Summary
Human sweet taste perception is mediated by the heterodimeric G-protein coupled receptor complex encoded by the TAS1R2 and TAS1R3 genes [1-7]. The extent of variation in these genes has been recently characterized [8], but the functional consequences of such variation are unknown. In this study we report that two C/T single nucleotide polymorphisms located at positions −1572 (rs307355) and −1266 (rs35744813) upstream of the TAS1R3 coding sequence strongly correlate with human taste sensitivity to sucrose, and explain 16% of population variability in perception. Individuals who carry T alleles display reduced sensitivity to sucrose compared to those who carry C alleles at these nucleotide positions. Using a luciferase reporter assay, we demonstrate that the T allele of each SNP results in reduced promoter activity in comparison to the C alleles, consistent with the phenotype observed in humans. We also found that the TAS1R3 promoter region extending from position −1700 to −1000 harbors a novel composite cis-acting element that has a strong silencing effect on promoter activity. We conclude that the rs307355 and rs35744813 SNPs affect gene transcription by altering the function of this regulatory element. A worldwide population survey reveals that the T alleles of rs307355 and rs35744813 occur at lowest frequencies in European populations. We propose that inherited differences in TAS1R3 transcription account for a substantial fraction of worldwide differences in human sweet taste perception.
Keywords: SYSNEURO
Results and Discussion
Measurements of Sucrose Sensitivity
In our study we analyzed threshold and suprathreshold sensitivity to sucrose by employing the novel use of signal detection analysis and R-index measures for this trait. The quantitative stimuli consisted of 9 different blinded sucrose solutions, which subjects sorted from least sweet to most sweet. For each pairwise sucrose concentration (e.g. 0−0.5%, 0.5−1%, etc) we estimated the classical signal detection measure of sensitivity P(A) - the area under a receiver operating characteristic (ROC) curve - by calculating the R-index [9, 10]. The area under an ROC curve, and hence the R-index, ranges from 0.5 (chance level discrimination) to 1 (perfect discrimination) and measures an individual's ability to discriminate between two stimuli. To obtain a singular measure of sensitivity across the entire sucrose concentration series, the pairwise R-indices were summed to create a concentration-response function for each subject and the area under this curve (AUC) was determined. The AUC, which can theoretically range from 0 (complete non-discrimination) to 9.25 (perfect discrimination) in this testing paradigm, was used as the dependent variable for assessing the effect of genotype on sucrose sensitivity.
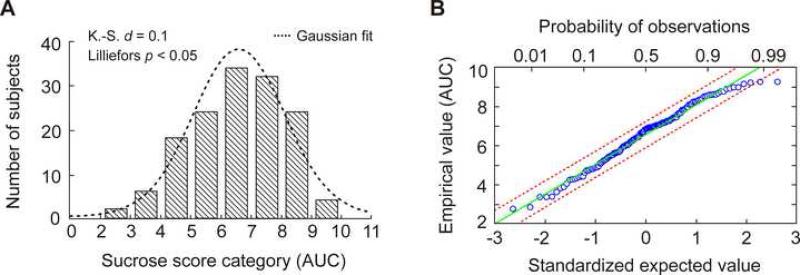
Our test population consisted of 144 unrelated individuals, who identified themselves as European (n = 92), Asian (n = 37), or African (n = 15). The distribution of AUC scores in the test sample population is shown in Figure 1A. Sensitivity scores were normally distributed in accordance with a Gaussian distribution (d = 0.1, Kolmogorov-Smirnov test; p < 0.05, Lilliefors test). Figure 1B shows a quantile-quantile plot for AUC scores determined in our subjects versus the expected values from a normal distribution. AUC scores follow a normal distribution, with the exception of modest deviation in the range of higher discrimination abilities.
Figure 1.
Sucrose sensitivity scores distributed normally in the population of 144 unrelated test subjects. (A) Frequency histogram showing the distribution of AUC scores. K.-S. d, Kolmogorov-Smirnov test for normality of empirical distribution. Lilliefors p, Lilliefors test for significance of K.-S. difference statistic. (B) Quantile-quantile plot of the empirical variable against the standardized normal variable. The vertical scale denotes the individual AUC values sorted in order from smallest to largest. The bottom axis shows a normally distributed variable. The top axis denotes probability of observations of a normally distributed variable. The regression line (green) is a reference demonstrating theoretically perfect fit between the empirical and theoretical distributions. Red dashed lines indicate 99% confidence intervals.
Sucrose Perception Genotype-Phenotype Relationships
Although individual variations in sweet taste perception have been associated with aging [11, 12], hormonal status [13, 14], sweet food intake [15], and anatomical factors [16], inherited factors clearly make important contributions to variability in this trait [17-19]. Human sweet perception is mediated by the products of the TAS1R2 and TAS1R3 genes, which encode the major carbohydrate sweet taste receptor [1-7]. An evolutionary genetic analysis revealed strong deviations from evolutionary neutral genetic drift for variation in the coding regions of TAS1R2 and, to a lesser extent, TAS1R3 [8]. Such deviations from neutrality suggested functional variations in sensitivity to sweet compounds.
Nucleotide sequence variations in the TAS1R2 and TAS1R3 genes in our subject group were assayed by complete DNA sequencing. TAS1R2 gene sequencing extended from 2553 bp upstream of the translation start site to 1366 bp downstream of the stop codon, spanning 23,981 bp of genomic DNA. TAS1R3 DNA sequencing extended from 3188 bp upstream of the translation start to 1828 bp downstream, comprising a total of 8135 bp of genomic DNA. Sequencing of the TAS1R2 gene and its flanking regions revealed 34 SNPs with minor allele frequencies (MAF) ≥ 0.05, including 5 exonic non-synonymous nucleotide changes (Table S1). Sequencing of the 8135 bp genomic region containing TAS1R3 revealed 20 SNPs with MAF ≥ 0.05, including two encoding protein amino acid replacements, as listed in Table S2.
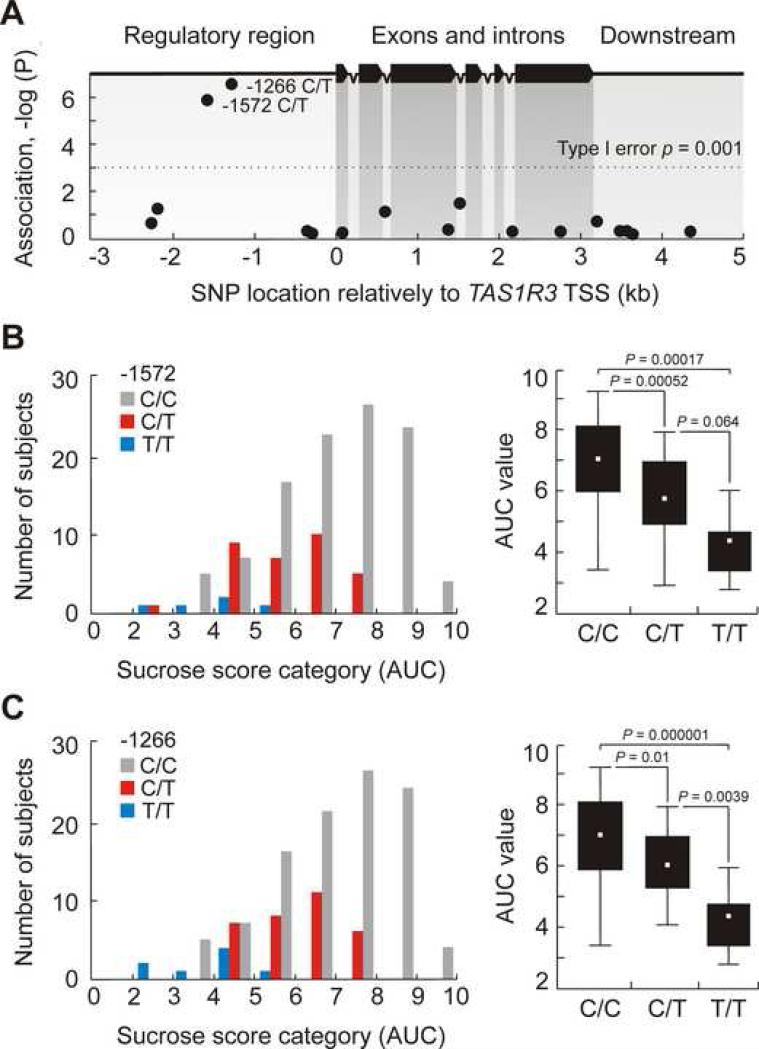
Putative effects of SNPs identified in the TAS1R2 and TAS1R3 genes were estimated using one-way ANOVA. No individual SNPs at the TAS1R2 locus displayed significant association with phenotype. However, we identified two, rs307355 (−1572 C/T) and rs35744813 (−1266 C/T) at the TAS1R3 locus that demonstrated strong association with decreased sucrose AUC scores (Bonferroni corrected p = 1.08×10−4 and 2×10−5, respectively) (Fig. 2 and Table 1). Determination coefficients calculated as the ratio SSTAS1R3 genotype/SStotal were 0.15 for rs307355 and 0.16 for rs35744813. Thus, together these SNPs account for ∼16% of the observed trait variability.
Figure 2.
Rs307355 (−1572) and rs35744813 (−1266) T alleles are significantly associated with decreased sucrose sensitivity scores. (A) The results of ANOVA association analysis of TAS1R3 SNP polymorphisms (MAF > 0.05) with phenotypes. Vertical scale is a probability of association, −log10 transformed. Horizontal axis indicates SNP positions in kb relatively to TAS1R3 translation start site, with bp 0 indicating the A of the ATG initiation codon. Dashed line on the graph indicates Type I error threshold, Bonferroni adjusted. (B and C) Frequency histograms and Box-Whisker plots demonstrate the distributions of individual AUC scores among rs307355 and rs35744813 genotypic groups. Black bars on Box-Whisker plots correspond to 25% and 75% percentiles, vertical lines indicate AUC value ranges. White rectangles inside of black bars are within group means. P, statistical significance of among group mean differences (Bonferroni post-hoc test).
Table 1.
Descriptive statistics for SNPs rs307355 and rs35744813
| Characteristic | SNP |
|||||
|---|---|---|---|---|---|---|
| rs307355 | rs35744813 | |||||
| Genotype | C/C | C/T | T/T | C/C | C/T | T/T |
| n in selection | 107 | 32 | 5 | 103 | 32 | 9 |
| Trait mean | 6.89 | 5.80 | 4.25 | 6.90 | 6.07 | 4.36 |
| +95% CI (AUC)* | 7.16 | 5.86 | 6.25 | 7.18 | 6.46 | 5.33 |
| −95% CI (AUC)* | 6.61 | 5.36 | 2.63 | 6.61 | 5.68 | 3.40 |
| SD* | 1.44 | 1.23 | 1.30 | 1.47 | 1.08 | 1.26 |
| Levene’ p** | 0.6 | 0.2 | ||||
| Fisher F | 14.5 | 16.4 | ||||
| P association*** | 1.08×10−4 | 2×10−5 | ||||
| R2, % | 14.9 | 16 | ||||
- CI, confidence intervals for the mean; SD, standard deviation
- Levene’ test p value for homogeneity of variances
- ANOVA probability of association, Bonferroni adjusted
Estimation of linkage disequilibrium between SNP polymorphisms identified in the TAS1R3 gene locus in our subject population showed that rs307355 and rs35744813 SNPs reside within the same haplotype block (r2 = 0.8). We observed only 6 individuals in which rs307355 and rs35744813 SNPs did not co-segregate. This number was insufficient for statistical analysis of a functional interaction between these SNPs. Population-based patterns of linkage disequilibrium available from the HapMap data base (http://snp.cshl.org/index.html) show that no other SNPs from the neighboring genes exist in linkage disequilibrium with rs307355 and rs35744813 (data not shown). This indicates that correlation of sequence variations in genes adjacent to TAS1R3 with AUC scores is unlikely in our sample populations.
Figures 2B and C illustrate that carriers of the C/T alleles exhibit a ∼25% decrease in sucrose sensitivity whereas homozygotes for the T/T alleles were more profoundly affected, as evidenced by a 50% decrease in their sucrose AUC scores. We observed reduced taste sensitivity to sucrose associated with T alleles in each of the three sample populations used in our study (Figure S1, Tables S3 and S4). These data suggest that this association is common to human populations worldwide.
Figure S2 shows examples of ROC-curves for the three groups of test subjects, who were C/C homozygotes for rs307355 and rs35744813 SNPs, C/T heterozygotes or T/T homozygotes for both SNPs. The presence of the T alleles of rs307355 and rs35744813 was associated with the reduced ability of subjects to perform fine discriminations in sucrose sweetness. However, discriminations at lower sucrose concentrations were performed by C/T heterozygote and T/T homozygote subjects with approximately equal aptitude as C/C homozygotes. At each sucrose concentration interval, the C/T heterozygotes and T/T homozygotes had cumulative R-indices that deviated further from the sensitivities of C/C homozygotes resulting in non-parallel concentration-response curves (Figure S2). This non-parallel shift between curves suggests that sensitivity of C/T heterozygotes and T/T homozygotes was decreased along the whole sucrose concentration range tested. Estimating the entire effect of such non-parallel shifting (AUC scores) produced statistically significant differences between the sucrose sensitivities of test subjects. However, sensitivity scores calculated at each particular sucrose concentration interval were less or not informative in genotype-phenotype association analysis (data not shown).
Distal Region of the Human TAS1R3 Promoter Harbor a Novel Cis-acting Element
Our findings suggested that SNPs rs307355 and rs35744813 affect the function of the TAS1R3 regulatory region. Quantitative variations of phenotypic traits in physiology, morphology, and disease susceptibility often approximate a statistical normal distribution [20, 21]. The molecular mechanism of such variations has been explained in a number of instances by differences in gene expression levels [22-24].
Toyono et al. have shown that the TAS1R3 core promoter is located upstream of the exon 1 and contains two potential transcription initiation sites, located 67 bp and 176 bp upstream of the translation start site [25]. However, we found that a transcription initiation site located 176 bp upstream of the ATG has a weak activity (see Supplemental Results and Discussions).
Our genotype-phenotype association data suggest that the sequence regions harboring SNPs rs307355 and rs35744813 and located 5’ upstream of the TAS1R3 core promoter also have a role in transcriptional regulation. To investigate the effects of corresponding nucleotide sequences on promoter activity, we created a series of luciferase reporter plasmids containing 5’-truncated variants of TAS1R3 regulatory region (constructs 1−12, Table S5). Promoter variants were assayed in two TAS1R3-expressing cell lines, NCI-H716 and HuCCT1. The NCI-H716 cell line was derived from enteroendocrine L cells of the human colon [26]. Jang et. al. have previously shown that enteroendocrine L cells express sweet taste receptors and other components of taste transduction pathway [26]. The HuCCT1 cell line was isolated from a human intrahepatic bile duct carcinoma [25]. Using immunohistochemical analysis, Toyono et. al. demonstrated that cholangiocytes in the intrahepatic bile duct express sweet taste receptors, indicating a role in chemical sensation in these tissues [25].
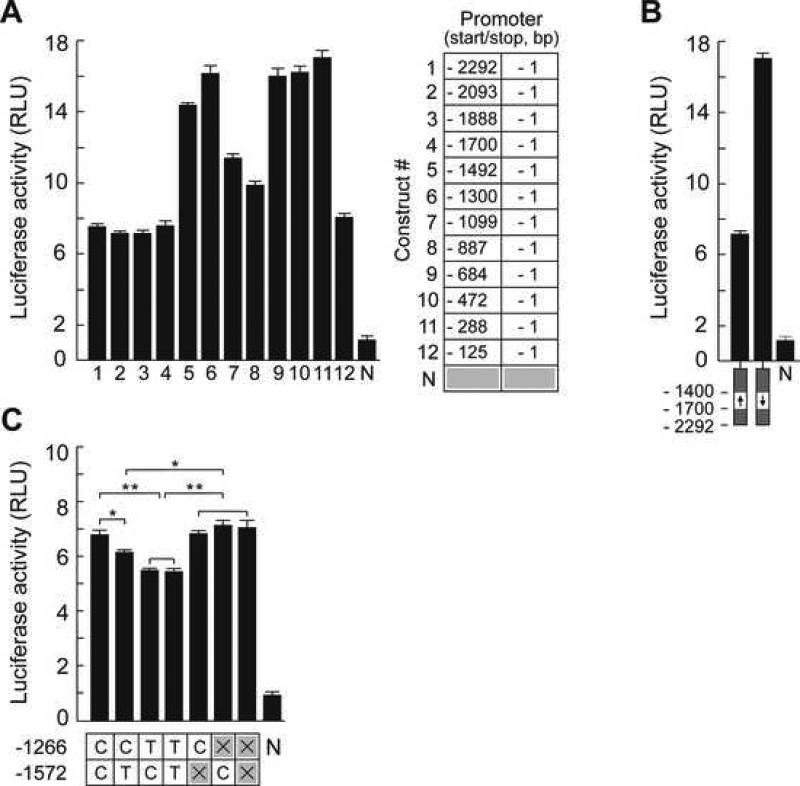
The results of reporter expression analysis in NCI-H716 cell line are shown on Figure 3A and B. Inclusion of the TAS1R3 promoter region from −2000 to −1500 bp in reporter constructs is associated with more than 50% reduction of the transcriptional level of the reporter gene (Fig. 3A, lines 1−4). In addition, a promoter variant created by inverting the nucleotide sequence between nt −1700 to −1400 demonstrated no reduction in activity (Fig. 3B). Another region suppressing transcription was observed between nt −1099 and −887 (Fig. 3A, lines 7 and 8). In contrast, the nucleotide sequences located between these two regions were associated with normal transcription (Fig. 3A, lines 5 and 6). These data demonstrate that the TAS1R3 regulatory region has a functionally modular structure in this model system.
Figure 3.
The results of ex vivo reporter analyses of TAS1R3 promoter variants in NCI-H716 cell line. (A) Reporter expression analyses of 5’-truncated promoter variants (table on the right and Table S5). Each black bar corresponds to individual assay and represents mean value of firefly luciferase activity normalized against the Renilla luciferase activity. Error bars show value ranges. RLU, relative light units. N, promoter-less negative control. (B) Reporter expression analysis of the intact promoter compared to the promoter variant with inverted sequence region located between nt −1700 and −1400. Black arrows below of the graph indicate normal orientation (arrow oriented up) and reverse orientation (arrow oriented down) of −1700 bp to −1400 bp sequence region. (C) Reporter expression analyses of promoter variants differing by genotypes at nt positions −1266 bp and −1572 bp. Table below of the graph shows specific genotypes at each of the two nucleotide positions. Symbols × in the table indicate eliminated E-box TFBS sequences. Asterisks, significance level of among group mean differences (unpaired t-test): one asterisk, P < 0.05; two asterisks, P < 0.01.
Similar results of reporter expression analysis were obtained in HuCCT1 cell line (Figure S3). However, the silencing effect of sequences located between nt −1099 and −887 on promoter activity was not observed in this cell line. This could be due to differences in the specific features of transcriptional regulation in NCI-H716 and HuCCT1 cell lines.
Repeat Masker software (http://www.repeatmasker.org) revealed the presence of a repetitive element in the promoter region extending from nt −1800 to −1100 (data not shown). Dot plot matrix alignment of TAS1R3 promoter sequence against itself identified multiple divergent duplications in this region indicating that this repetitive element belongs to the class of simple repeats (Figure S4). Simple repetitive elements account for ∼3% of the human genome [27, 28] and in some cases modulate transcription of housekeeping genes [29-31].
The DNA sequence of the TAS1R3 distal promoter region in higher primates shows no significant similarity to that of rodents (Figure S5). Furthermore, the ∼600 bp region containing SNPs rs307355 and rs35744813 is either deleted or shows no sequence similarity in other species, suggesting that this regulatory element has a unique role in humans in regulation of TAS1R3 gene expression. Although the TAS1R3 promoters of most vertebrates lack this regulatory region, a conservation track generated by Human Genome Browser (http://genome.ucsc.edu) revealed a high level of conservation with sequences located elsewhere in the genomes (data not shown), indicating that the similar sequence elements may be involved in transcriptional regulation of other genes.
Functional Effects of rs307355 and rs35744813 Alleles on Transcription
Examination of the TAS1R3 promoter sequence for statistically overrepresented transcription factor binding sites (TFBSs) revealed multiple E-box TFBSs in the region from nt −2000 to −1000 (Figure S6). SNPs rs307355 and rs35744813 are located within and downstream of the E-box core motif CANN(C/T)G(C/T), respectively (Figure S6). E-boxes serve as recognition sites for transcription factors of the bHLH (helix-loop-helix) class [32].
In order to further investigate the effects of rs307355 and rs35744813 alleles on transcription, we created sequence variants of the TAS1R3 promoter containing C and/or T alleles at nt positions −1572 and −1266, or which eliminated E-boxes at these positions (Table S5). We then used these to drive luciferase expression in reporter constructs transfected into NCI-H716 cell line. The results are shown in Figure 3C. Consistent with the reduced sucrose sensitivity in human subjects, the T alleles at each position individually significantly reduced luciferase transcription in comparison to the C alleles. However, no cumulative effect on transcription was observed when both T alleles were present in promoter simultaneously. Eliminating the E-boxes from our reporter constructs had the same effect as the C alleles. We conclude that the E-box TFBSs located at these positions are functional, and that naturally-occurring nucleotide variations in these regions affect transcription in our model cell system.
Worldwide Distribution of rs307355 and rs3574481 Alleles
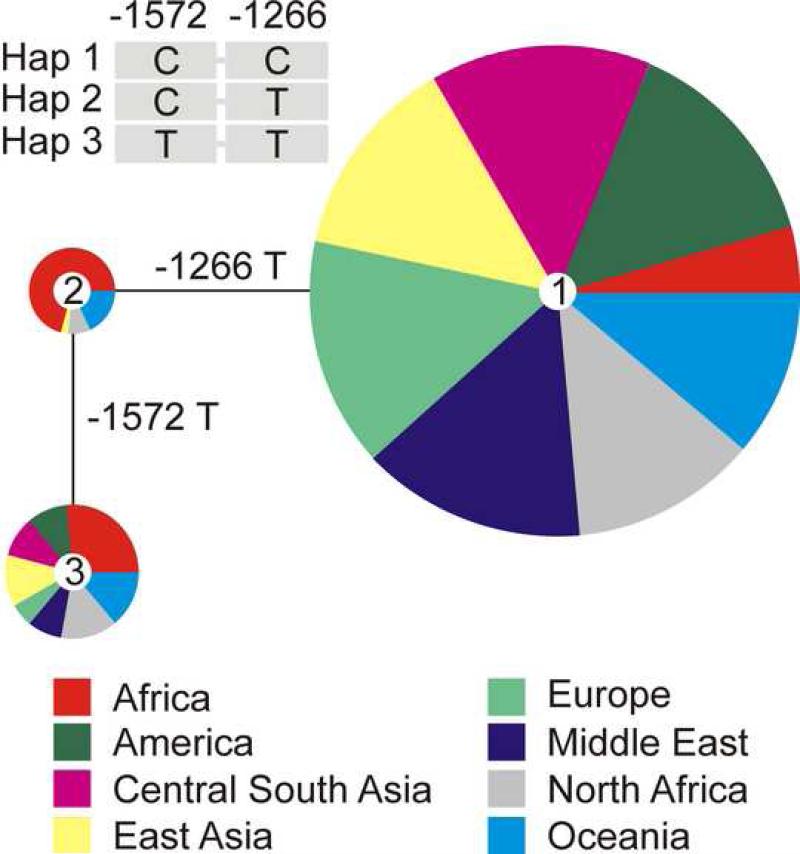
Figure S7 shows the distribution of rs307355 and rs3574481 alleles worldwide based on genotyping of 1062 DNA samples from 52 human populations in the HGDP-CEPH reference panel [33]. The rs307355 and rs3574481 C alleles are more frequent in worldwide populations outside of Africa, while the T-alleles are most prevalent in sub-Saharan African populations. Figure 4 shows the frequencies of haplotypes of these two SNPs in various populations. Rs307355 and rs3574481 C alleles (C – C haplotype) represent the major haplotype in all geographical regions except Africa. The alternative haplotype T - T is also distributed across most geographical regions with approximately equal frequencies, although it is much more frequent in African populations. Haplotype C – T is relatively rare and found mainly in African populations, while the T – C haplotype was not observed in more than 2000 chromosomes from our worldwide sample.
Figure 4.
Median joining network of three haplotypes found in 1064 individuals from the HGDP-CEPH diversity panel, color coded according to regions of origin.
Sucrose and other simple carbohydrates (glucose, fructose, maltose, etc.) are the main sources of energy for the human body and are involved in a multitude of biochemical pathways. The most common sources of simple carbohydrates for humans are plants, e.g. sucrose derived primarily from sugar cane and sugar beets [34]. Initially, carbohydrate-rich plants grew mainly in tropical latitudes [34], and we hypothesize that the observed differences in distribution of low and high sensitivity alleles among human populations could reflect the adaptation of those populations to their habitat in distant past. We hypothesize that the ability to taste sugars at lower concentrations was one of the critical factors for human survival in cold geographical regions. Thus, an evolutionary pressure favoring high sweet sensitivity alleles in the non-tropical geographical regions drove such alleles to high frequency in these populations.
Concluding remarks
This is the first observation that non-coding SNPs can affect human taste perception. Our finding of no significant association between phenotypes and non-synonymous SNPs located in protein coding regions of these genes was unexpected since allelic variation of the human TAS1R genes is unusually large, especially in TAS1R2 [8], the sweet-specific component of the receptor [3]. On the other hand, absence of significant sweet taste associations with individual SNPs at the TAS1R2 locus could be due to small sample size (n = 144) and the exclusion of rare variants (MAF < 0.05) from the statistical analysis. However, our data indicate that the several common alleles of TAS1R2 do not specify significant differences in sweet sensitivity in vivo. Consistent with these findings in vivo, we have also observed that the TAS1R2 protein variants commonly occurring in human populations and differing by single amino acid changes do not show appreciable differences in sensitivity to multiple sweeteners based on in vitro assessment (unpublished observations).
The TAS1R3 protein also serves as a component, along with the product of the TAS1R1 gene, of the receptor for umami taste, a savory flavor exemplified by the taste of glutamate [1]. The T alleles of rs307355 and rs3574481 exhibit a gradient across Eurasia, with East Asian populations showing the highest frequencies and Western European populations showing the lowest. The umami taste modality was discovered in East Asia and umami flavor enhancers are widely used there [35]. In addition to specifying population-specific preferences in sweet taste, these variants may also affect umami taste, especially in East Asians.
These findings also illustrate that rodents, in which most molecular sweet taste studies have been performed, are unlikely to serve as a high fidelity models of human sweet perception. Similar to other mammals, the rodent genomes bear no similarity to humans in the region upstream of TAS1R3 that contains these two SNPs. Our studies add to the growing body of evidence that human complex traits are associated with variants that reside outside non-coding regions, and are likely to affect gene expression [36]. In contrast to many such findings, however, these two SNPs exhibit a relatively large effect on phenotype, and represent common, rather than rare variants in the population.
Supplementary Material
Acknowledgements
We thank the research volunteers for their participation and J. Kleinman and Lindsay Bellegia for assistance with administration of taste tests. This research was supported by NIDCD/NIH funding Z01-000046-09, and by CRADA DC-CR-06-01 from Givaudan Flavors Corporation. C.T.S. and J.P.S. note they are employees of Givaudan Flavors Corporation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sainz E, Cavenagh MM, LopezJimenez ND, Gutierrez JC, Battey JF, Northup JK, Sullivan SL. The G-protein coupling properties of the human sweet and amino acid taste receptors. Developmental neurobiology. 2007;67:948–959. doi: 10.1002/dneu.20403. [DOI] [PubMed] [Google Scholar]
- 3.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 4.Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, West DB, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chemical senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nature genetics. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 6.Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nature neuroscience. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- 7.Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Molecular genetic identification of a candidate receptor gene for sweet taste. Biochemical and biophysical research communications. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- 8.Kim UK, Wooding S, Riaz N, Jorde LB, Drayna D. Variation in the human TAS1R taste receptor genes. Chemical senses. 2006;31:599–611. doi: 10.1093/chemse/bjj065. [DOI] [PubMed] [Google Scholar]
- 9.Brown J. Recognition assessed by rating and ranking. British Journal of Psychology. 1974;65:13–22. [Google Scholar]
- 10.O'Mahony M. Understanding discrimination tests: a user-friendly treatment of response bias, rating, and ranking R-index tests and their relationship to signal detection. Journal of Sensory Studies. 1992;7:1–47. [Google Scholar]
- 11.Fukunaga A, Uematsu H, Sugimoto K. Influences of aging on taste perception and oral somatic sensation. The journals of gerontology. 2005;60:109–113. doi: 10.1093/gerona/60.1.109. [DOI] [PubMed] [Google Scholar]
- 12.Stevens JC, Cain WS. Changes in taste and flavor in aging. Critical reviews in food science and nutrition. 1993;33:27–37. doi: 10.1080/10408399309527609. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura Y, Sanematsu K, Ohta R, Shirosaki S, Koyano K, Nonaka K, Shigemura N, Ninomiya Y. Diurnal variation of human sweet taste recognition thresholds is correlated with plasma leptin levels. Diabetes. 2008;57:2661–2665. doi: 10.2337/db07-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodin J. Insulin levels, hunger, and food intake: an example of feedback loops in body weight regulation. Health Psychol. 1985;4:1–24. doi: 10.1037//0278-6133.4.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Keskitalo K, Silventoinen K, Tuorila H, Perola M, Pietilainen KH, Rissanen A, Kaprio J. Genetic and environmental contributions to food use patterns of young adult twins. Physiology & behavior. 2008;93:235–242. doi: 10.1016/j.physbeh.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller IJ, Jr., Reedy FE., Jr. Variations in human taste bud density and taste intensity perception. Physiology & behavior. 1990;47:1213–1219. doi: 10.1016/0031-9384(90)90374-d. [DOI] [PubMed] [Google Scholar]
- 17.Bretz WA, Corby PM, Melo MR, Coelho MQ, Costa SM, Robinson M, Schork NJ, Drewnowski A, Hart TC. Heritability estimates for dental caries and sucrose sweetness preference. Archives of oral biology. 2006;51:1156–1160. doi: 10.1016/j.archoralbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Keskitalo K, Knaapila A, Kallela M, Palotie A, Wessman M, Sammalisto S, Peltonen L, Tuorila H, Perola M. Sweet taste preferences are partly genetically determined: identification of a trait locus on chromosome 16. The American journal of clinical nutrition. 2007;86:55–63. doi: 10.1093/ajcn/86.1.55. [DOI] [PubMed] [Google Scholar]
- 19.Mizuta E, Kokubo Y, Yamanaka I, Miyamoto Y, Okayama A, Yoshimasa Y, Tomoike H, Morisaki H, Morisaki T. Leptin gene and leptin receptor gene polymorphisms are associated with sweet preference and obesity. Hypertens Res. 2008;31:1069–1077. doi: 10.1291/hypres.31.1069. [DOI] [PubMed] [Google Scholar]
- 20.Belmont JW, Leal SM. Complex phenotypes and complex genetics: an introduction to genetic studies of complex traits. Current atherosclerosis reports. 2005;7:180–187. doi: 10.1007/s11883-005-0004-6. [DOI] [PubMed] [Google Scholar]
- 21.Mackay TF. The genetic architecture of quantitative traits. Annual review of genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- 22.Frey UH, Lieb W, Erdmann J, Savidou D, Heusch G, Leineweber K, Jakob H, Hense HW, Lowel H, Brockmeyer NH, et al. Characterization of the GNAQ promoter and association of increased Gq expression with cardiac hypertrophy in humans. European heart journal. 2008;29:888–897. doi: 10.1093/eurheartj/ehm618. [DOI] [PubMed] [Google Scholar]
- 23.Pare G, Chasman DI, Parker AN, Nathan DM, Miletich JP, Zee RY, Ridker PM. Novel association of HK1 with glycated hemoglobin in a non-diabetic population: a genome-wide evaluation of 14,618 participants in the Women's Genome Health Study. PLoS genetics. 2008;4:e1000312. doi: 10.1371/journal.pgen.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeufer A, Jalilzadeh S, Perz S, Mueller JC, Hinterseer M, Illig T, Akyol M, Huth C, Schopfer-Wendels A, Kuch B, et al. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circulation research. 2005;96:693–701. doi: 10.1161/01.RES.0000161077.53751.e6. [DOI] [PubMed] [Google Scholar]
- 25.Toyono T, Seta Y, Kataoka S, Toyoshima K. CCAAT/Enhancer-binding protein beta regulates expression of human T1R3 taste receptor gene in the bile duct carcinoma cell line, HuCCT1. Biochimica et biophysica acta. 2007;1769:641–648. doi: 10.1016/j.bbaexp.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J, Kim HH, Xu X, Chan SL, Juhaszova M, et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Sharma VK, Brahmachari SK, Ramachandran S. (TG/CA)n repeats in human gene families: abundance and selective patterns of distribution according to function and gene length. BMC genomics. 2005;6:83. doi: 10.1186/1471-2164-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epplen JT, Kyas A, Maueler W. Genomic simple repetitive DNAs are targets for differential binding of nuclear proteins. FEBS letters. 1996;389:92–95. doi: 10.1016/0014-5793(96)00526-1. [DOI] [PubMed] [Google Scholar]
- 30.Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 31.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Molecular biology and evolution. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 32.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Molecular and cellular biology. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science (New York, N.Y. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 34.Hussain R, Kinghorn A, Soearto D. Sweetening Agents of Plant Origin: Literature Search for Candidate Sweet Plants. Economic Botany. 1988;42(2):267–283. [Google Scholar]
- 35.Lindemann B, Ogiwara Y, Ninomiya Y. The discovery of umami. Chemical senses. 2002;27:843–844. doi: 10.1093/chemse/27.9.843. [DOI] [PubMed] [Google Scholar]
- 36.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.