Abstract
Background
UDP-GlcNAc 2-epimerase/ManNAc 6-kinase, GNE, is a bi-functional enzyme that plays a key role in sialic acid biosynthesis. Mutations of the GNE protein cause sialurea or autosomal recessive inclusion body myopathy/Nonaka myopathy. GNE is the only human protein that contains a kinase domain belonging to the ROK (repressor, ORF, kinase) family.
Principal Findings
We solved the structure of the GNE kinase domain in the ligand-free state. The protein exists predominantly as a dimer in solution, with small populations of monomer and higher-order oligomer in equilibrium with the dimer. Crystal packing analysis reveals the existence of a crystallographic hexamer, and that the kinase domain dimerizes through the C-lobe subdomain. Mapping of disease-related missense mutations onto the kinase domain structure revealed that the mutation sites could be classified into four different groups based on the location – dimer interface, interlobar helices, protein surface, or within other secondary structural elements.
Conclusions
The crystal structure of the kinase domain of GNE provides a structural basis for understanding disease-causing mutations and a model of hexameric wild type full length enzyme.
Enhanced Version
This article can also be viewed as an enhanced version in which the text of the article is integrated with interactive 3D representations and animated transitions. Please note that a web plugin is required to access this enhanced functionality. Instructions for the installation and use of the web plugin are available in Text S1.
Introduction
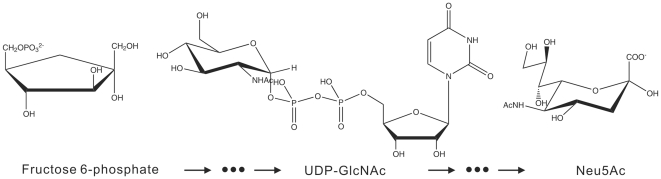
Sialic acids are N- or O- substituted terminal monosaccharides with a nine-carbon backbone highly expressed on eukaryotic cell surfaces [1]. Sialylation of glycoproteins and glycolipids modulates a wide range of biological and pathological events including early development [2], tumorigenesis [3], viral and bacterial infection, and immunity [4], [5]. In vertebrate systems, N-acetylneuraminic acid (Neu5Ac) is the metabolic precursor of all known naturally occurring sialic acids [6]. Neu5Ac is synthesized in the cytosol from UDP-N-acetylglucosamine (UDP-GlcNAc) by four consecutive reactions; and UDP-GlcNAc is a derivative of fructose-6-phosphate and the end-product of the hexosamine biosynthesis pathway (Figure 1).
Figure 1. Key sugar molecules in the sialic acid biosynthesis pathway.
The first two steps of the biosynthesis of Neu5Ac from UDP-GlcNAc are catalyzed by the bi-functional enzyme UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase (GNE). GNE contains an N-terminal epimerase domain and a C-terminal kinase domain [7]. The epimerase domain converts UDP-GlcNAc to N-acetylmannosamine (ManNAc), which is then phosphorylated at the 6 position by the kinase domain. GNE is feedback-inhibited by the activated form of Neu5Ac, i.e., cytidine-monophosphate N-acetylneuraminic acid (CMP-Neu5Ac). The kinase domain belongs to the ROK (Repressor, ORF, Kinase) family. The ROK family consists of a set of bacterial proteins that include repressors for sugar catabolic operons, and sugar kinases [8]. Gne is the only known gene in the entire human genome that encodes a ROK domain-containing protein.
Three protein isoforms have been described for human GNE, where isoform 1 is ubiquitously expressed and is believed to be responsible for the basic supply of sialic acids. Isoforms 2 and 3 are generated by alternative splicing and show tissue specific expression patterns. Isoforms 2 and 3 have reduced epimerase activities but almost intact kinase activities and may fine-tune the production of sialic acids [9]. Wild type GNE forms homo-hexamer in solution [10], and allosteric regulation of the epimerase and kinase activities of GNE is important for the normal function of the protein [10], [11]. Mutations in the epimerase domain lead to the rare congenital metabolism disorder sialurea, which results in the production of high levels of Neu5Ac due to loss of the allosteric feedback control of the UDP-GlcNAc 2-epimerase activity by CMP-Neu5Ac [12]. Late onset autosomal recessive inclusion body myopathy, which is also known as hereditary inclusion body myopathy (hereinafter referred to as HIBM), and allelic Nonaka myopathy are neuromuscular disorders that are caused by a number of different mutations within the gne gene. The mutations are located at either the epimerase domain or the kinase domain [13] and lead to hypoactivity of the enzyme [11]. Mutagenesis and enzymatic activity analysis revealed that the activities of the epimerase domain and the kinase domain are interrelated such that a single mutation in one domain could affect the activities of both domains [11]. Here, we solved the structure of the dimeric GNE kinase domain in the ligand-free state. The structure reveals the dimerization interface of the kinase domain and also suggests a possible hexameric assembly of the protein. Furthermore, the structure provides insights into the relationship between GNE mutations and GNE-related metabolism disorders.
Results and Discussion
Overview of the GNE kinase domain monomer
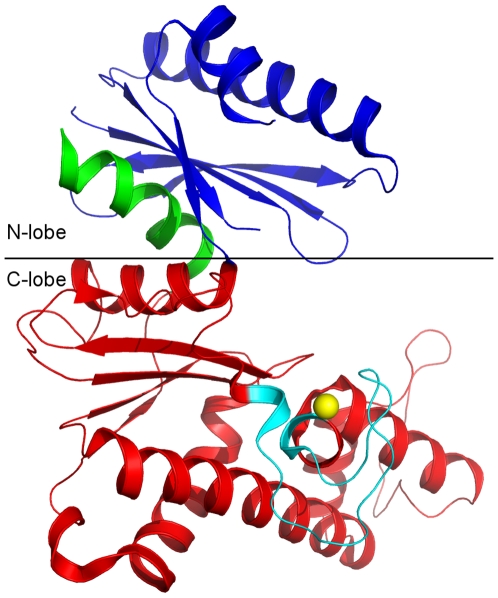
The overall structure adopts a typical bi-lobal kinase architecture. Both the N-lobe and the C-lobe have the α/β fold. Each lobe consists of a central β-sheet flanked by α-helices on both sides of the sheet. The last helix C-terminal to the C-lobe is part of the N-lobe and perpendicular to the interfacial helix of the C-lobe. Residues 475–498 of the N-lobe are invisible in the electron density map (Figure 2).
Figure 2. Overall structure of the kinase domain of GNE.
The C-terminal helix (residues 700–717, in green) is embedded into the N-lobe subdomain. The ROK family signature zinc-binding motif (cyan) is located on top of two parallel helices of the C-lobe. Residues 475–498 in the N-lobe are missing in the electron density map and are presented by dotted line (black).
The GNE kinase domain contains a type I zinc-binding motif GHx9–11CxCGx2G(C/H)xE, which forms an HC3 type zinc-finger with residues H569, C579, C581, C586. The zinc-binding motif is a characteristic feature for all ROK family members [14]. The kinase domain also contains a DxGxT type ATP-binding motif [15], [16]. The side chains of this ATP-binding motif residues point toward the cleft between the N-lobe and the C-lobe. Comparison with the actin/hexokinase/hsp70 ATPase domains suggests that the disordered residues 475–498 form part of the binding pocket for the adenosine moiety of ATP [17] and are located near the DxGxT ATP-binding motif. Taken together, these findings suggest that the ATP binding pocket of the GNE kinase domain is located in the cleft between the two lobes.
Oligomeric state of the GNE kinase domain
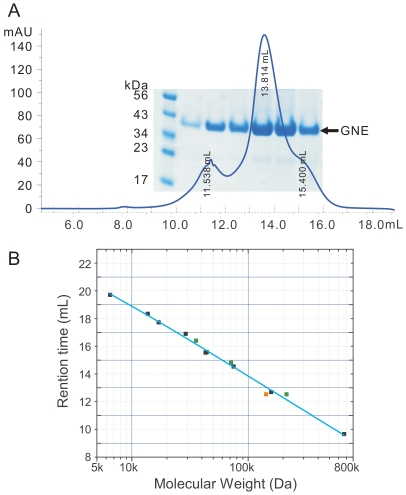
Previous deletion mutations study has suggested that the GNE kinase domain is responsible for dimerization, while a segment of residues between the epimerase and kinase domains, residues 360–382, is a potential site for trimerization [18]. Our gel filtration data (Figure 3) show that the kinase domain exists predominantly as a dimer in the solution, with small amounts of monomer and a higher order oligomer. The apparent molecular weight of the oligomer fits a hexamer of the kinase domain (see also below). However, the possibility of a tetramer [19] cannot be completely ruled out due to the low resolution of the gel-filtration column at this molecular size range (Figure 3b). The populations of the different oligomeric states of the protein are concentration dependant (data not shown), implying that the different oligomeric states exist in equilibrium.
Figure 3. Oligomeric state analysis of the GNE kinase domain.
Panel A: Superdex-200 10/300 (GE healthcare) size exclusion chromatography profile of GNE at 6 mg per mL concentration in 20 mM HEPES buffer, pH 7.5, 500 mM NaCl, 5 mM MgCl2, 1 mM tris-(2-carboxyethyl)phosphine hydrochloride (TCEP). The lanes on the SDS-PAGE gel correspond to different fractions on the gel filtration profile. Panel B: Size exclusion column calibration curve and measurement of GNE kinase domain apparent molecular weight. Standard protein samples (black data points) used include thyroglobulin (670 kDa), globulin (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), anhydrase (29 kDa), myoglobin (17 kDa), and ribonuclease (13.7 kDa). Green data points are values for GNE kinase domain in monomeric, dimeric and hexameric forms. The orange data point is for GNE kinase domain in putative tetrameric form.
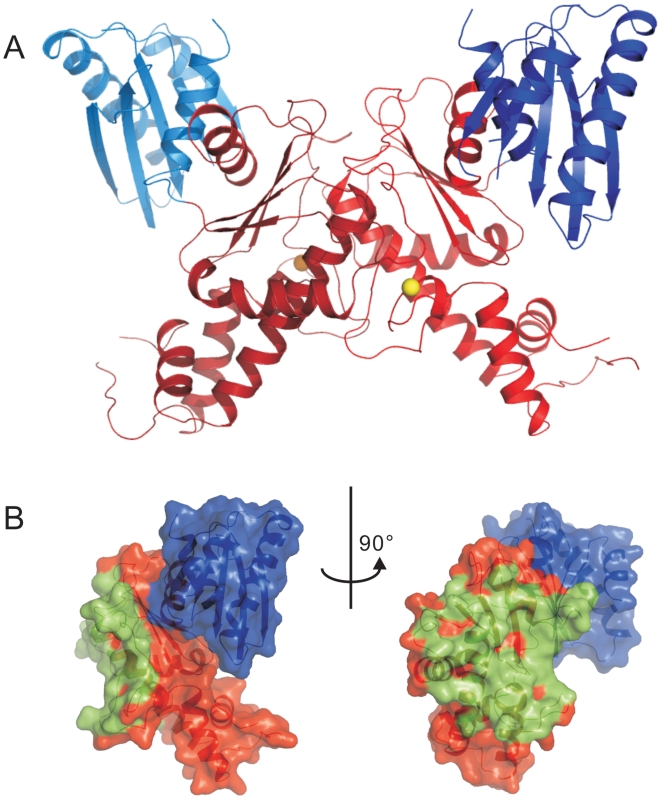
The GNE kinase domain was crystallized in space group P3121 with three molecules in the asymmetric unit. Protein interface and assembly analysis using the PISA server [20] suggests that two of the three molecules dimerize through the C-lobe with an average buried surface area of 1587 Å2 per molecule (Figure 4) whereas the third molecule dimerizes with a two-fold symmetry related molecule through the same C-lobe (Figure 5a). The solvation free energy gain upon formation of the interface, ΔiG, is −24.2 kcal·mol−1, indicating that the dimer interface is very stable and may not simply be a crystal packing artifact.
Figure 4. Dimeric GNE kinase domain.
Panel A: Ribbon representation of the dimeric structure of the kinase domain. The N-lobes are shown in blue colors, the C-lobes in red. Panel B: Orthogonal views of the protein surface of one of the monomers. Residues within 4 Å distance from the other monomer are colored green.
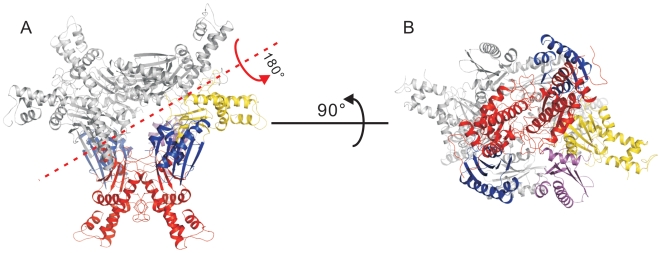
Figure 5. Crystallographic hexamer.
Panel A: A crystallographic hexamer is produced by applying a two-fold rotational symmetry operation (red axis) on the three molecules in the asymmetric unit. The N-lobes of the dimer in the biological unit are shown in blue while the C-lobe in red. The N-lobe of the third molecule that forms a dimer with a symmetry-related molecule is shown in magenta, and the C-lobe in yellow. The three symmetry-related molecules are in grey. Panel B: A bottom-up view of the hexamer by rotating the hexamer in panel A over 90° along x-axis. The N-lobes of each dimer are located on the opposite sides of the hexamer.
A crystallographic hexamer can be produced when a two-fold rotational symmetry operation is applied to the three molecules (one and a half dimers) in the asymmetric unit (Figure 5). In this hexamer, the N-lobes of three kinase molecules are pointing to the same side of the “hexamerization plane”, while the N-lobes of the other three molecules are pointing to the opposite side of the plane (Figure 5b). This assembly mode of the kinase domain allows locating the epimerase domain further away from the hexamerization plane and is consistent with the proposition that the interdomain segment (residues 360–382) is the site of trimerization [18].
Structure comparison with other ROK family members
Structural homology search of the GNE kinase domain using the FATCAT (Flexible structure AlignmenT by Chaining Aligned fragment pairs allowing Twists) server [21] revealed the top four non-redundant hits to be PDB codes 2aa4, 1xc3, 1z05 and 1z6r. All these structures contain the signature zinc-binding motif of the ROK family. The structure of E. coli putative ManNAc kinase (PDB 2aa4) was the top hit with twist-adjusted r.m.s. deviation (opt-rmsd value) of 1.94 Å. The structure of a putative sugar kinase from Bacillus subtilis was the second best hit (PDB 1xc3). The other two homologous structures were transcription repressors that belong to the ROK family (PDB: 1z05 and 1z6r). Vibrio cholerae transcriptional regulator (PDB: 1z05) is a homolog of the E. coli Mlc protein (PDB: 1z6r). The latter is a transcriptional repressor that controls the expression of malT, the central transcription activator of the E. coli maltose system [22]. The structure of the GNE kinase domain aligns well with the E. coli Mlc structure: the N-lobe of GNE kinase domain aligns to the E-domain of Mlc and the C-lobe aligns to the O-domain of Mlc. It is interesting to note that the Mlc O-domain is responsible for the oligomerization of Mlc protein [22] in a way similar to the dimerization of GNE kinase through the C-lobe. However, these four structures do not contain sugar ligands that would help inform on a substrate binding mode for GNE.
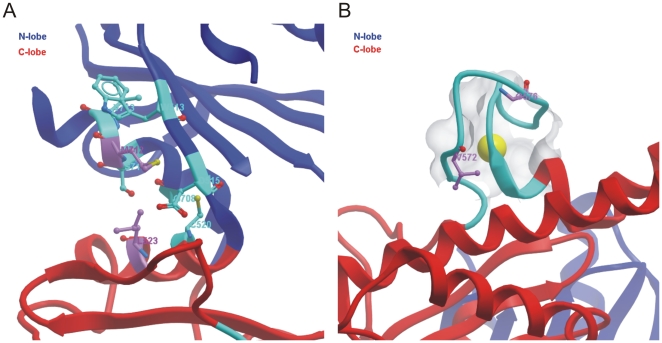
To evaluate the putative sugar binding site, the sequence of the GNE kinase domain was aligned with that of E. coli glucokinase complexed with glucose (PDB: 1sz2) [23], which is the closest homologous structure containing a bound substrate currently available in the PDB data bank. The five residues involved in sugar binding are conserved in GNE (N516, D517, E566, H569, E588, GNE numbering). These five residues are arranged to accommodate the sugar substrate (Figure 6). Two residues, H569 and E588, are located in the ROK family zinc-binding signature motif and H569 directly coordinates the zinc ion. This finding suggests that zinc may play a catalytic role in sugar substrate binding, as well as a structural role.
Figure 6. Putative sugar binding residues in the GNE kinase domain.
Panel A: structural alignment of GNE kinase (red, 3eo3, only key residues putatively involved in sugar binding are shown as sticks) with E. coli glucokinase + glucose complex (grey, PDB 1sz2). Panel B: Structure-based alignment of the two proteins, only the segment involved in sugar binding is shown. The key residues are marked using green triangles. Residues involved in zinc-binding are marked using blue triangles. HIBM-related mutation sites are marked using red triangles.
Structural mapping of disease-related GNE mutations
Since the identification of the relationship between gne mutations and HIBM [13], more than 60 mutations have been found to be associated with HIBM [24]. Among these mutations, 25 missense mutations at 23 unique sites are located in the kinase domain of the GNE protein. These 23 mutation sites can be classified into 4 different groups based on their solvent accessibility, and their locations (Table 1).
Table 1. Structural analysis of HIBM-associated missense mutations in the GNE kinase domain.
| Missense Mutationa | Epimerase Activity (%)b | Kinase Activity (%)b | Oligomeric Statec | Relative Accessibility in monomerd | Relative Accessibility in dimerd | Secondary structure elemente | Mutation site feature | Mutation group |
| V421A | 0.007 | 0.007 | β-strand | Buried | IV | |||
| A460V | 0.018 | 0.018 | Helix | Buried | IV | |||
| I472T | 50 | 5 | 0.000 | 0.000 | β-strand | Buried | IV | |
| P511H | 0.416 | 0.416 | Loop | Exposed | III | |||
| P511L | 0.416 | 0.416 | Loop | Exposed | III | |||
| N519S | 40 | 20 | hexameric | 0.027 | 0.027 | Helix | Interlobar | II |
| A524V | 5 | 50 | 0.000 | 0.000 | Helix | Interlobar | II | |
| F528C | 70 | 35 | hexameric | 0.140 | 0.140 | helix end | Interlobar | II |
| F537I | 45 | 60 | 0.000 | 0.000 | β-strand start | Buried | IV | |
| I557T | 0.297 | 0.005 | Loop | Dimer | I | |||
| G559R | 0.470 | 0.229 | Loop | Dimer | I | |||
| V572L | 70 | 10 | 0.221 | 0.000 | Loop | Dimer | I | |
| G576E | 15 | 15 | trimeric | 0.254 | 0.254 | Loop | Dimer | I |
| I587T | 55 | 35 | 0.000 | 0.000 | helix start | Buried | IV | |
| A591T | 0.009 | 0.009 | Helix | Buried | IV | |||
| A600T | 0.000 | 0.000 | Helix | Buried | IV | |||
| A630T | 80 | 40 | 0.000 | 0.000 | Helix | Buried | IV | |
| A631T | 80 | 75 | hexameric | 0.109 | 0.109 | Helix | Buried | IV |
| A631V | 70 | 65 | hexameric | 0.109 | 0.109 | Helix | Buried | IV |
| I656N | 0.038 | 0.005 | Helix | Buried | IV | |||
| Y675H | 0.000 | 0.000 | Helix | Buried | IV | |||
| V679G | 0.000 | 0.000 | Helix | Buried | IV | |||
| V696M | 0.020 | 0.020 | β-strand | Buried | IV | |||
| G708S | 45 | 5 | 0.000 | 0.000 | β-strand | Interlobar | II | |
| M712T | 100 | 70 | 0.155 | 0.155 | Helix | Interlobar | II |
UDP-GlcNAc epimerase and ManNAc kinase activities are percentage values relative to the corresponding activities of the full length wild type GNE. Data extracted from reference [11].
Oligomeric state of the full length mutant GNE. Data extracted from reference [11].
Relative solvent accessibility of the residue calculated using the DSSP program [30] and normalized according to values in reference [31]. A value of 1 means full exposure of the residue while a value of 0 means the residue is fully buried.
The type of the secondary structure element the residue is located at was assigned using the DSSP program [30].
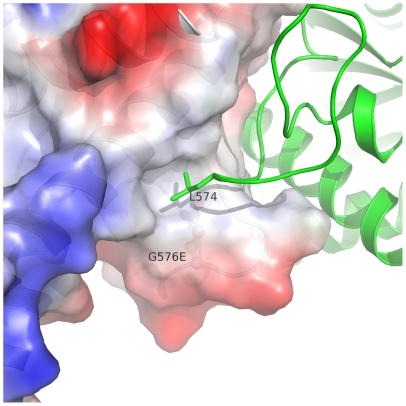
The first group of residues I557, G559, V572, and G576 is located at the dimerization interface of the C-lobe and mutation of these residues may interfere with dimerization of the kinase domain. It is noteworthy that kinase domain dimerization does not affect the solvent accessibility of G576 (Table 1), indicating that G576 is not directly involved in dimerization. The amino acid side chain of a mutant at this position would point into a hydrophobic niche that also accommodates the side chain of L574 from another monomer. The G576E mutation would exert both charge and space hindrances on the side chain of L574 and thus disrupt the dimerization (Figure 7), consistent with the previous observation that the G576E mutant of the full length GNE remains as a trimer [11].
Figure 7. Dimer interface around residue G576.
Surface charge of one molecule is shown around residue G576 (red for negatively charged surface, blue for positively charged surface). Side chain of L574 from another molecule is buried into the hydrophobic patch around G576. The side chain of the glycine residue is mutated to that of a glutamic acid residue and shown in ball-stick mode.
The location of this group of residues is also close to the residues involved in sugar substrate binding (Figure 6b). Residues V572 and G576 are located on the zinc-binding signature motif of the ROK family (Figure 6b, 8b), which could play both a functional and a structural role. Mutations of these residues could thus also affect the sugar substrate binding affinity of the kinase domain indirectly.
Figure 8. Disease-related mutations in the GNE kinase domain.
Panel A: M712 and its surrounding environment. M712 is within 4 Å with the side chain of L523 and forms hydrophobic interaction. Panel B: Disease-related mutations at residues V572 and G576 are located in the zinc-binding motif.
The second group of residues includes those located at the interfacial helices between the N-lobe and the C-lobe, i.e. N519, A524, F528, G708, and M712. Since the interlobar cleft is the site of ATP and carbohydrate binding as well as where phosphoryl transfer occurs, mutation of these residues could change the interlobar movement during catalysis and thus affect the kinase activity of the protein. For example, the first identified HIBM-related mutation, M712T [13], would likely abolish the hydrophobic interaction of the side chain of M712 with that of L523 from the C-lobe helix (Figure 8a). In the previous study [11], the M712T mutation has been shown to cause a 30% reduction in the kinase activity without affecting the epimerase activity of full length GNE. On the contrary, mutations of other residues in this second group reduce not only the kinase activity but also the epimerase activity of the full length protein ([11], Table 1). This suggests that the kinase domain is allosterically coupled to the epimerase domain. The structure of the full length GNE is needed to fully understand the coupled effects of the kinase and epimerase domains.
The third group currently includes residue P511. P511 has the highest relative solvent accessibility (>40%) among the 23 mutation sites and is located on a loop region of the structure. The underlying mechanism for the association of P511H and P511L mutations with HIBM is elusive without further data, but mutation of a proline to any other residue type will inevitably change the flexibility of the loop region around this residue and could thus change the allostery of full length GNE in the higher-order oligomeric state.
The fourth group of residues includes all the rest of mutation sites in Table 1. All residues have hydrophobic side chains and low solvent accessibilities, and are located within secondary structural elements. Mutations of these residues may disrupt the secondary structural elements at given mutation sites, and could interfere with the hydrophobic interactions of the secondary structure elements that stabilize the protein quaternary structure.
Summary
We show here the 3D structure of the N-acetylmannosamine kinase domain of GNE, the only ROK family kinase encoded in the human genome. The kinase domain dimerizes through an interface at the C-lobe. This is consistent with mutagenesis data from other groups on the full length GNE protein [11], [18]. The crystallographic hexamer, which consists of a trimer of kinase dimers, could serve as a prototype of a proposed full length GNE hexamer. Structure comparison of the GNE kinase domain with previously studied proteins revealed potential substrate binding sites at the interlobar cleft and also the structural and functional importance of the signature zinc-binding motif of the ROK family. Four groups of missense mutations associated with hereditary inclusion body myopathy are classified and their effects on the enzymatic activity can mostly be explained by the structure model.
Materials and Methods
DNA cloning, protein expression and purification
The cDNA template encoding the kinase domain of GNE was codon optimized for overexpression in E. coli and synthesized commercially (Codon Devices, Inc.) The DNA fragment encoding GNE residues 406–720 was PCR amplified and subcloned into the pET28-MHL vector (gi:134105571) using an In-Fusion dry-down PCR cloning kit (ClonTech). Protein was overexpressed in E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene) grown in terrific broth medium. The culture was grown at 37°C in a LEX bubbling system (Harbinger Biotech. & Engineering Corp.) until OD600 reached 3.0. The temperature of the culture was then lowered to 15°C and the cells were induced with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside and allowed to grow further overnight. Cells were harvested by centrifugation and flash frozen in liquid nitrogen and stored at −80°C until purification. Frozen cells were thawed and resuspended in 10 mM HEPES buffer (pH 7.5) containing 500 mM sodium chloride, 5% glycerol, 2 mM β-mercaptoethanol, and supplemented with 5 mM imidazole, and mechanically lysed using a microfluidizer (Microfluidics, model M-110EH) at 1,000 bar pressure. The lysate was clarified by centrifugation. GNE protein was bound with nickel-nitrilotriacetic acid (Ni-NTA) beads (Qiagen) at a ratio of 2.5 mL 50% Ni-NTA flurry per litre of cell culture. The bound protein was washed twice with the same HEPES buffer containing 30 mM or 75 mM imidazole, and finally eluted with the HEPES buffer supplemented with 300 mM imidazole. The elutant containing the GNE protein was further purified by Supderdex-75 size exclusion chromatography (GE Healthcare). The eluted fractions were pooled, concentrated to a final concentration of 40 mg per mL, and stored in a buffer containing 10 mM HEPES, pH 7.5, 500 mM sodium chloride, 5% glycerol and 5 mM dithiothreitol. The purity of the protein was better than 95% judging from SDS-PAGE gel.
Selenomethionine (SeMet) labelling of the protein was carried out using prepacked M9 SeMet growth media kit (Medicilon) following manufacturer's instructions.
Crystallization and structure determination
The ligand-free form crystals were grown at room temperature in sitting drops. A final concentration of 5 mM ADP, 1∶100 chymotrypsin (w/w) were added into the protein stock solution and 0.5 µL protein solution was mixed immediately with 0.5 µL well solution containing 15% polyethylene glycol (PEG) 4000, 0.2 M ammonium acetate, 0.1 M sodium citrate, pH 5.6 and set up for vapour diffusion crystallization. The SeMet crystal used for structure determination was grown in 14.55% PEG4000, 0.2 M ammonium acetate, 0.1 M sodium citrate, pH 6.0, with 1∶100 chymotrypsin (w/w) and 5 mM ADP in a sitting drop setup. Crystals grew to a mountable size within 24 hours. Paratone oil was used to cryo-protect the crystals.
Diffraction data of a selenomethionyl derivative of the GNE kinase domain were collected at beamline 19ID of the Advanced Photon Source (APS) at a wavelength of 0.9792 Å. Initial phases were obtained by single wavelength anomalous diffraction with SOLVE and density modification with RESOLVE [25]. For model building, the phases were combined with data collected at APS beamline 23ID-B at a wavelength of 0.9793 Å (see Table 2). The refined model of the target resulted from iterative application of density modification with DM and RESOLVE, interactive model building with COOT [26], coordinate and B-factor refinement with REFMAC [27] and PHENIX [28], and geometry validation with MOLPROBITY [29]. Diffraction data and refinement statistics are summarized in Table 2. The current model was deposited at the Protein Data Bank with PDB ID 3EO3.
Table 2. Crystallographic data and refinement statistics.
| SeMet | Native | |
| Diffraction data a | ||
| Space group | P3121 | P3121 |
| Cell dimensions: a, c (Å) | 128.88, 126.66 | 127.95, 127.25 |
| Resolution (Å) | 20.00−2.80 (2.90−2.80)b,c | 30.00−2.84 (2.94−2.84) |
| Unique HKLs | 27,466 (984) | 29,072 (2,860) |
| Completeness (%) | 90.7 (32.8) | 100.0 (100.00) |
| Rsym d (%) | 14.7 (>1) | 8.9 (97.8) |
| <I/σI> | 20.5 (0.4) | 40.0 (3.0) |
| Redundancy | 8.1 (1.5) | 11.1 (10.5) |
| Refinement | ||
| Initial phasing mean FOMe | - | 0.30 |
| No. of atoms: protein/others | - | 5,885/3 |
| Rwork/Rfree f (%) | - | 20.5/24.5 |
| Coordinate errorg Rwork/Rfree/max likelihood (Å) | - | 0.71/0.32/0.29 |
| RMSD bond length (Å)/angle (°) | - | 0.012/1.1 |
| Mean B-factor (Å2) | - | 73.8 |
| Ramachandran plot favored/outliers (%) | - | 96.1/0.0 |
Data reduced with DENZO and SCALEPACK.
Numbers in parentheses are for outer shell.
Phasing data were complete only to 3.15 Å, but were not re-scaled to that resolution limit.
Rsym = Σ|I-<I>|/ΣI.
FOM: Figures of merit.
Rwork = Σ||Fo|−|Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated as Rwork by using 3.8% of the data selected in thin resolution shells with SFTOOLS.
Estimated standard coordinate uncertainty.
Supporting Information
Standalone iSee datapack - contains the enhanced version of this article for use offline. This file can be opened using free software available for download at http://www.molsoft.com/icm_browser.html.
(ICB)
Instructions for installation and use of the required web plugin (to access the online enhanced version of this article).
(PDF)
Acknowledgments
We thank Dr. Linda McBroom-Cerajewski for editing the English of the manuscript and Dr. Wen Hwa Lee for preparing the iSee datapack version of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust. Diffraction data were collected at GM/CA-CAT (NCI Y1-CO-1020, NIGMS Y1-GM-1104) and Structural Biology Center at the Advanced Photon Source. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Traving C, Schauer R. Structure, function and metabolism of sialic acids. Cell Mol Life Sci. 1998;54:1330–1349. doi: 10.1007/s000180050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, et al. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyagi T, Wada T, Yamaguchi K, Hata K. Sialidase and malignancy: a minireview. Glycoconj J. 2004;20:189–198. doi: 10.1023/B:GLYC.0000024250.48506.bf. [DOI] [PubMed] [Google Scholar]
- 4.Adderson EE, Takahashi S, Bohnsack JF. Bacterial genetics and human immunity to group B streptococci. Mol Genet Metab. 2000;71:451–454. doi: 10.1006/mgme.2000.3025. [DOI] [PubMed] [Google Scholar]
- 5.Paltrinieri S, Gelain ME, Ceciliani F, Ribera AM, Battilani M. Association between faecal shedding of feline coronavirus and serum alpha1-acid glycoprotein sialylation. J Feline Med Surg. 2008;10:514–518. doi: 10.1016/j.jfms.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 7.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, et al. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 8.Titgemeyer F, Reizer J, Reizer A, Saier MH., Jr Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology. 1994;140 (Pt 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 9.Reinke SO, Eidenschink C, Jay CM, Hinderlich S. Biochemical characterization of human and murine isoforms of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE). Glycoconj J. 2009;26:415–422. doi: 10.1007/s10719-008-9189-6. [DOI] [PubMed] [Google Scholar]
- 10.Effertz K, Hinderlich S, Reutter W. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J Biol Chem. 1999;274:28771–28778. doi: 10.1074/jbc.274.40.28771. [DOI] [PubMed] [Google Scholar]
- 11.Penner J, Mantey LR, Elgavish S, Ghaderi D, Cirak S, et al. Influence of UDP-GlcNAc 2-epimerase/ManNAc kinase mutant proteins on hereditary inclusion body myopathy. Biochemistry. 2006;45:2968–2977. doi: 10.1021/bi0522504. [DOI] [PubMed] [Google Scholar]
- 12.Seppala R, Lehto VP, Gahl WA. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 14.Schiefner A, Gerber K, Seitz S, Welte W, Diederichs K, et al. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. J Biol Chem. 2005;280:29073–29079. doi: 10.1074/jbc.M504215200. [DOI] [PubMed] [Google Scholar]
- 15.Larion M, Moore LB, Thompson SM, Miller BG. Divergent evolution of function in the ROK sugar kinase superfamily: role of enzyme loops in substrate specificity. Biochemistry. 2007;46:13564–13572. doi: 10.1021/bi700924d. [DOI] [PubMed] [Google Scholar]
- 16.Holmes KC, Sander C, Valencia A. A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol. 1993;3:53–59. doi: 10.1016/0962-8924(93)90161-s. [DOI] [PubMed] [Google Scholar]
- 17.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blume A, Weidemann W, Stelzl U, Wanker EE, Lucka L, et al. Domain-specific characteristics of the bifunctional key enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biochem J. 2004;384:599–607. doi: 10.1042/BJ20040917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghaderi D, Strauss HM, Reinke S, Cirak S, Reutter W, et al. Evidence for dynamic interplay of different oligomeric states of UDP-N-acetylglucosamine 2-epimerase/N-acetyalmannosamine kinase by biophysical methods. J Mol Biol. 2007;369:746–758. doi: 10.1016/j.jmb.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam TW, Jung HI, An YJ, Park YH, Lee SH, et al. Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration. Proc Natl Acad Sci U S A. 2008;105:3751–3756. doi: 10.1073/pnas.0709295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunin VV, Li Y, Schrag JD, Iannuzzi P, Cygler M, et al. Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose. J Bacteriol. 2004;186:6915–6927. doi: 10.1128/JB.186.20.6915-6927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huizing M, Krasnewich DM. Hereditary inclusion body myopathy: a decade of progress. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbadis.2009.07.001. doi: 10.1016/j.bbadis.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad S, Gromiha M, Fawareh H, Sarai A. ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Standalone iSee datapack - contains the enhanced version of this article for use offline. This file can be opened using free software available for download at http://www.molsoft.com/icm_browser.html.
(ICB)
Instructions for installation and use of the required web plugin (to access the online enhanced version of this article).
(PDF)