, n = 6). These findings indicate that YFP-ND1 and the plasma membrane Src-CFP are in close proximity, suggesting a direct interaction between these two proteins. Finally, when cell lysates were immunoprecipitated by an anti-α1 antibody, we found that YFP-ND1 was co-precipitated (Fig. 3B). Thus, we concluded that YFP-ND1 is most likely capable of interacting with the Na/K-ATPase-associated Src.
FIGURE 3.
Targeting of YFP-ND1 to the Na/K-ATPase/Src complex in live cells. A, localization of YFP-ND1 in LLC-PK1 cells. LLC-PK1 cells were transfected with pEYFP-ND1 and immunostained with anti-Na/K-ATPase α1 antibody. Localization of YFP-ND1 (green) and Na/K-ATPase α1 (red) was visualized using a Leica DMIRE2 confocal microscope. Arrow indicated the co-localization of YFP-ND1 and Na/K-ATPase α1 on the plasma membrane. The scale bar represents 5 μm. B, lysates from transfected cells were immunoprecipitated with anti-Na/K-ATPase α1 antibody and then analyzed by Western blot using anti-GFP antibody and anti-Na/K-ATPase α1 antibody. A representative Western blot of three separate experiments is shown.
Development of NaKtide as a Specific Src Inhibitor
Because the above data suggest that ND1 binds and inhibits Src, we synthesized four 20-mer peptides that cover the entire ND1 domain to identify those which were capable of inhibiting Src (Fig. 4A). Based on the crystal structure of the Na/K-ATPase (4), peptides 1 and 2 are un-structured whereas peptides 3 and 4 may form an α-helix. As depicted in Fig. 4B, 1 μm peptide 3 caused almost a 100% inhibition of Src while peptide 4 produced a partial inhibition. On the other hand, both peptides 1 and 2 showed no effect. When the dose-response curve was constructed, we observed that peptide 3 was quite potent in inhibiting Src with an IC50 of about 70 nm (Fig. 4C), comparable to that of GST-ND1. As expected, peptide 1 had no effect. Because peptide 3 is derived from the Na/K-ATPase, we named it NaKtide. Because peptide 1 has no effect on Src, it was used as a control. Thus, we named it C1.
FIGURE 4.
Effect of ND1-derived peptides on Src activity. A, sequences of different ND1-derived peptides. B, each peptide (1 μm) was incubated with recombinant Src (4.5 units) for 15 min and then assayed for Src pY418. C, dose-dependent inhibition of Src by the P3. Curve fit analysis was performed with GraphPad Prism 5. D, effects of P3 on Lyn kinase activity. Recombinant Lyn was incubated with indicated amount of P3 for 15 min and then assayed for pY396 by Western blot. Curve fit analysis was done with GraphPad Prism 5. E, effects of P3 on kinase activity of PKC mixture. Quantitative data are presented as mean ± S.E. of at least three independent experiments. **, p < 0.01 compared with control.
To probe whether NaKtide is relatively specific to Src, we measured its dose-dependent effect on another Src family kinase member, Lyn. As shown in Fig. 4D, NaKtide produced a dose-dependent inhibition of Lyn. However, the IC50 was about 2.5 μm, ∼40 times higher than that for Src inhibition.
To test whether NaKtide affects serine/threonine kinases, we incubated a PKC family kinase mixture containing primarily PKC α and β isoforms with NaKtide and C1, and then measured the kinase activity. As shown in Fig. 4E, unlike staurosporine (a nonspecific PKC family kinase inhibitor), NaKtide as well as C1 showed no effect at concentrations up to 10 μm.
To verify that NaKtide does not act as an ATP analog as a generic Src inhibitor PP2, we measured its effect on Src in the presence of different concentrations of ATP. We found that changes in ATP concentration from 0.1 to 2 mm did not affect NaKtide-induced Src inhibition (data not shown).
Development of a Cell-permeable NaKtide as a Ouabain Antagonist
Recent studies have demonstrated that the coupling of biological molecules to a number of positively charged cell-penetrating peptides (CPPs) can facilitate their uptake into cultured mammalian cells as well as animal tissues (33, 34). Accordingly, we synthesized HIV-Tat-NaKtide (pNaKtide) (see Fig. 5A). HIV-Tat-C1 (pC1) was also synthesized and used as a control. To compare, we also synthesized penetratin-NaKtide (AP-NaKtide). In vitro kinase assays showed that pNaKtide was a highly potent Src inhibitor while the control pC1 was inactive (Fig. 5B). Interestingly, addition of HIV-Tat actually increased the potency of NaKtide in this in vitro assay. Specifically, the IC50 was decreased from 70 nm to about 4 nm. This was also true when AP-NaKtide was assessed (data not shown).
FIGURE 5.
Properties of cell-permeable peptides. A, sequence information of different peptides. B, dose-dependent inhibition of Src by pNaKtide and control pC1. Curve fit analysis was performed by GraphPad Prism 5. C, cell-loading analyses of pNaKtide and AP-NaKtide in LLC-PK1 cells. Cells were serum-starved for 12 h and then exposed to 1 μm FITC-pNaKtide or FITC-AP-NaKtide at 37 °C for 60 min. Cells were washed twice with PBS, and analyzed by confocal imaging. The scale bar represents 5 μm.
To assess the permeability of pNaKtide and AP-NaKtide, we labeled peptides with FITC. As depicted in Fig. 5C, confocal imaging analysis of live cells indicated that pNaKtide was cell-permeable. Maximal loading was achieved after 30 to 60 min of incubation, and labeled pNaKtide could be seen in virtually every LLC-PK1 cell examined. Moreover, unlike YFP-ND1, a large portion of pNaKtide resided in the plasma membrane (left panel in Fig. 5C). On the other hand, when the same experiments were conducted to assess AP-NaKtide, we found that most of AP-NaKtide was in intracellular vesicles. Unlike pNaKtide, very few of the labeled AP-NaKtide molecules were detected in the plasma membrane (right panel in Fig. 5C). Thus, different permeability tags are likely to affect the destination of the NaKtide fusion molecule within the cell.
We have demonstrated that the Na/K-ATPase and Src interact in the plasma membrane to form a functional receptor. Binding of ouabain to this receptor complex changes the conformation of Na/K-ATPase, resulting in a release and activation of the associated Src (23). Thus, pNaKtide, binding to Src from the cytosolic side, could function as an effective ouabain antagonist by blocking the formation of Na/K-ATPase/Src complex or by inhibiting ouabain-induced activation of Src. To test this postulate, we first performed FRET analysis to determine the effect of pNaKtide on the formation of Na/K-ATPase/Src receptor complex in the plasma membrane. LLC-PK1 cells were co-transfected with YFP-α1 and Src-CFP, and then exposed to different concentrations of pNaKtide. We focused on pNaKtide because this conjugate resided mainly in the plasma membrane where the receptor Na/K-ATPase/Src resides. As depicted in Fig. 6A, both YFP-α1 and Src-CFP were targeted to the plasma membrane. Moreover, a significant FRET was detected in control LLC-PK1 cells as we previously reported (23). Addition of pNaKtide produced a dose-dependent reduction in overall FRET efficiency (Fig. 6B) as well as the percentage of cells that exhibited detectable FRET (Fig. 6C), indicating that pNaKtide is effective as YFP-ND1 in interacting with the plasma membrane pool of Src, thus blocking the formation of a stable Na/K-ATPase/Src complex.
FIGURE 6.
Effects of pNaKtide on the formation of the Na/K-ATPase/Src complex. A, LLC-PK1 cells were transfected with EYFP-rat α1 (yellow) and Src-ECFP (cyan). FRET analysis was performed as described under “Experimental Procedures.” The Region of Interest 1 (Boxed area marked by ROI 1) was photobleached and analyzed for FRET. The same measurement was done in ROI 2 that was not photobleached. B and C, same FRET analyses were conducted in transfected cells pretreated with 1 μm pC1 or different concentrations of pNaKtide for 1 h. Average FRET efficiency (B) and percentage of cells with FRET (cut-off value is 4.0%) were calculated. At least 20 cells from three experiments were measured for each condition.
It is important to note that although the IC50 of pNaKtide as a Src inhibitor is about 4 nm when assayed in the test tube, the IC50 for blocking the formation of Na/K-ATPase/Src complex is about 1 μm. This decrease in potency may be because of its limited cell permeability and accessibility to the plasma membrane Src. Thus, 1 μm pNaKtide was used in the following four sets of experiments to test its effectiveness and specificity as an ouabain antagonist.
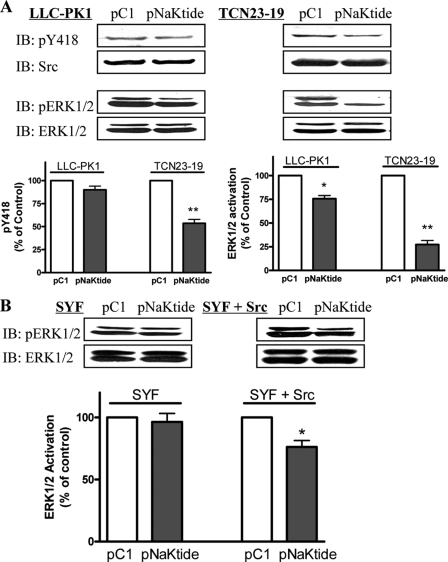
First, to assess whether pNaKtide acts as a nonspecific Src inhibitor, we exposed LLC-PK1 and the Na/K-ATPase-knockdown TCN23-19 cells to 1 μm pC1 or pNaKtide for different times. Cell lysates were then subjected to Western analysis of active Src and ERK1/2 (ERK1, upper band; ERK2, lower band in Figs. 7 and 8). We showed that about 25% of cellular Src interacts with the Na/K-ATPase in LLC-PK1 cells (23). TCN23-19 cells were derived from LLC-PK1 cells (24). Knockdown of the Na/K-ATPase in these cells reduces the pool of Src-interacting Na/K-ATPase and thus increases the basal Src and ERK1/2 activity (24). As depicted in Fig. 7A and Table 1, pNaKtide inhibited 10% of basal Src activity in LLC-PK1 cells. However, this inhibition was not statistically significant (p = 0.07). On the other hand, it caused a significant inhibition (over 20%) of basal ERK1/2. This is not surprising because ERK1/2 is a downstream effector of Src. Interestingly, the effects of pNaKtide on Src and ERK1/2 in TCN23-19 cells were much more than that seen in LLC-PK1 cells (Fig. 7A). Thus, we suggest that pNaKtide may selectively target the Na/K-ATPase-interacting pool of Src.
FIGURE 7.
Effect of pNaKtide on Src and Src-mediated signaling pathway. A, LLC-PK1 and TCN23-19 cells were serum-starved for 12 h and were exposed to 1 μm pC1 or pNaKtide for 1 h. Cell lysates were assayed for active Src (pY418) and ERK1/2 (pERK1/2) by Western blot as described under “Experimental Procedures.” n = 3. *, p < 0.05; **, p < 0.01 compared with control pC1. B, SYF and SYF + Src cells were exposed to 1 μm pC1 or pNaKtide. Cell lysates were analyzed by Western blot. n = 3. *, p < 0.05 compared with control pC1.
FIGURE 8.
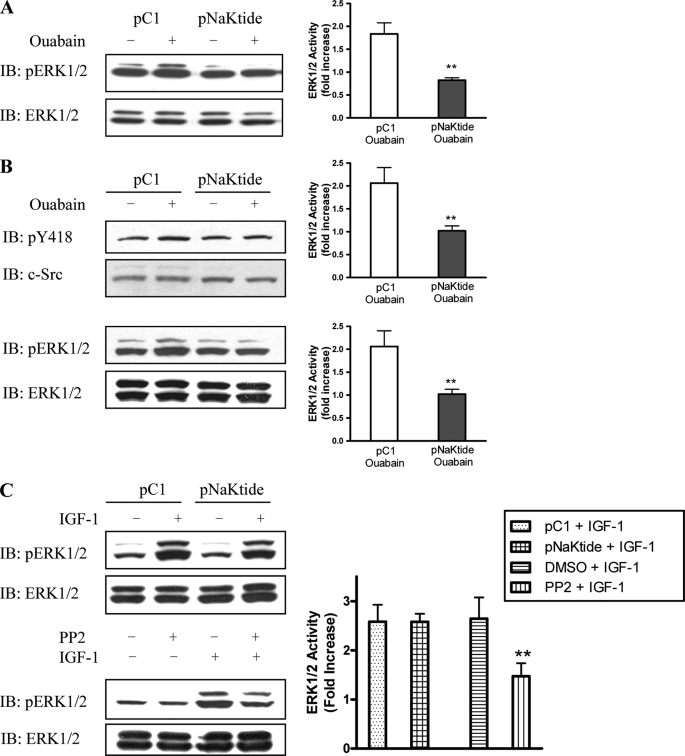
Effect of pNaKtide on ouabain-induced signal transduction. LLC-PK1 (A) and primary cultured cardiac myocytes (B) were preincubated with 1 μm peptides for 1 h and then exposed to 100 nm (LLC-PK1) or 100 μm (myocytes) ouabain. Cell lysates were analyzed by Western blot. n = 3. **, p < 0.01 compared with control peptide. C, primary cultures of cardiac myocytes were preincubated with 1 μm peptides for 1 h or PP2 for 30 min and then exposed to 20 ng/ml IGF-1 for 5 min. n = 3. **, p < 0.01.
TABLE 1.
Effects of ND1, NaKtide, pNaKtide, and PP2 on Src
| ND1 | pNaKtide | PP2 | NaKtide | |
|---|---|---|---|---|
| IC50in vitro | 50 nma | 4 nm | 5 nm | 70 nm |
| LLC-PK1 | 36 ± 11b | 90 ± 6c | 45 ± 7%d | 40 ± 9%e |
| Neonatal cardiac myocytes | NAf | 93 ± 9c | 42 ± 7%d | NA |
a GST-ND1 was used in vitro for Src activity assay.
b Cells were transfected with either YFP as control or YFP-ND1 for 24 h, or pretreated.
c Pretreated with 1 μm pNaKtide for 1 h.
d Pretreated with 1 μm PP2 for 30 min.
e LLC-PK1 cells were permeabilized with saponin, and then exposed to 1 μm NaKtide for 1 h. Src pY418 in lysates was analyzed, and data are means ± S.E., % of control, n = 3 to 5.
f NA, Not Available.
To verify that the effect of pNaKtide on ERK1/2 is due to its inhibition of Src, we repeated the above experiments in SYF + Src and SYF cells. As shown in Fig. 7B, pNaKtide had no effect on ERK1/2 in SYF cells where Src family kinases (Src, Yes, and Fyn) were knocked out. However, pNaKtide reduced ERK1/2 activity in Src-rescued SYF cells (SYF + Src).
To test whether the plasma membrane residence makes pNaKtide a relative specific inhibitor of the Na/K-ATPase-associated Src pool, we loaded LLC-PK1 cells with NaKtide in the presence of saponin. As shown in Table 1, NaKtide, like expression of YFP-ND1, caused a 60% inhibition of basal Src activity when loaded by saponin. Interestingly, when the effect of AP-NaKtide on basal Src was measured, like pNaKtide, it barely affected the basal Src activity in LLC-PK1 cells.
Second, we measured the effect of pNaKtide on ouabain-induced activation of ERK in LLC-PK1 cells. As expected, we found that 1 μm pNaKtide completely abolished ouabain-induced ERK1/2 activation in LLC-PK1 cells (Fig. 8A). To be sure that this is not a cell-specific effect, we repeated the same experiments in primary cultures of cardiac myocytes. As shown in Fig. 8B, ouabain-induced activation of Src and ERK1/2 was also blocked by pNaKtide.
Third, we compared pNaKtide and PP2, a generic Src inhibitor. As shown in Table 1, both pNaKtide and PP2 have a similar IC50 on Src kinase. However, PP2 produced more inhibition on basal Src activity than that of pNaKtide in both LLC-PK1 and cardiac myocytes (Table 1). Moreover, when cardiac myocytes were stimulated by IGF, PP2, but not pNaKtide, caused a significant inhibition of ERK activation (Fig. 8C).
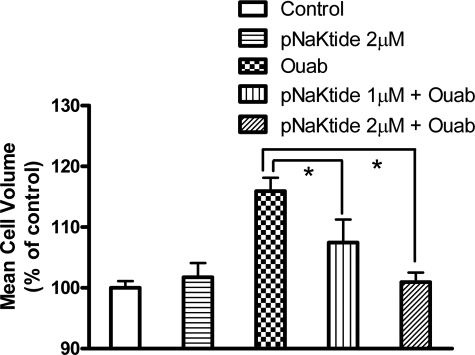
Finally, we assessed the effect of pNaKtide on ouabain-induced hypertrophy of neonatal cardiac myocytes. The primary cultures of cardiac myocytes were first exposed to pNaKtide for 1 h and then treated with 100 μm ouabain for 48 h. As expected, pNaKtide blocked ouabain-induced cell volume increase in a dose-dependent manner (Fig. 9). Furthermore, ouabain-induced hypertrophy was completely abolished by pNaKtide at the dose of 2 μm.
FIGURE 9.
Effect of pNaKtide on ouabain-induced hypertrophy in the neonatal cardiac myocytes. Primary cardiac myocytes were exposed to pNaKtides for 1 h and then incubated with 100 μm ouabain for 48 h. Mean cell volume was assessed by a Z2 Coulter Counter. n = 4. *, p < 0.05.
DISCUSSION
In this report, we have identified the specific substructure of the Na/K-ATPase α1 subunit that interacts and inhibits Src. Based on these data, we have further engineered a novel peptide Src inhibitor that can target the Na/K-ATPase/Src receptor complex and thus function as an effective ouabain antagonist in cultured cells. These and other important issues are further discussed in the following paragraphs.
Identification of NaKtide as a New Class of Src Inhibitor
The CD3 of Na/K-ATPase α1 subunit consists of both N and P domains. We showed that CD3 binds the Src kinase domain and inhibits Src kinase activity in vitro (23). Based on the newly published crystal structure of Na/K-ATPase, the N domain is exposed, whereas the P domain is relatively close to the membrane (4). Consistently, we found that the N domain binds and inhibits Src. Interestingly, it is known that the less structured N terminus of SR Ca2+-ATPase N domain interacts with phospholamban (32). Similarly, we have demonstrated that the ND1 (Fig. 2), the first 50 amino acid residues of the α1 N domain, inhibits Src. However, further mapping analyses reveal that the corresponding phospholamban-binding domain in ND1 (peptide 2, see Fig. 4A) actually had no effect on Src kinase activity. Instead, peptides 3 and 4 showed strong inhibitory effect on Src. Based on the crystal structure as well as NMR data (4, 30), both peptides possess a helix structure, suggesting that helix/helix interaction between the ND1 and the Src kinase may be responsible for the Na/K-ATPase-induced Src inhibition. A literature review reveals that several endogenous proteins interact and inhibit Src. Noteworthy are RACK1 and Wiscott-Aldrich syndrome protein. While RACK1 inhibits Src via its interaction with the SH2 domain (35), WASP does so by binding to the Src kinase domain (18). However, no detailed structural information is available for further comparison.
Like inhibitors of other tyrosine kinases, most Src inhibitors are ATP mimetics (36). When the ATP-dependence was assessed, we found that changes in ATP concentration did not affected NaKtide-induced Src inhibition. Moreover, it is unlikely that NaKtide acts as a substrate Src inhibitor (37) since the peptide does not contain Tyr residue. Thus, NaKtide represents a novel class of Src inhibitor. As a Src inhibitor, both ND1 and NaKtide are potent (, B and C). The IC50 is close to 50 nm, comparable to most of reported Src inhibitors. Finally, when the effects of NaKtide on other kinases were assessed, we found that NaKtide appears to be relatively specific to Src (Fig. 4D). It showed no effect on PKC family of kinases (Fig. 4E). Its effect on ERK1/2 depends on the expression of Src (Fig. 7B), indicating that it is not an ERK inhibitor but can affect ERK signaling by inhibiting Src. This is consistent with the fact that ERK1/2 are well-known effectors of Src kinase.
Development of pNaKtide as a Specific Ouabain Antagonist
Transient transfection assays indicate that YFP-ND1 resided as a soluble protein and inhibited basal Src as effective as PP2 (Fig. 3A and Table 1). Moreover, a small fraction of YFP-ND1 was detected in the plasma membrane. Both FRET and immunoprecipitation assays showed that this plasma membrane-targeted YFP-ND1 disrupted the interaction between the Na/K-ATPase and Src (Fig. 3B). Taken together, these findings suggest that ND1 and its derivative NaKtide may be used as an effective Src inhibitor or a relatively specific ouabain antagonist depending on its cellular distribution. This postulation is supported by the following experimental evidence. First, we found that loading NaKtide by saponin into LLC-PK1 cells caused about 60% inhibition of basal Src activity as did by PP2 or expression of YFP-ND1. Second, we demonstrated that tagging a positively charged leader peptide such as HIV-Tat to NaKtide made it readily cell permeable. Moreover, confocal imaging analyses showed that a majority of pNaKtide resided in the plasma membrane and had much less effect on basal Src activity in both LLC-PK1 and cardiac myocytes (data not shown). Interestingly, when pNaKtide was applied to TCN23-19 cells where the pool of Src-interacting Na/K-ATPase is depleted, in contrast to that in LLC-PK1 cells, pNaKtide caused about 50% inhibition of cellular Src activity. These findings suggest that the plasma membrane-targeted pNaKtide may have a relatively specific effect on the Na/K-ATPase-interacting pool of Src. Third, we observed that pNaKtide was very effective in blocking the formation of Na/K-ATPase/Src receptor complex (Fig. 6). In accordance, ouabain-induced activation of ERK1/2 in LLC-PK1 cells was completely abolished by 1 μm pNaKtide (Fig. 8A). Moreover, pNaKtide was effective in inhibiting ouabain-induced activation of ERK1/2 (Fig. 8B) and hypertrophy in cardiac myocytes (Fig. 9). Finally, the specificity of pNaKtide toward the Na/K-ATPase/Src complex was further demonstrated by experiments showing that PP2, but not pNaKtide, caused a significant inhibition of IGF-induced ERK1/2 activation (Fig. 8C).
Limitations
Needless to say, several important issues remain to be resolved. First, we have not identified key amino acid residues in NaKtide that bind and inhibit Src and do not know where in the kinase domain NaKtide binds to and how the binding inhibits Src. Resolution of these issues will reveal the molecular mechanism of Na/K-ATPase-mediated Src regulation. Second, we found that AP-NaKtide resided mainly in intracellular vesicles. Like pNaKtide, it also had almost no effect on basal Src activity. Although it is unlikely, it will be of interest to test whether AP-NaKtide can block ouabain-induced ERK1/2 activation. Moreover, AP-NaKtide may have a relative specific effect on endocytosis, exocytosis, or vesicle recycling because Src is known to play a role in these events. Finally, we have not assessed the ability of pNaKtide as an ouabain antagonist in intact animals or isolated organs.
Acknowledgment
We thank Martha Heck for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-36573 from NHLBI and GM-78565 from NIGMS.
- A domain
- actuator domain
- CD2
- second cytosolic domain
- CD3
- third cytosolic domain
- CTS
- cardiotonic steroids
- ECFP
- enhanced cyan fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- ERK
- extracellular signal-regulated protein kinase
- FRET
- fluorescence resonance energy transfer
- GST
- glutathione S-transferase
- IGF-1
- insulin-like growth factor 1
- N domain
- nucleotide-binding domain
- P domain
- phosphorylation domain
- PKC
- protein kinase C
- PLC
- phospholipase C
- PP2
- 4-amino-5-[4-chlorophenyl]-7-[t-butyl]pyrazolo[3,4-d]pyrimidine
- ROI
- region of interest
- PBS
- phosphate-buffered saline
- FITC
- fluorescein isothiocyante
- SR
- sarcoplasmic reticulum.
REFERENCES
- 1.Skou J. C. (1957) Biochim. Biophys. Acta. 23, 394–401 [DOI] [PubMed] [Google Scholar]
- 2.Kaplan J. H. (2002) Annu. Rev. Biochem. 71, 511–535 [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen P. L., Hakansson K. O., Karlish S. J. (2003) Annu. Rev. Physiol. 65, 817–849 [DOI] [PubMed] [Google Scholar]
- 4.Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sorensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 5.Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima C., Nomura H. (2002) Nature 418, 605–611 [DOI] [PubMed] [Google Scholar]
- 7.Jordan C., Püschel B., Koob R., Drenckhahn D. (1995) J. Biol. Chem. 270, 29971–29975 [DOI] [PubMed] [Google Scholar]
- 8.Barwe S. P., Anilkumar G., Moon S. Y., Zheng Y., Whitelegge J. P., Rajasekaran S. A., Rajasekaran A. K. (2005) Mol. Biol. Cell 16, 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K., Jung J., Kim M., Guidotti G. (2001) Biochem. J. 353, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Z., Cai T., Tian J., Ivanov A. V., Giovannucci D. R., Xie Z. (2005) Mol. Biol. Cell 16, 4034–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S., Malmersjö S., Li J., Ando H., Aizman O., Uhlén P., Mikoshiba K., Aperia A. (2006) J. Biol. Chem. 281, 21954–21962 [DOI] [PubMed] [Google Scholar]
- 12.Yudowski G. A., Efendiev R., Pedemonte C. H., Katz A. I., Berggren P. O., Bertorello A. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 14.Yeatman T. J. (2004) Nat. Rev. Cancer 4, 470–480 [DOI] [PubMed] [Google Scholar]
- 15.Boyce B. F., Xing L., Yao Z., Yamashita T., Shakespeare W. C., Wang Y., Metcalf C. A., 3rd, Sundaramoorthi R., Dalgarno D. C., Iuliucci J. D., Sawyer T. K. (2006) Clin. Cancer Res. 12, 6291s–6295s [DOI] [PubMed] [Google Scholar]
- 16.Chong Y. P., Ia K. K., Mulhern T. D., Cheng H. C. (2005) Biochim. Biophys. Acta. 1754, 210–220 [DOI] [PubMed] [Google Scholar]
- 17.Miller L. D., Lee K. C., Mochly-Rosen D., Cartwright C. A. (2004) Oncogene 23, 5682–5686 [DOI] [PubMed] [Google Scholar]
- 18.Schulte R. J., Sefton B. M. (2003) Biochemistry 42, 9424–9430 [DOI] [PubMed] [Google Scholar]
- 19.Li S., Couet J., Lisanti M. P. (1996) J. Biol. Chem. 271, 29182–29190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Z., Cai T. (2003) Mol. Interv. 3, 157–168 [DOI] [PubMed] [Google Scholar]
- 21.Tian J., Gong X., Xie Z. (2001) Am. J. Physiol. Heart Circ. Physiol. 281, H1899–1907 [DOI] [PubMed] [Google Scholar]
- 22.Aydemir-Koksoy A., Abramowitz J., Allen J. C. (2001) J. Biol. Chem. 276, 46605–46611 [DOI] [PubMed] [Google Scholar]
- 23.Tian J., Cai T., Yuan Z., Wang H., Liu L., Haas M., Maksimova E., Huang X. Y., Xie Z. J. (2006) Mol. Biol. Cell 17, 317–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang M., Cai T., Tian J., Qu W., Xie Z. J. (2006) J. Biol. Chem. 281, 19709–19719 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Cai T., Wang H., Li Z., Loreaux E., Lingrel J. B., Xie Z. (2009) J. Biol. Chem. 284, 14881–14890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y. C., Huang J., Ali S., Lowry W., Huang X. Y. (2000) Cell 102, 635–646 [DOI] [PubMed] [Google Scholar]
- 27.Peng M., Huang L., Xie Z., Huang W. H., Askari A. (1996) J. Biol. Chem. 271, 10372–10378 [DOI] [PubMed] [Google Scholar]
- 28.Haas M., Wang H., Tian J., Xie Z. (2002) J. Biol. Chem. 277, 18694–18702 [DOI] [PubMed] [Google Scholar]
- 29.Johnson J. A., Gray M. O., Karliner J. S., Chen C. H., Mochly-Rosen D. (1996) Circ. Res. 79, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 30.Hilge M., Siegal G., Vuister G. W., Güntert P., Gloor S. M., Abrahams J. P. (2003) Nat. Struct. Biol. 10, 468–474 [DOI] [PubMed] [Google Scholar]
- 31.Imagawa T., Kaya S., Taniguchi K. (2003) J. Biol. Chem. 278, 50283–50292 [DOI] [PubMed] [Google Scholar]
- 32.Toyofuku T., Kurzydlowski K., Tada M., MacLennan D. H. (1994) J. Biol. Chem. 269, 22929–22932 [PubMed] [Google Scholar]
- 33.Shokolenko I. N., Alexeyev M. F., LeDoux S. P., Wilson G. L. (2005) DNA Repair 4, 511–518 [DOI] [PubMed] [Google Scholar]
- 34.Mäe M., Langel U. (2006) Curr. Opin. Pharmacol. 6, 509–514 [DOI] [PubMed] [Google Scholar]
- 35.Chang B. Y., Chiang M., Cartwright C. A. (2001) J. Biol. Chem. 276, 20346–20356 [DOI] [PubMed] [Google Scholar]
- 36.Sawyer T. K. (2007) in Topics in Medicinal Chemistry: Cancer ( Bradbury R. ed) Vol. 1, pp. 383–405, Springer, Berlin Heidelberg, Germany [Google Scholar]
- 37.Lou Q., Leftwich M. E., McKay R. T., Salmon S. E., Rychetsky L., Lam K. S. (1997) Cancer Res. 57, 1877–1881 [PubMed] [Google Scholar]