Abstract
The Paramyxoviridae are enveloped, negative-stranded RNA viruses, some of which recognize sialic acid-containing receptors, while others recognize specific proteinaceous receptors. The major cytopathic effect of paramyxovirus infection is membrane fusion-induced syncytium formation. Paramyxoviruses are unusual in that the receptor-binding and fusion-promoting activities reside on two different spike structures, the attachment and fusion glycoproteins, respectively. For most paramyxoviruses, this distribution of functions requires a mechanism by which the two processes can be linked for the promotion of fusion. This is accomplished by a virus-specific interaction between the two proteins. An increasing body of evidence supports the notion that members of this family of viruses utilize this glycoprotein interaction in different ways in order to mediate the regulation of the fusion protein activation, depending on the type of receptor utilized by the virus.
Keywords: attachment protein, fusion protein, glycoprotein interaction, membrane fusion, paramyxovirus, receptor binding
Paramyxoviridae
The Paramyxoviridae are a family of enveloped viruses that contain a negative-sense, nonsegmented, single-stranded RNA genome [1]. The family includes viruses that cause disease in both humans and animals. Human pathogens in the subfamily Paramyxovirinae include measles virus (MV), mumps virus and the various human parainfluenza viruses (hPIVs; types 1–4), while those in subfamily Pneumovirinae include respiratory syncytial virus (RSV) and human metapneumovirus. MV can cause severe CNS disease and remains a leading killer of children worldwide despite successful vaccination programs in industrialized countries [2]. RSV and hPIV1–3 have long been recognized as causative agents of croup and as important respiratory pathogens, especially in infants and children [3]. RSV is known to cause bronchiolitis and pneumonia in infants and children, and human metapneumovirus causes both upper and lower respiratory infections in children and adults [4].
Important animal viruses in the family include Newcastle disease virus (NDV), Sendai virus (SV), parainfluenza virus 5 (PIV5) and canine distemper virus (CDV). NDV is an important avian pathogen which, owing to its potential as an agent of agricultural bioterrorism, has been classified as a select agent. In recent years, NDV has also gained importance for its ability to selectively kill human tumor cells and for its use as an oncolytic agent [5–7] and vaccine vector [8–11].
The relatively recently emerged henipaviruses, Hendra (HeV) and Nipah (NiV), are also members of the Paramyxovirinae subfamily. These viruses are unique among the paramyxoviruses in being capable of causing severe encephalitis and high mortality rates in both animals and humans [12,13]. Based on their highly infectious nature and virulence, they are potential agents of bioterrorism and are considered to be Risk Group 4 Overlap Select Agents.
Paramyxovirus fusion
The surfaces of paramyxovirions and infected cells possess two types of spikes, comprised of the attachment and fusion (F) proteins. Paramyxoviruses gain entry into cells by promoting the direct fusion of the viral and cellular membranes. The hallmark cytopathic effect of the infection of cells by paramyxoviruses is the formation of syncytia, which is mediated by membrane fusion induced by the two viral glycoproteins expressed on the surface of infected cells.
Porotto et al. developed a novel assay to dissect the individual steps in fusion [14] based on the idea that the association of cells expressing hPIV3 hemagglutinin–neuraminidase (HN) and F protein with receptor-bearing target cells have three possible fates. First, the target cells can be released by neuraminidase. Second, the target cells can be bound reversibly and released by a neuraminidase inhibitor. Third, the target cells can be bound irreversibly by insertion of F protein into the membrane. Using this assay, it was shown that HN’s capacity to promote fusion depends on a balance between receptor-binding avidity, neuraminidase and F protein triggering, and each has the ability to independently affect HN’s role in fusion. Thus, syncytium formation may play a role in optimizing the surface:volume ratio for viral replication (a cytoplasmic or ‘volume’ event) and viral assembly (a membrane or ‘surface’ event). With a few exceptions, the promotion of paramyxovirus fusion is tightly regulated through a virus-specific interaction between the two viral surface glycoproteins. In addition, in the context of MV infection, it has been shown that the matrix (M) protein can negatively regulate cell–cell fusion by promoting formation of virus particles [15].
Paramyxovirus attachment proteins & their receptors
All paramyxovirus attachment proteins are type II homotetrameric membrane glycoproteins, although they differ in the types of receptors they recognize (Table 1). The attachment proteins of the avulaviruses, rubulaviruses and respiroviruses (e.g., NDV, PIV5 and hPIV3) mediate binding to sialic acid-containing receptors and possess neuraminidase activity, the ability to cleave this same component. Neuraminidase is thought to be involved in preventing the self-aggregation of viral particles during budding from the host-cell membrane. Owing to the fact that these proteins are capable of agglutinating red blood cells, they are designated as HN proteins [1].
Table 1.
Some paramyxoviruses and their receptors.
| Virus | Attachment protein | Receptor(s) |
|---|---|---|
| Genus: Avulavirus | ||
| Newcastle disease virus | HN | Sialylated proteins and lipids |
| Genus: Respirovirus | ||
| Human parainfluenza viruses 1 and 3 | HN | Sialylated proteins and lipids |
| Sendai virus | HN | Sialylated proteins and lipids |
| Bovine parainfluenza virus 3 | HN | Sialylated proteins and lipids |
| Genus: Rubulavirus | ||
| Mumps virus | HN | Sialylated proteins and lipids |
| Parainfluenza virus 5 | HN | Sialylated proteins and lipids |
| Genus: Morbillivirus | ||
| Measles virus | H | CD150 (SLAM) and/or CD46* |
| Genus: Henipavirus | ||
| Hendra virus | G | EphrinB2 and B3‡ |
| Nipah virus | G | EphrinB2 and B3‡ |
EphrinB2 and ephrinB3, ligands for the Eph family of receptor tyrosine kinases, have been identified as cellular receptors recognized by the G glycoproteins of Hendra and Nipah [24–26].
G: Glycoprotein; H: Hemagglutinin; HN: Hemagglutinin–neuraminidase; SLAM: Signaling lymphocyte-activation molecule.
The attachment proteins of other members of the family do not recognize sialic acid, but rather, specific protein receptors [16]. The attachment protein of MV mediates binding to either CD46 or CD150 (signaling lymphocyte-activation molecule [SLAM]) [17–21]. Since it agglutinates primate erythrocytes and lacks neuraminidase activity, this protein is called hemagglutinin (H).
The attachment glycoproteins of henipa-viruses differ from the H/HN proteins in that they have neither hemagglutination nor neuraminidase activity [22,23]. For this reason, they are called G proteins. EphrinB2 and ephrinB3 have been identified as cellular receptors for the henipaviruses [24–26].
Similar to the henipaviruses, the pneumo-virus, RSV, has an attachment (G) protein that does not cause hemagglutination of erythrocytes. Receptor binding by RSV is not completely understood but is thought to involve interactions with heparan sulfate, a glucos-aminoglycan that is part of the extracellular matrix [27]. Interestingly, αvβ1-integrin was recently identified as a functional receptor for human metapneumovirus. However, this receptor appears to be unique in being recognized by the viral F protein rather than the G protein [28].
Structure of the globular head of the HN protein
The ectodomains of all paramyxovirus attachment proteins consist of a stalk and a globular head, in which the receptor binding and antigenic sites reside. The structures of the globular heads of several paramyxovirus attachment proteins have now been solved by x-ray crystallography. The initial structures solved were those of NDV HN, both unliganded and complexed with either sialic acid or a neuraminidase inhibitor [29]. The structures revealed a typical neuraminidase active site within a β-sheet propeller motif, although with significantly different conformations. Based on these structures, it was concluded that this protein is very dynamic, with both receptor binding and neuraminidase activities mediated by a single active site that is able to switch between two very different states through a conformational change, which drastically alters the dimer interface. Subsequently, this same group identified a second site on NDV HN composed of residues from each monomer at the membrane distal end of the dimer interface [30]. This second site is capable of binding to sialic acid analogs; however, it lacks enzymatic activity and most of the interactions with the ligand involve backbone atoms.
The structures of two other HN proteins have also been solved. The structures of HN from hPIV3, both unliganded and with several different ligands, were determined at pH 7.5 [31]. The structure is similar to that of NDV HN, but with a single flexible site that mediates both receptor binding and neuraminidase by a structural change limited to the active site, with no alteration of the dimer interface. There is some evidence to suggest the existence of a second sialic acid binding site in hPIV3 HN [32], and loss of an N-linked glycan at residue 173 in hPIV1 HN is proposed to expose a second binding site in that protein [33], although neither has been confirmed crystallographically. However, the structure of PIV5 HN identifies a single site, but with no changes in the positions of active site residues [34]. The PIV5 crystal structure is of the entire ectodomain, but the stalk was not visible, indicating that it is unstructured or may adopt multiple conformations in the crystal. In any event, the proposed drastic conformational change in the dimer interface of NDV HN would be unique among the HN proteins.
Structures of measles virus H & Nipah G protein
The structures of both the MV H [35,36] and NiV/HeV G protein [37,38] monomeric globular domains complexed with their respective receptors were recently solved. Both MV H and NiV G protein retain the β-sheet propeller motif characteristic of neuraminidase molecules, even though they lack neuraminidase activity and do not bind sialic acid. In MV H, the vestigial neuraminidase site has been inactivated by point mutations that abolish catalysis. The area of the molecule corresponding to the second sialic acid binding site present in NDV is blocked by the addition of a glycosylation site. In NiV G protein, the supposed active site is also inactivated by mutations; however, the binding site for the ephrinB2/B3 is still located at the pocket opening into the β-sheet propeller motif. This means that the binding site of this attachment protein to its protein receptor is positioned, relative to the β-sheet propeller motif, in a very similar location to the neuraminidase site on parmyxovirus HN proteins.
The actual orientation of the β-sheet propeller motif in the globular head of NiV G protein is still unknown owing to the lack of data for the dimeric structure of the molecule. Attempts at modeling a NiV G protein dimeric structure from the crystallographic data for the monomer have led to several different proposed dimer structures [37–39]. Thus, the actual dimeric structure of this molecule remains to be determined. The orientation of the β-sheet propeller motif in the dimeric structure of MV H, another paramyxovirus with a protein receptor, is proposed to be significantly different from the orientation characteristic of paramyxovirus HNs with their sialic acid receptors [36]. The axes of the two MV H β-sheet propellers are at very oblique angles, almost parallel to the membrane, and are directed away from each other. This orientation is quite similar to one of the proposed dimer structures for NiV G protein [38].
Structure & function of the F glycoprotein
The F protein must be proteolytically cleaved to promote fusion
All paramyxovirus F proteins are class I fusion proteins similar to the influenza HA, HIV gp41 and the Ebola virus GP2 [40]. The F protein is a trimeric, type I membrane glycoprotein. It is produced as an inactive precursor, F0, which must be cleaved by a cellular protease to produce an active, disulfide-linked F1–F2 complex and a new hydrophobic C-terminus of F1, called the ‘fusion peptide’. For most paramyxoviruses, the F protein is cleaved by a furin-like protease with susceptibility dependent on the number of basic residues in the cleavage site. However, the henipavirus F protein is very different in this regard. It was recently shown that cleavage of this protein is mediated by cathepsin L in acidic, endosomal compartments following a complex trafficking pathway [41–44]. This mechanism appears to be unique among the paramyxoviruses and orthomyxoviruses.
F protein heptad repeats
Two stretches of amphipathic α-helical structure are conserved among F proteins. They are heptad repeats (HRs), with every seventh residue being L, I or V. HR-A is adjacent to the fusion peptide at the N-terminus of F1. HR-B is located in the 40 membrane proximal residues of the stalk. Upon triggering, HR-A forms a triple-stranded coiled-coil hairpin for the insertion of the fusion peptide into the membrane. After a conformational change, HR-B fits into the grooves between the HR-A domains in antiparallel fashion to form a six-helix bundle (6-HB). HR-A and HR-B peptides block 6-HB formation and fusion [45].
Crystal structure of F protein in its pre- & postfusion conformations
The structures of soluble forms of the NDV [46] and hPIV3 [47] F proteins have been solved. The F protein trimer is organized into head, neck and stalk regions. These structures contain the 6-HB and constitute the trimeric, postfusion state into which the secreted, soluble form spontaneously folds. A major advance in our understanding of paramyxovirus fusion was the solution of the structure of the metastable, prefusion conformation of the PIV5 F protein [48]. Although the anchorless form of the protein does not trimerize, these investigators noted that it does assemble into trimers when appended with a trimeric coiled-coil domain at HR-B. These trimers were crystallized, making it possible to solve the structure of the prefusion form of the F protein. There are significant changes in the secondary and tertiary structures of the protein in the transition to the postfusion state, involving a melting of the HR-B stalk, formation of the extended HR-A coiled coil and insertion of the fusion peptide. HR-B is then repositioned to form the final 6-HB structure with HR-A, which completes the change leading to membrane merger and pore formation [48].
Attachment protein-independent fusion
Among the paramyxoviruses, there are several examples of membrane fusion induced by expression of only the fusion protein. The best-characterized attachment protein-independent F protein is that of the W3A strain of PIV5. Based on sequence differences with the F protein of the highly homologous WR strain, which does require HN for fusion, it was shown that several amino acids, specifically, residues 22, 132, 416 and 443, are responsible for the HN-independent phenotype [49,50]. It is thought that prolines at residues 22 and 443 may destabilize the F protein by decreasing the energy required to effect the transition to the fusion-active conformation. Other examples of attachment protein-independent fusion include the F proteins of several viruses in the Pneumovirus genus, most notably RSV, and that of peste des petits ruminants virus, a morbillivirus. RSV F protein can bind cellular heparin and heparan sulfate receptors to trigger a conformational change, leading to extensive fusion in the absence of the viral attachment protein [51,52].
Attachment protein-independent fusion has also been induced by site-directed mutagenesis in important domains of various F proteins. In this way, it was shown that mutations in the cytoplasmic domain of the SER virus F protein rescue syncytium formation and eliminate the HN protein requirement for membrane fusion [53]. It was also shown that the F protein of NDV can be rendered HN-independent for fusion by the introduction of an L289A mutation [54]. However, it should be noted that the fusion activity of each these attachment protein-independent F proteins is significantly enhanced by coexpression of the homologous attachment protein.
Attachment protein-dependent fusion
It is clear that the fusion-promoting activities of the vast majority of paramyxovirus F proteins are dependent not only on the attachment protein, but specifically on the homologous attachment protein. More than 15 years ago, Hu et al. initially established that there is a virus-specific aspect to the attachment protein-dependent mode of fusion [55]. With the exceptions of NiV/HeV [56], hPIV1/SV (not reciprocal) [57] and some MV/CDV combinations [58], the glycoproteins of which share significant homology, the attachment and F proteins derived from heterologous paramyxoviruses fail to result in syncytium formation. Hu et al. concluded from their results that the promotion of fusion requires a specific interaction between the homologous attachment and F protein that is mediated by specific complementary domains [55], which has now clearly been shown to be the case.
Detection of complexes between homologous attachment & F protein
The initial demonstration of a complex between paramyxovirus attachment and F protein was accomplished in the Compans laboratory (Emory University School of Medicine, GA, USA) by the coimmunoprecipitation (co-IP) of radiolabeled, surface biotinylated hPIV2 HN and F protein with an antibody to only one of the proteins [59]. Several laboratories have since been able to detect paramyxovirus glycoprotein complexes in cells using the approach of immunoprecipitation with an antibody to one protein and detection of the second coprecipitated protein by western blot. This includes NDV HN and F protein [60], MV H and F protein [58,61] and NiV/HeV G and F protein [39,62–64]. The approach described by Yao et al. [59] has been used to examine NDV HN–F protein [65–68] and MV H–F protein [69,70] complexes at the cell surface.
Determinants of the interaction
Chimera studies indicate that the stalk regions of several HN proteins determine specificity for the homologous F protein
Chimeric HN proteins with segments derived from heterologous paramyxoviruses have been used to identify the F protein-specific region(s) on the HN proteins of several viruses, including hPIV3, NDV, SV and hPIV2 [71–75]. In all of these studies, it was demonstrated that chimeras having N-terminal segments extending to near the top of the stalk region that are derived from the HN protein of a particular virus fuse with the F protein from that virus, regardless of the viral origin of the HN globular domain. These studies all indicate that specificity for the homologous F protein is determined by the stalk of HN, although portions of the transmembrane anchor and regions in the globular head may also contribute to F protein specificity in some cases.
Peptide studies indicate that a domain at the stalk–head interface of NDV HN interacts with HR-B in the homologous F protein
Based on the assumption that HR-B in F protein mediates the interaction with the homologous HN protein and that a peptide mimicking this domain will bind specifically to a peptide containing the F protein-interactive domain on HN, a 20-mer peptide spanning the NDV F protein HR-B was tested for binding to peptides mimicking various segments of the NDV HN stalk [76]. The HR-B peptide bound to a peptide mimicking HN amino acids 124–152, suggesting that this region defines the F protein-interactive site on HN. This approach is consistent with the complementary domains on the HN and F proteins localizing to the head–stalk transition region and HR-B, respectively.
Chimeric proteins suggest that the HR-B & cysteine-rich regions of F protein are involved in HN(H) specificity
Two groups have taken advantage of the close homology between the MV and CDV glycoproteins to try to map the H-specific domain on the F protein. Ten F protein ectodomain cysteines are highly conserved with eight clustered in a cysteine-rich domain, which begins 75 residues from the transmembrane. Wild et al. reported that replacement of only the 44 amino acid N-terminal half of this region from CDV F protein with the corresponding domain from MV F protein confers on CDV F protein the ability to fuse with MV H [77]. However, this conclusion is weakened by the lack of quantitative data on the extent of fusion and the efficiency of cleavage of the chimeras. Evaluation of hPIV2/SV-41 F protein chimeras has also identified regions clustered near the cysteine-rich domain and HR-B as determinants of HN specificity [78].
Residues in MV H & F protein that interdependently contribute to the formation of functional H–F protein complexes
A more complete study by Lee et al. took advantage of a pair of CDV F proteins that share more than 95% homology, but differ in their ability to promote fusion with the MV attachment protein [58]. This group identified complementary residues on the H and F proteins that contribute to their reciprocal specificity. This approach has identified CDV F protein residues 219 and 233 as contributors to the interaction with H. Conversely, the introduction of a five-residue domain (residues 110–114) from the CDV H stalk into the MV H stalk confers specificity for the CDV F protein. It was concluded that these residues in the two glycoproteins interdependently contribute to the formation of functional H–F protein complexes. The study is strengthened by the evaluation of the avidity of the interaction between the H and F proteins and by the close apposition of F protein residue 233 and H stalk residues 110–114 in structural models of the two proteins. Contrary to the peptide studies described previously, these models predict that it is the globular head of MV F protein that interacts with the stalk of H. Interestingly, these regions of the PIV5 HN and F protein spikes also align with each other based on biochemical data [79] and modeling of the structure of HN [Lamb, Jardetzky, Pers. Comm.]
Mutations in the NDV HN stalk specifically abolish fusion & the HN–F protein interaction
A series of N-linked glycosylation sites was introduced at several positions in the stalk region of HN and the effect on fusion and the HN–F protein interaction was determined [68]. As controls, hemadsorption (HAd), neuraminidase activities and antigenic structure were monitored. All the introduced sites are utilized and essentially abolish fusion. Glycosylation sites at residues 69 and 77 specifically abolish fusion, which correlates with a loss of the ability of the HN protein to interact with F protein in the co-IP assay. Thus, the presence of supernumerary N-glycans in the stalk of NDV HN specifically abolishes both fusion and the HN–F protein interaction, consistent with the stalk mediating the interaction with F protein.
An examination of the HN stalk identified a partially conserved motif that could mediate the HN–F protein interaction, which has enough sequence heterogeneity to account for its virus specificity. This is a stretch of almost 40 residues, 74–110 in NDV HN, which includes two partially conserved HR-like domains. Analyses of HN proteins carrying substitutions for the heptadic residues show that they are important for fusion, but not for attachment, although most also decrease levels of neuraminidase and alter the structure of the protein [80,81].
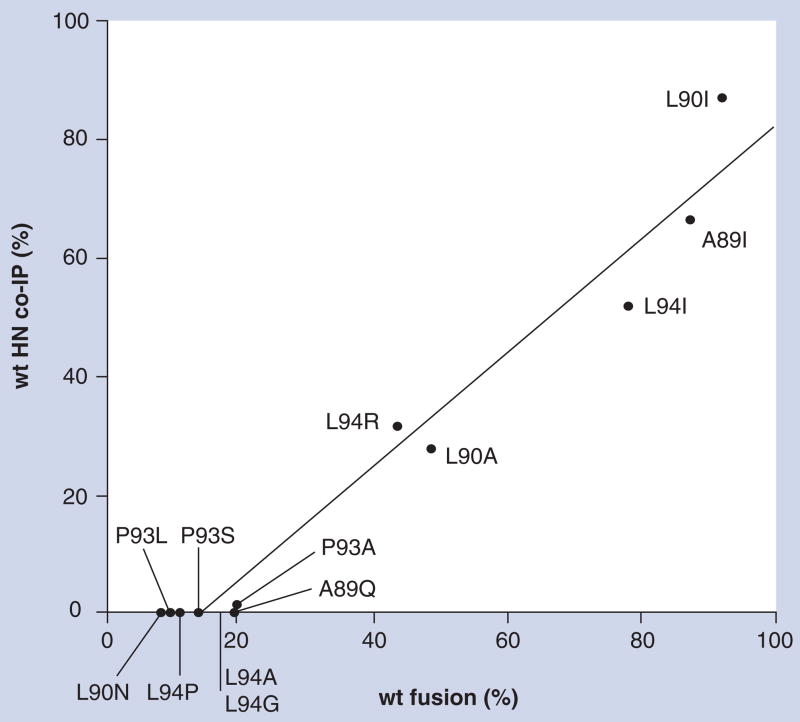
Between the two HR-like domains is a seven amino acid-intervening region defined by residues 89–95 [67]. Substitutions for A89, L90 and L94 in this region decrease fusion to less than 20% of wild-type (wt) and eliminate the HN–F protein interaction; however, they exhibit no discernable effect on other HN functions or structure [67]. Interestingly, some substitutions at these positions, specifically A89I, L90A and L94I-R, result in intermediate levels of fusion with no decrease in neuraminidase or HAd and no detectable effect on antigenic structure. Most importantly, the ability of each of these mutated proteins to co-IP with the homologous F protein correlates with the extent to which they promote fusion, down to approximately 20% of wt fusion (Figure 1). With fusion below this amount, no HN–F protein interaction is detectable, indicating the limits of the co-IP assay. Apparently, it is unable to detect extremely weak interactions that are still of sufficient strength to result in a low level of fusion. Nonetheless, even with this limitation, the data suggest that the co-IP assay accurately reflects the NDV HN–F protein interaction taking place at the cell surface and that the interaction is directly proportional to the extent of fusion.
Figure 1. A linear correlation between the level of fusion promotion and the extent of complex formation at the cell surface between Newcastle disease virus HN and fusion proteins.
Several individual point mutations were introduced for residues A89, L90, P93 and L94 in a short domain in the stalk of the Newcastle disease virus HN protein. The mutated proteins were coexpressed with fusion (F) protein in baby hamster kidney cells using the vaccinia-T7 expression system. The abilities of the mutated HN proteins to complement the homologous F protein in the promotion of fusion were determined in a content-mixing assay. The amounts of the mutated HN proteins associated with a cleavage-site mutant form of the F protein (csmF) at the cell surface were determined using a co-IP assay involving radiolabeling and biotinylation of the two proteins and co-IP of HN with a monoclonal antibody to the F protein. The rationale for using csmF in these studies was to make it possible to compare complex formation in two nonfusing monolayers, thus separating complex formation from the possible dissolution of the complex following F protein activation and fusion. The percentage of mutated HN that co-IPs with the F protein relative to that of the wt protein is graphed versus the percentage of wt fusion. Each data point is labeled with the amino acid substitution of the mutated HN protein and represents the mean of at least four independent determinations of fusion by the content-mixing assay and at least two independent determinations of the amount of HN immunoprecipitated by an anti-F protein monoclonal antibody in the co-IP assay. The graph establishes a linear correlation between the extent of Newcastle disease virus HN–F protein complex formation at the cell surface and the extent of fusion. The failure to detect co-IP of HN at fusion levels less than 20% of wt indicates the detection limit of the co-IP assay.
co-IP: Coimmunoprecipitation; HN: Hemagglutinin–neuraminidase; wt: Wild-type. Data taken from [67].
One other caveat must be considered with respect to this approach. Since the assay specifically examines the association of HN and F protein at the cell surface and apparently cannot detect weak interactions between the two proteins, one cannot rule out the possibility of an interaction between HN and F protein that takes place intracellularly and is too weak to detect in this assay and, for that matter, may not even be relevant to fusion.
Association versus dissociation models for the mechanism of paramyxovirus fusion
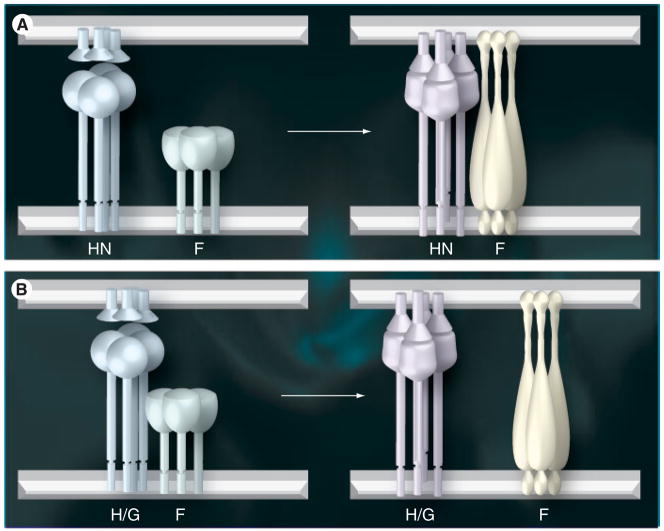
It is uniformly accepted in the paramyxovirus field that it is the recognition of receptors by the attachment protein that regulates the timing and site of activation of the homologous F protein. However, there are essentially two models for the mechanism by which this regulation is accomplished, as recently reviewed by White et al. [40]. The ‘association model’ predicts that the two glycoproteins arrive at the surface independent of each other. Receptor binding by the attachment protein induces a conformational change in that protein, which leads to complex formation with F protein. This complex between the two glycoproteins, which may be quite transient, then triggers the fusion activation of the F protein (Figure 2A).
Figure 2. Two models for the promotion of paramyxovirus fusion.
(A) The association model shows the attachment protein homotetramer and the metastable prefusion form of the F homotrimer existing separately at the surface prior to receptor binding (left panel). The attachment protein spike is depicted as being significantly taller than that of the prefusion F spike based on modeling studies [Lamb, Jardetzky, Pers. Comm.]. This aligns the head of F with the stalk of the attachment protein. The attachment protein is labeled as an HN protein in this figure based on the available data, suggesting that this mode of fusion promotion is specific to viruses that recognize sialic acid-containing receptors. Attachment of HN to receptors results in a conformational change in the protein that induces an interaction with the F protein, which, in turn, triggers the latter into its fusion-active form (right panel). This results in the insertion of the previously sequestered fusion peptide into the target membrane. In this model, the interaction between the two proteins may be quite transient, although this has not been proven.
(B) The dissociation model proposes that the attachment protein and the metastable prefusion form of F are associated in a complex on the surface prior to receptor binding (left panel). The association with the attachment protein presumably serves to maintain F in its metastable, prefusion conformation. The attachment protein in this figure is labeled as H/G because the available data suggest that this mode of fusion promotion is specific to viruses whose attachment proteins, specifically, measles virus H and Nipah/Hendra G, recognize specific protein receptors. Attachment of H/G to protein receptors results in a conformational change in the protein that releases F to assume its fusion-active form (right panel).
F: Fusion; G: Glycoprotein; H: Hemagglutinin; HN: Hemagglutinin–neuraminidase. Adapted from [40].
Alternatively, the ‘dissociation model’ postulates that the two glycoproteins associate with each other during transport and reach the surface as a complex. This complex serves to hold the F protein in its metastable prefusion state. Once the attachment protein binds to receptors on the target cell, the attachment protein undergoes a conformational change that triggers release from the F protein thereby allowing it to assume its fusion-active form (Figure 2B).
Both models predict that activation of the F protein is regulated by receptor binding by the viral attachment protein and both establish a link between receptor binding by one protein spike and fusion promotion by the other. However, they do so by different mechanisms. Various aspects of each of these models have been tested in different laboratories. Support exists for both models, although in many aspects it appears to depend on the particular virus studied.
Dimer interface model for NDV fusion
The structures obtained for the NDV HN protein with and without bound ligand [29] led the authors to propose a detailed version of the ‘dissociation model’ for NDV fusion based on the prediction that there is a significant conformational change in the dimer interface upon ligand binding. HN is thought to change from a structure having a minimal dimer interface to one with a much more extensive interface, with this drastic conformational change being integral to HN’s role in the fusion process. This model was subsequently modified to account for the second sialic acid binding site in NDV HN [30]. It predicts that the HN and F proteins may be associated at the surface through hydrophobic residues with the active site off and the fusion peptide sequestered. HN then binds to receptors via its active site, inducing neuraminidase activity that releases the sialic acid. Receptor binding causes a change in HN from the minimal interface structure to the extensive interface conformation. Hydrophobic residues predicted to interact with F protein to maintain its prefusion state now become sequestered in the extended interface. F protein is released and assumes its fusion-active conformation. The transformation to the extensive interface structure also creates the second binding site in HN, which the protein then uses to reattach to the cell surface to hold the target cell membrane in place as fusion proceeds [30].
Tetramer interface model for PIV5 fusion
This model is based on the structure of the PIV5 HN tetramer [34], which does not support the dimer interface model proposed for NDV HN. Instead, it proposes that binding to receptors by the receptor-binding sites on the four heads in the HN tetramer triggers the opening of the tetrameric head by perturbing relatively weak interactions between the globular heads. This opening of the head is predicted to result in changes in the HN stalk and the interaction with the F protein, thus activating it for fusion.
Peptide-based model for NDV fusion
The identification of a domain at the stalk–head transition region of NDV HN and HR-B in the stalk of F protein as the complementary interacting domains constitutes another variation of the dissociation model, as it is predicted that these two domains associate with each other to hold F protein in its prefusion state until receptor binding by HN releases F protein to promote fusion [76].
Experimental approaches to test the models
Endoplasmic reticulum co-IP & coretention studies
Based on co-IP and cross-linking studies in infected cells, evidence was presented for an interaction between the NDV HN and F proteins in the endoplasmic reticulum (ER) [82]. However, only minimal amounts of co-IP were detected in NDV-infected cells, and it could not be duplicated in transfected cells, making it impossible to include essential controls for nonspecific binding of the heterologous protein to the precipitating antibody. Another approach utilized by several laboratories is based on the idea that ER retention of one of the proteins may result in co-retention of the other. hPIV2 and hPIV3 F proteins carrying a C-terminal ER retention signal (KDEL) downregulate surface expression of the homologous HN [83,84], providing support for the existence of an intracellular complex between the two proteins. However, this conclusion is complicated by the finding that heterologous H/HN proteins are also co-retained with ER-retained hPIV2/3 F protein. By contrast, SV5 and hPIV3 F and HN proteins tagged for ER retention with, respectively, a C-terminal KK motif and multiple N-terminal arginines, do not affect transport of the other protein [85]. The latter results argue against the existence of an intracellular HN–F protein complex for these viruses, although the presence of an interaction between the two proteins that is too weak to result in co-retention cannot be ruled out.
Efficient retention of the MV H or F proteins in the ER results in co-retention of the other protein [61], consistent with oligomerization of the two proteins in the ER. Moreover, cotransfection of the retained proteins with transport-competent MV proteins results in a dominant negative effect on fusion. Futhermore, the addition of epitope tags to the MV H tail weakens the H–F protein interaction; however, it also increases fusion [86]. This study indicates that the MV H and F proteins not only interact in the ER, but also that fusion is inversely related to the strength of that interaction.
Relationship between receptor binding & the HN/H/G–F protein complex
Depending on the site of the HN–F protein interaction, formation of the complex may be differentially affected by a lack of receptor-binding activity. According to the association model, one would expect there to be little or no complex formation between F protein and a homologous attachment protein that lacks receptor-binding activity. By contrast, in the dissociation model, the complex would presumably form independently of receptor binding. In fact, because receptor binding is predicted to promote the dissociation of the preformed complex, diminished ability to bind receptors might be expected to result in an increase in the amount of the glycoprotein complex detectable at the cell surface relative to the two wt proteins.
Receptor-binding-deficient NDV HN cannot be detected in a complex with F protein at the cell surface
Substitutions for any of five residues in the neuraminidase site of NDV HN result in proteins that lack detectable neuraminidase, attachment and fusion. An I175E, D198R, K236R, Y526L or E547Q mutation abolishes all activities of HN, despite efficient expression at the cell surface and no detectable effect on protein conformation, as determined with a panel of conformation-specific monoclonal antibodies [87]. Zero or negligible interaction can be detected between the F protein and NDV HN carrying a D198R, K236R, Y526L or E547Q substitution, arguing that, in the absence of receptor binding, the HN and F proteins do not interact at the surface in a complex of sufficient avidity for detection. This is consistent with receptor binding by HN triggering the HN–F protein interaction at the cell surface, that is, NDV HN and F proteins promoting fusion according to the association model.
Although it also lacks all HN functions, I175E-mutated NDV HN does interact with F protein at the surface, albeit less efficiently than wt HN. Accordingly, this would seem to argue that the HN and F proteins can interact in the absence of receptor binding. However, this mutated protein is apparently a special case, since it may alter the dimer interface of the protein [88].
Of course, one cannot eliminate the possibility that the HN active-site mutations that do eliminate the HN–F protein interaction at the surface do so because they alter the structure of HN in ways that cannot be detected with the panel of monoclonal antibodies employed, or that these HN mutants do form a complex with F protein, but one that is of insufficient avidity to be detected by the surface co-IP protocol.
Interaction of NDV HN & F protein under conditions of receptor depletion
The relationship between NDV HN and F protein was examined under conditions in which receptor was depleted on target cells by treatment with exogenous neuraminidase [60]. This study led to the conclusion that the HN–F protein complex forms in the absence of receptor and is dissociated by attachment. It was shown that the amount of the HN–F protein complex decreases if neuraminidase-treated HN–F protein expressing effector cells are mixed with untreated target cells to stimulate attachment. This was used to argue that the complex is preformed and dissociated by attachment. However, the experiment involved comparison of nonfusing (without target cells) and fusing (with target cells) monolayers. The decreased amount of the complex in the presence of the added target cells could be due to the dissociation of the HN–F protein complex as fusion proceeds. This experiment would be more properly performed under conditions that separate receptor binding and complex formation from the subsequent steps in the fusion process.
MV H & HeV G proteins with impaired receptor-binding activity exhibit enhanced ability to interact with the homologous F protein
Contrary to the findings with receptor-binding-deficient NDV HN, mutations that severely compromise the ability of MV H to bind CD46 do not result in a reduction in the amount of the MV H–F protein complex at the surface, but rather, significantly increase it. For example, a mutated H protein in which amino acids 473–477 are changed to alanine exhibits only 1% of wt HAd activity and no detectable fusion-promoting activity, yet it co-IPs with the F protein from the cell surface 38% more efficiently than the wt H protein [70]. This is in sharp contrast to the results obtained with NDV HN proteins deficient in receptor binding [66] and argues strongly that NDV HN, which recognizes sialic acid-containing receptors, and MV H, which recognizes specific proteins as receptors, utilize receptor binding in different ways to regulate the fusogenic activation of their homologous F proteins.
Prior to the solution of the structure of the HeV G protein, Bishop et al. targeted several residues in MV H known to be involved in receptor binding for mutagenesis [62]. In the G protein, seven point mutations were identified to have significantly impaired binding to both ephrinB2 and ephrinB3 and, similarly, decreased fusion when coexpressed with F protein. Interestingly, all of these attachment/fusion-deficient proteins retained the ability to interact with F protein and the majority of them actually interacted more efficiently with F protein in co-IP assays, often as much as twice as efficiently compared with the wt G protein. The authors concluded that their data support a mechanism for the promotion of fusion in which HeV G and F proteins interact prior to receptor binding. This appears to be consistent with the dissociation model for fusion.
Tests of the dimer-interface model for the role of HN in NDV fusion
Mutation of purported F protein-interactive residues in the HN globular head
The dimer-interface model [29] contends that hydrophobic residues exposed in the minimal interface structure may mediate an interaction with F protein to maintain it in its metastable prefusion conformation. Upon binding to the receptor, the HN protein is proposed to undergo a significant conformational change, resulting in the sequestration of these residues in the dimer interface and the release of F protein.
Takimoto et al. tested this model by mutating several hydrophobic residues that are exposed in the minimal interface form, but sequestered in the extensive interface form [89]. They demonstrated that these mutations reduce fusion to as little as 5–10% of the wt level. The authors also reported that HAd and neuraminidase activity were not significantly affected by the mutations, although HAd was measured in the cold. On this basis, they concluded that these residues may be part of the fusion-promotion domain in HN.
However, these studies predated the identification of the second sialic acid-binding site at the membrane distal end of the dimer interface [30]. Some of the mutated residues, for example, R516 and F553, are actually located in the second site. For others, for example, F220 and L224, it was subsequently demonstrated that the diminished fusion induced by their mutation correlates with a temperature-dependent defect in attachment [90]. Although the mutants exhibit at least 60% of wt activity in the cold, HAd is severely reduced at 37°C, the temperature at which fusion normally occurs. Proteins carrying mutations for these residues elute much faster than wt HN from erythrocytes, even though neuraminidase is unaffected. Thus, the fusion deficiency of these mutated proteins correlates with instability of receptor binding, which could be related to the disruption of intermonomeric hydrogen bonds across the dimer interface. Elimination of these dimer contacts could disrupt the intermonomeric second sialic acid binding site and decrease fusion-promoting activity. These data call into question the idea that the dimer-interface residues in HN mediate an interaction with F protein and suggest that the fusion deficiency of these interface mutants could be the result of a perturbation of the second sialic acid binding site, which results in severely compromised attachment at 37°C.
Intermonomer disulfide bonds across the dimer interface of NDV HN do not impair fusion
The dimer-interface model predicts that fusion promotion is dependent on the minimal interface form of NDV HN holding F protein in its metastable prefusion state until receptor binding triggers conversion of HN to the extensive interface form, releasing F protein to promote fusion. The proposed requirement for the minimal interface form of HN for fusion was tested by the introduction of two cysteine mutations in the dimer interface of the globular domain of an otherwise nondisulfide-linked NDV HN protein [91]. These cysteine residues mediate the formation of a pair of intermonomeric disulfide bonds across the dimer interface. These bonds preclude formation of the minimal interface structure and effectively lock the protein into its extensive interface form. It was also shown that the disulfide bonds form intra-cellularly and are independent of receptor binding, suggesting that HN assumes the extensive interface form before reaching the cell surface and binding receptors. The mutated protein also exhibits an elevated level of fusion relative to the wt proteins. These results strongly imply that neither the minimal interface form of HN nor the proposed drastic conformational change in the protein is required for syncytium formation, arguing against this model for fusion.
An N-glycan in the HN domain 124–152 does not abolish fusion, disputing the peptide-based model
The peptide-based model for NDV fusion proposes that a domain defined by HN residues 124–152 and situated at the stalk–head transition mediates the interaction with F protein [76]. However, HN mutated to carry an N-glycan in this domain is efficiently expressed and retains most of its fusion activity, which is inconsistent with an involvement of this region in mediating the interaction with F protein [68]. In retrospect, an interaction between these two domains is difficult to reconcile on several accounts. Residues 124–152 are positioned at the stalk–head transition in the HN spike and one might question how this region on HN could physically interact with HR-B on F protein, which is situated close to the membrane. In addition, hPIV3-NDV HN chimeras with NDV HN residues 124–152 complement hPIV3 F protein, not NDV F protein [71]. Since this domain is not conserved between the two HN proteins, it seems unlikely that it could be the F protein-interactive site on HN.
Relationship between the extent of fusion & the strength of the glycoprotein complex
Deletion of N-glycans from the NiV F protein enhances fusion, but decreases the strength of the G–F protein interaction
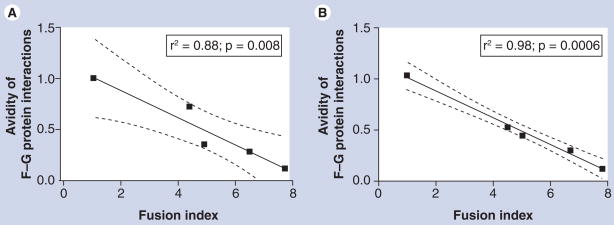
Whereas N-glycans on most paramyxovirus F proteins are required for proper folding and function, it has been shown that deletion of these moieties from the NiV F protein enhances fusion by as much as fivefold [63]. Interestingly, this marked increase in fusion correlates with a decrease in the relative avidity of the association of F with the G protein. In other words, the extent of fusion and the strength of the G–F protein interaction are inversely related (Figure 3). While the increased fusion is probably due to enhanced 6-HB formation, this inverse relationship beween fusion and glycoprotein complex avidity is consistent with the dissociation model for fusion.
Figure 3. Nipah virus fusion is inversely correlated with the avidity of the F–G protein interaction.
Reciprocal coimmunoprecipitations were performed on cell lysates of 293T cells transfected with wild-type Nipah virus (NiV) G protein and either wild-type NiV F or mutated forms of F protein lacking one or more N-glycans. The deletion of the N-glycans increases the extent of fusion, yet decreases the avidity of the complex between the F and G proteins. (A) The G protein was immunoprecipitated with anti-NiV-G-specific antisera and the samples were western blotted for NiV F protein. (B) The F protein was immunoprecipitated and the samples were western blotted for the G protein. The F–G protein interaction avidities were calculated and are graphed here against the fusion indices. A Pearson correlation analysis is included for each graph. The graph shows that the fusogenicity of the F protein mutants inversely correlates with the avidity of the complex formed with the wild-type G protein.
F: Fusion; G: Glycoprotein.
Reproduced with permission from [63].
Mutations for conserved isoleucines in the stalk of HeV G convert the protein to a postattachment conformation
Several conserved isoleucine residues in the stalk region of the HeV G protein have been targeted for mutagenesis to investigate their role in the promotion of fusion [92]. Although these mutated proteins were efficiently expressed, maintained their oligomeric structure and were recognized by a panel of conformation-specific antibodies, they were completely defective in promoting fusion. Based on their recognition by a panel of antibodies that preferentially recognize G protein in a receptor-bound conformation, it was concluded that the I–A mutations in the stalk convert G protein to a receptor-bound conformation in the absence of a receptor. This defect correlated with the appearance of a slightly altered glycosylated form of the protein, which did not interact with the F protein at the cell surface. From this, the authors concluded that there is a direct correlation between the inability of G protein to interact with the F protein and loss of fusion-promoting activity. They proceeded to argue that the promotion of fusion by he HeV G and F proteins is analogous to that of the NDV HN and F proteins. However, it should be noted that the predominantly intracellular form of these mutants that comigrates with the wt G protein retains its ability to interact with the F protein, which is consistent with the two proteins interacting intracellularly.
Receptor-induced dissociation of NiV G & F proteins induced by receptor binding
A monoclonal antibody, whose binding to NiV G protein is enhanced upon the binding of the protein to ephrinB2, has been identified [39]. This antibody inhibits viral entry and, by several measures, appears to recognize an epitope on the G protein that is involved in triggering the F protein. Interestingly, a mutated G protein, which is nonfusogenic and does not exhibit the ephrinB2-induced activation of antibody binding, does not dissociate from F protein, whereas the wt G protein does so quite efficiently. This correlation between fusion deficiency and the inability of G to release from F is consistent with the two proteins being associated in a preformed complex, the dissociation of which is triggered by receptor binding.
Mutations in the stalk of MV H block fusion & enhance the H–F protein interaction
Similar to the studies with NDV HN [67], mutations in an HR-like domain in the stalk of the MV H protein impair fusion [69]. However, unlike the situation with NDV, these mutations do not interfere with the interaction with the MV F protein. Indeed, they apparently act by stabilizing the H–F protein complex. For example, mutations of I84A and I98A, which eliminate fusion, increase the amount of H that is detectable in a complex with F protein at the cell surface by two- and threefold, respectively. Several other mutations have a similar effect, albeit to a lesser extent. Thus, for MV, blocking fusion apparently correlates with a stabilization of the H–F protein complex at the cell surface.
Conclusion & future perspective
The paramyxovirus receptor-binding and fusion-promotion activities reside on different glycoprotein spikes. Whereas the latter is dependent on the former, this requires a mechanism by which the two processes can be linked. There appears to be an increasing body of evidence to support the notion that paramyxoviruses regulate fusion by different mechanisms, depending on the type of receptor they recognize [16]. The extent of fusion promoted by NDV HN, which recognizes sialic acid-containing receptors, appears to directly correlate with the avidity of the HN–F protein complex at the cell surface. In addition, HN proteins that lack receptor-binding and fusion-promoting activities fail to interact with F protein in a complex of sufficient avidity to be detected at the cell surface. Finally, for two similar viruses, hPIV3 and PIV5, the ER retention of either HN or F protein does not affect the transport of the other protein. All of these findings are consistent with the complex between the two proteins being formed at the cell surface triggered by receptor binding, that is, the association model for fusion.
However, the possibility that HN and F proteins are associated intracellularly prior to receptor binding in a complex that is of insufficient avidity to be detected efficiently at the cell surface and may, in fact, not even be relevant to fusion, cannot be excluded. Binding to receptors would then induce the formation of a tighter, possibly transient, complex between the two proteins that triggers the activation of the F protein. In this scenario, fusion would still ultimately be regulated by the ability of the two proteins to associate in this higher avidity complex.
Similar approaches for viruses such as MV, NiV and HeV, which recognize specific protein receptors, lead to a very different conclusion. For NiV, increased fusion results in decreased avidity of the G–F protein complex, indicating that fusion and complex avidity are inversely related. MV H and HeV G proteins with impaired receptor binding activity exhibit enhanced ability to interact with the homologous F protein. In addition, MV H and F proteins interact in the ER, and fusion is inversely related to the strength of this interaction. All of this strongly suggests that the promotion of fusion by these viruses is regulated by the dissociation of a previously formed complex between the fusion and G proteins triggered by the binding of the latter to receptors (the dissociation model for fusion).
However, this proposed model of fusion promotion by the henipaviruses must be reconciled with the complex-processing pathway exhibited by the F proteins of these viruses. As discussed previously, the NiV and HeV F proteins are unique among those of the paramyxoviruses in that they are expressed in an uncleaved form at the cell surface prior to recycling into endosomes, cleavage by cathepsin L into the fusion-active form and retrafficking to the surface [41–44]. The most straightforward interpretation of this finding is that the G–F protein complex does not form intra-cellularly, but rather, at the cell surface subsequent to this complex sequence of events and is most likely triggered by receptor binding. Alternatively, it seems possible that the G–F protein complex may form subsequent to the endosomal cleavage of F protein and prior to receptor binding, with the latter triggering dissociation of the complex and fusion. It may be that the complex processing pathway of the henipavirus F protein will translate into an equally complex relationship between it and the homologous G protein during transport to the surface, especially in comparison to that of the MV H and F proteins.
The idea that viruses that bind sialic acid- containing receptors and those that bind proteinaceous receptors might regulate fusion by different mechanisms was originally proposed by Plemper et al. [61], who attributed it to the abundance of sialic acid in the cell relative to the protein receptors. However, the data in support of the distinction between the two mechanisms of regulation are predominantly biochemical in nature. Other approaches are needed to either confirm or dispute this hypothesis. In the future, the task will be to compare the glycoprotein interactions for the different types of viruses by more direct methods, especially in the living cell.
In addition, several other aspects of the process of paramyxovirus fusion remain to be elucidated. Foremost among these is the definitive identification of the complementary domains on each of the attachment and fusion proteins that mediate the interaction between them responsible for linking receptor binding to F protein triggering. A major advance in this regard has been made recently for MV H and F proteins, indicating that the stalk of H interacts with a complementary domain in the globular head of the prefusion F protein [58]. Most of the available evidence indicates that the stalk domain of HN mediates the interaction with the homologous F protein. However, the complementary domain on the homologous F protein remains to be identified.
Is an interaction between a domain in the stalk of the attachment protein and one in the head of the F protein physically possible? Based on biochemical data [79] and modeling, Lamb and Jardetzsky propose that the stalk of PIV5 HN is 50–80 residues of helix, which serves to extend most, if not all, of its head above the prefusion F protein spike [Pers. Comm.]. If this is the case, then one can easily envision an interaction taking place between the stalk of the HN/H/G protein and the head of F protein. Indeed, modeling of MV H and F protein by Lee et al. aligns the complementary regions they had identified in these two regions of the respective proteins [58].
All of the studies of paramyxovirus glycoprotein interactions discussed in this article focus on cell–cell fusion in infected or transfected cells. It must be stressed that the results for this mode of fusion cannot be extrapolated to virus–cell fusion or virus entry. This is apparent from the properties of monoclonal antibodies to one antigenic site on the NDV HN protein, which neutralize at a post-attachment step. Antibodies to this site inhibit virion-induced fusion-from-without (a model for virus–cell fusion), but not fusion-from-within, which is mediated by the glycoprotein-laden infected cell surface [93]. This suggests that the requirement for HN may be different for the two modes of fusion. Indeed, little is known about the relationship between the attachment and fusion spikes on virions. If the distinction between the two classes of paramyxoviruses proposed here holds true, it will be interesting to compare the relationship between the HN and F proteins and the H/G and F proteins on virions. It may be that the two classes of viruses may also exhibit distinctly different spike interactions on the virion.
The F protein-interactive domain on the henipavirus G protein has not yet been identified. In fact, unlike NDV HN, no mutation in either MV H or NiV/HeV G protein has been identified, to date, which eliminates its interaction with the homologous F protein. This could mean that the F protein-interactive site in these proteins is quite complicated and multiple mutations are required to fully disrupt the interaction. Another possibility is that there may be more than one set of interactive sites on the MV and henipavirus proteins.
The kinetics of fusion promotion by the two groups of viruses has not been compared under the same conditions. One could speculate that viruses, which are proposed to promote fusion by the dissociation of a preformed complex, might exhibit quite different kinetics than viruses in which the promotion of fusion requires the formation of a complex between the attachment and F protein triggered by receptor engagement. This remains to be determined.
Another question is how the signal is transduced from the receptor-binding site in the attachment protein to the domain within it that mediates the interaction with F protein. What changes does each attachment protein undergo during the triggering process? Are the changes conformational and limited to each monomer, or do they involve more significant changes in the oligomeric structure as was initially proposed for NDV HN [29]? It will be interesting to see if the changes are different depending on whether receptor binding triggers the formation or dissociation of the glycoprotein complex. A recent monoclonal antibody-based study has provided some insight into the changes that NiV G protein undergoes upon receptor binding [39]. The results from another study suggest that the stalk of HeV G protein is altered upon receptor recognition [92]. The significance of these studies will become clearer when the F protein-interactive site(s) on the G protein is identified.
Several questions also remain to be answered with respect to the involvement of the F protein in the regulation of fusion. Are the F proteins structurally different in ways that account for some to be associated with the homologous attachment protein prior to receptor recognition while others are apparently able to maintain the metastable prefusion state without binding to the attachment protein? How does the association between the attachment protein and F protein maintain the latter in its prefusion state (dissociation model) or actively trigger its conversion to an active state (association model)? The answers to these questions will undoubtedly become clearer as we learn exactly how each F protein interacts with its attachment protein and when the structures of the MV and henipavirus F protein become available.
Paramyxovirus fusion and entry are complex processes, requiring a virus-specific interaction between glycoprotein spike structures. Understanding how this interaction promotes the fusion of the viral or cell membrane with that of the target cell could identify new and potent antiviral strategies to interfere with the infection cycles of these important pathogens.
Conclusion
Evidence indicates that the stalk domain of HN mediates the interaction with the homologous F protein. Similar findings have now been reported for MV H;
It is likely that there is an interaction between the stalk of the HN/H and the head of F protein;
The results with NDV, MV and NiV/HeV reviewed here indicate that the mode of regulation of fusion utilized by paramyxoviruses may be related to the type of receptor they recognize;
Viruses that recognize sialic acid-containing receptors appear to use receptor binding to induce the association, or at least a more avid association, between the attachment and F protein at the cell surface. Viruses that recognize specific proteins as receptors appear to use receptor binding to promote the dissociation of a previously formed glycoprotein complex. The biological significance of the possible existence of the different mechanisms of fusion is a matter of speculation. However, as originally proposed by Plemper et al., it may be based on the relative abundances of the two types of receptors [61]. The less abundant protein receptors therefore may be less available for premature fusion than ubiquitous sialic acid-containing receptors.
Executive summary
Paramyxovirus fusion usually requires the attachment protein
The structures of the attachment proteins for several paramyxoviruses have been solved. In addition, the structure of the fusion (F) protein has been solved in both its pre- and postfusion states.
Some paramyxovirus attachment proteins recognize sialic acid-containing receptors while others recognize specific proteinaceous receptors.
There is a virus-specific interaction between the two glycoproteins
The glycoprotein complex can be detected by coimunoprecipitation assays.
Chimera studies indicate that the stalk regions of several hemagglutinin–neuraminidase (HN) proteins and of measles virus (MV) hemagglutinin (H) determine specificity for the homologous F protein.
Mutations in the Newcastle disease virus (NDV) HN stalk specifically abolish fusion and the HN–F protein interaction.
Receptor binding by the attachment protein is thought to trigger the activation of the fusion protein
Some models implicate receptor binding as the cause of the dissociation of a previously formed protein complex. Other models propose that receptor binding induces the formation of a complex between the two proteins that leads to fusion.
MV H and Nipah (NiV) attachment (G) proteins with impaired receptor binding activity exhibit enhanced ability to interact with the homologous F protein.
Most NDV HN mutants deficient in receptor binding and fusion-promoting activity fail to interact with the F protein at the cell surface.
There is a relationship between the extent of fusion & the strength of the glycoprotein complex
Deletion of N-glycans from the NiV F protein enhances fusion, but decreases the strength of the G–F protein interaction, indicating that the fusogenicity of this virus inversely correlates with the avidity of the G–F protein interaction.
The extent of NDV fusion for a series of HN stalk mutants is proportional to the extent of HN–F protein complex formation at the cell surface.
Acknowledgments
The authors thank Benhur Lee and the American Society for Microbiology for permission to utilize Figure 3. We also thank Robert Lamb and Theodore Jardetzky for helpful information pertinent to the structures of the paramyxovirus attachment and fusion protein spikes.
Financial & competing interests disclosure
The authors acknowledge ongoing financial support of the work performed in this laboratory by grant AI-49268 from the NIH (awarded to Ronald M Iorio), as well as previous financial support from a U19 grant (U19 AI057319) awarded by the NIH to the UMass Center for Translational Research on Human Immunology and Biodefense (Alan Rothman, PI). Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the US Army. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Ronald M Iorio, Program in Immunology & Virology, University of Massachusetts Medical School, Worcester, MA 01655, USA and Department of Molecular Genetics & Microbiology, University of Massachusetts Medical School, 55 Lake Avenue North, Worcester, MA 01655, USA, Tel.: +1 508 856 5257, Fax: +1 508 856 5920, ronald.iorio@umassmed.edu.
Vanessa R Melanson, United States Army Medical Research, Institute of Infectious Diseases, Division of Virology, 1425 Porter Street, Frederick, MD 21702, USA, Tel.: +1 301 619 1009, Fax: +1 301 619 2290, vanessa.melanson@us.army.mil.
Paul J Mahon, Department of Molecular Genetics & Microbiology, University of Massachusetts Medical School, Worcester, MA 01655, USA, Tel.: +1 508 769 3698, Fax: +1 508 767 7241, pmahon@assumption.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Lamb RA, Parks GD. In: Fields Virology. 5. Knipe DM, Howley PM, editors. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 1449–1496. [Google Scholar]
- 2.Elliman D, Sengupta N. Measles. Curr Opin Infect Dis. 2005;18(3):229–234. doi: 10.1097/01.qco.0000168383.93647.47. [DOI] [PubMed] [Google Scholar]
- 3.Moscona A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J Clin Invest. 2005;115:1688–1698. doi: 10.1172/JCI25669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams JV, Harris PA, Tollefson SJ, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350(5):443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elankumaran S, Rockemann D, Samal SK. Newcastle disease virus exerts oncolysis by both intrinsic and extrinsic caspase-dependent pathways of cell death. J Virol. 2006;80(11):5145–5155. doi: 10.1128/JVI.00241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vigil A, Martinez O, Chua MA, Garcia-Sastre A. Recombinant Newcastle disease virus as a vaccine vector for cancer therapy. Mol Ther. 2008;16(11):1883–1890. doi: 10.1038/mt.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamarin D, Vigil A, Kelly K, Garcia-Sastre A. Genetically engineered Newcastle disease virus for malignant melanoma therapy. Gene Ther. 2009;16(6):796–804. doi: 10.1038/gt.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiNapoli JM, Yang L, Suguitan A, Jr, et al. Immunization of primates with a Newcastle disease virus-vectored vaccine via the respiratory tract induces a high titer of serum neutralizing antibodies against highly pathogenic avian influenza virus. J Virol. 2007;81(21):11560–11568. doi: 10.1128/JVI.00713-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Q, Park MS, Palese P. Expression of transgenes from Newcastle disease virus with a segmented genome. J Virol. 2008;82(6):2692–2698. doi: 10.1128/JVI.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veits J, Romer-Oberdorfer A, Helferich D, et al. Protective efficacy of several vaccines against highly pathogenic H5N1 avian influenza virus under experimental conditions. Vaccine. 2008;26(13):112–121. doi: 10.1016/j.vaccine.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Carnero E, Li W, Borderia AV, Moltedo B, Moran T, Garcia-Sastre A. Optimization of human immunodeficiency virus gag expression by Newcastle disease virus vectors for the induction of potent immune responses. J Virol. 2009;83(2):584–597. doi: 10.1128/JVI.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray K, Selleck P, Hooper P, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 13.Chua KB, Bellini WJ, Rota PA, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 14.Porotto M, Murrell M, Greengard O, Doctor L, Moscona A. Influence of the human parainfluenza virus 3 attachment protein’s neuraminidase activity on its capacity to activate the fusion protein. J Virol. 2005;79(4):2383–2392. doi: 10.1128/JVI.79.4.2383-2392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spielhofer P, Bachi T, Fehr T, et al. Chimeric measles viruses with a foreign envelope. J Virol. 1998;72(3):2150–2159. doi: 10.1128/jvi.72.3.2150-2159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio RM, Mahon PJ. Paramyxoviruses: different receptors – different mechanisms of fusion. Trends Microbiol. 2008;16(4):135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for MV (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 18.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 19.Erlenhoefer C, Wurzer WJ, Loffler S, Schneider-Schaulies S, ter Meulen V, Schneider-Schaulies J. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J Virol. 2001;75(10):4499–4505. doi: 10.1128/JVI.75.10.4499-4505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol. 2001;75(9):4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masse N, Ainouze M, Neel B, Wild TF, Buckland R, Langedijk JP. Measles virus (MV) hemagglutinin: evidence that attachment sites for MV receptors SLAM and CD46 overlap on the globular head. J Virol. 2004;78(17):9051–9063. doi: 10.1128/JVI.78.17.9051-9063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M, Hansson E, Langedijk JP, Eaton BT, Wang LF. The attachment protein of Hendra virus has high structural similarity but limited primary sequence homology compared with viruses in the genus. Paramyxovirus Virology. 1998;251(2):227–233. doi: 10.1006/viro.1998.9302. [DOI] [PubMed] [Google Scholar]
- 23.Harcourt BH, Tamin A, Tsiazek TG, et al. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology. 2000;271(2):334–349. doi: 10.1006/viro.2000.0340. [DOI] [PubMed] [Google Scholar]
- 24.Bonaparte MI, Dimitrov AS, Bossart KN, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negrete OA, Levroney EL, Aguilar HC, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 26.Negrete OA, Wolf MC, Aguilar HC, et al. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah. PLoS Pathog. 2006;2(2):e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 1999;74(14):6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cseke G, Maginnis MS, Cox RG, et al. Integrin αvβ1 promotes infection by human metapneumovirus. Proc Natl Acad Sci USA. 2009;106(5):1566–1571. doi: 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29▪▪.Crennell S, Takimoto T, Portner A, et al. Crystal structure of the multifunctional paramyxovirus hemagglutinin–neuraminidase. Nat Struct Biol. 2000;7(11):1068–1074. doi: 10.1038/81002. First crystal structure of a paramyxovirus attachment protein. [DOI] [PubMed] [Google Scholar]
- 30▪.Zaitsev V, von Itzstein M, Groves D, et al. Second sialic acid binding site in Newcastle disease virus hemagglutinin–neuraminidase: implications for fusion. J Virol. 2004;78:3733–3741. doi: 10.1128/JVI.78.7.3733-3741.2004. Only crystallographic evidence for a second sialic acid binding site in an hemagglutinin–neuraminidase (HN) protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence MC, Borg NA, Streltsov VA, et al. Structure of the haemagglutinin–neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 32.Porotto M, Fornabaio, Kellogg GE, Moscona A. A second receptor binding site on human parainfluenza type 3 hemagglutinin–neuraminidase contributes to activation of the fusion mechanism. J Virol. 2007;81:3216–3228. doi: 10.1128/JVI.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alymova IV, Taylor G, Mishin VP, et al. Loss of the N-linked glycan at residue 173 of human parainfluenza virus type 1 hemagglutinin–neuraminidase exposes a second receptor-binding site. J Virol. 2008;82:8400–8410. doi: 10.1128/JVI.00474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34▪.Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, Jardetzky TS. Structural studies of the parainfluenza virus 5 hemagglutinin–neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. First crystallization data for an HN tetramer. [DOI] [PubMed] [Google Scholar]
- 35.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 36.Hashiguchi T, Kajikawa M, Maita N, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;104 (49):19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, Stuart DI. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 38.Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. Host–cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguilar HC, Ataman ZA, Aspericueta V, et al. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F) J Biol Chem. 2009;284(3):1628–1635. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deiderich S, Moll M, Klenk HD, Maisner A. The nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem. 2005;280(33):29899–298903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 42.Meulendyke KA, Wurth MA, McCann RO, Dutch RE. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79 (20):12643–12649. doi: 10.1128/JVI.79.20.12643-12649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deiderich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375(2):391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell CJ, Kantor KL, Jardetzky TS, Lamb RA. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J Cell Biol. 2003;163:363–374. doi: 10.1083/jcb.200305130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Gorman JJ, McKimm-Breschkin J, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9(3):255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 47.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102(26):9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪▪.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. First structure of a paramyxovirus fusion protein in its prefusion conformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270(1):17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 50.Ito M, Nishio M, Kawano M, Komada H, Ito Y, Tsurudome M. Effects of multiple amino acids of the parainfluenza virus 5 fusion protein on its haemagglutinin–neuraminidase-independent fusion activity. J Gen Virol. 2009;90(Pt 2):405–413. doi: 10.1099/vir.0.006437-0. [DOI] [PubMed] [Google Scholar]
- 51.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74(14):6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barretto N, Hallak LK, Peeples ME. Neuraminidase treatment of respiratory syncytial virus-infected cells or virions, but not target cells, enhances cell–cell fusion and infection. Virology. 2003;313(1):33–43. doi: 10.1016/s0042-6822(03)00288-5. [DOI] [PubMed] [Google Scholar]
- 53.Seth S, Vincent A, Compans RW. Mutations in the cytoplasmic domain of a paramyxovirus fusion glycoprotein rescue syncytium formation and eliminate the hemagglutinin–neuraminidase protein requirement for membrane fusion. J Virol. 2003;77(1):167–178. doi: 10.1128/JVI.77.1.167-178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sergel T, McGinnes LW, Morrison TG. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol. 2000;74(11):5101–5107. doi: 10.1128/jvi.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu X, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin–neuraminidase of human parainfluenza viruses. J Virol. 1992;66(3):1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bossart KN, Wang LF, Flora MN, et al. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J Virol. 2002;76(22):11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bousse T, Takimoto T, Gorman WL, Takahasi T, Portner A. Regions on the hemagglutinin–neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology. 1994;204(2):506–514. doi: 10.1006/viro.1994.1564. [DOI] [PubMed] [Google Scholar]
- 58▪▪.Lee JK, Prussia A, Paal T, White LK, Snyder JP, Plemper RK. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283(24):16561–16572. doi: 10.1074/jbc.M801018200. Identification of complementary domains on homologous attachment and fusion proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59▪.Yao Q, Hu X, Compans RW. Association of the parainfluenza virus fusion and hemagglutinin–neuraminidase glycoproteins on cell surfaces. J Virol. 1997;71(1):650–656. doi: 10.1128/jvi.71.1.650-656.1997. Initial detection of the HN–fusion glycoprotein complex at the cell surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGinnes LW, Morrison TG. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J Virol. 2006;80(6):2894–2903. doi: 10.1128/JVI.80.6.2894-2903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61▪▪.Plemper RK, Hammond AL, Cattaneo R. MV envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 276(47):44239–44246. doi: 10.1074/jbc.M105967200. 2001. First suggestion that paramyxoviruses that bind specific protein receptors may regulate fusion by a different mechanism than viruses that bind sialic acid-containing receptors. [DOI] [PubMed] [Google Scholar]
- 62.Bishop KA, Stantchev TS, Hickey AC, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81(11):5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63▪.Aguilar HC, Matreyek KA, Filone CM, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80(10):4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. Evidence that strength of the Nipah attachment–fusion protein interaction is inversely related to the extent of fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bossart KN, Tachedjian M, McEachern JA, et al. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372(2):357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Deng R, Wang Z, Mahon PJ, Marinello M, Mirza AM, Iorio RM. Mutations in the NDV HN protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology. 1999;253(1):43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 66.Li J, Quinlan E, Mirza A, Iorio RM. Mutated form of the Newcastle disease virus hemagglutinin–neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J Virol. 2004;78(10):5299–5310. doi: 10.1128/JVI.78.10.5299-5310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67▪.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78(23):13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. Evidence for a direct correlation between the level of fusion and the extent of HN–fusion protein complex formation at the cell surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80(2):623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corey EA, Iorio RM. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology. 2009;383(1):1–5. doi: 10.1016/j.virol.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71▪.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209(2):457–469. doi: 10.1006/viro.1995.1278. First demonstration that the stalk region of an HN protein determines its specificity for the homologous fusion protein. [DOI] [PubMed] [Google Scholar]
- 72.Tsurudome M, Ito M, Nishio M, et al. Identification of regions on the hemagglutinin–neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology. 1995;213(1):190–203. doi: 10.1006/viro.1995.1559. [DOI] [PubMed] [Google Scholar]
- 73.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: Identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70(9):6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng R, Mirza AM, Mahon PJ, Iorio RM. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch Virol Suppl. 1997;13:115–130. doi: 10.1007/978-3-7091-6534-8_12. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Mirza AM, Li J, Mahon PJ, Iorio RM. An oligosaccharide at the C-terminus of the F-specific domain in the stalk of the human parainfluenza virus 3 hemagglutinin–neuraminidase modulates fusion. Virus Res. 2004;99(2):177–185. doi: 10.1016/j.virusres.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Gravel K, Morrison TG. Interacting domains of the HN and F proteins of Newcastle disease virus. J Virol. 2003;77(20):11040–11049. doi: 10.1128/JVI.77.20.11040-11049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wild TF, Fayolle Beauverger P, Buckland R. Measles virus fusion: role of the cysteine-rich region of the fusion glycoprotein. J Virol. 68(11):7546–7548. doi: 10.1128/jvi.68.11.7546-7548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsurudome M, Ito M, Nishio M, et al. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemmagglutinin-neuraminidase proteins to promote cell fusion. J Gen Virol. 1998;79(Pt 2):279–289. doi: 10.1099/0022-1317-79-2-279. [DOI] [PubMed] [Google Scholar]
- 79.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin–neuraminidase (HN) protein. Virology. 2008;378(2):282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stone-Hulslander JT, Morrison TG. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J Virol. 1999;73(5):3630–3637. doi: 10.1128/jvi.73.5.3630-3637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Iorio RM. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J Gen Virol. 1999;80(Pt 3):749–753. doi: 10.1099/0022-1317-80-3-749. [DOI] [PubMed] [Google Scholar]
- 82.Stone-Hulslander J, Morrison TG. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J Virol. 1997;71(9):6287–6295. doi: 10.1128/jvi.71.9.6287-6295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tanaka Y, Hemingway BR, Galinski MS. Down-regulation of paramyxovirus hemagglutinin–neuraminidase glycoprotein surface expression by a mutant fusion protein containing a retention signal for the endoplasmic reticulum. J Virol. 1996;70(1):580–584. doi: 10.1128/jvi.70.8.5005-5015.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tong S, Compans RW. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J Gen Virol. 1999;80(Pt 1):107–115. doi: 10.1099/0022-1317-80-1-107. [DOI] [PubMed] [Google Scholar]
- 85.Paterson RG, Johnson ML, Lamb RA. Paramyxovirus fusion (F) protein and hemagglutinin–neuraminidase (HN) protein interactions: Intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology. 1997;237(1):1–9. doi: 10.1006/viro.1997.8759. [DOI] [PubMed] [Google Scholar]
- 86.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol. 2002;76(10):5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iorio RM, Field GM, Sauvron JM, et al. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin–neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J Virol. 2001;75(4):1918–1927. doi: 10.1128/JVI.75.4.1918-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Connaris H, Takimoto T, Russell R, et al. Probing the sialic acid binding site of the hemagglutinin–neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J Virol. 2002;76(4):1816–1824. doi: 10.1128/JVI.76.4.1816-1824.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A. Role of the hemagglutinin–neuraminidase protein in the mechanism of paramyxovirus–cell membrane fusion. J Virol. 2002;76:13028–13033. doi: 10.1128/JVI.76.24.13028-13033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Corey EA, Mirza AM, Levandowsky E, Iorio RM. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin–neuraminidase is due to a temperature-dependent defect in receptor binding. J Virol. 2003;77(12):6913–6922. doi: 10.1128/JVI.77.12.6913-6922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin–neuraminidase protein: implications for the mechanism of fusion promotion. J Virol. 2008;82(21):10386–10396. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bishop KA, Hickey AC, Khetawat D, et al. Residues in the stalk domain of the hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82(22):11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iorio RM, Glickman RL, Sheehan JP. Inhibition of fusion by neutralizing monoclonal antibodies to the haemagglutinin–neuraminidase glycoprotein of Newcastle disease virus. J Gen Virol. 1992;73:1167–1176. doi: 10.1099/0022-1317-73-5-1167. [DOI] [PubMed] [Google Scholar]