Abstract
Background
Studies of the bacterial communities of the gut microbiota have revealed a shift in the ratio of Firmicutes and Bacteroidetes in obese patients. Determining the variations of microbial communities in feces may be beneficial for the identification of specific profiles in patients with abnormal weights. The roles of the archaeon Methanobrevibacter smithii and Lactobacillus species have not been described in these studies.
Methods and Findings
We developed an efficient and robust real-time PCR tool that includes a plasmid-based internal control and allows for quantification of the bacterial divisions Bacteroidetes, Firmicutes, and Lactobacillus as well as the methanogen M. smithii. We applied this technique to the feces of 20 obese subjects, 9 patients with anorexia nervosa, and 20 normal-weight healthy controls. Our results confirmed a reduction in the Bacteroidetes community in obese patients (p<0.01). We found a significantly higher Lactobacillus species concentration in obese patients than in lean controls (p = 0.0197) or anorexic patients (p = 0.0332). The M. smithii concentration was much higher in anorexic patients than in the lean population (p = 0.0171).
Conclusions
Lactobacillus species are widely used as growth promoters in the farm industry and are now linked to obesity in humans. The study of the bacterial flora in anorexic patients revealed an increase in M. smithii. This increase might represent an adaptive use of nutrients in this population.
Introduction
Obesity disorders affect many developed countries and result from an imbalance in energy (input versus output) with serious heath consequences, including cardiovascular disease, type II diabetes mellitus, and colon cancer [1], [2]. The role of gut microbiota in obesity is a current public heath problem. The gut environment likely contributes to the energy balance disequilibrium because of its involvement in energy intake, conversion, and storage. The impact of the gut microbiota composition on body weight gain or host adiposity has been supported by reports involving mouse models with a genetic tendency for obesity [3]–[5].
Advances in high-throughput sequencing methods have recently allowed for better characterization of the activities and composition of the complex gut microbial community in both healthy patients and those in disease states [6]–[10]. Large-scale clonal Sanger sequencing of the 16S rRNA genes of gut microbiota [7], [11] have contributed to our understanding of their diversity and ecology. These methods have allowed the identification of a vast number of new and uncultivated species [12]. A recent 16Sr RNA sequencing-based investigation showed that the Firmicutes/Bacteroidetes (F/B) ratio differed in obese and lean human subjects, mainly due to a reduced Bacteroidetes proportion in obese subjects. This finding was unexpected, because the Bacteroidetes member Bacteroides thetaiotaomicron is associated with host adiposity [3]. Using a weight loss program, Ley and colleagues [7] demonstrated that a decrease in the F/B ratio in obese individuals correlated with weight loss over time and suggested that modulating the abundance of specific bacterial communities might be beneficial in the treatment of obesity. The growth promoters such as Lactobacillus species could be one of these specific bacterial communities since its impact on food conversion and weight gain has been demonstrated in farm animals [13].
The characterization and the relative abundance of higher taxonomic orders of gut microbiota have been investigated using multiple methods, including metagenomics, microarrays, and sequencing of large clone libraries. Although these methods are powerful in terms of data production, they involve time-consuming, costly, and tedious data analysis.
In the present work, we identified and used the genomic signature of Lactobacillus, Methanobrevibacter smithii, Bacteroidetes, and Firmicutes divisions. We rapidly assessed their relative abundance in the microbiota of obese subjects, lean subjects, and patients with anorexia nervosa using a real-time PCR assay. This real-time PCR assay is a practical, low cost clinical method for monitoring the variations of bacterial phyla of the gut. The genus, species, and divisions we focused on may play a role in the disorder of obesity.
Methods
Bacterial Sequences and System Assessment
The sequences of archaea domain, M. smithii species, bacterial phyla and genus were downloaded from the ribosomal database Project II (RDP-II) using an online hierarchical browser tool. “Isolates” and “uncultured” sequences that were labeled as “good quality” and had a length greater than 1,200 bp were selected. The Firmicutes, Bacteroidetes and M. smithii systems (the nucleotide sequence “forward primer-probe-reverse primer”) were built as a nucleotide pattern. The pattern was used to search against sequences of phyla and genera using the DREG program of the EMBOSS package.
Statistical Tests
The Bacteroidetes copy number values were converted into logarithm values in order to equalize the standard deviations. The normal distribution was checked using the Shapiro-Wilk test. One-way ANOVA (P value for the Bartlett's test) with the Tukey-Kramer post-test was performed on the Bacteroidetes values for the anorexic, obese, and lean groups. A Fisher exact test was performed on a contingency table for the Lactobacillus copy number.
An unpaired t-test was used for the comparison of M. smithii quantification between the anorexic or the obese group and the lean control group. The statistics were performed using GraphPad Prism version 5 software.
Patient Recruitment
All the study was approved by the local ethics committee “Comité d'éthique de l'IFR 48, Service de Médecine Légale”, (Faculté de Médecine, 27 Bld Jean Moulin, 13005 Marseille, FRANCE) under the accession number N°08-003, January the 21st, 2009. All patients were recruited at University Hospital (CHU) of Marseille. Only verbal consent was necessary from patients for this study. This is relied on the French bioethics decree N° 2007-1220 published in the official journal of the French Republic. Normal weight control subjects were anonymously recruited from individuals working at the laboratory (or belonging to their family) (n = 20; 13 to 68 years-old; BMI = 20.68 kg/m2±2.014). No several members of the same family have been included in the study. Patients were enrolled in inpatient and outpatient clinics at the Department of Nutrition (University Hospital of Marseille, France). Obese patients were selected from patients attending the clinic for surgical treatment of excessive body weight (n = 20, 17 to 72 years-old; BMI = 47.09 kg/m2±10.66) and had not taken probiotics before the stool samples were collected. Subjects with anorexia nervosa (defined by DSM-4 criteria) were recruited from patients who were recently hospitalized in the department (n = 9, 19 to 36 years-old; BMI = 12.73 kg/m2 ±1.602). The body mass index (BMI) values were represented as mean values±SD.
Fecal DNA Isolation
Approximately 1 g of sample (two spoonfuls) was suspended in 10 ml of saline buffer and filtered into a Fecal Specimen Filtration Vial (Orion Diagnostica–Fumouze-Division Diagnostics, Levallois Perret, France). A total of 250 µl of the filtrate was transferred in a sterile screw-cap Eppendorf tube containing 0.3 g acid-washed glass beads (Sigma, Saint Quentin Fallavier, France). The tube was shaken in a FastPrep instrument BIO 101 (Qbiogene, Strasbourg, France) at level 6.5 (full speed) for 90 s to accomplish mechanical lysis. The supernatant was collected and incubated overnight at 56°C with 180 µl of the T1 Buffer from NucleoSpin® Tissue Mini Kit (Macherey Nagel, Hoerdt, France) and 25 µl of proteinase K (20 mg/ml). After a second cycle of mechanical lysis, the samples were incubated for 15 min at 100°C. Next, the DNA was extracted using the NucleoSpin® Tissue Mini Kit according to the manufacturer's procedure. Finally, DNA was eluted in 100 µl of elution buffer and stored at −20°C until use. An extraction negative control with 250 µl of sterile water was introduced in each series of the DNA extraction.
Real-Time Quantitative PCR Assays
The targeted genes, probe sequences, primer sequences (Eurogentec, Seraing, Belgium), and amplicon sizes for the two real-time PCR assays used in this study are summarized in Table 1. The MGB probes and primers of the Bacteroidetes and the Firmicutes divisions were designed on the basis of the genomic DNA barcodes previously described [14]. The probe and primers of the Lactobacillus genus used in this study were previously reported [15]. The M. smithii probe and primers (Table 1) were designed using Primer3 v0.4.0 software [16]. Real-time assays were performed in the MX3000™ system (Stratagene Europe, Amsterdam, The Netherlands) using the QuantiTect PCR mix (Qiagen, Courtaboeuf, France). Included in the samples were 5 pmol of primers and probes labeled with FAM or VIC. A total of 5 µl of DNA extracted from stools or a bacterial strain was diluted to 1/10, 1/100, or 1/1000. The dilutions were added to a final volume of 25 µl. The real-time PCR programs for Bacteroïdetes detection included 95°C for 15 min and 45 cycles (95°C 30 s, 48°C 45 s, 72°C 1 min). For Firmicutes and Lactobacillus detections, the programs included 95°C for 15 min and 45 cycles (95°C 30 s, 60°C 1 min). The specificity of the real-time PCR was tested against DNA from 108 bacterial strains. The results of the comparisons are reported in Supplementary Table S1. The detection and quantification of Lactobacillus were performed as previously reported [15]. The Bacteroïdetes and Firmicutes samples were quantified using a quantification plasmid which was constructed as previously described [15].
Table 1. Bacteroidetes and Firmicutes Real-Time PCR System.
| Target phylum | Target gene | Amplicon length (bp) | Primer and probe sequences |
| Lactobacillus | Tuf | 90 | FP: TACATYCCAACHCCAGAACG |
| RP: AAGCAACAGTACCACGACCA | |||
| Probe: AAGCCATTCTTRATGCCAGTTGAA | |||
| Firmicutes | 16S rRNA | 179 | FP: GTCAGCTCGTGTCGTGA |
| RP: CCATTGTAKYACGTGTGT | |||
| Probe: GTCAANTCATCATGCC | |||
| Bacteroidetes | 16S rRNA | 184 | FP: AGCAGCCGCGGTAAT |
| RP: CTAHGCATTTCACCGCTA | |||
| Probe: GGGTTTAAAGGG | |||
| M. smithii | 16S rRNA | 123 | FP: CCGGGTATCTAATCCGGTTC |
| RP: CTCCCAGGGTAGAGGTGAAA | |||
| Probe: CCGTCAGAATCGTTCCAGTCAG |
FP indicates the forward primer, and RP indicates the reverse primer.
Quantification Plasmid
Chimeric fragments were used for the quantification of Bacteroïdetes, Firmicutes, and M. smithii.
agcagccgcggtaatACGGAGGATCCGAGCGTTATCCGGATTTATTgggtttaaagggAGCGTAGGTGGACTGGTAAGTCAGTTGTGAAAGTTTGCGGCTCAACCGTAAAATTGCAGTTGATACTGTCAGTCTTGAGTACAGTAGAGGTGGGCGGAATTCGTGGTgtagcggtgaaatgcttaggtcagctcgtgtcgtga GATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATTGTTAGTTGCCATCATTTAGTTGGGCACTCTAGCGAGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGAC gtcaaatcatcatgcc CCTTATGACCTGGGCTacacacgtgctacaatgg ccgggtatctaatccggttcGCGCCCCTAGCTTTCGTCCCTCAccgtcagaatcgttccagtcagACGCCTTCGCAACAGGCGGTCCTCCCAGGATTACAGAAtttcacctctaccctgggag .
The fragment length is 485 bp. The Bacteroïdetes targeted sequence is in regular style, the Firmicutes targeted sequence is in bold style, and the M. smithii targeted sequence is in underlined style. The primer sequences are in lowercase and are underlined; the probe sequences are in lowercase and are in italic.
Reproducibility Assay
The fidelity of the DNA extraction and microbial quantifications were assessed. Three samples of feces from one obese patient (Ob10), one anorexic patient (Ano1), and one control patient (lean11) were defrosted. For each trial, about 1 g of fecal sample was suspended in 5 ml of 0.05 M Tris, pH 7.3. A volume of 250 µL of the suspension was used for the DNA extraction as described in the fecal DNA isolation section. This operation was performed five times and generated five independent DNA extracts per sample. Each DNA extract was tested five times by real-time PCR to determine the quantity of Firmicutes, Bacteroidetes, and Lactobacillus.
Results
Sensitivity and Specificity of the “Forward primer-probe-reverse primer” Systems
Oligonucleotide primers and probe sequences from Bacteroidetes, Firmicutes, Lactobacillus and M. smithii are shown in Table 1. The sensitivity and the specificity of the Bacteroidetes, Firmicutes and M. smithii systems were assessed in silico using the ribosomal RDP-II database [17]. The Bacteroidetes system is highly sensitive (89.89%), since it has a perfect match with 30,237 out of the 33,639 16S rRNA sequences of the RDP-II Bacteroidetes phylum. The Firmicutes system is also highly sensitive (88.94%), because it has a perfect match with 83,576 out of 93,969 of 16S rRNA sequences of the RDP-II Firmicutes phylum. The M. smithii system has a sensitivity of 88.89% (8 matches out of 9 sequences).
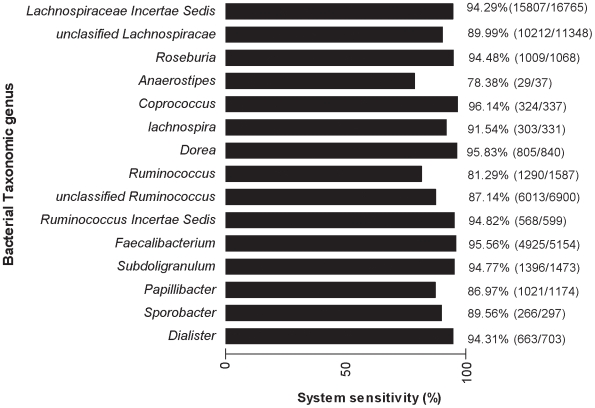
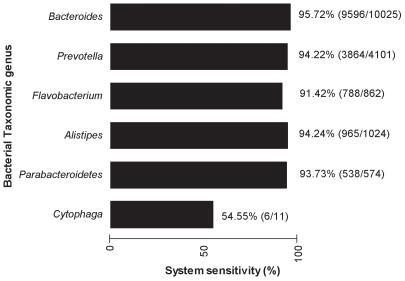
The assessment of the system's sensitivity was refined at the genus taxonomic level for Firmicutes and Bacteroidetes. Although Anaerostipes is the least sensitive genus (sequence match % <80) with the Firmicutes system (Figure 1), the most abundant genera, including Lachnospiraceae, Faecalibacteriun, and Subdoligranulum, are highly detected (sequence match % >90 for each genus). The 16S rRNA sequences of the Bacteroidetes genus were also efficiently recovered by the Bacteroidetes system (sequence match % >90), except for the Cytophaga genus (Figure 2).
Figure 1. Firmicutes system sensitivity at genus taxonomic level.
Scores in parentheses indicate the number of sequences matching the system out of the total sequence number of a given genus.
Figure 2. Bacteroidetes system sensitivity at genus taxonomic level.
Scores in parentheses indicate the number of sequences matching the system out of the total sequence number of a given genus.
The specificity (or the false discovery rate) of the two systems was evaluated by looking for perfect matches with sequences outside of the Firmicutes and Bacteroidetes phyla. Scanning all the RDP-II 16S rRNA sequences outside of the Firmicutes or the Bacteroidetes phyla, the false-discovery rate for the Firmicutes and Bacteroidetes phyla is 0.83% and 0.01%, respectively. As for the sensitivity, these results suggest a high specificity for the two systems. Finally, the false discovery rate of the M. smithii system reaches 0.025% for the archaea domain (2 matches out of 7,873 sequences) and is of 0% for the bacteria domain (no match out of 322,863 sequences).
Assessment of the Detection of Cultivated Strains from Bacteroidetes and Firmicutes Phyla
A large collection of strains inhabiting the gut microbiota that were taxonomically assigned to the Bacteroidetes and the Firmicutes bacterial phyla were tested by real-time PCR (Supplementary Table S1). The Bacteroidetes system (Supplementary Table S1, column A) successfully detected all Bacteroidetes strains, including Prevotella, Bacteroidetes, Parabacteroidetes, Captocytophaga, and Alistipes. A few false-positives were identified, including three strains that were assigned to the Firmicutes phylum (two Streptococcus and one Enterococcus). However, the detection of these strains is extremely weak due to the threshold cycle (CT) values, which were close to 40. With regard to the assessment of the Firmicutes system (Supplementary Table S1, column B), all the Firmicutes strains except Clostridium perfringens were identified. In addition, no false-positive results were obtained, suggesting a high specificity for the system. Finally, Fusobacterium strains were detected as false-positives by the Firmicutes probe system. The detection efficiency of the Lactobacillus system has been previously reported using a broad range of Lactobacillus strains [15].
Bacterial Quantification of Different Human Populations
Quantification of the bacterial groups using real-time PCR was performed on three distinct populations, including normal-weight healthy controls (lean), patients exhibiting obesity (BMI≥30 kg/m2), and patients suffering from anorexia nervosa (defined by DSM-4 criteria). These populations differed in terms of their body mass indices. The bacterial quantification results for the individuals are showed in Table 2.
Table 2. Bacteroidetes, Firmicutes, Lactobacillus and M. smithii Quantifications.
| Sample | Bacteroidetes | Firmicutes | Lactobacillus | M. smithii |
| Lean1 | 3.26E+10 | 4.68E+10 | 0.00E+00 | 8.16E+07 |
| Lean2 | 5.08E+10 | 4.60E+10 | 0.00E+00 | 4.56E+08 |
| Lean3 | 2.91E+10 | 5.40E+10 | 0.00E+00 | 1.93E+04 |
| Lean4 | 1.62E+10 | 1.52E+10 | 0.00E+00 | 2.55E+08 |
| Lean5 | 2.87E+09 | 9.00E+09 | 0.00E+00 | 0.00E+00 |
| Lean6 | 6.12E+09 | 1.49E+10 | 0.00E+00 | 2.77E+03 |
| Lean7 | 1.02E+10 | 1.45E+10 | 9.64E+05 | 6.72E+07 |
| Lean8 | 6.80E+09 | 1.52E+10 | 0.00E+00 | 0.00E+00 |
| Lean9 | 7.88E+09 | 1.72E+10 | 0.00E+00 | 8.80E+08 |
| Lean10 | 5.20E+09 | 3.46E+10 | 5.44E+07 | 5.72E+02 |
| Lean11 | 3.18E+10 | 2.15E+10 | 4.80E+04 | 7.24E+05 |
| Lean12 | 1.58E+09 | 2.14E+09 | 1.08E+04 | 2.96E+06 |
| Lean13 | 9.44E+09 | 1.72E+10 | 0.00E+00 | 0.00E+00 |
| Lean14 | 1.67E+10 | 4.32E+10 | 0.00E+00 | 0.00E+00 |
| Lean15 | 3.05E+10 | 3.37E+10 | 0.00E+00 | 2.02E+08 |
| Lean16 | 2.15E+09 | 1.70E+10 | 0.00E+00 | 0.00E+00 |
| Lean17 | 2.03E+09 | 4.16E+09 | 0.00E+00 | 8.64E+06 |
| Lean18 | 4.72E+09 | 1.39E+10 | 0.00E+00 | 6.68E+02 |
| Lean19 | 3.33E+09 | 1.06E+10 | 0.00E+00 | 2.53E+06 |
| Lean20 | 2.06E+08 | 4.68E+08 | 2.37E+04 | 9.16E+04 |
| Lean Mean | 1.35E+10 | 2.16E+10 | 2.77E+06 | 9.78E+07 |
| Ob1* | 1.93E+10 | 3.82E+10 | 4.36E+08 | 3.62E+06 |
| Ob2 | 3.85E+09 | 5.84E+10 | 3.36E+06 | 0.00E+00 |
| Ob3* | 1.77E+08 | 3.80E+09 | 1.29E+06 | 1.96E+02 |
| Ob4 | 4.92E+09 | 9.80E+09 | 0.00E+00 | 6.16E+08 |
| Ob5 | 2.01E+10 | 2.44E+10 | 0.00E+00 | 0.00E+00 |
| Ob6 | 5.44E+08 | 4.44E+09 | 0.00E+00 | 5.48E+03 |
| Ob7 | 1.05E+10 | 1.50E+10 | 0.00E+00 | 3.15E+02 |
| Ob8 | 1.63E+09 | 6.04E+09 | 4.28E+07 | 0.00E+00 |
| Ob9 | 1.94E+09 | 7.20E+09 | 3.61E+08 | 4.68E+03 |
| Ob10 | 2.68E+08 | 1.14E+10 | 2.21E+07 | 0.00E+00 |
| Ob11 | 9.08E+08 | 8.84E+09 | 0.00E+00 | 1.01E+09 |
| Ob12 | 4.76E+07 | 2.20E+09 | 5.28E+06 | 2.76E+04 |
| Ob13 | 2.14E+09 | 1.12E+10 | 0.00E+00 | 1.40E+04 |
| Ob14 | 1.34E+08 | 1.30E+10 | 0.00E+00 | 6.76E+02 |
| Ob15 | 6.20E+09 | 2.14E+10 | 0.00E+00 | 5.44E+02 |
| Ob16 | 7.20E+08 | 2.79E+09 | 3.96E+06 | 3.88E+02 |
| Ob17 | 6.60E+08 | 6.44E+09 | 0.00E+00 | 1.14E+03 |
| Ob18 | 2.96E+08 | 5.52E+09 | 1.96E+05 | 1.45E+09 |
| Ob19 | 3.37E+08 | 6.28E+09 | 0.00E+00 | 2.82E+08 |
| Ob20 | 5.80E+08 | 1.74E+10 | 0.00E+00 | 6.96E+05 |
| Ob Mean | 3.76E+09 | 1.37E+10 | 4.38E+07 | 1.68E+08 |
| Ano1 | 5.60E+08 | 5.44E+09 | 5.56E+05 | 5.08E+03 |
| Ano2 | 4.72E+08 | 6.72E+09 | 0.00E+00 | 1.17E+04 |
| Ano3 | 2.84E+09 | 1.36E+10 | 2.30E+04 | 4.48E+08 |
| Ano4 | 2.44E+10 | 1.98E+10 | 0.00E+00 | 3.82E+08 |
| Ano5 | 7.12E+09 | 1.42E+10 | 0.00E+00 | 3.19E+03 |
| Ano6 | 3.59E+10 | 1.48E+10 | 0.00E+00 | 9.40E+08 |
| Ano7 | 6.24E+09 | 1.26E+10 | 0.00E+00 | 7.12E+02 |
| Ano8 | 1.09E+10 | 2.04E+10 | 0.00E+00 | 8.84E+08 |
| Ano9 | 6.60E+09 | 7.24E+09 | 1.45E+05 | 2.08E+09 |
| Ano Mean | 1.06E+10 | 1.28E+10 | 8.04E+04 | 5.26E+08 |
Scores are bacteria/archaea copies per g of feces. Ano indicates anorexic individuals and Ob indicates obese individuals. Lean indicates normal weight individuals. The diabetic patients are mentioned by an asterisk (*).
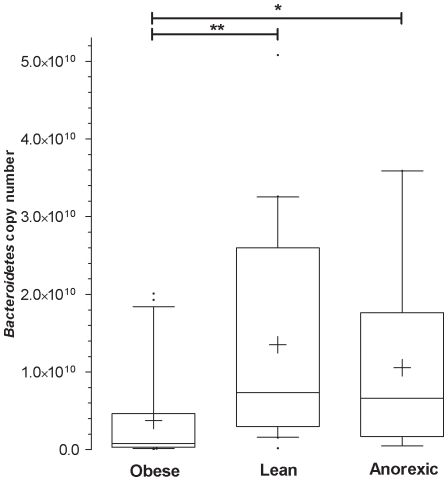
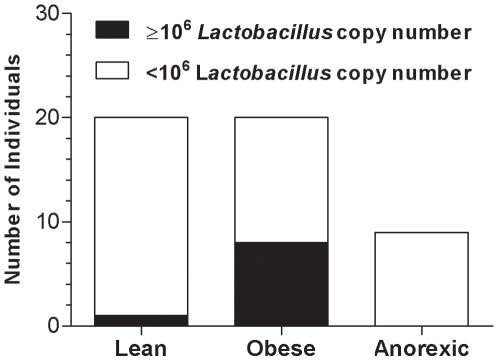
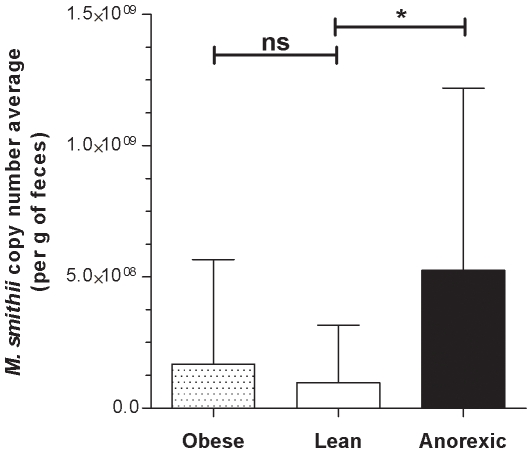
The Firmicutes data was similar in the three categories (data not shown). Compared to the lean and anorexic groups, the Bacteroidetes data from the obese group had a smaller mean, reduced median and smaller scatter (Figure 3). The differences in Bacteroidetes abundance between the obese group and the lean (** p<0.01) or anorexic groups (*p<0.05) were statistically significant (Figure 3). Although the average copy number of Lactobacillus was higher in the obese group than the other groups, the difference was not significant. However, at the individual level, the Lactobacillus copy number was generally much greater in the obese patients than in the lean or anorexic patients. A contingency bar graph using a threshold of 106 Lactobacillus copy numbers confirmed this observation (Figure 4). Based on this threshold, the difference in Lactobacillus concentration was significant when obese patients were compared with lean (p = 0.0197) and anorexic (p = 0.0332) patients. The average copy number of the archaeon M. smithii is indicated in Figure 5. The M. smithii quantification average was slightly higher for the obese group than the lean group as indicated by an obese/lean ratio of 1.72. In addition, the quantification average of M. smithii for the anorexic group was much greater than the lean and obese groups by factors of five and three, respectively. Overall, the obese group had a bacterial profile poor in Bacteroidetes and rich in Lactobacillus. No correlation was found between the quantification of bacterial and archaeal groups and the diabetic state of some obese patients. The anorexic bacterial profile is similar to the lean control group for the Firmicutes, Bacteroidetes, and Lactobacillus but different for M. smithii (anorexic group had higher quantification).
Figure 3. The Bacteroidetes quantification.
Box and whisker: 10–90 percentiles. Outliers are plotted as a black bubble, means are plotted as a plus, and medians are the black lines in the boxes. P value <0.05 is represented as * and P value <0.01 is represented as ** on the graph.
Figure 4. Distribution of high Lactobacillus concentrations.
The number of individuals having a high Lactobacillus concentration was 1 out of 20, 8 out of 20, and 0 out of 9 for the lean, obese, and anorexic groups, respectively.
Figure 5. Quantification of the archaeon M. smithii species.
Values are means±SD. P value <0.05 is represented as * and P value > 0.05 is indicated by “ns” for not significant.
Discussion
The molecular biology method developed in this study aimed to produce a large quantity of data concerning the composition of human gut microbiota. The data was used to analyze bacterial community variations and their correlation with body weight. Using in silico primers and probes designed by a pattern identification method [14], our method utilized highly sensitive and specific tests for Bacteroidetes and Firmicutes. In silico and experimental tests demonstrated the efficiency of our real-time PCR assay. To our knowledge, no such benchmarking of the detection quality of the Firmicutes and Bacteroidetes members has been carried out. The bacterial groups that belong to the Firmicutes and Bacteroidetes division and the Lactobacillus genus were amplified and identified by our method. The system's sensitivity and specificity of the detection of this last bacterial group was previously assessed by Menard et al. [15]. The reproducibility of the quantification results obtained from the different samples demonstrates the robustness of the extraction and quantification steps (Table 3). The analysis of five-fold independent DNA extracts indicated low inter-extract variability, with a coefficient of variation ranging from 0.52 to 6.08% for Bacteroidetes, from 0.98 to 4.81% for Firmicutes, and from 0.64 to 2.67% for Lactobacillus (Table 3). In addition, a quantification plasmid allowed for quantification of the bacteria copy number. This method was previously performed in the quantitative evaluation of bacteria genes associated with bioterrorism [18] and vaginosis [15]. The development of our real-time PCR system allowed us to test similar conditions to those described by the Gordon team [4]–[7]. To our knowledge, our study is the largest human study concerning obesity and gut microbiota to date. The 20 obese and 20 lean individuals in this study varied in Bacteroidetes number by a magnitude similar to that described by Gordon [4], [7] (Figure 3; p<0.01). This comparison validates our method and reveals that the Bacteroidetes profile of French obese patients is similar to that of American obese patients. Nine anorexics patients were also investigated in our study. The results of this group were similar to the lean control group for Bacteroidetes (Figure 3), Firmicutes (Table 2), and Lactobacillus (Table 2 and Figure 4). A major difference was the relative increase in M. smithii in anorexic patients compared to the lean control group (Figure 5; p = 0.0171). Accumulation of H2 in the human gut reduces the efficiency of microbial fermentation and thereby, the yield of energy. Methanogenic archaea are important since they are involved in the removal of H2 excess from the human gut [19]. The most common methanogenic archaeon found in the gut microbiota is M. smithii which can reduce CO2 with H2 to methane. The methanogen identification by cultivation based studies (in human faeces) has been hampered by the strict anaerobic nature of these archaea. The use of culture-independent methods has revealed the high proportion of methanogens [11] that may comprise up to 10% of all anaerobes in the colons of healthy adults [3], [11], [20], [21]. In addition, although methanogens are usually found in 30–50% of Western populations [19], [22], we believe that these proportions are underestimated. Besides, using our method, M. smithii was detected in 75% (15 out of 20), 80% (16 out of 20), and 100% (9 out of 9) of the lean, obese, and anorexic faeces, respectively. Methanobrevibacter associated with Bacteroides thetaiotaomicron led to weight gain in gnotobiotic mice [3]. An increase in Methanobrevibacter in obese patients was recently revealed in a study of obese patients before and after gastric bypass by comparing them to lean controls [21]. Our results were in agreement with these previous reports, since our obese group exhibited more Methanobrevibacter than our lean group (1.72-fold increase). However, the difference was not significant in our study (p = 0.0883). Surprisingly, the anorexic population showed significantly more Methanobrevibacter than lean patients (p = 0.0171). One explanation for this result may be that the development of Methanobrevibacter in anorexia nervosa patients might be associated with an adaptive attempt towards optimal exploitation of the very low caloric diet absorbed by these patients. M. smithii recycles hydrogen in methane, allowing for an increase in the transformation of nutrients into calories [3]. For anorexic patients, the increase of Methanobrevibacter can lead to optimization of food transformation in very low calorie diets. Another explanation for the increase in Methanobrevibacter is that it could be related to constipation, which is a common phenomenon in anorexia nervosa patients. An increase in methane producing-bacteria has been demonstrated in patients who suffer from constipation or diverticulosis [23], [24].
Table 3. Reproducibility Assay.
| Fecal Sample | Bacterial group | Assay extract1 | Assay extract2 | Assay extract3 | Assay extract4 | Assay extract5 | |
| Obese | Firmicutes | mean (Ct) | 17.6 | 17.28 | 17.12 | 17.56 | 18.55 |
| SD | 0.68 | 0.33 | 0.32 | 0.61 | 0.58 | ||
| CV (%) | 3.86 | 1.91 | 1.87 | 3.47 | 3.13 | ||
| Obese | Bacteroidetes | mean (Ct) | 16.46 | 16.22 | 15.62 | 16.79 | 18.35 |
| SD | 0.36 | 0.78 | 0.77 | 0.47 | 1.35 | ||
| CV (%) | 2.19 | 4.81 | 4.93 | 2.8 | 7.36 | ||
| Obese | Lactobacillus | mean (Ct) | 24.15 | 24.49 | 24.25 | 24.47 | 25.08 |
| SD | 0.32 | 0.42 | 0.28 | 0.29 | 0.67 | ||
| CV (%) | 1.33 | 1.71 | 1.15 | 1.19 | 2.67 | ||
| Anorexic | Firmicutes | mean (Ct) | 15.84 | 16.07 | 16.66 | 16.37 | 15.18 |
| SD | 0.43 | 0.6 | 0.28 | 0.16 | 0.73 | ||
| CV (%) | 2.71 | 3.73 | 1.68 | 0.98 | 4.81 | ||
| Anorexic | Bacteroidetes | mean (Ct) | 16.34 | 15.06 | 16.12 | 16.27 | 16.22 |
| SD | 0.64 | 0.15 | 0.98 | 0.73 | 0.47 | ||
| CV (%) | 3.92 | 1.00 | 6.08 | 4.49 | 2.9 | ||
| Anorexic | Lactobacillus | mean (Ct) | 26.05 | 26.14 | 26.34 | 26.25 | 25.14 |
| SD | 0.3 | 0.22 | 0.31 | 0.27 | 0.16 | ||
| CV (%) | 1.15 | 0.84 | 1.18 | 1.03 | 0.64 | ||
| Lean | Firmicutes | mean (Ct) | 17.94 | 17.85 | 18.12 | 18.26 | 17.75 |
| SD | 0.35 | 0.37 | 0.38 | 0.3 | 0.32 | ||
| CV (%) | 1.95 | 2.07 | 2.1 | 1.64 | 1.8 | ||
| Lean | Bacteroidetes | mean (Ct) | 15.23 | 15.33 | 15.71 | 14.73 | 15.27 |
| SD | 0.32 | 0.22 | 0.27 | 0.39 | 0.08 | ||
| CV (%) | 2.1 | 1.44 | 1.72 | 2.65 | 0.52 |
SD indicates Standard Deviation, means are the Cycle Threshold values (Ct), and CV indicates the Coefficient of Variation. Each assay extract used independent DNA extracts from a given fecal sample. The Ct mean of an assay extract is the result of five replicate real-time PCR measures.
The Lactobacillus population inhabiting the gut microbiota was also investigated. In a recent study, we focused on the potential role of Lactobacillus in animal weight gain [13]. Recently, child growth was suggested to be associated with Lactobacillus intake [25], [26]. Obese populations typically have higher concentrations of Lactobacillus than lean (p<0.05) or anorexic (p<0.05) populations (Figure 4). Due to the high inter-subject variability in Lactobacillus copy number, only a fraction (8 out of 20) of the obese population had a higher Lactobacillus copy number than the lean (1 out of 20) or anorexic (0 out of 9) population. This observation reinforces the possibility of a role for Lactobacillus in weight gain. This phenomenon was experimentally confirmed by iterated use of a growth promoter. Recent work by the Gordon team showed that specific enzymatic activities of obese individuals were found in Gram-positive bacteria of the Firmicutes phylum (of which Lactobacillus belong to) rather than in Bacteroidetes [6].
Interestingly, no or few Lactobacillus were retrieved from the databank of the analyzed bacteria [7]. The difference in these results can be explained by the difference of bacterial detection sensitivity of the methodologies as well as amplification or cloning bias. Our real time PCR tool is highly sensitive with a bacterial detection threshold of 102 bacteria copies/g of feces. Owing to the relative paucity of the fragments sequenced from studies that use clonal Sanger sequencing of 16S rRNA genes [11], the evaluation of the approximately 1011 bacteria copies/g of feces remains superficial by this sequencing method. Therefore, a range of 187 to 1100 fragments sequenced per obese [7] probably does not allow the detection of bacteria in concentrations lower than 107 copies/g of feces (such as the Lactobacillus). A recent study found major differences in the gut microbiota of Chinese people compared to American people [27]. Therefore, it is possible that our findings reflect a specific European or French diet.
Conclusion
This is the first study on the human gut microbiota that shown an increase of Lactobacillus in some obese individuals and an increase of M. smithii in anorexic patients. Based on these important findings, the use of microarrays for transcriptomic data comparison of Lactobacillus and M. smithii gene pool between the different populations could be an interesting challenge for the understanding of metabolic activities. However, the purpose of this work was to provide a useful tool for medical monitoring and diagnostic. We developed a molecular biology tool for quantifying the Bacteroidetes, Firmicutes, M. smithii, and Lactobacillus microbial groups. This tool allowed for the identification of specific patient microbial communities and the confirmation of a specific profile related to obesity. Patients suffering from anorexia nervosa had a profile similar to that of lean individuals. However, the M. smithii species quantification was higher in anorexic patients. Overall, this study provides preliminary data that links Lactobacillus levels with obesity.
Supporting Information
Sensitivity and Specificity of the Detection of Firmicutes and Bacteroidetes Strains by Quantitative PCR
(0.14 MB DOC)
Footnotes
Competing Interests: The « Université de la Mediterranée » deposed a patent on this study for which the authors Armougom F., Henry M., Drancourt M., and Raoult D. are the inventors.
Funding: The authors have no support or funding to report.
References
- 1.DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, et al. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. 2008;83:460–469. doi: 10.4065/83.4.460. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D. Obesity pandemics and the modification of digestive bacterial flora. Eur J Clin Microbiol Infect Dis. 2008;27:631–634. doi: 10.1007/s10096-008-0490-x. [DOI] [PubMed] [Google Scholar]
- 3.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 8.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 13.Khan M, Raoult D, Richet H, Lepidi H, La SB. Growth-promoting effects of single-dose intragastrically administered probiotics in chickens. Br Poult Sci. 2007;48:732–735. doi: 10.1080/00071660701716222. [DOI] [PubMed] [Google Scholar]
- 14.Armougom F, Raoult D. Use of pyrosequencing and DNA barcodes to monitor variations in Firmicutes and Bacteroidetes communities in the gut microbiota of obese humans. BMC Genomics. 2008;9:576. doi: 10.1186/1471-2164-9-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menard JP, Fenollar F, Henry M, Bretelle F, Raoult D. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis. 2008;47:33–43. doi: 10.1086/588661. [DOI] [PubMed] [Google Scholar]
- 16.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–366. [DOI] [PubMed] [Google Scholar]
- 17.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrel RN, La SB, Raoult D. Multi-pathogens sequence containing plasmids as positive controls for universal detection of potential agents of bioterrorism. BMC Microbiol. 2004;4:21. doi: 10.1186/1471-2180-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker A. Say hello to our little friends. Nat Rev Microbiol. 2007;5:572–573. doi: 10.1038/nrmicro1720. [DOI] [PubMed] [Google Scholar]
- 20.Miller TL, Wolin MJ. Methanogens in human and animal intestinal tracts. Systematic and applied microbiology. 1986;7:223–229. [Google Scholar]
- 21.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker A, Cerdeno-Tarraga A, Bentley S. Faecal matters. Nat Rev Microbiol. 2006;4:572–573. doi: 10.1038/nrmicro1483. [DOI] [PubMed] [Google Scholar]
- 23.Fiedorek SC, Pumphrey CL, Casteel HB. Breath methane production in children with constipation and encopresis. J Pediatr Gastroenterol Nutr. 1990;10:473–477. doi: 10.1097/00005176-199005000-00010. [DOI] [PubMed] [Google Scholar]
- 24.Weaver GA, Krause JA, Miller TL, Wolin MJ. Incidence of methanogenic bacteria in a sigmoidoscopy population: an association of methanogenic bacteria and diverticulosis. Gut. 1986;27:698–704. doi: 10.1136/gut.27.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchida K, Takahashi T, Inoue M, Morotomi M, Otake K, et al. Immunonutritional effects during synbiotics therapy in pediatric patients with short bowel syndrome. Pediatr Surg Int. 2007;23:243–248. doi: 10.1007/s00383-006-1866-6. [DOI] [PubMed] [Google Scholar]
- 26.Kanamori Y, Sugiyama M, Hashizume K, Yuki N, Morotomi M, et al. Experience of long-term synbiotic therapy in seven short bowel patients with refractory enterocolitis. J Pediatr Surg. 2004;39:1686–1692. doi: 10.1016/j.jpedsurg.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Wang B, Zhang M, Rantalainen M, Wang S, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity and Specificity of the Detection of Firmicutes and Bacteroidetes Strains by Quantitative PCR
(0.14 MB DOC)