Abstract
Background/aim
Alkali generation by oral bacteria plays a key role in plaque pH homeostasis and may be a major impediment to the development of dental caries. To determine if the capacity of oral samples to produce ammonia from arginine or urea was related to caries experience, the arginine deiminase system (ADS) and urease activity in saliva and dental plaque samples were measured in 45 adult subjects.
Methods
The subjects were divided into three groups according to caries status; 13 caries-free (CF) individuals (decayed, missing, and filled teeth = 0); 21 caries-active (CA) individuals (decayed teeth ≥ 4); and 11 caries-experienced (CE) individuals (decayed teeth = 0; missing and filled teeth > 0). Real-time polymerase chain reaction was used to quantify the proportion of certain acid- or alkali-producing organisms in the samples.
Results
The amount of ammonia generated from the test substrates by plaque samples was generally higher than that produced by salivary samples in all groups. Significantly higher levels of salivary ADS activity and plaque urease activity were observed in CF subjects compared to CA subjects (P = 0.0004 and P = 0.014, respectively). The proportions of Streptococcus mutans from saliva and dental plaque of CA subjects were significantly higher than those from the CF group (P = 0.0153 and P = 0.0009, respectively). In the CA group, there was an inverse relationship between urease activity and the levels of S. mutans (P < 0.0001).
Conclusion
This study supports the theory that increased caries risk is associated with reduced alkali-generating capacity of the bacteria colonizing the oral cavity; providing compelling evidence to further our understanding of oral alkali-generation in health and disease.
Keywords: alkali, arginine, caries, plaque, saliva, urea
Oral bacteria that colonize the teeth form dental plaque, a biofilm community that exists in dynamic equilibrium with host defenses and is generally compatible with the integrity of the tooth tissues (15, 16, 47). The transition from oral health to oral diseases, such as dental caries and periodontal disease, is characterized by compositional and metabolic changes in the complex communities of oral biofilms (16, 63). In the case of caries, frequent acidification of dental plaque favors the emergence of an acidogenic and acid-tolerant (aciduric) microflora, enriched for mutans streptococci and Lactobacillus spp., that is capable of rapidly fermenting dietary carbohydrates and lowering the pH to the extent where significant amounts of tooth demineralization can occur (12, 15, 16, 39, 63, 64). Although production of acid by dental plaque is the direct cause of dental caries, it is noteworthy that increases in the proportions of aciduric organisms appear to occur at the expense of species that are less aciduric and generally associated with dental health (1, 10, 12, 13, 24); including Streptococcus sanguinis and Streptococcus gordonii. Some of the less aciduric organisms associated with dental health derive protection from plaque acidification by hydrolyzing urea or arginine to ammonia, either by expressing a urease enzyme or by the arginine deiminase system (ADS), respectively. Production of ammonia by oral bacteria can positively influence the balance between remineralization and demineralization of the tooth and may help to prevent the emergence of a cariogenic microflora (26, 27, 34, 58, 59). Therefore, the capacity of oral biofilms to generate alkali appears to be a major caries-inhibiting factor (18).
The potential for alkali generation by oral bacteria as a means to prevent dental caries is supported by an extensive amount of evidence from in vitro studies (28, 32, 34, 35, 43, 44, 57, 60–62), as well as by some indirect clinical observations (26, 43, 49, 56, 65). For example, urea and arginine can be rapidly metabolized by oral bacteria to elicit a rise in environmental pH (19, 20, 48, 68, 69). Also, chronic renal failure patients, who have salivary urea levels as much as 50-times greater than healthy subjects, rarely develop caries, despite consuming a diet that is predominated by carbohydrates (29, 49, 55). A strong correlation between elevated levels of free arginine in saliva and caries resistance has also been revealed (65). In addition, dental plaque of caries-resistant individuals has been shown to have higher pH values compared to the plaque of caries-susceptible individuals (44, 61, 62), and in part the increased pH has been correlated with elevated ammonia levels. Notably, when rats were infected with a recombinant, urease-producing strain of Streptococcus mutans, a strong inhibitory effect on the development of caries was observed (22, 23). Recently, it was found that urease activity in the dental plaque of caries-free subjects was about three-fold higher than in caries-active subjects (56). Finally, the testing of formulations that contain arginine bicarbonate have revealed a profound anti-caries effect of this compound, presumably partly because of the ability of arginine to serve as a substrate for ammonia generation by plaque bacteria (3, 4). Collectively, a substantial body of evidence has accumulated to confirm that the modulation of the alkalinogenic potential of oral biofilms may be a promising strategy for caries control. Despite the fact that efforts are underway to modulate alkali generation in human oral biofilms, a detailed understanding of the role of alkali production in microbial ecology, and oral health and disease in humans, is only beginning to develop.
Arginine is found in saliva in micromolar concentrations, but is abundant in salivary peptides and proteins (65). Arginine entering the mouth is catabolized primarily by the ADS, which yields ornithine, carbon dioxide and ammonia, with the concurrent generation of adenosine triphosphate. The ADS is present in a variety of bacteria that colonize the teeth and soft tissues of the mouth in high numbers, including Streptococcus gordonii, Streptococcus sanguinis, Streptococcus parasanguis, and certain Lactobacillus species (18, 45, 52). Urea, the other major source of alkali in the mouth, is delivered in all salivary gland secretions at concentrations ranging from 3 to 10 mM in healthy individuals (6, 30). Urea entering the mouth is hydrolyzed to carbon dioxide and ammonia by bacterial ureases. Streptococcus salivarius is perhaps the most ureolytic oral organism, although Actinomyces naeslundii and oral haemophili, are also ureolytic (21, 54, 59). Even though other systems for the generation of alkali from arginine and urea may exist in the oral cavity, the majority of the ammonia produced from these substrates appears to come from the ADS and urease activity, respectively (17, 18, 33). Ammonia production from arginine and urea metabolism has been identified as a mechanism by which oral bacteria (i) are protected against acid killing (18, 19); (ii) maintain a relatively neutral environmental pH that may suppress the emergence of a cariogenic microflora (18, 45); and (iii) derive bioenergetic advantages, including increasing ΔpH and, for arginine specifically, synthesizing adenosine triphosphate (18, 51).
Although considerable progress has been made in dissecting the role and regulation of ureases and the ADS in oral bacteria, there remain serious deficiencies in our understanding of the importance of alkali generation in the ecology of human oral biofilms and the relationship of these activities to oral health. It is clear from the study by Shu et al. (56) that the ability to produce base in the oral environment can vary substantially within humans and that there is a correlation with caries status; but to date, no similar studies have been conducted to evaluate arginine catabolic potential or to begin to disclose the microbiological, biochemical, or molecular basis for heterogeneity in plaque alkali generation. Here, we compared the levels of the ADS and urease activities in saliva and dental plaque collected from caries-free (CF), caries-active (CA), and caries-experienced (CE) subjects, and quantified selected alkali-producing and acid-producing bacterial species. The information obtained is crucial to understand the influence of arginine and urea catabolism on plaque ecology and virulence, as well as for the development of novel diagnostic and caries risk assessment strategies and for conceiving novel preventive therapies based on enzymatic modulation of plaque pH.
Materials and methods
Study group
A total of 45 adult subjects (27 females and 18 males; mean age 33 years) with good periodontal and general health were recruited for this study from the predoctoral and faculty-practice clinics of the University of Florida. The subjects were organized into three groups consisting of 13 caries-free (CF) individuals with no clinical evidence of caries experience [decayed, missing and filled teeth (DMFT) = 0]; 21 caries-active (CA) individuals with at least four active, cavitated, and unrestored carious lesions (DT ≥ 4), independently of the number of missing and filled teeth (MFT ≥ 0); and 11 (caries-experienced (CE) individuals with previous experience of caries and absence of active carious lesions (DT = 0; MFT > 0). The DT mean was 8.8 and the MFT mean was 8.3 for the CA group. The selection process excluded subjects younger than 18 years of age and those with fewer than 20 teeth. Exclusion criteria also included subjects with systemic diseases; those treated with antibiotics, steroids or any medication known to cause xerostomia in the last 3 months; and subjects making use of removable or fixed dental appliances. Informed consent was obtained from each volunteer under a protocol reviewed and approved by the University of Florida Health Science Center Institutional Review Board (#226-2006).
Sampling
Subjects refrained from oral hygiene procedures and had fasted for 16 h overnight before sample collection. Whole unstimulated saliva was collected between 09.00 and 11.00 a.m. by asking the patients to expectorate into a chilled sterile plastic tube (Falcon 2070, Becton Dickinson and Company, Franklin Lakes, NJ, USA). After saliva collection, supragingival plaque was collected from all smooth dental surfaces of incisors and molar regions using sterile periodontal curettes (GR 4 and 5, Gracey finishing curettes, Ransom and Randolph, Toledo, OH, USA). Each portion of plaque scraped from surfaces of the teeth was transferred to sterile, chilled microcentrifuge tubes containing 10 mM sodium phosphate buffer (pH 8.0) (56). The samples were immediately transported on ice to the laboratory to be analyzed or, if necessary, snap-frozen by emersion in a carbon dioxide–ethanol bath, and stored at −80°C until the day of analysis. Previous work had shown that snap-freezing of oral samples does not adversely affect arginine or urea hydrolysis (M. M. Nascimento, V. V. Gordan, C. W. Garvan, C. M. Browngardt, R. A. Burne; data not published).
Biochemical assays: ADS and urease activity levels
Before the enzymatic assays, dental plaque and saliva samples were dispersed by external sonication (Heat Systems, Ultrasonics, Farmingdale, NY) for two cycles of 15 s, with cooling on ice during the interval. Plaque samples were also washed once with 10 mM Tris–maleate (pH 7.0) and resuspended in 500 μl of the same buffer. ADS activity was measured by quantification of the ammonia generated from the incubation of dental plaque (25 μl) and saliva samples (5 μl) in a mixture containing 50 mM arginine–HCl (Sigma-Aldrich Canada, Oakville, Ontario, Canada) and 0.5 mM Tris–maleate buffer (pH 6.0) for 90 min at 37°C. The ammonia produced was detected by Nessler’s reagent (Sigma-Aldrich, St. Louis, MO, USA) using ammonium sulfate as the standard. Each sample was assayed in triplicate, and controls for background and interference were always included. Urease activity was determined by quantification of ammonia produced from 50 mM urea (Fisher Scientific, Pittsburgh, PA, USA). ADS and urease activities were normalized to protein content and defined as μmol ammonia liberated [minute × (mg of protein)]−1.
Protein determination
To measure the amount of protein present in the dental plaque and saliva samples, 200 μl of the clinical samples were mixed with 200 μl of glass beads (0.1-mm, BioSpec Products, Inc., Bartlesville, OK). The samples were then homogenized in a Bead Beater using two cycles of 30 s, with cooling on ice during the interval. The samples were centrifuged for 5 min at 13,000 g in a refrigerated microcentrifuge (Labnet International, Woodbridge, NJ, USA). Protein concentration of the supernatant fluid was determined by the method of Bradford (14) using the BioRad Protein assay reagent (BioRad Laboratories, Hercules, CA, USA) with bovine serum albumin as the standard.
Quantitative real-time polymerase chain reaction
To obtain genomic DNA, the samples were dispersed as described previously and precipitated by centrifugation for 2 min at 13,000 g in a refrigerated microcentrifuge (Labnet International). The supernatant fluids were discarded and the cells were suspended in 480 μl of 50 mM ethylenediaminetetraacetic acid. Genomic DNA was then isolated from the samples using the Wizard DNA Purification kit (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. A total of 10 ng genomic DNA from each clinical sample was used in every real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) run. Oligonucleotide primers used in this study are listed in Table 1 and were designed using DNA MFOLD (http://www.bioinfo.rpi.edu/) and Beacon Designer 2.0 (Premier Biosoft International, Palo Alto, CA, USA), as described elsewhere (5). Species-specific primers were designed from the gtfB gene of S. mutans (70), the arginine deiminase genes of S. gordonii (arcA) and S. sanguinis (sagP), and urease subunit genes from A. naeslundii (ureA), and S. salivarius (ureC). The universal oligonucleotide primers for a broad range of bacterial 16S ribosomal RNAs (rRNAs) were also utilized (53). To test the specificity of the primers, genomic DNA from a battery of reference strains representing oral bacterial species and DNA isolated from pooled, human, whole saliva were used as templates for PCR. Standard curves for each gene were prepared as described by Yin et al. (71) and used in every real-time quantitative RT-PCR run. A range of 101–108 copies was found to be adequate for all the genes examined. Real-time PCRs were carried out in an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories) using iQSYBR green supermix (BioRad Laboratories).
Table 1.
Real-time polymerase chain reaction primers
| Bacterial strain | Target | Sequence | Product size (bp) |
|---|---|---|---|
| Universal for bacteria | 16S rRNA | F-ACT ACG TGC CAG CAG CC | 296–300 |
| R-GGA CTA CCA GGG TAT CTA ATC C | |||
| Streptococcus mutans | gtfB | F-AGC CAT GCG CAA TCA ACA GGT T | 415 |
| R-CGC AAC GCG AAC ATC TTG ATC AG | |||
| Streptococcus gordonii | arcA | F-GCT ATT CGT GAG TTG CTT CAA GG | 107 |
| R-TTT GCT GCT TCT GGA ATT TCT GG | |||
| Streptococcus sanguinis | sagP | F-GTG GTG GTG GCA ATA TCG TAG | 178 |
| R-CGA CCT CGA ACC AAT TCA CTT CC | |||
| Streptococcus salivarius | ureC | F-AGG TTC AGG TGG TGG ACA TGC | 98 |
| R-TTG TGG TGT ATG GGT TGA TTG GG | |||
| Actinomyces naeslundii | ureA | F-ACG AAG ACG CAA GGA CAG AGG | 118 |
| R-GTA GGC CAT GAG ATC CGT GAC C |
F, forward primer (5′ to 3′) and R, reverse primer (3′ to 5′).
Data analysis
Descriptive statistics and graphical displays including means, standard deviations, ranges, and box and whiskers plots were used to identify outliers, summarize the data, and check for distributional forms. To determine whether there were associations between ADS and/or urease activity levels in dental plaque and/or saliva and dental caries status (categorized as CF, CA, and CE), an analysis of variance (ANOVA) testing of outcome measures was applied. Tukey’s multiple comparison procedure was used to identify significant pairwise differences. To determine whether ADS and/or urease activity levels were associated with the numbers of selected acid-producing and alkali-producing bacterial species in plaque and/or saliva, three variables were constructed: (i) total number of acid-producing bacteria per ml of clinical sample (A/ml), (ii) total number of alkali-producing bacteria per ml of clinical sample (B/ml), and (iii) the ratio of A: B. Finally, the Spearman’s correlation was used to test the association between ADS and/or urease activity levels and the three constructed variables. SAS version 9.1 (SAS Institute, Cary, NC, USA) was used for all data management and statistical analyses.
Results
Alkali-generating activity of plaque and salivary bacteria
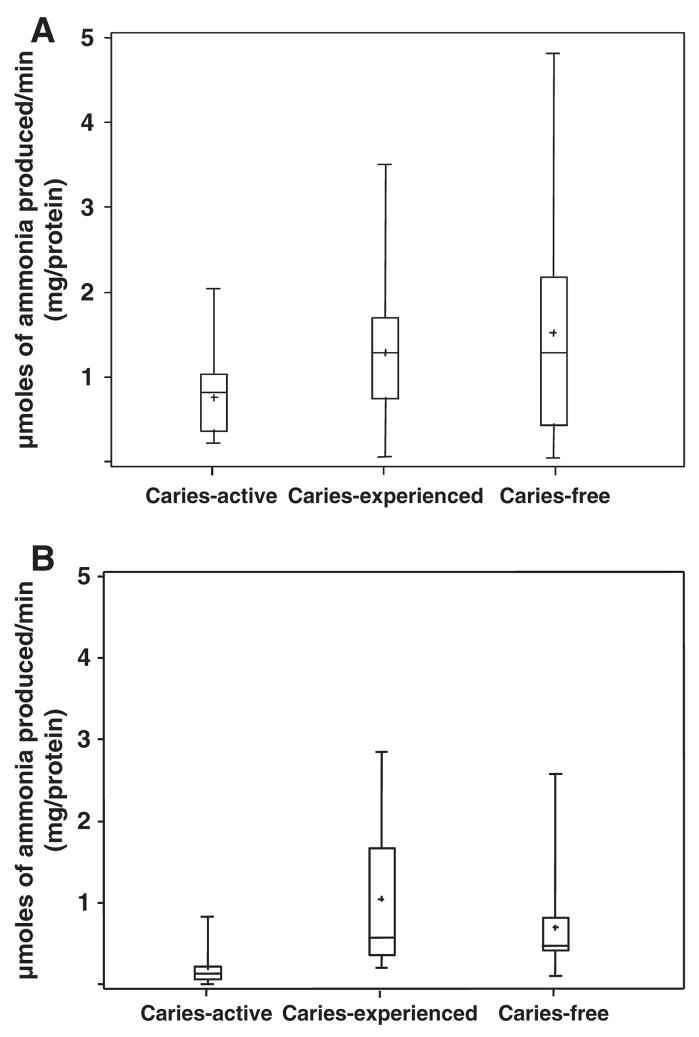
ADS activity levels in supragingival dental plaque and whole unstimulated saliva samples collected from the caries-status groups are presented in Fig. 1A and B and in Table 2. Both plaque and salivary ADS activity distributions were considerably asymmetric between individuals within all groups. Salivary ADS activity levels were generally lower than plaque ADS levels in all groups and within samples from the same individual. As shown in Table 2, the average ADS activity in the salivary samples was approximately five-fold higher in the CF group compared to the CA (P = 0.0004). Although plaque ADS activity clearly tended to be higher in CF and CE subjects compared to CA subjects, the differences between the groups were not statistically significant.
Fig. 1.
Arginine deiminase system activity in dental plaque A and saliva B.
Table 2.
Study variables in the caries-free, caries-active, and caries-experienced groups
| Caries-free | Caries-active | Caries-experienced | ||
|---|---|---|---|---|
| (n = 13) ± SD/Mean | (n = 21) ± SD/Mean | (n = 11) ± SD/Mean | ||
| SE2 (min–max) | SE2 (min–max) | SE2 (min–max) | P value3 | |
| Saliva AD1 | 1.08 ± 0.17 (0.73–1.42) | 0.21 ± 0.13 (0.005–0.48) | 0.69 ± 0.18 (0.33–1.07) | 0.0004 |
| Plaque AD | 1.36 ± 0.25 (0.85–1.87) | 0.74 ± 0.20 (0.34–1.15) | 1.41 ± 0.28 (0.85–1.96) | >0.05 |
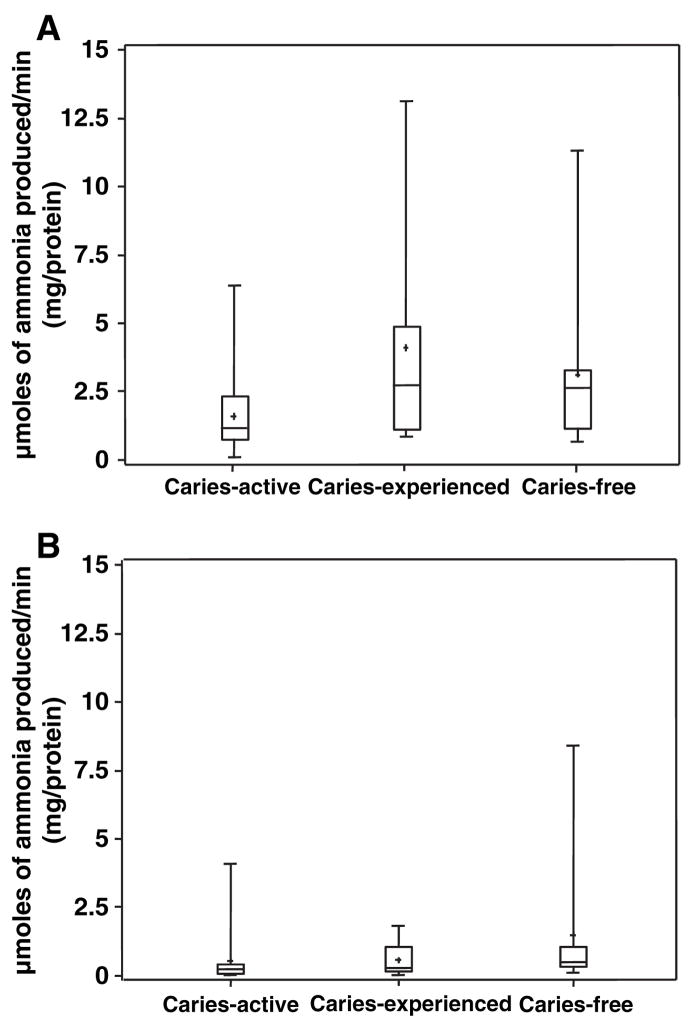
| Saliva urease* | 0.60 ± 0.39 (0.018–1.38) | 0.52 ± 0.31 (0.009–1.14) | 1.3 ± 0.42 (0.45–2.16) | >0.05 |
| Plaque urease | 4.13 ± 0.77 (2.58–5.68) | 1.51 ± 0.60 (0.29–2.73) | 3.22 ± 0.84 (1.53–4.91) | 0.014 |
Arginine deiminase system (ADS) and urease activity levels: μmol ammonia liberated min−1 (mg protein); SD, standard deviation.
Original values.
Log-transformation is used in the data analysis comparing caries-free to caries-active subjects.
Urease activity in dental plaque and saliva from each group of subjects is represented in Fig. 2A and B and in Table 2. As observed with arginine metabolism, urease activity in the saliva samples was generally lower than plaque urease activity in all groups. Dental plaque urease activity of CF and CE subjects was 2.7-fold and 2.1-fold higher, respectively, than that of CA subjects (P = 0.014 and P = 0.029), whereas salivary urease levels did not differ significantly (P > 0.05) with caries-status.
Fig. 2.
Urease activity in dental plaque A and saliva B.
Oral bacteria quantified by quantitative RT-PCR
Real-time PCR was used to quantify selected acid- and alkali-producing organisms in oral samples. Species-specific oligonucleotides were used to enumerate S. mutans, one of the most acidogenic species of the MS group; two arginolytic strains, S. sanguinis and S. gordonii; and two ureolytic strains, S. salivarius and A. naeslundii. The latter four organisms were selected based on their consistent ADS- or urease-positive phenotypes and the availability of DNA sequence for the genes of interest. ADS gene-specific primers were employed to identify strains in saliva samples based on the fact that statistically significant differences were observed in ADS activity only in the saliva samples between the caries groups (Table 2). Likewise, urease gene-specific primers were used only in plaque samples because of the differences shown in plaque urease activity between the groups (Table 2). Previously, the number of arginolytic and ureolytic strains was investigated in both dental plaque and saliva samples of a small group of subjects from the CA (n = 2) and CF (n = 4) and no significant differences were found between the oral samples and the caries groups (data not shown). S. mutans primers were used in both saliva and dental plaque samples. The proportions of each strain were obtained by normalization to total bacteria present in the same clinical sample as determined using 16S rRNA-specific primers (Table 3). Where applicable, the information derived from the PCR-based enumeration was found to be consistent with the data obtained by plating (data not shown), although, not surprisingly, higher numbers of total bacteria were routinely detected using the PCR-based method.
Table 3.
Percentage of salivary and dental plaque bacteria of the caries-free, caries-active and caries-experienced groups as determined by quantitative reverse transcription–polymerase chain reaction and normalized by total bacteria
| Caries-free | Caries-active | Caries-experienced | |||
|---|---|---|---|---|---|
| (n = 13) ± SD/Mean | (n = 21) ± SD/Mean | (n = 11) ± SD/Mean | |||
| SE (min–max) | SE (min–max) | SE (min–max) | Mean | P value1 | |
| Salivary Streptococcus gordonii | 0.32 ± 0.35 (0.016–0.95) | 0.65 ± 0.73 (0.034–2.16) | 0.70 ± 1.06 (0.029–3.00) | 0.55 | >0.05 |
| Salivary Streptococcus sanguinis | 0.060 ± 0.073 (0.005–0.26) | 0.078 ± 0.14 (0.0005–0.54) | 0.027 ± 0.045 (0.003–0.12) | 0.056 | >0.05 |
| Salivary Streptococcus mutans | 0.0018 ± 0.0029 (0.0005–0.0086) | 0.050 ± 0.094 (0.001–0.28) | 0.0012 ± 0.0012 (0.00026–0.0034) | 0.018 | 0.0153 |
| Plaque Streptococcus salivarius | 0.026 ± 0.053 (0.0001–0.18) | 0.52 ± 1.33 (0.00002–5.44) | 0.063 ± 0.16 (0.0007–0.44) | 0.20 | >0.05 |
| Plaque Actinomyces naeslundii | 0.317 ± 0.63 (0.00003–2.15) | 0.16 ± 0.27 (0.00003–0.89) | 0.076 ± 0.092 (0.00002–0.25) | 0.18 | >0.05 |
| Plaque S. mutans | 0.016 ± 0.041 (0.00026–0.14) | 0.051 ± 0.085 (0.00029–0.28) | 0.0012 ± 0.0012 (0.00027–0.0038) | 0.023 | 0.0009 |
Log-transformation is used in the data analysis comparing CF to CA subjects.
Among the three bacterial strains analyzed in the saliva samples of all groups, S. gordonii was detected in the highest proportions, followed by S. sanguinis and S. mutans (Table 3). A. naeslundii showed the highest proportion among the three bacterial strains examined in plaque samples of CF and CE subjects, followed by S. salivarius and S. mutans, although S. salivarius was detected in higher proportions in the plaque of CA subjects compared to the other organisms. There was no significant correlation between the levels of an individual arginolytic or ureolytic species and the associated alkali-producing enzyme activity detected in the groups of subjects. Also, when the proportions of S. sanguinis and S. gordonii were combined as the arginolytic group and the proportions of S. salivarius and A. naeslundii as the ureolytic group, no significant associations were found between the related enzyme activity or caries-status and the pairs of alkali-producing organisms.
The proportions of S. mutans from saliva and dental plaque of CA were significantly higher than those from the CF group (P = 0.0153 and P = 0.0009, respectively). At the time of collection, subjects with active caries lesions harbored higher levels of S. mutans than did CE or CF subjects. Interestingly, a negative correlation was found between the levels of S. mutans and alkali-producing enzyme activities, both ADS (P = 0.0023) and urease activity (P = 0.20); although the latter was not statistically significant when the Spearman’s rank correlation was used. Notably, an inverse relationship was also found between the levels of urease activity and S. mutans in the CA group (P < 0.0001).
Discussion
Ammonia production by oral bacteria is believed to have a major impact on oral microbial ecology and to be intimately intertwined with oral health and diseases. The nitrogenous salivary substrates that contribute significantly to base formation in the oral cavity are arginine and urea (18). In the present study, the rate of ammonia production from arginine and urea by clinical samples from individuals with no evidence of caries experience was greater than that of individuals with active caries lesions. More specifically, higher levels of salivary ADS activity and plaque urease activity were observed in CF individuals. Although not statistically significant, the higher levels of plaque ADS activity and saliva urease activity verified in CF and CE subjects may be biologically significant if they are able to impact the acid–base balance in the mouth. In fact, in an in vitro study where the levels of urease produced by a recombinant S. mutans strain could be manipulated, comparatively small increases in urease activity resulted in significant decreases in environmental acidification by cells co-metabolizing urea and an excess of glucose (22). It also appears from that same study that it is necessary to reach some threshold of ammonia-generating capacity before any substantial effects are seen on the pH profile of cells metabolizing glucose and urea.
There are a number of possible explanations for the observed differences among individuals from the caries-status groups in their capacity to generate ammonia from arginine and urea. The simplest, and the most consistent with the ecological plaque hypothesis for caries (47), is that alkali-generating enzymes are more abundant in CF individuals; which reflects differences in the proportions of alkali-producing microflora (36, 68). It is also well established that the expression of bacterial ureases and the ADS is subject to tight genetic control. Consequently, the levels of induction or repression of genes encoding alkali-generating enzymes may be affected by host factors, including diet and salivary molecules. Finally, it is also possible that the enzyme activity measured in salivary and plaque samples from the caries groups is influenced by the presence of factors that have an effect on the ability of cells to internalize the substrates or on the urease or ADS enzymes themselves (9, 25, 40, 41, 50).
Even though the function and diversity of the oral microflora associated with health, disease, and the transition from health to disease is only beginning to be characterized in detail (1, 24, 42), it is clear that organisms capable of maintaining plaque pH homeostasis are of crucial importance to oral health (17, 46). However, based on the results presented in Tables 2 and 3, a correlation between the presence of four specific species, or combinations of these species, and the alkalinogenic potential of plaque was not evident; suggesting that it may not be a simple change in the proportions of ADS- or urease-positive bacteria that is associated with caries development; at least in terms of the species examined in this study. However, these results emphasize just how little is known about the phenotypic capabilities of individual strains of oral bacteria and, in general, about the functions fulfilled by members of the oral microbiome. The selected probes for the AD and urease genes yielded melting curves in real-time PCR that were consistent with the detection of a single species so it is possible that the particular species we selected based on available sequence information and their established ability to express ADS or urease, are not the species responsible for the bulk of oral alkali generation. An alternative explanation, given the remarkable heterogeneity of individual species of oral bacteria studied so far, is that known ADS- or urease-positive bacteria differ widely in their capacity to break down arginine or urea. In fact, this idea is supported by the early work from the Sissons Laboratory that attempted to identify the principal urea-metabolizing organisms in plaque and saliva (58, 59). This heterogeneity among strains may be the result of (i) a constitutional difference in the expression of the genes for the enzymes, (ii) the differential sensitivity of the ADS or urease genes to induction or repression, or (iii) an inherent characteristic of the enzymes involved in urea or arginine metabolism, for example, differences in pH optima, Km or turnover rates. Along these lines, dental plaque samples generally had a higher capacity to produce ammonia than did the salivary samples, after normalization to the protein concentration of the samples. Even though salivary bacteria are, in part, derived from shedding of the microbiota colonizing the different oral niches, differences in the composition of the microbial populations in plaque and saliva are well-established (42). These results again highlight the paucity of information available on the microbiology of oral arginine and urea metabolism in humans and the relevance of investigating alkali production by salivary and plaque bacteria because of its affects on saliva and plaque pH and the cariogenic potential of the populations in these environments. More detailed microbiological, genetic and biochemical analyses of the oral microbiome are underway to disclose the basis for the substantial differences in the alkalinogenic potential of subjects.
Abelson and Mandel (2) demonstrated that the differences in plaque acidogenesis between caries-free and caries-susceptible individuals are no longer observed if salivary contact with dental plaque is blocked, which emphasizes the crucial role of saliva in the acid–base cycling of the oral cavity. The complexity of salivary components and the microbial populations has made it difficult to confirm specific associations between host factors and disease resistance, although there has been recent progress in associating specific salivary factors with caries susceptibility (7, 8, 31, 66, 72). Future investigations on functional properties of saliva and its protective components could focus on the ability of host factors to foster the colonization, growth and persistence of alkalinogenic species. For example, does heterogeneity in ADS activity in humans arise because of primary sequence heterogeneity in the polypeptides that fuel the ADS? Clearly, it will eventually be necessary to study microbial composition and behavior in the context of host factors to derive a comprehensive picture of how the composition and function of the oral microbiome is modulated.
Numerous studies have demonstrated an association between the proportions of mutans streptococci (MS), mostly S. mutans, in plaque and saliva and the caries status of populations, and to a minor extent to the caries status of an individual (11, 64). Consistent with this knowledge, S. mutans was detected in significantly greater proportions in clinical samples collected from CA subjects as verified by real-time PCR. One particularly interesting finding from this study was the observation that there was a negative correlation between the proportions of S. mutans and alkali-producing activity, mainly regarding urease activity in the dental plaque of CA subjects. A key virulence attribute of S. mutans that gives it a selective advantage over many other plaque bacteria is its ability to grow and continue to produce acid at low pH (38). Indeed, S. mutans competes comparatively weakly with organisms associated with a healthy flora at neutral pH values in vitro (15, 16), so the elevated pH that may be associated with increased urease activity would tend to select against aciduric species like S. mutans. An alternative theory, albeit not one that is mutually-exclusive, is that certain alkali-producing organisms suppress the growth and the expression of virulence genes of S. mutans. Such inhibition has been shown to occur through a variety of mechanisms, including bacteriocin production (67) and production of hydrogen peroxide (37).
The fact that dental caries still constitutes a major clinical problem mandates that oral health researchers explore new strategies for caries prevention and treatment therapy, as well as for assessment of caries risk. The findings from the present study suggest that an increased caries risk is associated with reduced alkali-generating capacity of the microbial populations colonizing the oral cavity. Studies are ongoing to identify novel bacterial strains and/or microbial associations capable of contributing to total arginolysis and ureolysis in the oral cavity. Likewise, additional investigations are being initiated to establish the nature and function of the factors controlling the alkali-generating activity of the oral microbiome.
Acknowledgments
This study was supported by DE10362 from the National Institute of Dental and Craniofacial Research.
References
- 1.Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abelson DC, Mandel ID. The effect of saliva on plaque pH in vivo. J Dent Res. 1981;60:1634–1638. doi: 10.1177/00220345810600090101. [DOI] [PubMed] [Google Scholar]
- 3.Acevedo AM, Machado C, Rivera LE, Wolff M, Kleinberg I. The inhibitory effect of an arginine bicarbonate/calcium carbonate CaviStat-containing dentifrice on the development of dental caries in Venezuelan school children. J Clin Dent. 2005;16:63–70. [PubMed] [Google Scholar]
- 4.Acevedo AM, Montero M, Rojas-Sanchez F, et al. Clinical evaluation of the ability of CaviStat in a mint confection to inhibit the development of dental caries in children. J Clin Dent. 2008;19:1–8. [PubMed] [Google Scholar]
- 5.Ahn SJ, Lemos JA, Burne RA. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol. 2005;187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Nowaiser A, Roberts GJ, Trompeter RS, Wilson M, Lucas VS. Oral health in children with chronic renal failure. Pediatr Nephrol. 2003;18:39–45. doi: 10.1007/s00467-002-0999-7. [DOI] [PubMed] [Google Scholar]
- 7.Anderson LC, Mandel ID. Salivary protein polymorphisms in caries-resistant adults. J Dent Res. 1982;61:1167–1168. doi: 10.1177/00220345820610101101. [DOI] [PubMed] [Google Scholar]
- 8.Ayad M, Van Wuyckhuyse BC, Minaguchi K, et al. The association of basic proline-rich peptides from human parotid gland secretions with caries experience. J Dent Res. 2000;79:976–982. doi: 10.1177/00220345000790041401. [DOI] [PubMed] [Google Scholar]
- 9.Barboza-Silva E, Castro AC, Marquis RE. Mechanisms of inhibition by fluoride of urease activities of cell suspensions and biofilms of Staphylococcus epidermidis, Streptococcus salivarius, Actinomyces naeslundii and of dental plaque. Oral Microbiol Immunol. 2005;20:323–332. doi: 10.1111/j.1399-302X.2005.00228.x. [DOI] [PubMed] [Google Scholar]
- 10.Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beighton D, Hellyer PH, Lynch EJ, Heath MR. Salivary levels of mutans streptococci, lactobacilli, yeasts, and root caries prevalence in non-institutionalized elderly dental patients. Community Dent Oral Epidemiol. 1991;19:302–307. doi: 10.1111/j.1600-0528.1991.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 12.Bowden GH. Possibilities for modifying the caries attack by altering the oral microflora. J Can Dent Assoc. 1984;50:169–172. [PubMed] [Google Scholar]
- 13.Bowden GHW, Ellwood DC, Hamilton IR. Microbial ecology of the oral cavity. Adv Microb Ecol. 1979;3:135–217. [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Bradshaw DJ, Marsh PD. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 1998;32:456–462. doi: 10.1159/000016487. [DOI] [PubMed] [Google Scholar]
- 16.Burne RA. Oral streptococci…products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 17.Burne RA, Chen YY. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2:533–542. doi: 10.1016/s1286-4579(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 18.Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 19.Casiano-Colon A, Marquis RE. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54:1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YY, Clancy KA, Burne RA. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect Immun. 1996;64:585–592. doi: 10.1128/iai.64.2.585-592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YY, Weaver CA, Burne RA. Dual functions of Streptococcus salivarius urease. J Bacteriol. 2000;182:4667–4669. doi: 10.1128/jb.182.16.4667-4669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clancy A, Burne RA. Construction and characterization of a recombinant ureolytic Streptococcus mutans and its use to demonstrate the relationship of urease activity to pH modulating capacity. FEMS Microbiol Lett. 1997;151:205–211. doi: 10.1111/j.1574-6968.1997.tb12571.x. [DOI] [PubMed] [Google Scholar]
- 23.Clancy KA, Pearson S, Bowen WH, Burne RA. Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect Immun. 2000;68:2621–2629. doi: 10.1128/iai.68.5.2621-2629.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corby PM, Lyons-Weiler J, Bretz WA, et al. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–5759. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curran TM, Ma Y, Rutherford GC, Marquis RE. Turning on and turning off the arginine deiminase system in oral streptococci. Can J Microbiol. 1998;44:1078–1085. doi: 10.1139/cjm-44-11-1078. [DOI] [PubMed] [Google Scholar]
- 26.Dawes C, Dibdin GH. Salivary concentrations of urea released from a chewing gum containing urea and how these affect the urea content of gel-stabilized plaques and their pH after exposure to sucrose. Caries Res. 2001;35:344–353. doi: 10.1159/000047473. [DOI] [PubMed] [Google Scholar]
- 27.Dibdin GH, Dawes C. A mathematical model of the influence of salivary urea on the pH of fasted dental plaque and on the changes occurring during a cariogenic challenge. Caries Res. 1998;32:70–74. doi: 10.1159/000016432. [DOI] [PubMed] [Google Scholar]
- 28.Dibdin GH, Shellis RP. Physical and biochemical studies of Streptococcus mutans sediments suggest new factors linking the cariogenicity of plaque with its extracellular polysaccharide content. J Dent Res. 1988;67:890–895. doi: 10.1177/00220345880670060101. [DOI] [PubMed] [Google Scholar]
- 29.Epstein SR, Mandel I, Scopp IW. Salivary composition and calculus formation in patients undergoing hemodialysis. J Periodontol. 1980;51:336–338. doi: 10.1902/jop.1980.51.6.336. [DOI] [PubMed] [Google Scholar]
- 30.Golub LM, Borden SM, Kleinberg I. Urea content of gingival crevicular fluid and its relation to periodontal diseases in humans. J Periodontal Res. 1971;6:243–251. doi: 10.1111/j.1600-0765.1971.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Ruan M, Wang Q. A study of parotid salivary proteins from caries-free and caries-active people by high performance liquid chromatography. Zhonghua Kou Qiang Yi Xue Za Zhi. 1997;32:95–98. [PubMed] [Google Scholar]
- 32.Imfeld T, Birkhed D, Lingstrom P. Effect of urea in sugar-free chewing gums on pH recovery in human dental plaque evaluated with three different methods. Caries Res. 1995;29:172–180. doi: 10.1159/000262065. [DOI] [PubMed] [Google Scholar]
- 33.Kanapka JA, Kleinberg I. Catabolism of arginine by the mixed bacteria in human salivary sediment under conditions of low and high glucose concentration. Arch Oral Biol. 1983;28:1007–1015. doi: 10.1016/0003-9969(83)90055-9. [DOI] [PubMed] [Google Scholar]
- 34.Kleinberg I. Effect of urea concentration on human plaque pH levels in situ. Arch Oral Biol. 1967;12:1475–1484. doi: 10.1016/0003-9969(67)90183-5. [DOI] [PubMed] [Google Scholar]
- 35.Kleinberg I. Prevention and dental caries. J Prev Dent. 1978;5:9–17. [PubMed] [Google Scholar]
- 36.Kleinberg I, Jenkins GN, Chatterjee R, Wijeyeweera L. The antimony pH electrode and its role in the assessment and interpretation of dental plaque pH. J Dent Res. 1982;61:1139–1147. doi: 10.1177/00220345820610100601. [DOI] [PubMed] [Google Scholar]
- 37.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- 39.Lingstrom P, van Ruyven FO, van Houte J, Kent R. The pH of dental plaque in its relation to early enamel caries and dental plaque flora in humans. J Dent Res. 2000;79:770–777. doi: 10.1177/00220345000790021101. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Hu T, Jiang D, Zhang J, Zhou X. Regulation of urease gene of Actinomyces naeslundii in biofilms in response to environmental factors. FEMS Microbiol Lett. 2008;278:157–163. doi: 10.1111/j.1574-6968.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 41.Ma Y, Rutherford GC, Curran TM, Reidmiller JS, Marquis RE. Membrane locus and pH sensitivity of paraben inhibition of alkali production by oral streptococci. Oral Microbiol Immunol. 1999;14:244–249. doi: 10.1034/j.1399-302x.1999.140408.x. [DOI] [PubMed] [Google Scholar]
- 42.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 43.Margolis HC, Duckworth JH, Moreno EC. Composition and buffer capacity of pooled starved plaque fluid from caries-free and caries-susceptible individuals. J Dent Res. 1988;67:1476–1482. doi: 10.1177/00220345880670120701. [DOI] [PubMed] [Google Scholar]
- 44.Margolis HC, Duckworth JH, Moreno EC. Composition of pooled resting plaque fluid from caries-free and caries-susceptible individuals. J Dent Res. 1988;67:1468–1475. doi: 10.1177/00220345880670120601. [DOI] [PubMed] [Google Scholar]
- 45.Marquis RE, Bender GR, Murray DR, Wong A. Arginine deiminase system and bacterial adaptation to acid environments. Appl Environ Microbiol. 1987;53:198–200. doi: 10.1128/aem.53.1.198-200.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh PD. Dental plaque as a biofilm and a microbial community – implications for health and disease. BMC Oral Health. 2006;6(Suppl 1):S14. doi: 10.1186/1472-6831-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 48.Morou-Bermudez E, Burne RA. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect Immun. 1999;67:504–512. doi: 10.1128/iai.67.2.504-512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson S, Woodhead J, Crall J. Caries resistance in children with chronic renal failure: plaque pH, salivary pH, and salivary composition. Pediatr Res. 1985;19:796–799. doi: 10.1203/00006450-198508000-00003. [DOI] [PubMed] [Google Scholar]
- 50.Phan TN, Buckner T, Sheng J, Baldeck JD, Marquis RE. Physiologic actions of zinc related to inhibition of acid and alkali production by oral streptococci in suspensions and biofilms. Oral Microbiol Immunol. 2004;19:31–38. doi: 10.1046/j.0902-0055.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 51.Poolman B, Driessen AJ, Konings WN. Regulation of arginine–ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol. 1987;169:5597–5604. doi: 10.1128/jb.169.12.5597-5604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers AH. Utilization of nitrogenous compounds by oral bacteria. Aust Dent J. 1990;35:468–471. doi: 10.1111/j.1834-7819.1990.tb05432.x. [DOI] [PubMed] [Google Scholar]
- 53.Rupf S, Merte K, Eschrich K. Quantification of bacteria in oral samples by competitive polymerase chain reaction. J Dent Res. 1999;78:850–856. doi: 10.1177/00220345990780040501. [DOI] [PubMed] [Google Scholar]
- 54.Salako NO, Kleinberg I. Incidence of selected ureolytic bacteria in human dental plaque from sites with differing salivary access. Arch Oral Biol. 1989;34:787–791. doi: 10.1016/0003-9969(89)90029-0. [DOI] [PubMed] [Google Scholar]
- 55.Shannon IL, Feller RP, Eknoyan G, Suddick RP. Human parotid saliva urea in renal failure and during dialysis. Arch Oral Biol. 1977;22:83–86. doi: 10.1016/0003-9969(77)90082-6. [DOI] [PubMed] [Google Scholar]
- 56.Shu M, Morou-Bermudez E, Suarez-Perez E, et al. The relationship between dental caries status and dental plaque urease activity. Oral Microbiol Immunol. 2007;22:61–66. doi: 10.1111/j.1399-302X.2007.00325.x. [DOI] [PubMed] [Google Scholar]
- 57.Sissons CH, Cutress TW. pH changes during simultaneous metabolism of urea and carbohydrate by human salivary bacteria in vitro. Arch Oral Biol. 1988;33:579–587. doi: 10.1016/0003-9969(88)90133-1. [DOI] [PubMed] [Google Scholar]
- 58.Sissons CH, Hancock EM, Cutress TW. The source of variation in ureolysis in artificial plaques cultured from human salivary bacteria. Arch Oral Biol. 1988;33:721–726. doi: 10.1016/0003-9969(88)90005-2. [DOI] [PubMed] [Google Scholar]
- 59.Sissons CH, Hancock EM, Perinpanayagam HE, Cutress TW. The bacteria responsible for ureolysis in artificial dental plaque. Arch Oral Biol. 1988;33:727–733. doi: 10.1016/0003-9969(88)90006-4. [DOI] [PubMed] [Google Scholar]
- 60.Stephan RM. Changes in hydrogen-ion concentration on tooth surfaces and in carious lesions. J Am Dent Assoc. 1940;27:718–723. [Google Scholar]
- 61.Stephan RM. Intra-oral hydrogen-ion concentration associated with dental caries activity. J Dent Res. 1944;23:257–266. [Google Scholar]
- 62.Turtola LO, Luoma H. Plaque pH in caries-active and inactive subjects modified by sucrose and fluoride, with and without bicarbonate-phosphate. Scand J Dent Res. 1972;80:334–343. doi: 10.1111/j.1600-0722.1972.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 63.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73:1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 64.van Ruyven FO, Lingstrom P, van Houte J, Kent R. Relationship among mutans streptococci, ‘low-pH’ bacteria, and lodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J Dent Res. 2000;79:778–784. doi: 10.1177/00220345000790021201. [DOI] [PubMed] [Google Scholar]
- 65.van Wuyckhuyse BC, Perinpanayagam HE, Bevacqua D, et al. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res. 1995;74:686–690. doi: 10.1177/00220345950740021001. [DOI] [PubMed] [Google Scholar]
- 66.Vieira AR, Marazita ML, Goldstein-McHenry T. Genome-wide scan finds suggestive caries loci. J Dent Res. 2008;87:435–439. doi: 10.1177/154405910808700506. [DOI] [PubMed] [Google Scholar]
- 67.Wang BY, Kuramitsu HK. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol. 2005;71:354–362. doi: 10.1128/AEM.71.1.354-362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wijeyeweera RL, Kleinberg I. Arginolytic and ureolytic activities of pure cultures of human oral bacteria and their effects on the pH response of salivary sediment and dental plaque in vitro. Arch Oral Biol. 1989;34:43–53. doi: 10.1016/0003-9969(89)90045-9. [DOI] [PubMed] [Google Scholar]
- 69.Yaling L, Tao H, Jingyi Z, Xuedong Z. Characterization of the Actinomyces naeslundii ureolysis and its role in bacterial aciduricity and capacity to modulate pH homeostasis. Microbiol Res. 2006;161:304–310. doi: 10.1016/j.micres.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Yano A, Kaneko N, Ida H, Yamaguchi T, Hanada N. Real-time PCR for quantification of Streptococcus mutans. FEMS Microbiol Lett. 2002;217:23–30. doi: 10.1111/j.1574-6968.2002.tb11451.x. [DOI] [PubMed] [Google Scholar]
- 71.Yin JL, Shackel NA, Zekry A, et al. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol Cell Biol. 2001;79:213–221. doi: 10.1046/j.1440-1711.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- 72.Zakhary GM, Clark RM, Bidichandani SI, Owen WL, Slayton RL, Levine M. Acidic proline-rich protein Db and caries in young children. J Dent Res. 2007;86:1176–1180. doi: 10.1177/154405910708601207. [DOI] [PubMed] [Google Scholar]