Abstract
The RecQ family helicase BLM is critically involved in the maintenance of genomic stability and BLM mutation causes the heritable disorder, Bloom’s syndrome. Affected individuals suffer from a predisposition to a multitude of cancer types and an ill-defined immunodeficiency involving low serum antibody titers. To investigate its role in B cell biology, we inactivated murine Blm specifically in B lymphocytes in vivo. Numbers of developing B lymphoid cells in the bone marrow and mature B cells in the periphery were drastically reduced upon Blm-inactivation. Of the major peripheral B cell subsets, B1a cells were most prominently affected. In the sera of Blm-deficient naïve mice, concentrations of all Ig isotypes were low, particularly IgG3. Specific IgG antibody responses upon immunization were poor and mutant B cells exhibited a generally reduced antibody class switch-capacity in vitro. We did not find evidence for a crucial role of Blm in the mechanism of class switch recombination. However, a modest shift towards microhomology-mediated switch junction formation was observed in Blm-deficient B cells. Finally, a cohort of p53-deficient, conditional Blm knockout mice revealed an increased propensity for B cell lymphoma development. Impaired cell cycle progression and survival as well as high rates of chromosomal structural abnormalities in mutant B cell blasts was identified as the basis for the observed effects. Collectively, our data highlight the importance of BLM-dependent genome surveillance for B cell immunity by ensuring proper development and function of the various B cell subsets while counteracting lymphomagenesis.
Keywords: B cells, immunodeficiency diseases, transgenic/knockout mice, cell differentiation, cell proliferation
INTRODUCTION
Maintenance of genomic stability is fundamental for the faithful propagation of genomic information and, hence, for the vitality of every cell type. Genome integrity is constantly jeopardized by a diverse array of insults emanating from the organism’s interaction with the environment (e.g. ionizing radiation, chemicals), endogenous sources intrinsic to cellular physiology (e.g. free radicals, collapsed replication forks), or specialized developmental programs (e.g. somatic recombination in lymphocytes) (1). In this regard, B lymphocytes are particularly challenged. Not only are vast numbers of B cells continuously being produced during adult life but they also undergo three distinct modes of genetic alterations associated with DNA strand breaks: V(D)J recombination, antibody class switch recombination (CSR)3, and somatic hypermutation (SHM). If such damage is dealt with inappropriately, tumorigenesis or immune deficiency may ensue (2, 3).
A drastic example for an inherited genetic defect revealing this relationship is given by Bloom’s syndrome (BS), a rare autosomal recessive disorder in which the RecQ helicase BLM is mutated (4). Although its multiple functions remain incompletely characterized, BLM is believed to act as ‘roadblock remover’ during replication, chiefly through unwinding of secondary DNA structures detrimental to replication fork progression. It is also assumed to function as ‘anti-recombinase’, either through disruption of early intermediates of homologous recombination (HR) at sites of limited homology (discussed in ref. (5)) or through resolution of Holiday junctions yielding non-crossover products (6). Indeed, grossly elevated frequencies of sister chromatid exchanges are a typical feature for BS cells (7). The hallmark characteristics of BS patients are small stature, male infertility, a profound susceptibility to most cancer types, and immunodeficiency (7), with the latter two features accounting for early death (8). Among body tissues, the effect of BLM-deficiency on the immune system appears particularly severe as non-Hodgkin’s lymphomas and leukemias dominate the cancer spectrum (8) whereas impaired hypersensitivity and hypoglobulinemia imply a generally compromised adaptive immunity. Indeed, BS patients frequently develop serious ear and respiratory tract infections (7). Data from multiple studies point towards defects in the B cell lineage, exemplified by low concentrations of serum IgM (9, 10) or non-IgM isotypes (11, 12), compromised mitogen-induced B cell responses (10), or impaired generation of non-IgM-secreting antibody-forming cells (AFCs) in vitro (11). These findings are compatible with impaired B cell development or differentiation, including defective CSR.
During B cell development in the mouse bone marrow, B220+ progenitor B cells successively rearrange DH→JH and VH→DHJH gene segments at the Ig H chain locus to generate a productive VHDHJH exon. This VDJ-joint is subsequently expressed as class μ H chain in combination with a surrogate L chain forming the pre-BCR (Hardy-fractions (Fr.) A-C (13)). Pre-BCR-expression of precursor B cells triggers a proliferative burst (Fr. C’) followed by a resting phase (Fr. D), at which the cells rearrange Igκ or Igλ L chain genes to become IgM+ immature (Fr. E) B cells. Fr. E cells migrate to the spleen where they complete their maturation to join the pool of long-lived follicular (FO) or marginal zone (MZ) B cells, collectively referred to as B2 cells (14–16). FO cells, chiefly localized in the follicles of spleen and lymph nodes, participate in T cell-dependent (TD) antibody responses and occasionally re-circulate through the bone marrow where they appear as IgM+IgD+ Fr. F cells (17). MZ B cells, located in the marginal zone of the spleen, are able to rapidly respond to blood-borne pathogens through predominantly T cell-independent (TI) antibody responses (15). B1 cells, mainly populating peritoneal and pleural body serosa, are thought to produce the bulk of “natural antibodies” that are enriched in specificities to common pathogens. They thus function as a ‘first line of defense’ through rapid TI antibody responses. In contrast to B2 cells, B1 cells seem to be generated predominantly during fetal life and early ontogeny, after which they propagate themselves throughout adulthood. Furthermore, CD5-expression distinguishes CD5+ B1a and CD5− B1b cells in the peritoneal cavity (14, 18).
All four principal B cell subsets are capable of undergoing CSR to generate antibodies of the IgG, IgE, and IgA subclasses. During CSR, the VHDHJH-exon which by default forms a transcription unit with the Cμ/Cδ constant region genes is joined to a downstream CH region. This process involves the generation of double strand breaks (DSBs), deletion of the intervening DNA, and DSB-repair by general DNA response factors (19). CSR critically depends on the enzyme activation-induced cytidine deaminase (AID) (20) and prior transcription through the intronic switch-(S)-regions of the participating CH genes (21). Such transcription at a particular CH locus is widely believed to generate R-loop structures within the S-region, thereby exposing cytidines in the non-template strand as targets for AID-mediated deamination (19). Interestingly, due to the abundance of guanines in the non-template strand, S-regions likely adopt stable DNA secondary structures upon transcription such as G4 quartets (19, 22) that represent classical substrates for the unwinding activity of BLM (5, 23, 24). Also, BLM is known to directly interact with factors proven to participate in CSR such as the mismatch repair protein Mlh1 (25, 26). A direct or indirect role for BLM in the mechanism of CSR is therefore conceivable.
To investigate the role of BLM in B cell biology and circumvent the embryonic lethality in conventional Blm-knockout mice (27), we selectively ablated murine Blm in B-lineage cells. In accordance with and complimentary to our earlier report where we encountered an impaired T cell lineage in T cell-specific knockout mice (28), we describe here a dramatic effect of Blm-deficiency on B lymphocytes. In conditional knockout mice, we found compromised B cell development and maintenance, strongly impaired TD and TI antibody responses after immunization, and a propensity for developing B cell lymphomas. Our results from in vitro experiments are compatible with a minor yet non-essential role for Blm in CSR.
MATERIALS AND METHODS
Mice
The mutant Blm-alleles had been generated previously in our lab: The BlmΔ-allele (Blmtm1Ches) is rendered non-functional by disruption of exon 8 upstream of the helicase domain resulting from an insertion of a Neo resistance cassette (27). The Blmf-allele (Blmtm4Ches) contains a loxP-flanked (floxed) exon 8 (29) that is excised upon Cre-expression (Blmtm3Ches) causing a premature termination of Blm translation (30). Mb1-Cre mice expressing a humanized Cre-recombinase under the control of the Igα promoter/enhancer elements were described recently (31), and the Trp53−/− line (32) has been carried in this lab for years. All mice were housed in specific pathogen-free environments. Experimental procedures were approved by the institutional review board of the animal facility at the Harvard Medical School.
Tumor cohorts, histological, and immunohistochemical analysis of tumor material
Mice set up to allow for tumor development were immunized twice at the age of 9–12 and 14–17 weeks i.p. with 108 SRBCs (Cedarlane) in PBS, monitored twice per week, and sacrificed when overtly sick. Spleens and macroscopically cancerous organs were archived in Optimalfix (American Mastertech Scientific) and pathological examination was carried out on H&E-stained organ sections by Dr. Roderick Bronson at the Harvard Medical School Rodent Histopathology core.
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissue sections. After rehydration, slides were pre-treated with 10 mM citrate pH 6.0 (for anti-B220 and PNA), 1 mM EDTA pH 8.0 (for anti-CD3), and DAKO pH 9 buffer (DAKO; for anti-TdT) in a steam pressure cooker. After treatment with Peroxidase Block (DAKO), slides were incubated with either rat anti-murine B220 (RA3-6B2; BD Biosciences) followed by polyclonal rabbit anti-rat immunoglobulin antibody (DAKO), or polyclonal rabbit anti-CD3 (Cell Marque, Cat. #CMC363), or polyclonal rabbit anti-TdT (DAKO, Cat. #A3524), and detected with the anti-rabbit EnVision+ kit (DAKO) as per the manufacturer’s instructions. To reveal cytoplasmic immunoglobulin, no primary antibody was used. Instead, slides were only stained with the anti-mouse EnVision+ kit (DAKO). To detect peanut agglutinin-(PNA)-binding, slides were treated with biotinylated PNA (Vector Laboratories) followed by strepavidin-HRP. All immunoperoxidase stains were developed using a DAB chromogen (DAKO) and counterstained with hematoxylin.
Analysis of germinal center (GC) development
15–17 week old mice were immunized i.p. with 108 SRBCs in PBS and sacrificed after 8, 12, and 18 days. Spleens were snap-frozen, and tissue sections were cut on a cryostat microtome followed by formalin fixation. Antigen was retrieved by incubating slides in 1 mM EDTA (pH 7.5) for 30 min at 95°C. After treatment with the Avidin/Biotin Blocking kit (Vector Laboratories), GC B cells were identified by incubation with biotinylated PNA (Vector Laboratories) followed by avidin-coupled HRP (DAKO). Peroxidase activity was visualized employing the AEC peroxidase substrate kit (Vector Laboratories). GC and total section area measurements were performed on micrographs using Openlab software (Improvision).
Cell preparation and flow cytometry
Tibiae and peritoneal cavities were flushed with HBSS + 3% BSA to collect bone marrow and peritoneal exudate cells, respectively. Single-cell suspensions from spleens and thymi were prepared using the same buffer. Bone marrow and spleen preparations were subjected to erythrocyte lysis in 0.165 M NH4Cl (pH 7.5) solution and surface labeling was done using the following fluorescently conjugated antibodies: anti-B220 (RA3-6B2), anti-CD3 (145-2C11), anti-CD4 (L3T4), anti-CD5 (53-7.3), anti-CD8 (53-6.7), anti-CD11b (M1/70), anti-CD19 (MB19-1), anti-CD21/35 (7G6), anti-CD23 (B3B4), anti-CD43 (S7), anti-CD117 (ACK2), anti-Sca-1 (E13-161.7), anti-Gr1 (RB6-8C5), anti-Flt3 (A2F10), anti-IgG1 (A85-1), anti-IgM (II/41), anti-TER119 (TER119), AA4.1 (eBioscience, BD Biosciences, or Caltag), and polyclonal goat anti-IgM, anti-IgG2a, and anti-IgG3 (SouthernBiotech). To exclude dead cells, 1 µg/ml propidium iodide was added immediately prior to analysis. Flow cytometric analysis and cell sorting was performed on a FACSCalibur cytometer and FACSAria cell sorter (BD Biosciences), respectively, and data were analyzed using FlowJo software (Tree Star).
Blm deletion PCR
Blm-deletion on genomic DNA from FACS-purified lymphocyte populations was determined semi-quantitatively using a three-primer PCR assay in which a 330 bp- and a 120 bp-product is amplified from the floxed Blmtm4Ches and the deleted Blmtm3Ches allele, respectively. PCR and assessment of the approximate deletion-efficiencies was done exactly as described (28).
Induction of TI- and TD immune responses, ELISA, and ELISPOT
11–19 week old mice were immunized i.p. with 10 µg NP41-Ficoll (4-Hydroxy-3-nitrophenylacetyl-AECM-Ficoll) in PBS or 10 µg NP25-CGG (4-Hydroxy-3-nitrophenylacetyl chicken-γ-globulin) precipitated in alum, and bled from tail veins at regular intervals.
Serum Ig concentrations from 7–10 week old naive were measured by ELISA using the “SBA Clonotyping System” and the “Mouse Ig Isotype Panel” as reference standards (both from SouthernBiotech). Relative NP-specific Ig levels were determined similarly with the following exceptions: ELISA plates were coated with 10 µg/ml NP30-BSA in PBS and pools of reference sera from ten mice (all genotypes) that had been immunized using the same protocols served as standards.
Quantification of IgM-, IgG1-, and IgG2a-secreting AFCs was done by culturing bone marrow cells from NP-CGG-immunized mice on NP-BSA-coated PVDF membranes (Pall Corporation) for 18 h at 37°C. Bound NP-specific Igs were detected using the appropriate HRP-conjugated antibodies from the “SBA Clonotyping System” and visualized by chemiluminescence. All NP-conjugates were purchased from Biosearch Technologies.
BrdU labeling
Cohorts of (Trp53+/+ or Trp53+/−) conditional knockout and control mice were given 0.8 mg/ml BrdU (Sigma) in the drinking water for two weeks. Groups of 4+4 mice were sacrificed after 0, 8, 15, 22, 33 days following the pulse period. Bone marrow, spleen, and peritoneal exudate cells were surface-labeled and BrdU incorporation was determined utilizing the BrdU Flow Kit (BD Biosciences).
In vitro CSR assays
Resting splenic CD43− B cells were isolated by CD43-microbead-mediated negative selection (Miltenyi Biotec), labeled with 5 µM CFSE (Molecular Probes) for 10 min at 37°C, and cultured at 5 × 105 cells/ml in complete RPMI + 10% FCS for four days with 20 µg/ml LPS (Sigma), LPS + 50 ng/ml recombinant IFN-γ (Biosource), or LPS + 20 ng/ml recombinant IL-4 (eBioscience). To prepare RNA for real-time RT-PCR, a portion of the cells was harvested after two days, lysed in TRIZOL reagent (Invitrogen), and stored as lysate at −80°C until used for RT. On day four, cells were harvested, stained for surface expression of IgM, IgG1, IgG3, or IgG2a, and analyzed by cytometry in the presence of 1 µg/ml 7-AAD to discriminate dead cells.
Cell cycle and cytogenetic analysis
Splenic B cells were prepared and cultured as explained above with the modification that a combination of 1 µg/ml anti-CD40 antibody (HM40-3; eBioscience) and 25 ng/ml IL4 was used as stimulus. On day three, 25 ng/ml KaryoMax Colcemid (Invitrogen) was added for the last 90 min of culture. Subsequently, 2 × 106 cells were harvested and surface-labeled with anti-IgG1 antibody, followed by fixation and permeabilization using the BD Cytofix/Cytoperm and BD Perm/Wash buffers (BD Biosciences). Cells were incubated with 0.5 mg/ml RNase A (Roche) in PBS for 30 min at 37°C, after which 20 µg/ml propidium iodide was added for another 30 min-incubation on ice, followed by DNA content analysis at the cytometer.
For DNA damage analysis, Colcemid-treated cells were swelled in 0.075 M KCl solution for 12 min at 37°C and fixed with Methanol/Acetic acid (3:1). Metaphase spreads were prepared and stained with Giemsa-reagents following standard procedures.
Quantitative RT-PCR
Total RNA stored in TRIZOL (see above) was extracted according to the manufacturer’s recommendations and reverse transcribed with SuperScript III reverse transcriptase and oligo(dT)20 primers, followed by RNase H-treatment. Real-time PCR was done on an AB 7300 machine (Applied Biosystems) using the QuantiTect SYBR Green PCR system (Qiagen) according to the Qiagen protocol. Specificity of the primer pairs was ensured by analysis of dissociation curves and verification of expected fragment sizes by gel electrophoresis. Serially diluted cDNA samples were employed to ensure comparable amplification efficiencies of target and endogenous reference genes (β-actin). Relative mRNA abundance was calculated using the comparative CT method. Primers were: β-actin: 5’-CTTTTCCAGCCTTCCTTCTTGG-3’ and 5’-CAGCACTGTGTTGGCATAGAGG-3’; μ GL: 5’-CTCGGTGGCTTTGAAGGAAC-3’ and 5’-TGGTGCTGGGCAGGAAGT-3’; γ1 GL: 5’-TCGAGAAGCCTGAGGAATGTG-3’ and 5’-ATGGAGTTAGTTTGGGCAGCA-3’; γ2a GL: 5’-CTGGCAGTACCGATGCAGG-3’ and 5’-GCCAGTTGTATCTCCACACACAG-3’; γ3 GL: 5’-GCAAGATCTCTGCAGCAGAAATC-3’ and 5’-CCAGGGACCAAGGGATAGACA-3’; γ1 PS: 5’-TCTGGACCTCTCCGAAACCA-3’ and 5’-ATGGAGTTAGTTTGGGCAGCA-3’; γ2a PS: 5’-GACCTCTCCGAAACCAGGC-3’ and 5’-GGGCCAGTGGATAGACCGA-3’; γ3 PS: 5’-TCTGGACCTCTCCGAAACCA-3’ and 5’-CCAGGGACCAAGGGATAGACA-3’.
Analysis of Sμ-Sγ3 switch recombination junctions
400 ng of genomic DNA isolated from B cells stimulated with LPS for 4 days (see above) was subjected to PCR amplification using Expand HiFidelity Taq polymerase (Roche) and primers μ3-H3mod TGGCTTAACCGAGATGAGCC (modified from ref. (26)) and g3-2 TACCCTGACCCAGGAGCTGCATAAC (26). Six cycles of 94°C (1 min), 65–55°C (30 s; touch-down annealing) and extension at 72°C (2 min) were followed by 29 cycles of 94°C (30 sec), 55°C (30 sec), and 72°C (2 min). PCR products of three parallel reactions per sample were pooled, cleaned up with the QIAquick PCR purification Kit (Qiagen), and cloned into pCR2.1 using the TOPO TA Cloning Kit (Invitrogen). Plasmids from individual bacterial colonies were sequenced in the Biopolymers Facility at Harvard Medical School by high throughput colony sequencing. Switch junctions were identified using BLAST (Megablast). Only S-junctions longer than 100 bp were examined and clonally related sequences were excluded from the analysis. Invariably, the sequences producing the most significant alignments were the genomic contigs NT_114985.2 (strain 129/SvJ) or NT_166318.1 (strain C57BL/6J) reflecting the mixed background of our mice (129×B6). Alignment of all sequences obtained in the entire analysis using Lasergene software (DNASTAR) did not reveal evidence of further polymorphisms in the Sμ or Sγ3 GL segments of our mice.
Statistical analysis
To determine p-values, the 2-sample student’s t-test (unpaired, 2-tailed, assuming unequal variances) was employed (as calculated by Microsoft Excel software) unless otherwise noted. All other statistical and non-linear regression analyses were done with Prism 4 (GraphPad Software). The error bars in Fig. 4B accommodate the SDs of the respective subset cellularities from mutant and control mouse cohorts, and were derived as follows: Means (X̄) of mutant (Δ) cell populations are expressed as fractions of control means (C). Therefore, the SDs of the quotients are calculated as
where the coefficient of variation is defined as thus
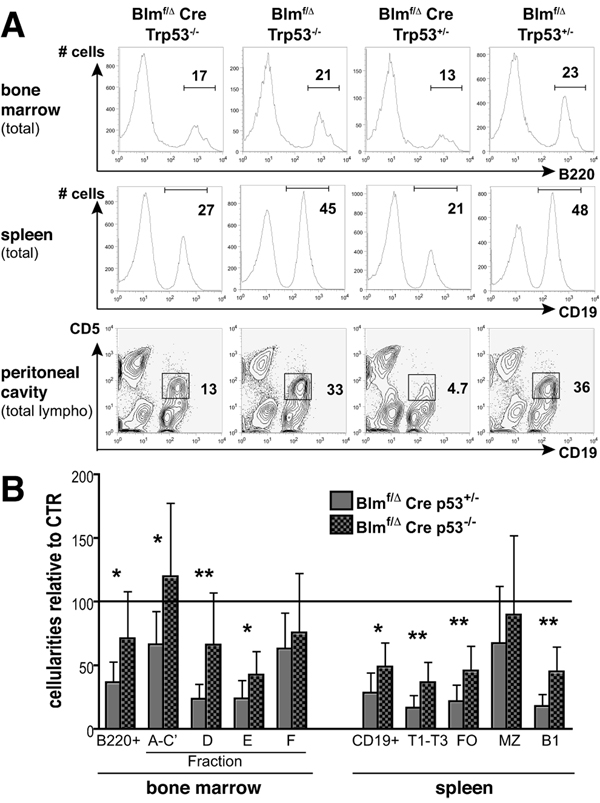
FIGURE 4. Partial rescue of B lineage compartments by p53-deficiency.
(A) Nucleated bone marrow, splenic, and peritoneal lavage cells from p53-proficient or p53-deficient Blmf/Δ Cre and littermate control mice were analyzed for B220, CD19, as well as CD19 and CD5 expression. A representative set of results is shown.
(B) Cellularities of the indicated B lineage populations from 7–11 week old Trp53+/− and Trp53−/− Blmf/Δ Cre mice are expressed as mean percentages (± SD) of the respective subsets from Trp53+/− and Trp53−/− control littermates (controls set to 100%, n = 5 or 6). Bone marrow populations were defined as in Fig. 2A and splenic compartments were: T1-T3: IgM+AA4.1+, FO: IgM+AA4.1−CD23hiCD21+, MZ: IgM+AA4.1−CD23loCD21hi and B1: IgM+AA4.1−CD23loCD21−. For calculation of SD values see the materials and methods section (*p < 0.05, **p < 0.005).
RESULTS
B lineage-specific ablation of Blm results in impaired B cell development and maintenance
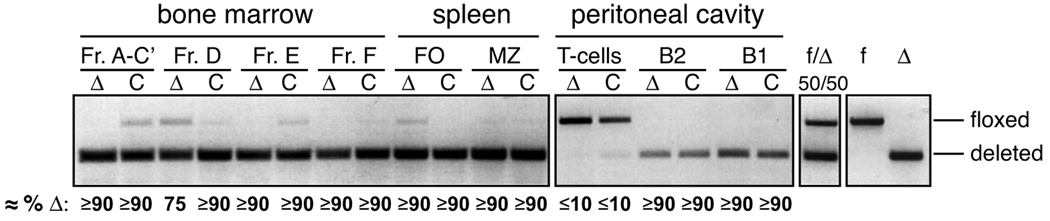
To delete Blm in B lineage cells, we bred mice harboring the constitutive null (Δ) (27) and floxed (f) Blm alleles (29) to mice expressing Cre recombinase under the mb1 regulatory elements (31). Deletion analysis of selected lymphocyte populations in bone marrow, spleen, and peritoneal cavity from Blmf/Δ mb1-Cre mice by semi-quantitative PCR revealed an almost complete loss of the floxed allele in all B cell populations analyzed (Fig. 1), as expected (31). In contrast, T lymphocytes found in the peritoneal cavity were virtually devoid of deleted cells (Fig. 1).
FIGURE 1. Efficiency of Blm-deletion in Blmf/Δ Cre mice.
Approximate fractions of cells deleted for Blm in selected lymphocyte populations from Blmf/Δ Cre (Δ) and Blmf/+ Cre control (C) mice as determined by semi-quantitative PCR. Bone marrow B220intCD43+IgM− Fr. A-C’, B220intCD43−IgM− Fr. D, B220intIgM+ Fr. E, and B220hiIgM+ Fr. F, splenic CD19+CD23hiCD21+ FO and CD19+CD23loCD21hi MZ cells, peritoneal IgM−CD5hi T, IgM+B220hiCD19lo B2 and IgM+B220loCD19hi B1 cells were sorted by cytometry and subjected to genomic PCR (pools of two mice per genotype). A 1:1 mixture of genomic DNA from cell lines harboring either only the floxed or the null Blm allele served as a reference (28). A repetition of the experiment yielded similar results.
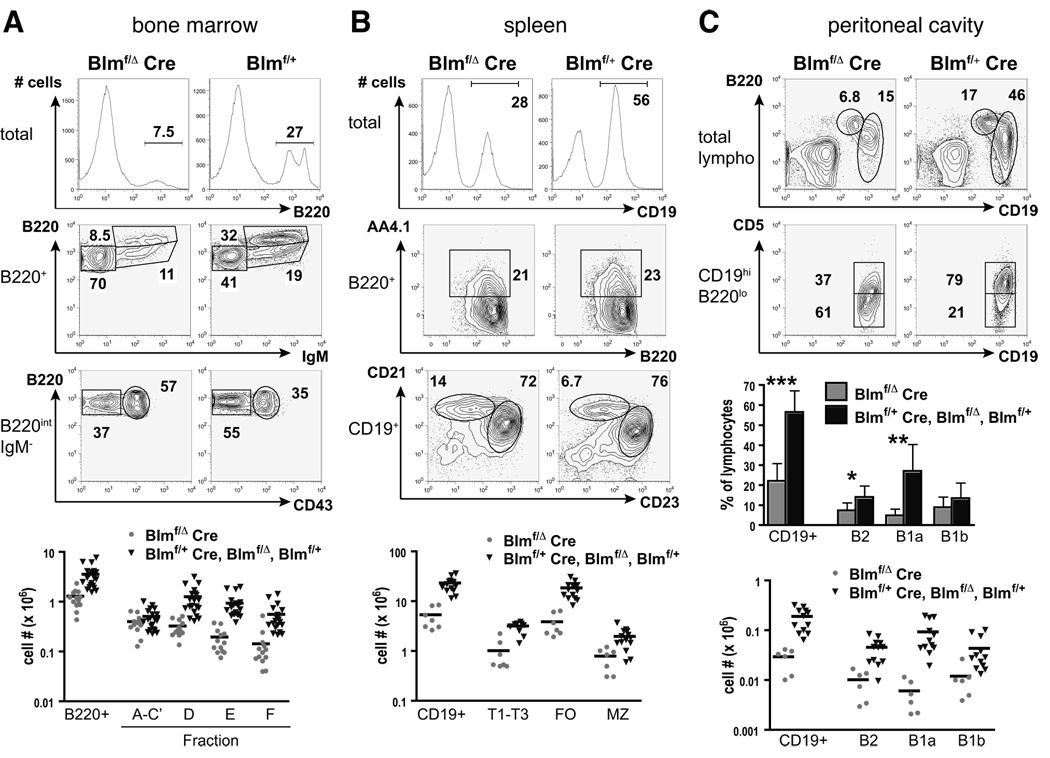
We analyzed select cell populations of bone marrow, spleen, and peritoneal lavage for phenotypic abnormalities by flow cytometry. Strikingly, B220+ B-lineage cells were dramatically underrepresented in the bone marrow of Blmf/Δ Cre mice compared to Blmf/+ Cre, Blmf/Δ, or Blmf/+ littermate controls (Fig. 2A). Absolute numbers of B220+ bone marrow cells accounted for only about a third of those seen in controls (Fig. 2A). Accordingly, B cell development was severely compromised. While Fr. A-C’ cells were relatively enriched, all later stages, i.e. Fr. D-F, were less frequent in mutant mice. Cellularities were significantly diminished in all subsets: Fr. A-C’ were reduced by a fifth, and Fr. D, E, and F by three quarters each (Fig. 2A). Of note, lineage−Sca1+CD117+ hematopoietic stem cells were present in normal frequencies and numbers in Blmf/Δ Cre mice (data not shown). These data thus indicate that B cell progenitors, shortly after commitment to the B cell lineage and subsequent loss of Blm function, start to exhibit an impaired developmental capacity, i.e. as soon as they engage in pre-BCR-mediated rapid proliferation following expression of a productive VHDHJH joint (Fr. C’ and early Fr. D) (14, 16).
FIGURE 2. Defective B cell development and reduced FO and B1 cell compartments in Blm-deficient mice.
Flow cytometric analysis of bone marrow cells, splenocytes, and peritoneal lavage recovered from 6–10 week old Blmf/Δ, Cre and control mice.
(A) Bone marrow cells from representative Blmf/Δ Cre and Blmf/+ mice were analyzed for expression of the indicated markers and percentages of cells within the respective gates are given. Total bone marrow cells are shown in the top, B220+ cells in the middle, and B220intIgM− cells in the bottom row panels. Absolute numbers of bone marrow B220+ and Fr. A-F cells are displayed as scatter plots with each triangle representing an individual mouse and horizontal bars denoting the mean for each group (B220+ bone marrow cells: p = 2 × 10−5, B220intCD43+IgM− Fr. A-C’: p = 0.14, B220intCD43−IgM− Fr. D: 4 × 10−5, B220intIgM+ Fr. E: 2 × 10−6, B220hiIgM+ Fr. F: 8 × 10−5; n = 13 for Blmf/Δ Cre mice and n = 20 for Blmf/+ Cre, Blmf/Δ and Blmf/+ controls).
(B) Splenocytes from representive Blmf/Δ Cre and Blmf/+ Cre mice were analyzed for expression of the indicated markers to differentiate CD19+ B lymphocytes (% of total, top row panels), B220+AA4.1+ transitional (T1-T3) B cells (% of B220+, middle panels), CD19+CD23hiCD21+ FO, and CD19+CD23loCD21hi MZ B cells (% of CD19+, lower panels). Scatter plots depict absolute numbers of these populations (CD19+ cells: p = 3 × 10−7, T1-T3: 10−4, FO: p = 10−6, MZ: 5 × 10−4; n = 7 or 8 for Blmf/Δ Cre mice and n = 9–14 for controls).
(C) Peritoneal exudate cells were analyzed to identify B220hiCD19lo B2 and B220loCD19hi B1 cells (upper contour plots). Analysis of CD5 expression on B220loCD19hi B1 cells revealed CD5int and CD5− B1a and B1b cells, respectively (lower contour plots). Bar diagrams show the average proportions (± SD) of total CD19+, B2, B1a, and B1b cells among peritoneal lymphocytes (*p < 0.05, **p < 0.005, ***p < 0.0005), and scatter plots display their absolute numbers (CD19+ cells: p = 10−4, B2: p = 4 × 10−4, B1a: p = 0.001, B1b: p = 0.009, n = 6 for Blmf/Δ Cre mice and n = 12 for controls).
Likewise, the sizes of peripheral B cell populations were dramatically reduced in Blm-deficient mice. Consistently smaller spleens (data not shown) corresponded to cellularities of total CD19+ and FO B cells of less than a quarter compared to littermate controls. Immature transitional B cells were reduced to a third and MZ B cells to 40% of control numbers (Fig. 2B). Strikingly and very consistently, the CD5+ B1a cell subset in the peritoneal cavities of mutant mice exhibited a severe atrophy. Peritoneal B1a cells accounted for only about 20% in frequency and 7% in absolute numbers compared to controls, whereas CD5− B1b were somewhat more numerous (27% of controls; Fig. 2C).
Taken together, the sizes of all major peripheral B cell subsets were drastically reduced in Blmf/Δ Cre mice and of those, splenic FO and peritoneal B1a cells were most dramatically affected by the loss of Blm. In contrast, thymic cellularities of CD4−CD8− double negative, CD4+CD8+ double positive, CD4+ and CD8+ single positive developing T cells were indistinguishable in mutant and control mice (data not shown). This underscores the B cell-intrinsic nature of the observed abnormalities. Since cohorts of Blmf/+ Cre, Blmf/Δ, and Blmf/+ control mice generally did not differ amongst each other phenotypically, we regarded all Blm-proficient mice as a single control group.
Compromised antibody responses in Blm-deficient mice
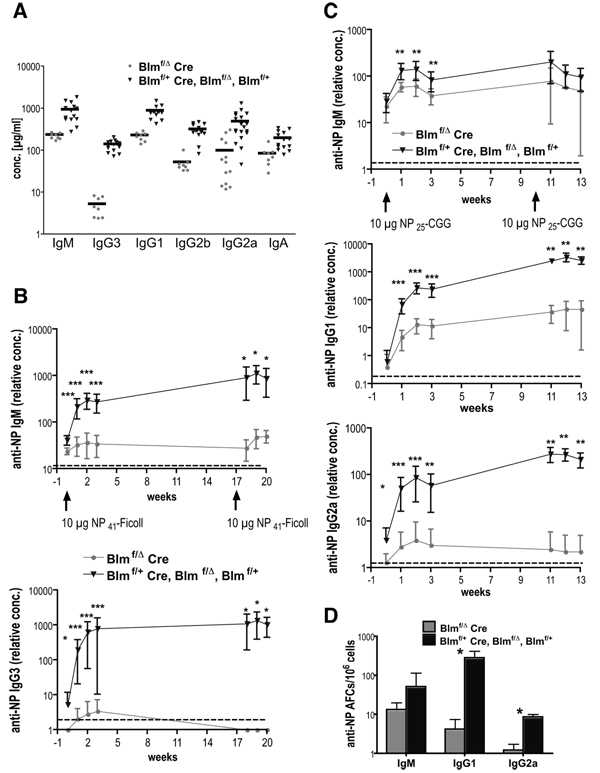
Even in naïve mice, antibodies of most Ig isotypes are detectable in considerable concentrations. Measuring steady-state antibody titers in non-immunized animals, we found serum levels of all Ig isotypes drastically diminished in naïve 7–10 weeks old mutant mice (Fig. 3A). While IgA was least affected (43% of controls), IgG1, IgM, IgG2a, and IgG2b concentrations were at 26%, 25%, 20%, and 16% of normal levels, respectively. Strikingly, IgG3 was consistently most dramatically reduced, present at only 4% of control titers (Fig. 3A).
FIGURE 3. Diminished immunoglobulin production in Blmf/Δ Cre mice.
(A) Serum Ig isotype concentrations from 7–10 week old naive Blmf/Δ Cre (n = 8–13) and littermate control mice (n = 14–19) were measured by ELISA. Each data point represents an individual animal and horizontal bars denote mean concentrations for each group and isotype (IgM: p = 10−4, IgG3: p = 2 × 10−8, IgG1: p = 10−6, IgG2b: p = 10−6, IgG2a: p = 10−4, IgA: p = 0.001).
(B) TI immune responses after primary and secondary immunizations. 12–17 week old Blmf/Δ Cre and control mice were intraperitoneally immunized with 10 µg NP41-Ficoll in PBS on weeks 0 and 17, and NP-specific serum IgM and IgG3 levels were determined after 0 (pre-immune), 1, 2, 3, 18, 19, and 20 weeks by ELISA. Average concentrations (± SD) for each group are expressed relative to a pool of reference sera that were immunized using the same protocol (n = 7–10 for sera collected after 0–3 and n = 4 for sera obtained after 18–20 weeks). Broken lines represent the detection limit of the ELISA (*p < 0.05, **p < 0.005, ***p < 0.0005 by Mann-Whitney test).
(C) TD immune responses after primary and secondary immunizations. Cohorts of 11–19 week old Blmf/Δ Cre and control mice immunized i.p. with 10 µg NP25-CGG in alum on weeks 0 and 10 were bled on weeks 0, 1, 2, 3, 11, 12, and 13. NP-specific serum IgM, IgG1, and IgG2a titers were determined by ELISA and analyzed as in (B). For each group, seven and five mice were analyzed on weeks 0–3 and 11–13, respectively.
(D) Frequencies (± SD) of NP-specific IgM-, IgG1-, and IgG2a-secreting AFCs in the bone marrow of four Blmf/Δ Cre and five control mice 12–13 weeks after secondary immunization with NP25-CGG as determined by ELISPOT. The mice were part of the cohort shown in (C). Statistical analysis was performed as in (B).
We then asked whether induced antibody responses would be similarly affected. Cohorts of Blmf/Δ Cre and littermate control mice were immunized with the TI-II immunogen NP-Ficoll, and NP-specific IgM and IgG3 responses were measured in weekly intervals by ELISA. Conditional knockout mice produced only 12–15% specific IgM compared to controls after one, two, and three weeks after immunization (Fig. 3B upper panel). Even more drastically, NP-specific IgG3 was detectable only in a few Blmf/Δ Cre mice at all, and those showed titers hardly above the detection limit of the assay. The averages consequently amounted to less than 1% of control levels (Fig. 3B lower panel). After re-challenge, the differences became even more dramatic. NP-specific IgM concentrations dropped to ≤5% of the controls, whereas specific IgG3 was undetectable (Fig. 3B and data not shown). Since challenge with TI-II antigens is not expected to produce large numbers of long-lived AFCs (33), these data suggest that an already small pool of specific B cell precursors present in naive Blmf/Δ Cre mice is further reduced in size by an inadequate and possibly abortive primary antibody response. It should be noted that, using an i.p. immunization protocol, our experiments chiefly tested MZ B cell responses (33).
TD B cell immunity was probed by immunization with NP-CGG followed by assessment of specific IgM-, IgG1-, and IgG2a antibody titers one, two, and three weeks after a primary and a secondary immunization. In mutant mice, NP-specific IgM was reduced by 50–60% throughout the experiment (Fig. 3C, upper panel). Concentrations of specific IgG1 and IgG2a, though, were as low as 5% after primary challenge and dropped to ≤1% upon recall immunization (Fig. 3C, middle and lower panels). To test whether the vestigial memory response in Blmf/Δ Cre mice was reflected by lower frequencies of long-lived plasma cells, we enumerated AFCs in the bone marrow (where they preferentially reside (34)) by ELISPOT. Numbers of NP-specific IgM-producing AFCs were diminished by approximately 75% and IgG1 and IgG2a-secreting plasma cells by ≥90% (Fig. 3D). The generation of long-lived AFCs and TD B cell memory is largely dependent on the GC reaction (35). To test whether the GC reaction requires Blm, we monitored the formation and maintenance of GCs over time. Groups of Blmf/Δ Cre and Blmf/+ Cre control mice were sacrificed 8, 12, and 18 days after immunization with SRBCs, and GC numbers and areas were determined in spleen sections by immunohistochemistry. As shown in Table I, we observed a tendency toward lower GC frequencies in conditional mutants (although this was not statistically significant). GC areas, however, were lower in Blmf/Δ Cre mice throughout the analysis, exemplified by reduced frequencies of large GCs. The differences between mutants and controls increased with time and by day 18 after immunization, the average frequency of large GCs in Blmf/Δ Cre mice was only 1/8 compared to control animals (Table I). We conclude that GC formation can occur robustly in the absence of Blm (acknowledging the lower numbers of splenic B cells) but that GC maintenance is impaired and the reaction dies off more rapidly providing a rationale for defective B cell memory and the low frequencies of long-lived AFCs in the bone marrow.
Table I.
GC frequencies and sizes in Blmf/Δ Cre and Blmf/+ Cre control mice after immunization with SRBCsa
| # of GCs / mm2 | # of large GCs / mm2b | total area examined [mm2] | ||
|---|---|---|---|---|
| DAY 8 | ||||
| Blmf/Δ Cre #1 | 1.9 | 0.6 | 8.0 | |
| Blmf/Δ Cre #2 | 2.6 | 0.9 | 6.4 | |
| Blmf/Δ Cre #3 | 2.0 | 1.1 | 18.0 | |
| average ± SD | 2.2 ± 0.4c | 0.9 ± 0.3d | ||
| control #1 | 4.4 | 2.0 | 6.4 | |
| control #2 | 4.6 | 1.9 | 9.2 | |
| control #3 | 2.9 | 1.9 | 11.0 | |
| average ± SD | 3.9 ± 0.9 | 1.9 ± 0.1 | ||
| DAY 12 | ||||
| Blmf/Δ Cre #1 | 2.2 | 0.7 | 10.1 | |
| Blmf/Δ Cre #2 | 2.1 | 0.3 | 14.0 | |
| Blmf/Δ Cre #3 | 1.2 | 0.3 | 18.7 | |
| average ± SD | 1.9 ± 0.5c | 0.4 ± 0.2d | ||
| control #1 | 5.7 | 1.8 | 10.7 | |
| control #2 | 3.8 | 1.4 | 11.2 | |
| control #3 | 2.8 | 1.2 | 17.0 | |
| average ± SD | 4.1 ± 1.5 | 1.5 ± 0.3 | ||
| DAY 18 | ||||
| Blmf/Δ Cre #1 | 1.1 | 0.3 | 16.0 | |
| Blmf/Δ Cre #2 | 0.6 | 0.0 | 16.1 | |
| Blmf/Δ Cre #3 | 1.9 | 0.1 | 9.7 | |
| average ± SD | 1.2 ± 0.7c | 0.1 ± 0.1d | ||
| control #1 | 1.5 | 0.8 | 14.5 | |
| control #2 | 1.8 | 0.9 | 15.2 | |
| control #3 | 1.9 | 0.7 | 10.1 | |
| average ± SD | 1.8 ± 0.2 | 0.8 ± 0.1 |
mice were sacrificed 8, 12, and 18 days after i.p. immunization with SRBCs; spleens were removed, fixed, and sectioned followed by immunohistochemistry; GCs were identified by PNA-binding; GC and total section areas were measured using Openlab software.
large GCs: > 9000 µm2
p > 0.05 (comparing mutant and corresponding control averages)
p < 0.05 (comparing mutant and corresponding control averages)
Taken together, the highly compromised short-term as well as long-term antibody responses mounted by mutant mice suggest as underlying cause a defective expansion and survival of antigen-specific B cells drawn from an already diminished pool of precursors. The observation that non-IgM (i.e. switched) isotypes were generally more severely affected raised the possibility of a dedicated role for Blm in CSR (see below).
Partial rescue of B cell subsets by p53-deficiency
To test whether p53-induced cell cycle arrest or apoptosis (36) is responsible for the small B cell compartments in Blm-deficient animals, we introduced a mutated Trp53 gene. P53-deficiency is known to substantially suppress the normally high apoptosis rates in Fr. A-C cells during B cell development in normal mice (37) and we had reported earlier a 55% rescue of thymocyte numbers in T cell-specific Blm knockout mice (28). Indeed, we observed a modest, but consistent proportional increase of B lymphoid cells in bone marrow and spleen in Blmf/Δ Cre mice upon p53-ablation (Fig. 4A, upper and middle panels). The size of the B1a subset in the peritoneal cavity benefited from p53-deficiency in particular (Fig. 4A, lower panels). As depicted in Fig. 4B, the absolute numbers of most B cell populations increased significantly as well, most notable were 2.8, 2.1, and 2.5-fold increases in bone marrow Fr. D, splenic FO, and splenic B1 cells, respectively. Serum Ig concentrations in naïve mice, however, remained unchanged (data not shown).
Impact of Blm on CSR
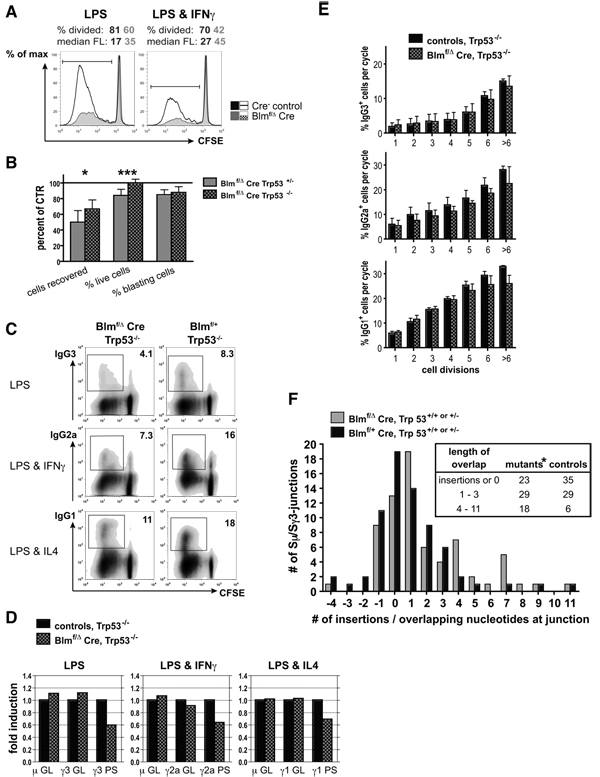
Given the selectively low IgG3 serum levels and the particularly dwarfed IgG antibody responses and AFC numbers upon antigen challenge in mutant animals, we investigated a potential involvement of Blm in the mechanism of CSR. To this end, CD43− splenocytes were labeled with CFSE to follow cell division and stimulated in vitro with the polyclonal mitogen LPS, either alone or in combination with IFNγ. FACS-analysis at day four revealed a prominent proliferative defect and excess cell death in mutant B cell cultures (Fig. 5A, and data not shown). As proliferation is a mandatory prerequisite for CSR (19) and hence for meaningful data interpretation, we used p53-deficient B cells instead. Indeed, similar to the beneficial effects of p53-deficiency on B cell cellularities in vivo, significantly higher absolute numbers of live cells were recovered after identical in vitro culture using cells from Blmf/Δ Cre Trp53−/− vs. Blmf/Δ Cre Trp53+/− mice (Fig. 5B). Interestingly, p53 deficiency rescued cell survival to control levels, but the proportions of cycling cells recovered at day four remained unchanged (Fig. 5B). This indicates that the absence of p53 suppresses apoptosis induction, but not cell cycle arrest in our system.
FIGURE 5. Impaired CSR in Blm-deficient B cells but no essential role for Blm in the mechanism of CSR.
(A) CD43-depleted splenocytes from Blmf/Δ Cre (filled histograms) and control mice (open histograms, pool of two mice per genotype) were labeled with CFSE and cultured in the presence of LPS (left) or LPS and IFNγ (right) for four days to induce proliferation (and CSR), followed by FACS-analysis to detect CFSE-dilution. The percentages of cells that had divided at least once and the median CFSE fluorescence values are shown. The results are representative of three independent experiments for each set of stimuli.
(B) Effects of p53-deficiency on cell survival and proliferative capacity of cultured B lymphocytes. Numbers of cells (as determined by enumeration of trypan blue excluding cells), fractions of live cells (as determined by cytometry and 7AAD exclusion, total events analyzed), and fractions of cell blasts (as determined by cytometric scatter analysis, gated on live cells) recovered after four days from Blmf/Δ Cre Trp53+/− and Blmf/Δ Cre Trp53−/− switch cultures are given as percentages (± SD) of controls. The graph includes data from eight to ten independent experiments in which B cells were stimulated with either LPS or LPS and IFNγ (*p < 0.05, ***p < 0.0005).
(C) Representative cytometric analyses of CD43-depleted and CFSE-labeled splenocytes from Blmf/Δ Cre Trp53−/− and Blmf/+ Trp53−/− mice after four days of in vitro culture to detect CFSE dilution and surface expression of IgG3, IgG2a, and IgG1 upon stimulation with LPS (top), LPS and IFNγ (middle), and LPS and IL4 (bottom), respectively. Gates outline the populations of switched cells and numbers denote their frequencies. Events that appear positive for the respective IgG isotype, but fall outside of the indicated gate were found to be false-positive as determined by isotype control analysis.
(D) Real-time RT-PCR for GL and PS sterile transcripts of S-regions in CD43-depleted B cells from Blmf/Δ Cre Trp53−/− and Trp53−/− control mice stimulated with LPS (μ and γ3, left), LPS and IFNγ (μ and γ2a, middle), and LPS and IL4 (μ and γ1, right) for two days. Mean expression levels in Blm-deficient B cells of three independent experiments are expressed as ‘fold induction’ relative to controls.
(E) Fractions of switched cells after a specific number of cell divisions as determined by surface staining for IgG3, IgG2a and IgG1 expression and CFSE-dilution after four days of appropriate stimulation. Mean values (± SEM) representing the same three experiments as in (D) are shown. None of the differences in Blmf/Δ Cre Trp53−/− vs. Trp53−/− control cultures reached a level of statistical significance.
(E) Microhomologies at switch recombination junctions from p53-proficient Blmf/Δ Cre and Blmf/+ Cre control mice. B cells were stimulated with LPS for four days as described in (C) and Sμ-Sγ3 junctions were amplified, subcloned, and sequenced. The number of nucleotides of uninterrupted Sμ donor/Sγ3 acceptor identity was determined by comparison to published Sμ and Sγ3 GL sequences. Sequences were compiled from two independent experiments employing Trp53+/+ and Trp53+/− B cells which yielded qualitatively similar results (n = 70 for each genotype). Negative numbers denote insertions at the breakpoint (*statistical significance for differences in distribution between mutants and controls in tabular insert: p = 0.01 by χ2 test).
Testing p53-deficient splenocytes (Fig. 5C), we consistently recovered substantially reduced proportions of switched B cells in Blm conditional knockout mice after four days of in vitro culture, i.e. IgG3+, IgG2a+, or IgG1+ cells upon stimulation with LPS alone, LPS and IFNγ, or LPS and IL4, respectively. Switching to IgG2a and IgG3 was generally more affected than to IgG1 (Fig 5C).
During CSR, transcription through the S regions of participating isotypes generates sterile germline (GL) transcripts consisting of the non-coding I exon spliced to the CH exons of the respective isotype. The transcriptional rates correlate with CSR potential (21). To test if the induction phase of CSR is compromised as a consequence of Blm-deficiency, we assessed the relative levels of GL transcripts by quantitative real-time RT-PCR after two days of in vitro culture. As depicted in Fig. 5D, average GL Iμ-Cμ and target GL Iγ3-Cγ3, Iγ2a-Cγ2a, and Iγ1-Cγ1 transcripts were expressed at similar levels in appropriately stimulated Trp53−/− Blm-deficient and Trp53−/− Blm-competent cells. We conclude that the CSR induction phase remains uncompromised in Blm-deficient B cells.
Iμ promoter-driven transcription continues beyond the CSR event generating post-switch (PS) transcripts with the Iμ exon spliced to the CH exons of the switched isotype (38). Compromised CSR is expected to yield lower than control levels of the respective PS transcripts. Indeed, we found the expression of Iμ-Cγ3, Iμ-Cγ2a, and Iμ-Cγ1 transcripts reduced to about 60–70% of control levels averaging three independent experiments (Fig. 5D).
However, since these effects were rather moderate and since Trp53-inactivation could not rescue the proliferation defect completely, we analyzed the switch efficiencies on a “per cell generation” basis, i.e. we compared the percentages of switched cells that had undergone a specific number of cell divisions. Consistently poorer switch efficiencies across all generations would be indicative of a prominent Blm-dependent CSR defect. However, averaging the same three experiments shown in Fig. 5D, we did not find such indication in either experimental setup (Fig. 5E). Thus, these data argue against an essential role for Blm in the mechanism of CSR.
To determine whether the CSR junctions are normal, we cloned and sequenced S-regions of p53-proficient B cells that had switched to IgG3 (i.e. Sμ-Sγ3 junctions) after LPS-stimulation for 4 days. We found similar mutation frequencies in the vicinity (i.e. within a 300 bp window) of the junctions in B cells from Blm-deficient and control mice (1.28 × 10−2 [Sμ; n = 49] and 1.5 × 10−3 [Sγ3; n = 45] for mutants and 1.33 × 10−2 [Sμ; n = 37] and 2.0 × 10−3 [Sγ3; n = 47] for controls; differences not significant). To see whether the absence of Blm influences the repair phase of CSR, we determined the extent of Sμ donor/Sγ3 acceptor homology at the junctions. The length of microhomology was shown to increase upon inactivation of certain DNA repair factors, such as Mlh1 or Pms2 (26, 39) and thus deviate from the situation in wild type mice whose S-S junctions display only little microhomology (40). In Blmf/Δ Cre B cells, a smaller number of Sμ-Sγ3 junctions showed no microhomology or small insertions whereas junctions with 4 – 11 bp of perfect donor/acceptor homology were slightly overrepresented (see insert in Fig. 5F). Accordingly, the average length of overlap was greater in Blm-deficient B cells (2.1 ± 0.3 bp SEM vs. 1.3 ± 0.3 bp in controls, n = 70 for each genotype; insertions counted as ‘no microhomolgy’; p = 0.03 by Mann-Whitney test). Taken together, although Blm appears to be dispensable for the CSR mechanism, these data suggest that DNA ends are processed somewhat differently during CSR in the absence of Blm.
Reduced proliferative capacity due to impaired genomic stability entails altered turnover-rates of several B-lineage subsets
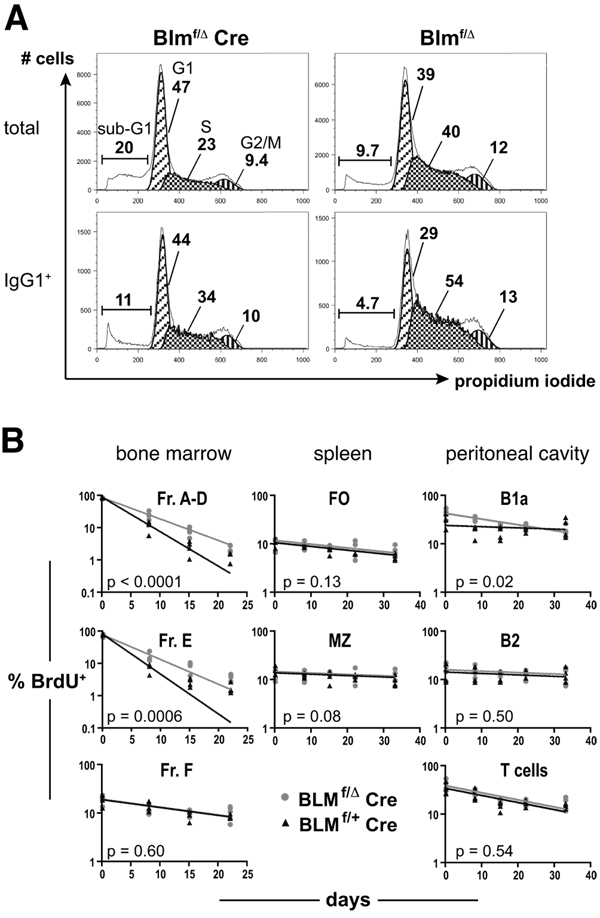
We sought to specify the underlying causes for the in vivo defects in Blmf/Δ Cre mice, especially for the remarkably contracted peritoneal B1a cell subset. To this end, we performed cell cycle analyses and examined metaphase spreads for the occurrence of chromosomal damage in class switch cultures. Upon stimulation of splenic B lymphocytes with anti-CD40 antibody and IL4 for three days, considerably fewer B cells were in the S or G2/M phases of the cell cycle in Blmf/Δ Cre vs. control cultures, whereas the sub-G1 fraction (i.e apoptotic cells and cell debris) was approximately twice as large (Fig. 6A upper panels). Together with the data shown in Fig. 5A and B, these data identify slowed (or suspended) cell cycle progression and increased apoptosis as the basis for the impaired in vitro and in vivo responses exhibited by mutant B cells. As depicted in Fig. 6A (lower panels), cell cycle analyses of exclusively IgG1+ events revealed similar differences between mutant and control B cells when compared to the un-fractionated populations. This suggests that switched cells did not display grossly more accentuated cell cycle defects than did cells that had not or not yet undergone CSR, arguing against CSR-induced DSBs as a major cause.
FIGURE 6. Impaired viability and proliferative capacity of Blm-deficient B cells in vitro and abnormal turnover rates of several B cell populations in vivo in Blmf/Δ Cre mice.
(A) Cell cycle analysis of in vitro activated B cells after CSR induction. CD43-depleted splenocytes from Blmf/Δ Cre and Blmf/Δ mice were stimulated with anti-CD40 antibody and IL4 for three days (with 50 ng/ml Colcemid added for the last 90 min of culture). The cells were subsequently stained for surface IgG1-expression and analyzed for determination of DNA content by flow cytometry. Histograms in the upper and lower rows depict PI incorporation of total and switched (IgG1+) cells, respectively. The results shown are representative of three independent experiments.
(B) In vivo turnover of selected lymphocyte populations. Blmf/Δ Cre and Blmf/+ Cre mice were administered 0.8 mg/ml BrdU via the drinking water for two weeks (pulse period). On days 0, 8, 15, 22, 33 of the chase period cohorts of four mice per genotype were sacrificed and bone marrow (0, 8, 15, and 22 d time points only), splenic, and peritoneal exudate cells were analyzed for expression of appropriate surface markers to identify individual lymphocyte populations and BrdU incorporation by cytometry. Each data point corresponds to an individual mouse and solid lines represent trend lines describing the loss of BrdU+ cells (as fitted by non-linear regression). P-values comparing the slopes of the fitted curves are given for each population analyzed. Cell populations were defined as: bone marrow B220intIgM− Fr. A-D, B220intIgM+ Fr. E, and B220hiIgM+ Fr. F, splenic B220+CD23hiCD21+ FO and B220+CD23loCD21hi MZ, and peritoneal B220intCD5int B1a, B220hiCD5− B2, and B220−CD5hi T cells.
When we analyzed metaphase spreads prepared from those cultures we encountered drastically elevated numbers of Blm-deficient cells that exhibited various manifestations of genomic instability (Table II). Not only were the frequencies of aneuploid cells increased 1.8–2.7 fold, but there was also ample evidence for aberrant replication amongst mutant B cell blasts. In three independent experiments, Blmf/Δ Cre cells displayed 41-, 6.6-, and 3.4 times as many abnormal DNA structures per chromosome than control cells (Table II). Thus, an inability to faithfully replicate genomic material upon rapid cell division provides the likely basis for the Blm-dependent compromised proliferative capacity.
Table II.
Genomic instability in proliferating B cellsa
| experiment | genotype | metaphases analyzed | # of aneuploid metaphases (%) | # of metaphases with chromosomal aberrations (%) | # of chromosomal aberrations (% per chromosome) |
|---|---|---|---|---|---|
| 1 | Blmf/Δ Cre | 48 | 17 (35%) | 30 (63%) | 84 (4.13%) |
| Blmf/Δ | 47 | 9 (19%) | 2 (4%) | 2 (0.10%) | |
| 2 | Blmf/Δ Cre | 42 | 19 (45%) | 16 (38%) | 27 (1.52%) |
| Blmf/+ | 52 | 9 (17%) | 5 (10%) | 5 (0.23%) | |
| 3 | Blmf/Δ Cre | 50 | 18 (36%) | 27 (54%) | 50 (2.42%) |
| Blmf/+ Cre | 58 | 11 (19%) | 8 (14%) | 17 (0.72%) |
Chromosome breaks, translocations, small marker chromosomes, double minutes, and prematurely condensed chromosomes were scored on Giemsa-stained metaphase spreads prepared from CD43− B cells that had been stimulated with anti-CD40 and IL4 for three days. Cells from one mouse per genotype were used for each experiment.
In naïve mice, we found Blm-deficiency to affect different B cell subsets and Ig isotype levels to various extents (Fig. 2 and Fig. 3) – findings that represent a mere snapshot of a steady state maintained by the interplay of multiple parameters in vivo. To establish how Blm-dependent genomic instability has shaped the phenotype observed in adult mice, we measured the turnover rates of select lymphocyte subsets from bone marrow, spleen, and peritoneal cavity. Cohorts of Blmf/Δ Cre and Blmf/+ Cre control mice were fed BrdU-containing drinking water for two weeks and groups of four mice per genotype were sacrificed after particular time intervals during a chase period of several weeks. We detected no differences in the long-lived and rather non-proliferative (41) subsets of bone marrow Fr. F, splenic FO, splenic MZ, and peritoneal B2 cells (Fig. 6B). The same was true for peritoneal T cells, which served as a Blm-proficient control population with no differential outcome expected (Fig. 6B). In contrast, short-lived early B lymphoid fractions in the bone marrow (41) up to the Fr. E stage showed significantly slower turnover rates in Blm-deficient animals. Whereas Fr. A-E cells from both groups incorporated BrdU to a similarly high degree (≥80%) during the two-week pulse period, Blmf/Δ Cre cells lost the label at a much slower rate (Fr. A-D: t1/2 = 4.5 days and 2.8 days, and Fr. E: t1/2 = 3.9 days and 2.4 days for mutant and control mice, respectively). A different situation emerged in the subset of slowly dividing peritoneal B1a cells. Mutants entered the chase period with a higher fraction of BrdU+ B1a cells (43.8 ± 5.3% vs. 28.8 ± 5.5% SEM) whose numbers, however, declined more rapidly than in controls, translating into calculated half lives of 26 days in Blmf/Δ Cre and 121 days in Blmf/+ Cre mice (Fig. 6B).
These data indicate that in situations where cells undergo multiple cell divisions in rapid succession (Fr. C’ and early Fr. D), Blm-dependent decelerated cell cycle progression delays the passage of B lymphoid cells through the early developmental stages. In turn, since Fr. A-D cells feed the subsequent stage of immature Fr. E cells (characterized by little proliferation and massive cell death in normal mice (16)) this subset, too, loses BrdU+ cells at a slow kinetics in Blmf/Δ Cre mice. In contrast, the faster turnover rates of mutant peritoneal B1a cells appear symptomatic in that an atrophic population of naturally self-renewing cells (18) undergoes slow but continuing homeostatic proliferation, but fails to fill up the compartment to wild type levels, likely due to excess apoptosis.
Development of B cell lymphomas in p53-deficient Blmf/Δ Cre mice
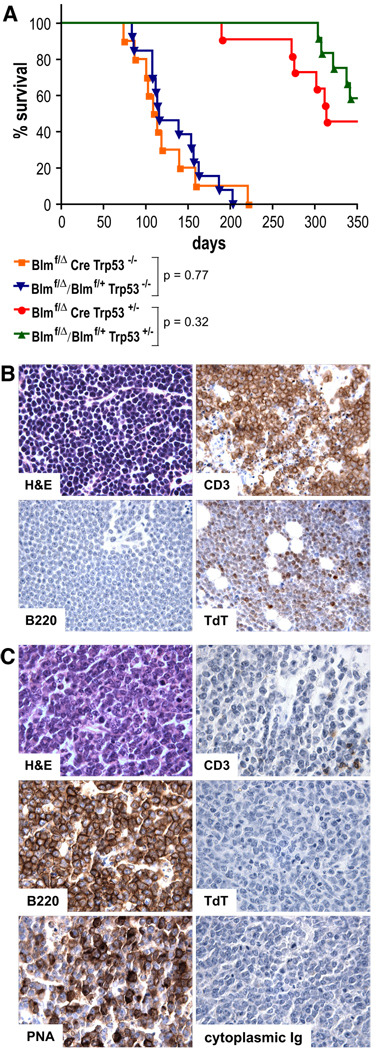
We monitored several cohorts of mice over a 14 month-period for the occurrence of B cell lymphomas. Since it is now well established that Blm-deficiency entails only a mild predilection for tumor-development in the mouse (28–30) we sought to facilitate tumor induction by analyzing cohorts of Blm-deficient and Blm-proficient mice on either Trp53-null- or Trp53-heterozygous backgrounds. Furthermore, to stimulate GC formation and thereby induce CSR-dependent DNA strand breaks, we immunized the mice twice with SRBCs. As evident from Fig. 7A, the survival curves of Blmf/Δ Cre and Cre− control mice did not differ significantly, irrespective of p53-proficiency.
FIGURE 7. Survival of mice as a function of Blm- and p53-deficiency.
(A) Kaplan-Meier survival plots representing percent survival of Blmf/Δ Cre Trp53−/− (n = 10), Cre− control Trp53−/− (n = 13), Blmf/Δ Cre Trp53+/− (n = 11), and Cre− control Trp53+/− (n = 12) mice vs. age. Adult mice were immunized with SRBCs twice at an interval of 4–5 weeks and sacrificed when in overtly sick condition. Each symbol represents an individual mouse at the time of its death and p values were calculated using the Mantel-Haenszel logrank test.
(B) and (C) Histopathology and immunohistochemistry of representative tumors (1000×). (B) Thymic T cell lymphoma from a Blmf/+ Trp53−/− and (C) splenic B cell lymphoma from a Blmf/Δ Cre Trp53−/− mouse. Sections were stained with H&E, anti-CD3, anti-B220, anti-TdT, PNA, and anti-cytoplasmic Ig as indicated. Positive cells or nuclei (anti-TdT only) appear brown in the immunohistochemically stained sections.
We nevertheless determined the tumor spectra in the carcasses. Necropsy revealed that thymic T cell lymphoma caused terminal disease in six out of 13 Trp53−/− control mice (47%), whereas none of them had developed a B cell malignancy (Fig. 7B and Table III). In contrast and unexpectedly, thymic lymphoma was found only in one of ten Blmf/Δ Cre Trp53−/− animals, while a B220+ B cell lymphoma was prevalent in six, as revealed by immunohistochemistry (Fig. 7C and Table III). The Blm-mutants developed B cell malignancies slightly earlier (113 ± 32 days) than did the controls succumb to thymic lymphoma (135 ± 39 days), but the difference was not significant. Of the five moribund Trp53+/− control mice encountered during the course of the experiment, none had developed either thymic or B cell lymphoma. Yet, of six terminally sick Blmf/Δ Cre Trp53+/− animals one and three came down with thymic and B cell lymphoma, respectively (Table III). Intriguingly, further immunohistochemical analysis revealed that all six B cell malignancies from Blmf/Δ Cre Trp53−/− mice were PNA+/TdT−/cytoplasmic Ig− (although PNA-binding was somewhat heterogeneous in five of them), suggesting their derivation from mature B cells (Table III and Fig. 7C). We conclude that B lineage-specific ablation of Blm promotes B cell lymphoma development. This effect, however, only becomes significant upon concomitant inactivation of the tumor suppressor p53.
Table III.
Tumors found in Blmf/Δ Cre Trp53−/−, Blmf/Δ Cre Trp53+/−, and control micea
| Tumor type |
Blmf/Δ Cre Trp53−/− (n=10)b |
Blmf/Δ/Blmf/+ Trp53−/− (n=13)c |
Blmf/Δ Cre Trp53+/− (n=11)d |
Blmf/Δ/Blmf/+ Trp53+/− (n=12)e |
|---|---|---|---|---|
| B220+/CD3−/PNA+/TdT−/cytoplasmic Ig− B cell lymphoma |
6 | |||
| B220+/CD3−/PNA−/TdT+/cytoplasmic Ig− B cell lymphoma |
3 | |||
| B220−/CD3+ thymic lymphoma | 1 | 6 | 1 | |
| Hemangiosarcoma | 4 | 7 | 1 | |
| Rhabdomyosarcoma | 1 | 1 | ||
| Medulloblastoma | 1 | |||
| Osteosarcoma | 1 | |||
| No tumors foundf | 1 | 5 |
mice were immunized twice with SRBCs, monitored for 350 days, sacrificed when in moribund condition (see Fig. 7A), and subjected to necropsy; spleens and macroscopically cancerous organs were examined histopathologically on H&E-stained tissue sections; cases with diagnosed lymphoma were subsequently classified by immunohistochemistry
two mice with B cell lymphoma and hemangiosarcoma, one mouse with rhabdomyosarcoma and hemangiosarcoma
two mice with thymic lymphoma and hemangiosarcoma
five mice alive after 350 days
seven mice alive after 350 days
four mice with neurological symptoms before sacrifice, such as ataxia, axial rotatory movement, hyperexcitability, and lethargy; all were lymphoma-free
DISCUSSION
We report a vital role for the Bloom’s syndrome helicase in all major aspects of the B cell life cycle, manifest in disturbed B cell immunity and tumor development in its absence. Since pleiotropic inactivation of the murine Blm gene confers embryonic lethality (27), we generated tissue-specific Blm knockout mice using the mb1-Cre strain that was shown to yield early and specific deletion of floxed target genes in the B cell lineage (31) – a result we could confirm using our Blmf-allele (Fig. 1). We thus conclude that the findings reported here are B cell-autonomous in nature.
In the bone marrow of Blm mutants, B220+ B lineage cells were dramatically underrepresented indicating impaired B cell development. This effect was primarily attributable to a severe attrition of Fr. D-F cells. Since rearrangement of the Ig H chain genes occurs during the Fr. A-C developmental stages (14) these findings could be explained by an impaired formation of functional VHDHJH joints and a subsequent failure of developing B cells to pass the pre-BCR checkpoint. However, a major role for Blm in the mechanism of VDJ-recombination has been ruled out before (28, 42, 43). Specifically, we previously reported normal VDJ-recombination efficiencies of Tcrb genes in Blm-deficient thymocytes and the inability of a rearranged TCR transgene to rescue atrophic thymi in T cell-specific Blm knockout mice (28). We therefore favor an alternative explanation. The proliferative burst triggered by pre-BCR-expression is considered substantial (≤ 6 cycles) (44, 45) and divisions occur in rapid succession (≥ 6.5 h/cycle) (44), imposing intense replicative stress on developing B cells. BS cells are well-known for their inability to cope with such stress as they exhibit delayed Okazaki fragment growth, accumulate abnormal replication intermediates, and are unable to efficiently restart replication following replication fork demise (46–48). Collapsing replication forks are thought to occur in every replicative cell cycle imposing a potentially serious problem for rapidly dividing cells (49). Blm is recruited to sites of damaged replication forks (50) where it can aid fork re-initiation through potentially several means, including the promotion of fork regression to allow lesion bypass of the leading strand (51) (for detailed discussion see (5, 49)). The helicase’s proposed function as a remover of bulky DNA secondary structures to avoid stalling and breakdown of replication forks in the first place (23, 24) was already mentioned. Thus, an inability of Blm-deficient developing B cells to suppress replication errors will manifest in delayed cell cycle progression (see the slow turnover rates of Blmf/Δ Cre Fr. A-E cells in Fig. 6B), likely accompanied by increased apoptosis rates (52) as observed in mature B cells (see below).
In the spleens of Blmf/Δ Cre mice, we observed greater attrition of the B2 than the MZ cell compartment (Fig. 2B and Fig. 4B). Most affected by the absence of Blm, however, were B1 cells. We encountered an at best rudimentary B1a cell compartment, amounting to <10% and <20% of control numbers in peritoneal cavity and spleen, respectively (Fig. 2C and Fig. 4B). The proliferative capacity of mutant B cells was remarkably impaired upon in vitro culture with several stimuli such as LPS or CD40-ligation. Increased global genomic instability was determined to be the underlying reason, manifesting in elevated levels of chromosomal structural abnormalities and chromosome loss during replication (Table II). This in turn led to elevated apoptosis rates (Fig. 5B and Fig. 6A) and a combination of cell cycle arrest and decelerated cell cycle progression (Fig. 5A, 5B and Fig. 6).
These findings are in line with earlier studies in humans (47, 48) and mice (28–30), and represent classical features of BLM-deficient cells (5, 7). B1a, B1b, FO, and MZ B lymphocytes might exhibit differential sensitivity to or suffer from variable degrees of replication stress during their respective life cycles – possibilities that cannot be excluded, but are hard to test for in our model. In the light of our findings, however, we consider the following scenario most pertinent. The MZ and B2 cell pools are constantly being replenished through bone marrow-derived precursors that are backed up by Blm-proficient HSCs. B1a cells, on the other hand, originate from fetal and postnatal precursors, yet are otherwise dependent on self-propagation throughout adulthood (16, 18). Ongoing self-replenishment is expected to be strongly handicapped as we detected near complete Blm-deletion in peritoneal B1a cells (Fig. 1). Concordantly, while no differences in the turnover rates of mature B2 and MZ cell subsets were seen in Blmf/Δ Cre vs. control mice, we found shorter half-lives of Blm-deficient B1a cells in the peritoneum (Fig. 6B), reflecting the strong but futile drive to replenish the main producers of “natural antibodies” (15).
BLM and p53 physically and functionally interact in vivo (50, 53). Although p53-dependent apoptosis was found deregulated in cell lines from BS patients (53, 54), BS cells were shown to undergo p53-dependent apoptosis at stalled replication forks and p53-deficiency prevents death of damaged BS cells (52). Also, Lu and Osmond reported a marked suppression of normally high apoptosis rates in early B cell development upon Trp53-inactivation (37). We found a general, modestly beneficial effect of p53-deficiency on the survival of B cells in the absence of Blm, e.g. a doubling of FO and B1a cell numbers in Blmf/Δ Cre Trp53−/− compared to Blmf/Δ Cre Trp53+/− mice (Fig. 5B). However, in a T cell-specific Blm knockout model, p53-inactivation had a stronger effect than seen here, as severely reduced thymic cellularities were rescued almost threefold (28). Furthermore, in Trp53−/− scid mice, T cell development was promoted while B cell hematopoiesis remained essentially unaffected (55). In this respect, the findings reported here are in line with the notion that p53-dependent cell survival plays a more significant role in the T cell vs. the B cell lineage (55, 56).
We found low levels of IgM, IgG1, IgG2b, IgG2a, and IgA, but extraordinarily reduced IgG3 concentrations in the sera of naïve mutant animals (Fig. 3A). Similar to our findings, certain gene mutations in mice were reported to produce a substantial depletion of the B1a cell compartment while less affecting conventional B2 or MZ cells. As expected, these strains exhibited marked IgM hypoglobulinaemia, along with comparably low concentrations of several (57) or all other isotypes (58). Our distinct Ig isotype pattern prompted us therefore to test whether Blm might be involved in CSR.
Because of the detrimental effect of Blm-ablation on proliferation and survival (Fig. 5A) we tested B cells doubly deficient for Blm and p53, whose proliferative capacity indeed proved to be more similar albeit not identical to control levels (Fig. 5B). We detected substantially reduced class switching to IgG1, IgG2a, and IgG3 in double-mutant splenic B cells, and the abundance of sterile PS Iμ-Cγ1, Iμ-Cγ2a, and Iμ-Cγ3 transcripts was reduced as well (Fig. 5C and 5D). However, PS transcripts were still at more than 50% of control levels, contrasting with other reports that showed obligatory roles for ATM and 53BP1 in CSR. Here, quantities of productive transcripts in stimulated mutant B cells were insignificant (59, 60). Most importantly, the frequencies of switched cells per generation were not significantly different between Blm/p53 double-deficient and control B cells across all generations and culture conditions. We thus ascribe the reduced switch efficiencies to the impaired proliferative capacity resulting from an inability of Blm-deficient B cells to cope with replicative stress. We did not find evidence for the possibility that the observed cell death was a consequence of the CSR process itself, i.e. that an excess number of induced DSBs remained unrepaired in switching Blm-deficient B cells. In fact, the impairment in CSR was consistently mildest in mutant B cells stimulated with LPS and IL4, while those cultures uniformly produced the highest yields of switched (i.e. IgG1+) cells compared to the other culture conditions. We add that a possible minor function of Blm in the early phase of CSR is probably undetectable using our system due to the obligatory role of proliferation in the CSR process. However, we observed a shift towards microhomology-mediated end joining in Sμ-Sγ3 junction sequences amplified from activated Blm-deficient B cells (Fig. 5F). This effect, although moderate, is generally compatible with a role for Blm in the resolution phase of CSR, i.e. the synapsis and/or joining of the participating S-regions. The relevance of this finding remains unclear at this point and its elucidation will be an interesting subject for future experiments. A possible direct role for Blm could include the disruption of DNA structures bearing only short stretches of homology, thereby disfavoring the generation of CSR joints that rely on microhomologies for their resolution. This would be reminiscent of the presumed role of RecQ helicases as fidelity regulators during HR (suppressing illegitimate recombination events (5)). Indirectly, a lack of Blm could interfere with the proper function of certain interaction partners, such as ATM (61) or the mismatch repair factor Mlh1 (25) that are known to be involved in CSR (26, 59, 62). For example, longer stretches of microhomology were also found in switched B cells deficient for Mlh1, albeit the effect was stronger (26).
BS patients develop tumors of various types at a high incidence prominently including B cell neoplasias (7, 8), yet most mouse models for BS exhibit only a mild propensity for cancerogenesis (28–30). The reason for this discrepancy is unclear, but a certain functional redundancy amongst the five mammalian RecQ family members (RECQ1, BLM, WRN, RECQ4, and RECQ5 in man), differentially accentuated in rodents and primates, is a possibility (63). Also, a role for Blm in telomere maintenance is emerging (64); an effect hard to model in mice due to the inherently longer telomeres of murine chromosomes. Indeed, in an attempt to faithfully model Werner syndrome, a premature aging disease caused by WRN-mutation, the full spectrum of the human disease (including an increased cancer incidence) was only unveiled after induced exhaustion of telomere reserves in engineered mice that lacked both, the Wrn gene and the telomerase RNA template Terc (65). In the Wrn−/−Terc−/−Blmtm3Brd−/− model described by Du et al. (64) cancer development was not investigated. But since this model used a hypomorphic Blm-allele (30, 66), it is tempting to speculate that Terc-deficiency in Blmf/Δ mb1-Cre mice could well accelerate the development of B cell lymphomas.
We used inactivation of the gatekeeper tumor suppressor p53 to facilitate lymphoma development in a small cohort of mice. P53-deficient mice are known to frequently develop lymphomas, though predominantly of thymic origin (32, 67). If ablation of Blm in the B cell lineage actually facilitated the development of B cell malignancies, we would expect a shift in the tumor spectrum favoring B over T cell-derived lymphomas. This is what we observed here. Although the life spans of Blmf/Δ Cre Trp53−/− and control Trp53−/− mice were similar (Fig. 7A) more than half of the double mutants exhibited B cell neoplasias at the time of death (Table III). We conclude that Blm-deficiency in B lymphocytes exerts a mild tumor-promoting effect in mice. This is in line with Luo et al. who reported late cancer development in 29% of mice homozygous for the hypomorphic Blmtm3Brd allele by the age of 20 months, including lymphomas (66). Interestingly, judging from their immunohistochemical phenotype, the six B cell lymphomas found in Blmf/Δ Cre Trp53−/− animals appear to have developed from GC or post-GC B cells. This finding is reminiscent of the situation in BS patients where Non-Hodgkin’s lymphomas dominate the spectrum of hematogenous malignancies (ref. (8), yet with unknown p53-status). Most Non-Hodgkin’s lymphomas are known to originate from GC or post-GC B cell precursors (68).
With the methodology employed, we cannot gauge to which extend unfaithfully repaired AID-dependent DNA strand breaks generated during CSR or SHM contribute to lymphoma development in immunized Blmf/Δ Cre Trp53−/− mice. Franco et al. (69) found dramatic levels of chromosomal instability in switching B cells deficient for histone H2AX in vitro, and those high levels were independent of p53-proficiency. In contrast to H2AX−/− mice, however, H2AX−/− Trp53−/− mice are dramatically cancer prone (70, 71), arguing that the generation of potentially oncogenic translocations can occur in the presence of p53 (similar to our findings, e.g. Table Table II). Defective p53-dependent checkpoints may, however, allow the propagation and outgrowth of post-GC B cells bearing them (69). It will be interesting to delineate to which extend the BCR loci are involved in oncogenic translocation events in lymphomas from Blmf/Δ Cre Trp53−/− animals. In addition to its function as roadblock remover during replication, facilitator of replication fork restarts, and anti-recombinase (as discussed above), the tumor suppressor functions exercised by Blm are thought to depend on its role as a suppressor of crossover formation (e.g. through double Holiday junction dissolution (6)). These functions in turn counteract the occurrence of LOH (66) and global genomic instability (29), crucial processes underlying tumor formation.
In conclusion, we generated a mouse model for the immunodeficiency in BS highlighting disturbances in key aspects of B cell biology. We propose that, together with a Blm-dependent T cell defect we described earlier (28), an impairment in both arms of the adaptive immune system accounts for the compromised immunity seen in affected individuals (72). But, besides the issues concerning embryonic lethality and tumor incidence mentioned above, we see further evidence for an accentuation of Blm function in mice vs. man. Although low Ig levels of one or several isotypes are typically found in most BS patients, B cell numbers are largely normal, though at the lower end of the spectrum (9, 11). Unfortunately, specific information on the distribution of different B cell subsets in affected individuals (e.g. size of the B1 cell compartment) is not available to extend the comparison. Nevertheless, it will be interesting to explore the impact of postulated backup mechanisms in mice that could fill in upon Blm-ablation, e.g. by a double knockout strategy. RECQ5, for instance, exhibits similar biochemical and functional properties (63), and a pan-Recql5-deficient mouse, although developmentally normal, revealed a prominent tumor suppressor function for this RecQ helicase as well (73).
ACKNOWLEDGEMENTS
We are grateful to Boris Reizis (Columbia University Medical Center, New York City, NY), Klaus Rajewsky (IDI, Harvard Medical School), and Nicholas Chester (Schering-Plough Research Institute, Cambridge, MA) for valuable suggestions and critically reviewing the manuscript. We thank Sven Kracker and Heide Christine Patterson (both IDI, Harvard Medical School) for helpful discussions and Natasha Barteneva (Flow Cytometry facility, IDI, Harvard Medical School) for technical assistance with cell sorting. We are further indebted to Roderick Bronson (Rodent Histopathology Core, Harvard Medical School) for providing expert advice with histopathology and the Biopolymers Facility at Harvard Medical School for high throughput colony sequencing service.
Footnotes
This work was supported in part by the Deutsche Forschungsgemeinschaft (SFB620, Teilprojekt B5) to M. R and grants from the NIH (JW).
Publisher's Disclaimer: Disclaimer:
"This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at http://www.jimmunol.org."
Abbreviations used in this paper:
CSR, class switch recombination; SHM, somatic hypermutation; BS, Bloom’s syndrome; AFCs, antibody-forming cells; Fr., Hardy-fraction; FO, follicular; MZ, marginal zone; TD, T cell-dependent; TI, T cell-independent; DSBs, double strand breaks; S-regions, switch-regions; AID, activation-induced cytidine deaminase; floxed, loxP-flanked; GC, germinal center; PNA, peanut agglutinin; GL, germline; PS, post-switch
DISCLOSURES
The authors have no financial conflict of interest.
REFERENCES
- 1.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 2.Gennery AR, Cant AJ, Jeggo PA. Immunodeficiency associated with DNA repair defects. Clin Exp Immunol. 2000;121:1–7. doi: 10.1046/j.1365-2249.2000.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 4.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 7.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 8.German J. Bloom's syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- 9.Kondo N, Motoyoshi F, Mori S, Kuwabara N, Orii T, German J. Long-term study of the immunodeficiency of Bloom's syndrome. Acta Paediatr. 1992;81:86–90. doi: 10.1111/j.1651-2227.1992.tb12088.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Kerckhove CW, Ceuppens JL, Vanderschueren-Lodeweyckx M, Eggermont E, Vertessen S, Stevens EA. Bloom's syndrome. Clinical features and immunologic abnormalities of four patients. Am J Dis Child. 1988;142:1089–1093. doi: 10.1001/archpedi.1988.02150100083032. [DOI] [PubMed] [Google Scholar]
- 11.Etzioni A, Lahat N, Benderly A, Katz R, Pollack S. Humoral and cellular immune dysfunction in a patient with Bloom's syndrome and recurrent infections. Journal of clinical & laboratory immunology. 1989;28:151–154. [PubMed] [Google Scholar]
- 12.Hutteroth TH, Litwin SD, German J. Abnormal immune responses of Bloom's syndrome lymphocytes in vitro. J Clin Invest. 1975;56:1–7. doi: 10.1172/JCI108058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 15.Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–335. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 16.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 17.Pillai S. The chosen few? Positive selection and the generation of naive B lymphocytes. Immunity. 1999;10:493–502. doi: 10.1016/s1074-7613(00)80049-7. [DOI] [PubMed] [Google Scholar]
- 18.Baumgarth N, Tung JW, Herzenberg LA. Inherent specificities in natural antibodies: a key to immune defense against pathogen invasion. Springer seminars in immunopathology. 2005;26:347–362. doi: 10.1007/s00281-004-0182-2. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 21.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends in immunology. 2002;23:31–39. doi: 10.1016/s1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- 22.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Jr, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 25.Pedrazzi G, Perrera C, Blaser H, Kuster P, Marra G, Davies SL, Ryu GH, Freire R, Hickson ID, Jiricny J, Stagljar I. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001;29:4378–4386. doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrader CE, Vardo J, Stavnezer J. Role for mismatch repair proteins Msh2, Mlh1, and Pms2 in immunoglobulin class switching shown by sequence analysis of recombination junctions. J Exp Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chester N, Kuo F, Kozak C, O'Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babbe H, Chester N, Leder P, Reizis B. The Bloom's syndrome helicase is critical for development and function of the alphabeta T-cell lineage. Mol Cell Biol. 2007;27:1947–1959. doi: 10.1128/MCB.01402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chester N, Babbe H, Pinkas J, Manning C, Leder P. Mutation of the murine Bloom's syndrome gene produces global genome destabilization. Mol Cell Biol. 2006;26:6713–6726. doi: 10.1128/MCB.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDaniel LD, Chester N, Watson M, Borowsky AD, Leder P, Schultz RA. Chromosome instability and tumor predisposition inversely correlate with BLM protein levels. DNA Repair (Amst) 2003;2:1387–1404. doi: 10.1016/j.dnarep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 33.Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- 34.Manz RA, Hauser AE, Hiepe F, Radbruch A. Maintenance of serum antibody levels. Annu Rev Immunol. 2005;23:367–386. doi: 10.1146/annurev.immunol.23.021704.115723. [DOI] [PubMed] [Google Scholar]
- 35.McHeyzer-Williams MG. B cells as effectors. Curr Opin Immunol. 2003;15:354–361. doi: 10.1016/s0952-7915(03)00046-3. [DOI] [PubMed] [Google Scholar]
- 36.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 37.Lu L, Osmond DG. Apoptosis and its modulation during B lymphopoiesis in mouse bone marrow. Immunol Rev. 2000;175:158–174. [PubMed] [Google Scholar]
- 38.Li SC, Rothman PB, Zhang J, Chan C, Hirsh D, Alt FW. Expression of I mu-C gamma hybrid germline transcripts subsequent to immunoglobulin heavy chain class switching. Int Immunol. 1994;6:491–497. doi: 10.1093/intimm/6.4.491. [DOI] [PubMed] [Google Scholar]
- 39.Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc Natl Acad Sci U S A. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forster I, Rajewsky K. The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc Natl Acad Sci U S A. 1990;87:4781–4784. doi: 10.1073/pnas.87.12.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsieh CL, Arlett CF, Lieber MR. V(D)J recombination in ataxia telangiectasia, Bloom's syndrome, and a DNA ligase I-associated immunodeficiency disorder. J Biol Chem. 1993;268:20105–20109. [PubMed] [Google Scholar]
- 43.Petrini JH, Donovan JW, Dimare C, Weaver DT. Normal V(D)J coding junction formation in DNA ligase I deficiency syndromes. J Immunol. 1994;152:176–183. [PubMed] [Google Scholar]
- 44.Osmond DG. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr Opin Immunol. 1991;3:179–185. doi: 10.1016/0952-7915(91)90047-5. [DOI] [PubMed] [Google Scholar]
- 45.Rolink AG, Winkler T, Melchers F, Andersson J. Precursor B cell receptor-dependent B cell proliferation and differentiation does not require the bone marrow or fetal liver environment. J Exp Med. 2000;191:23–32. doi: 10.1084/jem.191.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc Natl Acad Sci U S A. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer research. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 49.Wu L, Hickson ID. RecQ helicases and cellular responses to DNA damage. Mutation research. 2002;509:35–47. doi: 10.1016/s0027-5107(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 50.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. Embo J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 52.Davalos AR, Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. The Journal of cell biology. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang XW, Tseng A, Ellis NA, Spillare EA, Linke SP, Robles AI, Seker H, Yang Q, Hu P, Beresten S, Bemmels NA, Garfield S, Harris CC. Functional interaction of p53 and BLM DNA helicase in apoptosis. J Biol Chem. 2001;276:32948–32955. doi: 10.1074/jbc.M103298200. [DOI] [PubMed] [Google Scholar]
- 54.Spillare EA, Wang XW, von Kobbe C, Bohr VA, Hickson ID, Harris CC. Redundancy of DNA helicases in p53-mediated apoptosis. Oncogene. 2006;25:2119–2123. doi: 10.1038/sj.onc.1209242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nacht M, Strasser A, Chan YR, Harris AW, Schlissel M, Bronson RT, Jacks T. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes Dev. 1996;10:2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- 56.Marsden VS, Strasser A. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu Rev Immunol. 2003;21:71–105. doi: 10.1146/annurev.immunol.21.120601.141029. [DOI] [PubMed] [Google Scholar]
- 57.Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen PJ. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 1999;11:547–554. doi: 10.1016/s1074-7613(00)80130-2. [DOI] [PubMed] [Google Scholar]
- 58.Engel P, Zhou LJ, Ord DC, Sato S, Koller B, Tedder TF. Abnormal B lymphocyte development, activation, and differentiation in mice that lack or overexpress the CD19 signal transduction molecule. Immunity. 1995;3:39–50. doi: 10.1016/1074-7613(95)90157-4. [DOI] [PubMed] [Google Scholar]
- 59.Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC, 3rd, Gearhart PJ, Wynshaw-Boris A, Max EE, Hodes RJ. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–1121. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, Nussenzweig MC, Chen J. 53BP1 is required for class switch recombination. The Journal of cell biology. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- 62.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 66.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 67.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 68.Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. The New England journal of medicine. 1999;341:1520–1529. doi: 10.1056/NEJM199911113412007. [DOI] [PubMed] [Google Scholar]
- 69.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, Manis JP, Chen J, Carpenter PB, Alt FW. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 70.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 71.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.German J. The Immunodeficiency of Bloom syndrome. In: Ochs HD, Smith CIE, Puck J, editors. Primary immunodeficiency diseases : a molecular and genetic approach. New York: Oxford University Press; 1999. pp. 335–338. [Google Scholar]
- 73.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]