Abstract
Decapping is a critical step in the conserved 5′-to-3′ mRNA decay pathway of eukaryotes. The hetero-octameric Lsm1-7–Pat1 complex is required for normal rates of decapping in this pathway. This complex also protects the mRNA 3′-ends from trimming in vivo. To elucidate the mechanism of decapping, we analyzed multiple lsm1 mutants, lsm1-6, lsm1-8, lsm1-9, and lsm1-14, all of which are defective in decapping and 3′-end protection but unaffected in Lsm1-7–Pat1 complex integrity. The RNA binding ability of the mutant complex was found to be almost completely lost in the lsm1-8 mutant but only partially impaired in the other mutants. Importantly, overproduction of the Lsm1-9p- or Lsm1-14p-containing (but not Lsm1-8p-containing) mutant complexes in wild-type cells led to a dominant inhibition of mRNA decay. Further, the mRNA 3′-end protection defect of lsm1-9 and lsm1-14 cells, but not the lsm1-8 cells, could be partly suppressed by overproduction of the corresponding mutant complexes in those cells. These results suggest the following: (1) Decapping requires both binding of the Lsm1-7–Pat1 complex to the mRNA and facilitation of the post-binding events, while binding per se is sufficient for 3′-end protection. (2) A major block exists at the post-binding steps in the lsm1-9 and lsm1-14 mutants and at the binding step in the lsm1-8 mutant. Consistent with these ideas, the lsm1-9, 14 allele generated by combining the mutations of lsm1-9 and lsm1-14 alleles had almost fully lost the RNA binding activity of the complex and behaved like the lsm1-8 mutant.
Keywords: Lsm1-7–Pat1 complex, mRNA decay, Sm-like, decapping, oligo(A) tail
INTRODUCTION
Decapping is a key step in several mRNA decay pathways. In the 5′-to-3′ pathway, which is a major cytoplasmic mRNA decay pathway conserved in all eukaryotes, decay is initiated by poly(A) shortening, and decapping of the oligoadenylated mRNA permits the degradation of the message body by the 5′-to-3′ exonuclease Xrn1p (Coller and Parker 2004; Meyer et al. 2004; Wilusz and Wilusz 2004; Yamashita et al. 2005). On the other hand, the 5′-to-3′ exonucleolytic degradation of aberrant mRNAs is triggered by decapping independent of their A-tail length in the nonsense-mediated decay (NMD) pathway conserved in all eukaryotes (Muhlrad and Parker 1994; Beelman et al. 1996; He and Jacobson 2001; Lejeune et al. 2003; Unterholzner and Izaurralde 2004; Isken and Maquat 2007). Decapping is also a critical event in the miRNA-mediated mRNA decay (Behm-Ansmant et al. 2006a,b; Eulalio et al. 2007), mRNA decay stimulated by AU-rich elements (Gao et al. 2001; Wilusz and Wilusz 2004; Fenger-Gron et al. 2005; Lykke-Andersen and Wagner 2005; Stoecklin et al. 2006), decay of nonpolyadenylated histone mRNAs (Mullen and Marzluff 2008), and the initiation-mediated mRNA decay (Heikkinen et al. 2003). Finally, the nuclear decay of pre-mRNAs and nuclear-restricted mRNAs also seems to involve decapping of such transcripts (Kufel et al. 2004). A nuclear protein with decapping activity, Nudt16/X29, is conserved in several organisms (Taylor and Peculis 2008) and has the ability to cleave both m7-monomethyl and m2,2,7-trimethyl caps (characteristic of snRNAs and snoRNAs) of RNAs in vitro (Ghosh et al. 2004), implying that decapping may be a required step in the decay of several different RNA substrates in the nucleus.
The mRNA decapping enzyme consists of at least two subunits, Dcp1p and Dcp2p, which are conserved in all eukaryotes. While Dcp2p is the catalytic subunit, Dcp1p associates with Dcp2p and stimulates its activity (Parker and Song 2004; Liu and Kiledjian 2006; Simon et al. 2006; Deshmukh et al. 2008). Decapping is a precisely controlled rate-limiting step in the eukaryotic 5′-to-3′ mRNA decay pathway (Muhlrad et al. 1994, 1995; Tharun and Parker 1999; Coller and Parker 2004); however, the mechanism of decapping is poorly understood. Several observations suggest that decapping is a complex process comprised of multiple substeps, although the nature and order of such substeps in vivo are not clear. First, in the 5′-to-3′ mRNA decay pathway, decapping is influenced by the 3′-A-tail status of the mRNA such that oligoadenylated, but not polyadenylated, messages are selectively decapped (deadenylation-dependent decapping) (Coller and Parker 2004; Meyer et al. 2004), and the poly(A) binding protein is implicated in the inhibition of decapping of the polyadenylated messages in vivo (Caponigro and Parker 1995; Coller et al. 1998; Wilusz et al. 2001; Khanna and Kiledjian 2004). Second, multiple factors are known to affect the mRNA decapping rates in vivo, and these include both positive and negative regulators. While the translation initiation factors and the poly(A) binding protein are known to be inhibitory to decapping (Caponigro and Parker 1995; Coller et al. 1998; Schwartz and Parker 1999, 2000; Vilela et al. 2000; Wilusz et al. 2001; Ramirez et al. 2002; Khanna and Kiledjian 2004), several other factors like the Lsm1-7–Pat1 complex, Dhh1p, and the Edc proteins facilitate decapping (Bouveret et al. 2000; Tharun et al. 2000; Coller et al. 2001; Schwartz et al. 2003; Kshirsagar and Parker 2004; Parker and Song 2004; Fillman and Lykke-Andersen 2005; Tharun 2009). Third, multiple observations suggest that an mRNP rearrangement event involving the displacement of the translation initiation factors from the mRNA and recruitment of the decapping activators onto the mRNA is necessary for the decapping machinery to gain access to the cap (Schwartz and Parker 1999, 2000; Tharun et al. 2000; Tharun and Parker 2001; Andrei et al. 2005).
The Lsm1-7–Pat1 complex made up of Pat1p and the seven Sm-like proteins Lsm1p–Lsm7p is an activator of decapping required for the normal rates of decapping in vivo and is conserved in all eukaryotes (Salgado-Garrido et al. 1999; Bouveret et al. 2000; Tharun et al. 2000; Tharun 2009). Lsm1p is the key subunit that distinguishes this complex from the related U6 snRNP-associated nuclear complex, Lsm2p-8p, which is made of the seven Sm-like proteins Lsm2p–Lsm8p (Achsel et al. 1999; Mayes et al. 1999; Bouveret et al. 2000). The Lsm1-7–Pat1 complex preferentially associates in vivo with the oligoadenylated mRNPs targeted for decay rather than polyadenylated translating messages (Tharun et al. 2000; Tharun and Parker 2001; Tharun 2009). Consistently, the purified Lsm1-7–Pat1 complex exhibits a strong binding preference for oligoadenylated RNAs over polyadenylated and unadenylated RNAs in vitro (Chowdhury et al. 2007). Importantly, such ability of this complex to recognize the oligoadenylated status of the RNA is necessary for its mRNA decay function in vivo, because mutations in LSM1 impairing such ability result in mRNA decay defect in vivo (Chowdhury and Tharun 2008). These observations imply that the association of the Lsm1-7–Pat1 complex with the mRNA is a crucial event in the 5′-to-3′ mRNA decay pathway and that the preferential binding of the oligoadenylated mRNAs by this complex contributes to the deadenylation dependence of decapping in this pathway. Thus, the Lsm1-7–Pat1 complex and the poly(A) binding protein could play complementary roles in facilitating the selective decapping of oligoadenylated messages in this pathway.
Apart from being an activator of decapping, the Lsm1-7–Pat1 complex also serves to protect the 3′-ends of mRNAs from trimming in vivo as revealed by the accumulation of “3′-trimmed” forms of several mRNAs in the lsm1–lsm7 (but not lsm8) and pat1 loss of function mutants (Boeck et al. 1998; He and Parker 2001; Tharun et al. 2005). These trimmed species (generated by an unknown trimming nuclease) are truncated by ∼10 nucleotides (nt) at their (fully deadenylated) 3′-ends. The relationship between the mRNA decapping and 3′-end protection functions of the Lsm1-7–Pat1 complex is not understood.
In the present study, we have analyzed a set of lsm1 mutants to gain insight into the mechanism of decapping. The mutants, lsm1-6, lsm1-8, lsm1-9, and lsm1-14, are defective in mRNA decay and 3′-end protection (Tharun et al. 2005) but are able to assemble the Lsm1-7–Pat1 complex. However, while the RNA binding ability of the complex seems to be almost fully abolished in the case of the lsm1-8 cells, it is only partly affected for the other mutants. Interestingly, the lsm1-9 and lsm1-14 alleles but not the other alleles caused a dominant inhibition of mRNA decay when overexpressed in wild-type cells. The RNA binding activity of the complexes made with the Lsm1-9p and Lsm1-14p proteins seems to be important for the ability of the lsm1-9 and lsm1-14 alleles to cause such dominant inhibition, because the lsm1-9, 14 allele created by combining the mutations of the lsm1-9 and lsm1-14 alleles was unable to cause such inhibition and was severely impaired in the RNA binding ability of the complex. The mRNA decay defect of the lsm1-9, lsm1-14, lsm1-9, 14, and lsm1-8 mutants could not be suppressed by overexpression of the corresponding lsm1 alleles. However, a partial suppression of the 3′-end protection defect was observed upon such overexpression in the lsm1-9 and lsm1-14 mutants. These results suggest that although the Lsm1-7–Pat1-complex/mRNA interaction is very important for mRNA decapping, such interaction per se is not sufficient to support decapping.
RESULTS
The lsm1-6 and lsm1-8 mutants are able to assemble the Lsm1-7–Pat1 complex
We had earlier generated several lsm1 mutants that are defective in mRNA decay (Tharun et al. 2005). Analysis of multiple decay defective lsm1 mutants to understand the basis of the decay defect in each of them could provide more insight into the mechanism of decapping. To this end, we studied the lsm1-6, lsm1-8, lsm1-9, and lsm1-14 mutants, all of which are defective in both mRNA decay and 3′-end protection, although the defects are relatively less severe in lsm1-6 compared with the other three mutants (Tharun et al. 2005). The levels of the mutant Lsm1p proteins are normal in these mutants (compared with wild-type Lsm1p in wild-type cells) suggesting that insufficient accumulation of the mutant Lsm1p protein is not the cause of their decay and 3′-end protection defects (Tharun et al. 2005). We had shown earlier that, in the case of the lsm1-9 and lsm1-14 mutants, the Lsm1-7–Pat1 complex integrity is not affected and that the mutant complexes isolated from them are able to bind the RNA although with a lower affinity than the wild-type complex (Chowdhury and Tharun 2008). In order to determine if all the component proteins are present in the mutant complexes made in the lsm1-6 and lsm1-8 cells, we purified those mutant complexes following the strategy that we used earlier (Chowdhury et al. 2007; Tharun 2008). Sodium dodecyl sulfate–polyacrylamide gel electrophoretic (SDS-PAGE) analysis of the mutant complexes revealed a band pattern similar to that of the wild-type complex (Fig. 1A). The presence of all the component subunits in the mutant complexes was confirmed by mass spectrometry analysis based on the detection of at least one tryptic peptide with unambiguous sequence match for each protein (Supplemental Table 1).
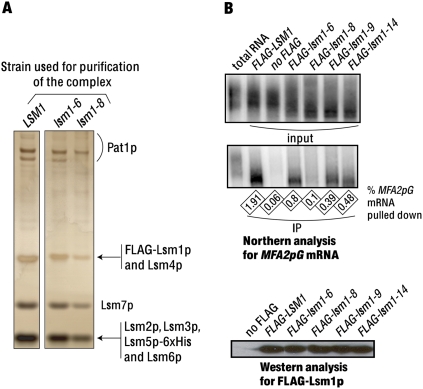
FIGURE 1.
(A) Lsm1-7–Pat1 complexes isolated from the lsm1-8 and lsm1-6 mutants contain all the component proteins. Complexes purified from the wild-type and mutant cells (indicated above the lanes) were subjected to SDS-PAGE, and the bands were visualized by silver staining. Identity of the proteins present in the various bands (revealed by mass spectrometry analysis) is indicated on the right. (B) Coprecipitation of mRNA with the mutant Lsm1p is almost completely abolished in the case of the lsm1-8 mutant but only partly impaired in the case of the lsm1-6, lsm1-9, and lsm1-14 mutants. Anti-FLAG antibody immunoprecipitations were carried out from the lysates of various strains (indicated above the lanes), after which the RNA extracted from the entire pull-down fraction (middle panel) or a small aliquot of the input lysate (top panel) were subjected to Northern analysis for MFA2pG mRNA. (bottom panel) Western analysis using anti-FLAG antibodies to visualize the FLAG-protein present in the pull-down fractions. The fraction of the lysate mRNA that got coprecipitated in each experiment is indicated as a percentage below the middle panel. Total RNA from wild-type yeast was loaded in the first lane (from left) in the top and middle panels to show the poly(A) tail distribution of the MFA2pG mRNA.
Mutant complex isolated from the lsm1-8 cells is severely defective in RNA binding in vitro
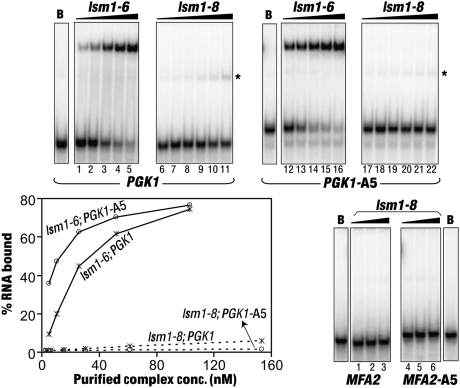
We had shown earlier that the purified wild-type Lsm1-7–Pat1 complex is able to bind the RNA directly in vitro and that it has a strong binding preference for RNAs carrying a 3′-oligo(A) tail over unadenylated RNAs (Chowdhury et al. 2007). In order to test the RNA binding properties of the mutant complexes purified from the lsm1-6 and lsm1-8 cells, we carried out gel shift assays using uniformly radiolabeled PGK1 and PGK1-A5 RNAs, which are the unadenylated and penta-adenylated forms of a short in vitro transcript carrying the 3′-most 42 nt of the yeast PGK1 mRNA preceded by two G's at the 5′-end (Chowdhury et al. 2007; Chowdhury and Tharun 2008). As shown in Figure 2, the complex isolated from the lsm1-6 cells binds RNA efficiently, with its affinity being only slightly lower than that of the wild-type complex (see Fig. 5 below) (PGK1 RNA is bound with apparent KD's of ∼35 nM and 22 nM by the complexes purified from the lsm1-6 and wild-type cells, respectively). Further, it shows higher affinity toward the oligoadenylated RNA (PGK1-A5) than the unadenylated RNA (PGK1) like the wild-type complex (Fig. 2). However, the complex isolated from the lsm1-8 cells exhibits barely detectable RNA binding ability with both the PGK1 and PGK1-A5 RNAs (Fig. 2). Such poor binding efficiency of this complex is not specific to the PGK1 RNA sequence, because similar results were also obtained when the gel shift assays were carried out using the MFA2 and MFA2-A5 RNAs (Fig. 2). Therefore, these observations indicate that the overall RNA binding ability of the Lsm1-8p-containing complex is in general very severely compromised. Weak gel shifted RNA bands of higher than expected mobility (marked by asterisks in Fig. 2) observed in the assays using the complex purified from the lsm1-8 cells are likely due to the dissociation of the RNPs during the gel run (Chowdhury et al. 2007).
FIGURE 2.
The Lsm1-7–Pat1 complex purified from the lsm1-8 cells has barely detectable RNA binding activity. Gel mobility shift assays were carried out using the PGK1, PGK1-A5, MFA2, or MFA2-A5 RNAs (indicated below the phosphorimages) with BSA (lanes marked as “B”) or increasing concentrations of the complexes purified from the lsm1-6 (5 nM, 10 nM, 26 nM, 52 nM, and 103 nM) or the lsm1-8 (3 nM, 6 nM, 15 nM, 31 nM, 61 nM, and 154 nM) cells (indicated above the lanes). Binding of the PGK1 and PGK1-A5 RNAs was quantitated using a PhosphorImager, and plots of the fraction of the RNA bound versus the concentration of the purified complex are shown on the bottom-left panel. Final concentrations of the Lsm1-8p-containing complex used in the assays with the MFA2 and MFA2-A5 RNAs (bottom right) are 31 nM, 61 nM, and 154 nM, respectively. Asterisks mark weak gel shifted RNA bands of higher mobility possibly resulting from dissociation of the RNPs during the gel run in assays carried out using the complex purified from the lsm1-8 cells.
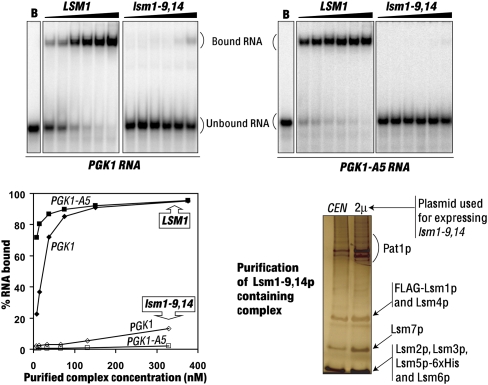
FIGURE 5.
The lsm1-9, 14 allele is severely impaired in the RNA binding ability of the complex. (Top) Gel mobility shift assays were carried out using the PGK1, and PGK1-A5 RNAs with BSA (lanes B) or increasing concentrations of the complexes purified from the wild-type (7.5 nM, 15 nM, 38 nM, 75 nM, 150 nM, and 376 nM) or the lsm1-9, 14 (6.6 nM, 13 nM, 33 nM, 66 nM, 131 nM, and 329 nM) cells (indicated above the lanes). Binding was quantitated using the PhosphorImager and plots of the fraction of the RNA bound versus the concentration of the purified complex are shown on the bottom-left panel. (Bottom right) The Lsm1-9, 14p-containing complex purified from cells expressing the lsm1-9, 14 allele from a CEN or a 2μ vector (indicated above the lanes) was subjected to SDS-PAGE analysis and bands were visualized by silver staining.
mRNA coprecipitation with Lsm1-8p from cell lysates is almost completely abolished
Our earlier studies showed that the pull-down of Lsm1p, Lsm5p, or Pat1p from the yeast cell lysates results in the coprecipitation of mRNA suggesting that the Lsm1-7–Pat1 complex interacts with the mRNA in vivo (Tharun et al. 2000; Tharun and Parker 2001). Given that the mutant complexes made in the lsm1-9 and lsm1-14 cells but not the lsm1-8 cells are able to bind RNA in vitro (Fig. 2; Chowdhury and Tharun 2008), we asked whether this difference is also reflected in the coprecipitation of mRNA with the corresponding mutant Lsm1p proteins from cell lysates. To this end, we made lysates of cells expressing the FLAG-tagged versions of the wild-type and the mutant Lsm1p proteins (as the only source of Lsm1p in the cell) and pulled down the FLAG-protein using anti-FLAG antibodies from such lysates. RNA made from the immunoprecipitates and the lysates were then subjected to Northern analysis to compare the fraction of MFA2pG mRNA in the lysate that coprecipitated with the FLAG–Lsm1p in each case. The immunoprecipitates were also subjected to Western analysis to determine the amount of FLAG–Lsm1p protein that was pulled down in each case. As shown in Figure 1B (top two panels), mRNA does get coprecipitated with the FLAG–Lsm1-6p, FLAG–Lsm1-9p, and FLAG–Lsm1-14p proteins from the corresponding mutant cell lysates, although the efficiency of such coprecipitation was decreased by ∼2.5-, five-, and fourfold, respectively, compared with the wild-type FLAG–Lsm1p pulled down from the wild-type cell lysate. However, the coprecipitation of mRNA with Lsm1-8p is close to background levels. The observed differences in the mRNA coprecipitation efficiencies are not a result of the differential abilities of the FLAG–Lsm1p proteins to interact with the anti-FLAG antibodies because the amounts of the wild-type and the mutant FLAG–Lsm1p proteins pulled down are comparable (Fig. 1B, bottom panel). These results suggest that the ability of the Lsm1-7–Pat1 complex to associate with the mRNA in vivo is severely impaired in the lsm1-8 mutants but only partially affected in the lsm1-6, lsm1-9, and lsm1-14 mutants. This is consistent with the in vitro RNA binding activities of the mutant complexes purified from these mutants (Fig. 2; Chowdhury and Tharun 2008).
Decay defect of the lsm1-9 and lsm1-14 mutants cannot be suppressed by overexpression of the corresponding lsm1 alleles
The lsm1-8 mutant exhibits a strong defect in mRNA decay and 3′-end protection in vivo (Tharun et al. 2005). Consistent with that, the mRNA coprecipitation experiments (Fig. 1B) and the in vitro assays using the purified complex (Fig. 2) reveal that the Lsm1-7–Pat1 complex assembled in this mutant is severely impaired in RNA binding. On the other hand, the other mutants, lsm1-6, lsm1-9, and lsm1-14, which are also defective in mRNA decay and 3′-end protection (Tharun et al. 2005), yield complexes that are capable of binding RNA in vitro (Fig. 2; Chowdhury and Tharun 2008). Furthermore, pull-down of the corresponding mutant Lsm1p proteins from cell lysates results in the coprecipitation of mRNA (although with lower efficiency compared with the wild-type Lsm1p) (Fig. 1B). This suggests that the nature of the block to mRNA decay is different in the various lsm1 mutants that we have studied. Therefore, to analyze our lsm1 mutants further, we asked whether their decay defects can be suppressed by overexpression of the corresponding lsm1 alleles. For these studies we utilized the MFA2pG reporter mRNA system. This mRNA decays by the 5′-to-3′ decay pathway (Muhlrad et al. 1994) and is expressed under the control of the GAL promoter from a chromosomal location in our strains (Hatfield et al. 1996). It contains a poly(G) insertion in its 3′-UTR that blocks Xrn1p in cis, resulting in the stable accumulation of a degradation intermediate called poly(G) fragment in vivo. The level of the poly(G) fragment relative to that of the full-length MFA2pG mRNA at steady state is a good indicator of the status of mRNA decay via the 5′-to-3′ pathway in any given strain, since the production of the poly(G) fragment is decreased upon impairment of decapping or Xrn1p function (Hatfield et al. 1996; Tharun et al. 2005).
lsm1Δ cells expressing different alleles of LSM1 from a CEN or 2μ vector were grown to log phase in galactose-containing medium, and RNA was prepared from them. Northern analysis of the RNA samples to visualize the MFA2pG mRNA and the poly(G) fragment revealed (Fig. 3) that, as expected, lsm1Δ cells expressing lsm1-6, lsm1-8, lsm1-9, or lsm1-14 from a CEN vector accumulate less poly(G) fragment (relative to the full-length MFA2pG mRNA) compared with those expressing LSM1 from a CEN vector [Fig. 3, lanes 1,4,7,10,13—the level of poly(G) fragment relative to that of the MFA2pG mRNA is expressed as “% poly(G) fragment”], consistent with the known mRNA decay defects of the lsm1-6, lsm1-8, lsm1-9, and lsm1-14 mutants (Tharun et al. 2005). However, importantly, lsm1Δ cells expressing lsm1-8, lsm1-9, or lsm1-14 from a 2μ vector did not show an increased accumulation of the poly(G) fragment compared with lsm1Δ cells expressing these alleles from a CEN vector, suggesting that the decay defect of the lsm1-8, lsm1-9, and lsm1-14 mutants cannot be rescued by overexpression of the corresponding lsm1 alleles (Fig. 3, lanes 7–15). However, a similar analysis revealed a moderate but reproducible increase in the poly(G) fragment levels in cells expressing the lsm1-6 allele from a 2μ vector compared with cells expressing the same allele from a CEN vector (Fig. 3, lanes 4–6).
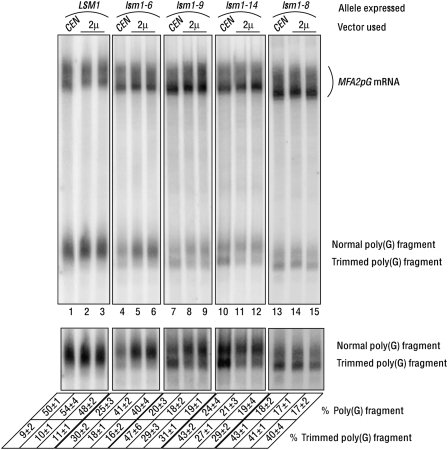
FIGURE 3.
mRNA 3′-end protection defect but not the mRNA decay defect of the lsm1-9 and lsm1-14 mutants can be partly suppressed by overexpression of the corresponding lsm1 alleles. lsm1Δ cells transformed with a CEN or a 2μ vector expressing the LSM1, lsm1-6, lsm1-9, lsm1-14, or the lsm1-8 allele (indicated above the lanes) were grown to log phase, and RNA was extracted from them. The RNA samples were then subjected to Northern analysis to visualize the MFA2pG mRNA and the normal and trimmed forms of the poly(G) fragment (indicated on the right), followed by quantitation of the levels of those species using a PhosphorImager. (Bottom) A longer exposure of the phosphorimage shown in the top panel [only the portion containing the poly(G) fragment is shown]. Percentage of the trimmed poly(G) fragment was determined by taking the total poly(G) fragment level (trimmed + normal) as 100%. Percentage of the poly(G) fragment was determined by taking the total signal [full-length mRNA + total poly(G) fragment] as 100%. Values are averages of at least two repeats for each strain and are shown at the bottom.
mRNA 3′-end protection defect of the lsm1-9 and lsm1-14 mutants, but not the lsm1-8 mutant, can be partially suppressed by overexpression of the corresponding lsm1 alleles
We had shown earlier that the lsm1-6, lsm1-8, lsm1-9, and lsm1-14 mutants are also defective in mRNA 3′-end protection and hence accumulate the 3′-trimmed forms of the full-length MFA2pG mRNA and the poly(G) fragment, while such trimmed species are hard to detect in wild-type cells (Tharun et al. 2005). Due to their smaller size, the trimmed and normal forms of the poly(G) fragment resolve better than the corresponding forms of the full-length mRNA during electrophoresis, and hence determination of the levels of the trimmed poly(G) fragment relative to that of the normal poly(G) fragment is a convenient way to determine the degree of 3′-end protection (Tharun et al. 2005). In order to determine the fraction of the poly(G) fragment accumulating in trimmed form in the lsm1Δ cells expressing the various lsm1 alleles from CEN and 2μ vectors, we quantitated the level of the trimmed poly(G) fragment in the corresponding RNA samples from the Northern blot shown in Figure 3 and calculated the percentage of the poly(G) fragment present in trimmed form (shown at the bottom of Fig. 3), taking the level of total poly(G) fragment (normal + trimmed) as 100%. Interestingly, we observed that in cells expressing the lsm1-9 or the lsm1-14 allele from a 2μ vector, a smaller fraction of the poly(G) fragment is present in trimmed form compared with the cells expressing the same allele from a CEN vector (Fig. 3, lanes 7–12), suggesting that the mRNA 3′-end protection defect of the lsm1-9 and lsm1-14 mutants can be rescued at least partially by overexpression of the corresponding lsm1 alleles. However, in the lsm1-8 cells the relative levels of the normal and trimmed poly(G) fragment were not affected significantly depending on whether the lsm1-8 allele was expressed from a CEN or a 2μ vector (Fig. 3, lanes 13–15).
Thus, the mRNA 3′-end protection defect but not the mRNA decay defect of the lsm1-9 and lsm1-14 mutants can be partly rescued by overexpression of the corresponding lsm1 alleles in those cells. On the other hand, neither the mRNA decay defect nor the mRNA 3′-end protection defect of the lsm1-8 cells could be suppressed to a significant extent by overexpression of the lsm1-8 allele.
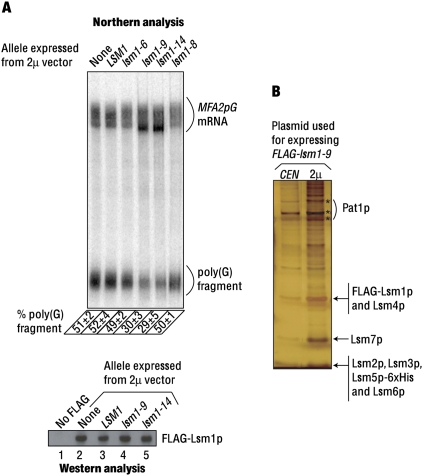
The lsm1-9 and lsm1-14 alleles can cause dominant inhibition of mRNA decay when overexpressed
The results discussed above (Fig. 3) suggest that the nature of the decay defect in the lsm1-8 mutant is different from that of the lsm1-9 and lsm1-14 mutants. Thus, they are in synchrony with the in vitro analysis of the mutant complexes purified from these cells (Fig. 2; Chowdhury and Tharun 2008) and the mRNA coprecipitation experiments (Fig. 1B). Therefore, to gain further insight into the basis of the decay defect in these mutants, we studied the effect of overexpression of these lsm1 alleles in wild-type background on the decay of the MFA2pG mRNA. We introduced 2μ vectors expressing different (untagged) lsm1 alleles (wild type or mutant) into the wild-type FLAG–LSM1 cells. After growing these transformants in galactose medium to log phase, the RNA samples made from them were subjected to Northern analysis to determine the relative levels of the poly(G) fragment and the full-length MFA2pG mRNA in each sample using a PhosphorImager. The results (Fig. 4A) revealed that expression from a 2μ vector of the lsm1-9 or the lsm1-14 allele (but not the LSM1, the lsm1-6, or the lsm1-8 allele) in wild-type background leads to a clear decrease in the accumulation of the poly(G) fragment (relative to the level of the full-length MFA2pG mRNA). Further, while the MFA2pG mRNA from wild-type cells bearing an empty 2μ vector or a 2μ vector expressing LSM1 (or lsm1-6 or lsm1-8) formed a uniform smear [representing a poly(A) tail length distribution of 10–70 A-residues] in the Northern blot as expected for the cells unaffected in the 5′-to-3′ decay, wild-type cells expressing lsm1-9 or lsm1-14 from a 2μ vector showed an accumulation of the oligoadenylated MFA2pG mRNA, which is typically seen in cells defective in decapping (Tharun et al. 2005). Thus, wild-type cells overexpressing lsm1-9 or lsm1-14 exhibit two important hallmarks of strains defective in decapping, namely, underaccumulation of the poly(G) fragment and overaccumulation of the oligoadenylated MFA2pG mRNA. Western analyses (Fig. 4A) revealed that the levels of the wild-type FLAG–Lsm1p are not affected in cells where the lsm1-9 or the lsm1-14 allele is overexpressed. Therefore, these results indicate that upon overexpression in wild-type background, the lsm1-9 and lsm1-14 alleles cause dominant inhibition of mRNA decay by affecting the function and not the levels of wild-type Lsm1p protein in the cell.
FIGURE 4.
(A) lsm1-9 and lsm1-14 alleles cause dominant inhibition of mRNA decay when overexpressed. Wild-type (FLAG-LSM1) cells transformed with 2μ vectors expressing different untagged alleles of LSM1 or an empty 2μ vector (indicated above the lanes in the top and bottom panels) were grown to log phase, after which RNA made from them was subjected to Northern analysis for MFA2pG mRNA (top) or lysates made from them were subjected to western analysis using anti-FLAG antibodies (bottom). The “% poly(G) fragment” values determined as described for Figure 3 and indicated at the bottom of the top panel are averages of at least two repeats. Positions of the MFA2pG mRNA and the poly(G) fragment are indicated on the right. In the bottom panel, lane 1 shows the western analysis of the lysate prepared from a wild-type strain that lacks a tag on Lsm1p. (B) Overexpression of the Lsm1p subunit results in an increase in the levels of the Lsm1-7–Pat1 complex. Lsm1-9p-containing complex purified from cells expressing lsm1-9 from a CEN or a 2μ vector (indicated above the lanes) was separated by SDS-PAGE and visualized by silver staining.
The inability of the lsm1-8 allele to inhibit mRNA decay upon expression from 2μ vectors in the wild-type cells is not because Lsm1-8p fails to accumulate to as high levels as Lsm1-9p or Lsm1-14p expressed similarly in those cells, because Western analysis of the lysates from wild-type cells (with untagged endogenous LSM1) expressing FLAG-tagged versions of lsm1-8, lsm1-9, and lsm1-14 from 2μ vectors reveal that these mutant proteins are expressed at comparable levels (Northern analysis of the RNA made from these cells for MFA2pG mRNA gave results similar to those shown in Fig. 4A) (Supplemental Fig. 1A; data not shown). This is consistent with our earlier studies, which also showed that the protein products of all four lsm1 alleles studied here accumulate as efficiently as the wild-type Lsm1p (Tharun et al. 2005). Experiments involving the pull-down of Lsm5p from the lysates of the wild-type and lsm1-8 cells revealed that the efficiency of coprecipitation of Lsm1p and Lsm1-8p are comparable, suggesting that the association of Lsm1-8p with the other subunits of the complex is not significantly impaired compared with the wild-type Lsm1p (Supplemental Fig. 1B). This suggests that the inability of the lsm1-8 allele to cause dominant inhibition of decay when overexpressed in wild-type cells is not due to its decreased ability to form the complex.
The above results (dominant inhibition of mRNA decay) and the results presented in Figure 3 (partial suppression of the 3′-end protection defect) show that the functions of the Lsm1-7–Pat1 complex can be impacted by overexpression of just the lsm1 allele. This suggested that the excess Lsm1p subunit produced upon overexpression is assembled into the complex elevating the levels of the complex. To test this, we purified the Lsm1-9p-containing complexes from the cells that express FLAG–lsm1-9 from either a CEN or a 2μ vector under identical conditions using the same number of cells as the starting material for the purification. SDS-PAGE analysis of the purified material revealed that indeed the yield of the complex is higher from the cells that express the FLAG–lsm1-9 allele from a 2μ vector compared with the cells expressing the same allele from a CEN vector (Fig. 4B). This observation suggests that the availability of the other subunits (Lsm2p–Lsm7p and Pat1p) for complex formation is not limiting in vivo under the conditions of our experiments. Similar experiments involving FLAG–lsm1-14 and FLAG–LSM1 gave analogous results (data not shown).
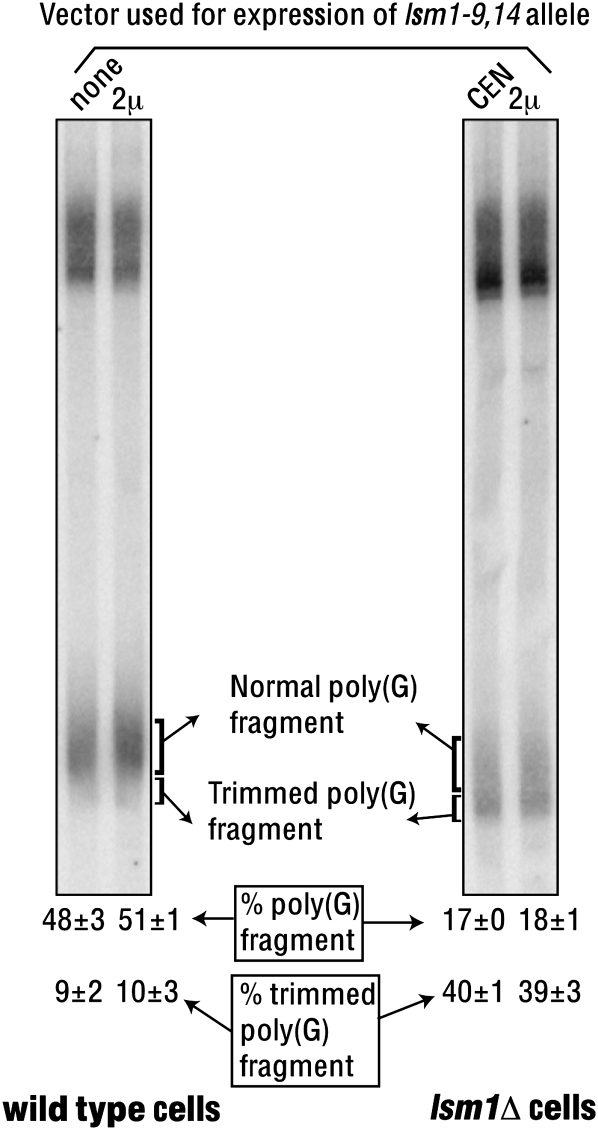
The lsm1-9, 14 allele is severely impaired in the RNA binding ability of the complex and is unable to cause dominant inhibition of mRNA decay
Our results (Figs. 1–4) suggest that the inability of the lsm1-8 allele to cause dominant inhibition of decay like the lsm1-9 and lsm1-14 alleles is due to the lack of RNA binding ability of the Lsm1-8p-containing complex. In order to test this idea, we created a new lsm1 allele (lsm1-9, 14) by combining the mutations of both the lsm1-9 and lsm1-14 alleles (Tharun et al. 2005). Purification of the complex from the lsm1-9, 14 mutant revealed that the complex integrity is not significantly affected in this mutant (Supplementary Table 1 and Fig. 5, bottom-right panel, left lane), similar to the lsm1-9 and lsm1-14 mutants. Further, the complex yield (during purification) from the lsm1-9, 14 mutant was comparable with that of the wild-type cells and the other mutants, suggesting that the accumulation of the Lsm1-9, 14p protein and the efficiency of its incorporation into the complex are not significantly affected. Experiments addressing the consequence of the overexpression of the lsm1-9, 14 allele (similar to those presented in Fig. 3) revealed that, although the lsm1-9, 14 mutant exhibits strong defects in mRNA decay and 3′-end protection (as expected) similar to the lsm1-9 and lsm1-14 mutants, neither of those defects could be suppressed by overexpression of the lsm1-9, 14 allele (Fig. 6). Gel shift assays carried out using the PGK1 and PGK1-A5 RNAs revealed that the mutant complex isolated from the lsm1-9, 14 cells has barely detectable RNA binding activity (Fig. 5, bottom-left and top panels). Importantly, overexpression of the lsm1-9, 14 allele in the wild-type cells failed to cause dominant inhibition of mRNA decay unlike the lsm1-9 and lsm1-14 alleles (Fig. 6). This is not due to an inability to elevate the levels of the Lsm1-9, 14p-containing complex upon such overexpression, because purification of the Lsm1-9, 14p-containing complex from cells expressing the lsm1-9, 14 allele from a 2μ vector revealed that the complex yield is indeed higher from such cells compared with the cells expressing the same allele from a CEN vector (Fig. 5, bottom-right panel). These results support the idea that the ability of the Lsm1-9p- and Lsm1-14p-containing complexes to bind RNA is essential for the ability of the lsm1-9 and lsm1-14 alleles to cause dominant inhibition of mRNA decay upon overexpression.
FIGURE 6.
Overexpression of the lsm1-9, 14 allele fails to suppress the mRNA decay and 3′-end protection defects of the lsm1-9, 14 cells and fails to cause dominant inhibition of mRNA decay in the wild-type cells. Wild-type and lsm1Δ cells (indicated below the lanes) expressing the lsm1-9, 14 allele from a CEN or a 2μ vector (indicated above the lanes) were grown to log phase, and RNA isolated from them was subjected to Northern analysis to visualize the MFA2pG mRNA. The “% poly(G) fragment” and “% trimmed poly(G) fragment” values (shown at the bottom) were determined as described for Figure 3 and are averages of at least two repeats.
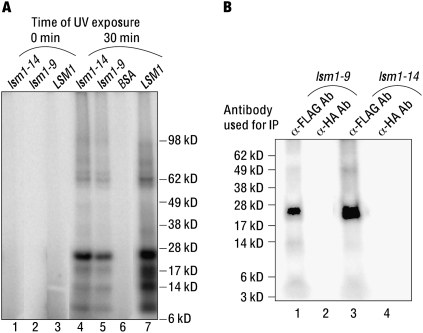
Mutant Lsm1p in the mutant complexes contacts RNA during RNA binding
Although the mutant complexes purified from the lsm1-9 and lsm1-14 cells are able to bind the RNA, they lack an important characteristic feature of the Lsm1-7–Pat1 complex, namely, the ability to recognize the 3′-oligo(A) tail of the RNA (Chowdhury and Tharun 2008). This raised the possibility that the manner in which these mutant complexes actually contact the RNA is different from that of the wild-type complex. Such an idea is consistent with our observation that the mRNA decay defect of these two mutants could not be suppressed by overexpression of the corresponding lsm1 alleles although such overexpression elevates the level of the mutant complex. Moreover, overexpression of the lsm1-9 and lsm1-14 alleles in wild-type cells results in a dominant inhibition of mRNA decay. Since the lsm1-9 and lsm1-14 alleles bear lesions in the predicted RNA binding surfaces of Lsm1p (Tharun et al. 2005), we asked if the mutant Lsm1-9p and Lsm1-14p subunits indeed make direct contacts with the RNA when the corresponding mutant complexes bind to the RNA or if the binding of these mutant complexes is primarily mediated by the other subunits (Lsm2p–Lsm7p and Pat1p) of the complex. To this end, we analyzed the nature of the interaction of the mutant complexes with RNA using UV-crosslinking assays. Lsm1-7–Pat1 complexes purified from the LSM1, lsm1-9, and lsm1-14 cells were bound to the radiolabeled MFA2 RNA and then UV irradiated. After ribonuclease treatment, the crosslinked proteins were analyzed by SDS-PAGE followed by autoradiography. As shown in Figure 7A, the band pattern of the proteins crosslinked to the RNA is very similar between the wild-type and the mutant complexes with an ∼23-kDa band being a major crosslinked species as we had observed earlier (Chowdhury et al. 2007). By detergent disruption followed by immunoprecipitation using anti-FLAG antibodies of the UV-crosslinked wild-type complex, we had shown earlier that this ∼23-kDa band indeed contains crosslinked Lsm1p, revealing that Lsm1p subunit makes direct contacts with the RNA when the wild-type complex binds RNA (Chowdhury et al. 2007). To determine whether the Lsm1-9p and Lsm1-14p proteins still contact the RNA in the mutant complexes, we extended a similar analysis to the mutant complexes. As shown in Figure 7B, the mutant Lsm1p proteins do contact RNA. This is consistent with the fact that in each of these mutants only a subset of the predicted RNA binding residues is changed and the observation that mutating both of the predicted RNA binding surfaces of Lsm1p (the lsm1-9, 14 allele) results in a greater impairment of the RNA binding ability of the Lsm1-7–Pat1 complex than mutating one of the two predicted RNA binding surfaces (the lsm1-9 and lsm1-14 alleles).
FIGURE 7.
Mutant Lsm1p subunit present in the complexes isolated from the lsm1-9 and lsm1-14 cells is able to contact RNA. (A) RNA binding reactions containing the MFA2 RNA (radiolabeled at U residues) were carried out with the complex isolated from the LSM1, the lsm1-9, or the lsm1-14 cells at a final concentration of 88 nM or with BSA (indicated above the lanes). Aliquots of the reaction mix were then exposed to UV irradiation for 0 or 30 min (indicated above the lanes) followed by ribonuclease treatment. The samples were then subjected to SDS-PAGE separation and autoradiography. (B) Samples (irradiated 30 min) of reactions with the complexes purified from the lsm1-9 and lsm1-14 cells were subjected to detergent treatment after ribonuclease treatment (to disrupt the protein complexes) and then immunoprecipitated using either the anti-FLAG antibody matrix or the unrelated anti-HA antibody matrix (as control) as indicated above the lanes before SDS-PAGE separation and autoradiography. Positions of the molecular weight markers are shown on the right and left in A and B, respectively.
DISCUSSION
Several studies suggest that decapping involves multiple steps, although the nature and order of such steps are not clear. Many observations made from the studies on the Lsm1-7–Pat1 complex suggest that the interaction of this complex with the mRNA is a critical event in the 5′-to-3′ mRNA decay pathway. First, mRNA coprecipitates with this complex from cell lysates and the purified complex is able to directly bind RNA in vitro (Tharun et al. 2000; Tharun and Parker 2001; Chowdhury et al. 2007). Second, mutations in the predicted RNA binding residues of Lsm1p lead to the inhibition of mRNA decay in vivo (Tharun et al. 2005). Third, consistent with the selective decapping of oligoadenylated mRNAs in the 5′-to-3′ pathway, the purified Lsm1-7–Pat1 complex exhibits an intrinsic binding preference for oligoadenylated RNAs over polyadenylated RNAs (Chowdhury et al. 2007), and mutations in LSM1 impairing such preferential binding ability result in the inhibition of mRNA decay in vivo (Chowdhury and Tharun 2008). Here we have analyzed a set of decay-defective lsm1 mutants to gain more insight into the process of decapping. Our studies suggest that while RNA binding by this complex is important for decapping, additional post-binding steps are also needed.
An interesting observation in our studies is that overexpression of the lsm1-9 and lsm1-14 alleles in wild-type cells results in dominant inhibition of mRNA decay. A simple model to explain this inhibition is that the large amounts of mutant complexes formed upon such overexpression bind to the mRNA and thereby decrease the availability of the mRNA to the wild-type complex, while the mRNPs formed by the mutant complexes themselves are unable to proceed through the downstream steps (post-binding steps) needed to support decapping. These events could ultimately result in decreased decapping rates. Several observations support this model. First, the ability of mutant complexes purified from the lsm1-9 and lsm1-14 cells to bind RNA (Chowdhury and Tharun 2008) and the coprecipitation of mRNA with the Lsm1-9p and Lsm1-14p proteins (Fig. 1B) suggest that the mutant complexes assembled in the lsm1-9 and lsm1-14 cells are able to bind mRNA in vivo. Second, the lsm1-8 and lsm1-9, 14 alleles that are severely impaired in the RNA binding ability of the Lsm1-7–Pat1 complex are also unable to cause dominant inhibition of mRNA decay when overexpressed in wild-type cells. Further, while the 3′-end protection defect (but not the mRNA decay defect) of the lsm1-9 and lsm1-14 mutants can be partly suppressed by overexpression of the corresponding lsm1 alleles, neither the mRNA decay defect nor the 3′-end protection defect of the lsm1-8 and lsm1-9, 14 mutants could be suppressed by such overexpression. These observations imply that the RNA binding ability of the (Lsm1-9p- and Lsm1-14p-containing) mutant complexes are important for the dominant inhibition of mRNA decay caused by the lsm1-9 and lsm1-14 alleles. Third, although the mutant complexes purified from the lsm1-9 and lsm1-14 cells retain considerable RNA binding activity, they are unable to recognize the oligo(A) tail, suggesting that the manner in which these complexes contact the RNA is different from that of the wild-type complex (Chowdhury and Tharun 2008). This is in synchrony with the idea that the mRNPs formed by the association of these mutant complexes with the mRNA in vivo are impaired in proceeding through the post-binding steps needed to support decapping because they are probably different from the “normal” mRNPs formed by the wild-type complex.
Overexpression of the lsm1-6 allele in wild-type cells does not cause inhibition of mRNA decay, although the Lsm1-7–Pat1 complex purified from the lsm1-6 mutant is able to bind RNA. This could be because the mRNPs formed in vivo by the Lsm1-6p-containing complex are able to go through the post-binding steps (needed to facilitate decapping) at least at a slower pace as suggested by the observation that the decay defect of the lsm1-6 mutant can be at least partly suppressed by overexpression of the lsm1-6 allele. Consistently, the mutant complex purified from lsm1-6 cells exhibits a binding preference for oligoadenylated RNA over unadenylated RNA similar to the wild-type complex (and unlike the mutant complexes, purified from the lsm1-9 and lsm1-14 cells).
An alternate model to explain the dominant inhibition of mRNA decay (caused by the lsm1-9 and lsm1-14 alleles) is that the mutant Lsm1p proteins (Lsm1-9p and Lsm1-14p) produced by the overexpression of the lsm1-9 and lsm1-14 alleles in wild-type cells sequester the other subunits of the Lsm1-7–Pat1 complex (e.g., Lsm2p–Lsm7p), making them unavailable to the wild-type Lsm1p and thereby reducing the level of the wild-type complex. However, such a model is not consistent with several observations. First, the lsm1-8 and lsm1-9, 14 alleles are unable to cause dominant inhibition of mRNA decay (upon overexpression in wild-type cells) although both of these mutants are not significantly affected in the complex integrity and their decay defects are not suppressible by overexpression of the corresponding lsm1 alleles (just like the lsm1-9 and lsm1-14 mutants). Second, the above model predicts that the availability of the subunits other than Lsm1p is limiting in vivo. However, the results presented in Figures 4B and 5 reveal that overexpression of Lsm1p is sufficient to elevate the level of the Lsm1-7–Pat1 complex, and therefore suggest that the other subunits of the Lsm1-7–Pat1 complex are not limiting under our experimental conditions. This is consistent with the observation that overexpression of Lsm1p leads to the mobilization of the other Lsm subunits (Lsm2p–Lsm7p) from the nucleus (at the expense of the Lsm2p-8p complex) to the cytoplasm (Spiller et al. 2007), suggesting that such mobilization could lead to an increase in the levels of the Lsm1-7–Pat1 complex in the cytoplasm. Further, studies in human cells reveal that overexpression of hLSM1 (also known as CaSm) alone is sufficient to cause malignant transformation (Kelley et al. 2003) by affecting the mRNA decay profile of the cells (Fraser et al. 2005; Streicher et al. 2007). Finally, none of the various mRNA decay defective lsm1 alleles that we have (Tharun et al. 2005) other than lsm1-9 and lsm1-14 are able to cause inhibition of mRNA decay when overexpressed in wild-type cells (data not shown).
We showed earlier that the mutant complexes purified from the lsm1-9 and lsm1-14 cells are unable to recognize the presence of the 3′-oligo(A) tail of the RNA (Chowdhury and Tharun 2008). Unlike the wild-type complex, these mutant complexes fail to show an enhanced affinity for the oligoadenylated RNAs over the unadenylated and polyadenylated RNAs. However, they do bind to the oligoadenylated RNA with a considerable affinity that is comparable to their affinity for the unadenylated and polyadenylated RNAs. Consistent with this, oligoadenylated mRNA does coprecipitate with Lsm1-9p and Lsm1-14p from the mutant cell lysates although at a lower efficiency compared with the coprecipitation with wild-type Lsm1p pulled down from the wild-type cell lysate (Fig. 1B). Thus, the ability of the mutant complexes to bind to the oligoadenylated mRNA is not completely abolished in the lsm1-9 and lsm1-14 cells. These observations and the results presented in Figures 3 and 4 therefore suggest that the major reason for the mRNA decay defect in the lsm1-9 and lsm1-14 mutants is the inability of the mutant Lsm1-7–Pat1 complexes in these cells to facilitate the post-binding events, although the decreased association of the mutant complexes with the oligoadenylated mRNA may also partly contribute to the mRNA decay defect. In any case, since most of the mRNA is present in polyadenylated form in the wild-type cell lysate but exists in oligoadenylated form in the lsm1-9 and lsm1-14 cell lysates (due to the block to decapping in these mutants), the coprecipitation of oligoadenylated mRNA with the wild-type Lsm1p, but not the mutant Lsm1-9p and Lsm1-14p proteins, is a likely reflection of the binding preference of the Lsm1-7–Pat1 complex for oligoadenylated messages (Fig. 1B).
The tertiary structure of yeast Lsm1p modeled using the homology of yeast Lsm1p with the other Sm-like proteins of known tertiary structures allowed the prediction of two sets of residues (one in each of the loops 3 and 5 of the modeled Lsm1p structure) as forming the RNA contacting surfaces of the Sm domain of Lsm1p. One of these two sets of residues is mutated in each of the lsm1-9 and lsm1-14 alleles (Tharun et al. 2005). The observation that the mutant Lsm1-7–Pat1 complexes of the lsm1-9 and lsm1-14 cells retain considerable RNA binding ability is therefore suggestive of the existence of some functional redundancy between these two RNA contacting surfaces (Chowdhury and Tharun 2008). Consistently, mutating both RNA binding surfaces (as in the lsm1-9, 14 allele) results in almost complete loss of RNA binding ability of the Lsm1-7–Pat1 complex. Interestingly, the mutant complex purified from the lsm1-8 cells is also severely impaired in RNA binding, although the predicted RNA binding residues are intact in this allele. This could be due to the mutation of the D72 residue in the lsm1-8 allele (Tharun et al. 2005). The corresponding aspartic acid residue in the human Sm-B protein and in the archaebacterial AF-Sm1 protein (D35 in both proteins) is involved in fixing the orientation of the highly conserved Asparagine of the loop 3 RNA binding surface (N39 in Sm-B and AF-Sm1 and N76 in yeast Lsm1p) (Kambach et al. 1999; Toro et al. 2001). Therefore, the inability of the Lsm1-8p-containing complex to bind RNA could at least partly be due to the misorientation of the RNA binding residues in three-dimensional space. On the other hand, it is also possible that the residues R69 and D72 (which are mutated in lsm1-8) play a more direct role in RNA binding.
The Lsm1-7–Pat1 complex is also involved in mRNA 3′-end protection in addition to mRNA decay. However, the relationship between these two functions is not known. Comparison of the various lsm1 mutants reveals that the severity of their defects in mRNA decay and 3′-end protection go hand in hand suggesting that the initial event(s), including the binding of the Lsm1-7–Pat1 complex, is (are) common in both processes (Tharun et al. 2005). In this regard, the studies on the lsm1-9 and lsm1-14 alleles presented here allow us for the first time to separate these two functions of this complex. As shown in Figure 3, only the 3′-end protection defect but not the mRNA decay defect of the lsm1-9 and lsm1-14 mutants could be partly suppressed by overexpression of the corresponding lsm1 alleles. Further, overexpression of lsm1-9 and lsm1-14 in wild-type cells inhibits mRNA decay but does not seem to impair 3′-end protection significantly (Fig. 4). These results suggest that 3′-end protection directly results from the binding of the Lsm1-7–Pat1 complex to the mRNA while activation of decapping requires such binding to be followed by additional post-binding events. Consistent with this, the 3′-end protection defect of the lsm1-8 and lsm1-9, 14 mutants, which are severely affected in the RNA binding activity of the Lsm1-7–Pat1 complex, is not suppressible by overexpression of the corresponding lsm1 alleles.
Overall, the observations presented here suggest that the activation of decapping by the Lsm1-7–Pat1 complex involves at least two steps, namely, the binding of the Lsm1-7–Pat1 complex to the mRNA and the facilitation of the post-binding events. However, the binding event per se may be sufficient for the protection of mRNA 3′-ends from trimming. Our results suggest that the lsm1-9 and lsm1-14 mutants are able to get past the first (binding) step (at least when the corresponding lsm1 alleles are overexpressed) but are blocked in the second step (post-binding). On the other hand, the lsm1-8 and lsm1-9, 14 mutants seem to be severely impaired in the first step itself. Thus, although both of these two pairs of mutants are defective in mRNA decay, they seem to be differently affected in the different steps of decapping. The mechanism by which the decapping enzyme is stimulated by the Lsm1-7–Pat1p complex is not understood and elucidation of the post-binding events could enable us reveal such mechanisms.
MATERIALS AND METHODS
Yeast strains and plasmids
Strains used in this study are in the genetic background of yRP841 (Hatfield et al. 1996). The lsm1Δ (yRP1365) strain has been previously described (He and Parker 2001). Strains needed for the purification of the different Lsm1-7–Pat1 complexes were generated by introducing the CEN or 2μ vectors carrying the FLAG-tagged versions of the different alleles of LSM1 into yST247 [MAT α, leu2, ura3, lys2, trp1, cup1ΔrLEU2(PM), lsm1ΔrTRP1, LSM5-6xHis-NEOr (Chowdhury et al. 2007)]. Such CEN vectors were pST17 (Chowdhury et al. 2007), pST66 (Chowdhury and Tharun 2008), pST63, pST65, and pST155 for the alleles LSM1, lsm1-9, lsm1-6, lsm1-8, and lsm1-9, 14, respectively. pST17 was used as the template for making pST63 and pST65 while pST66 served as the template for making pST155 via the QuikChange mutagenesis procedure (Stratagene). The 2μ vectors used for the purification of the Lsm1-9p- and Lsm1-9, 14p-containing complexes, pST196 and pST188, respectively, were made by transferring the NotI inserts from pST66 and pST155 into the NotI site of pRS426 (Sikorski and Hieter 1989). The 2μ plasmids expressing untagged LSM1, lsm1-6, lsm1-8, lsm1-9, and lsm1-14 alleles (pST70, pST54, pST56, pST57, and pST59, respectively) were also made in a similar way by transferring the NotI inserts from the corresponding previously described CEN vectors (Tharun et al. 2005) to pRS426. The wild-type FLAG–LSM1 strain used for the experiment in Figure 4 was made by transforming yRP1365 with pST122. This plasmid was generated by transferring the NotI-insert from pST17 (Tharun et al. 2005) to pRS317 (Sikorski and Hieter 1989). Strains needed for the immunoprecipitation experiment shown in Figure 1B were made by introducing CEN vectors expressing the different FLAG–lsm1 alleles into the lsm1Δ strain. The LSM1 alleles carried by all the plasmids used in this study contained the native flanking sequences of LSM1.
Protein purification, immunoprecipitation, and RNA analyses
Purification of the mutant and wild-type complexes was carried out as described before (Chowdhury et al. 2007; Tharun 2008). Radiolabeled RNA substrates for gel shift assays were made by in vitro transcription using T7 RNA polymerase, oligonucleotide templates, and [α-32P]UTP as described earlier (Chowdhury et al. 2007) with the help of Mirvana kit (Ambion). Purification of the substrate RNAs, RNA binding reactions, and gel shift analyses were done as described (Chowdhury et al. 2007). In all the experiments, substrate RNA was heat denatured before it was included in the binding reaction. RNA substrates whose binding affinities are compared in an experiment (e.g., PGK1 and PGK1-A5 RNAs) were prepared and tested together. BSA was used at final 147 nM concentration in the control reactions. Northern analyses, quantitation of the relative levels of the total and trimmed poly(G) fragment and full-length MFA2pG mRNA, Western analysis for FLAG–Lsm1p, and immunoprecipitations were done as described before (Tharun and Parker 1999, 2001; Tharun et al. 2000, 2005). Efficiency of coprecipitation of the MFA2pG mRNA with Lsm1p was worked out by comparing the amounts of MFA2pG mRNA (quantitated from the Northern blot using a PhosphorImager) in the input lysate and the immunoprecipitate (Tharun and Parker 2001).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by USUHS intramural grant (C071HJ) and NIH grant (GM072718) to S.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1650109.
REFERENCES
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006a;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006b;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Tharun S. lsm1 mutations impairing the ability of the Lsm1p-7p–Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA. 2008;14:2149–2158. doi: 10.1261/rna.1094208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p–Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes & Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fillman C, Lykke-Andersen J. RNA decapping inside and outside of processing bodies. Curr Opin Cell Biol. 2005;17:326–331. doi: 10.1016/j.ceb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Watson PM, Fraig MM, Kelley JR, Nelson PS, Boylan AM, Cole DJ, Watson DK. CaSm-mediated cellular transformation is associated with altered gene expression and messenger RNA stability. Cancer Res. 2005;65:6228–6236. doi: 10.1158/0008-5472.CAN-05-0650. [DOI] [PubMed] [Google Scholar]
- Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Peterson B, Tomasevic N, Peculis BA. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol Cell. 2004;13:817–828. doi: 10.1016/s1097-2765(04)00127-3. [DOI] [PubMed] [Google Scholar]
- Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen HL, Llewellyn SA, Barnes CA. Initiation-mediated mRNA decay in yeast affects heat-shock mRNAs, and works through decapping and 5′-to-3′ hydrolysis. Nucleic Acids Res. 2003;31:4006–4016. doi: 10.1093/nar/gkg474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kelley JR, Fraser MM, Hubbard JM, Watson DK, Cole DJ. CaSm antisense gene therapy: A novel approach for the treatment of pancreatic cancer. Anticancer Res. 2003;23:2007–2013. [PubMed] [Google Scholar]
- Khanna R, Kiledjian M. Poly(A)-binding–protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5′ degradation in yeast require the Lsm2-8p complex. Mol Cell Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Kiledjian M. Decapping the message: A beginning or an end. Biochem Soc Trans. 2006;34:35–38. doi: 10.1042/BST20060035. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Ramirez CV, Vilela C, Berthelot K, McCarthy JE. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol. 2002;318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E, Camier S, Seraphin B. New insights into the control of mRNA decapping. Trends Biochem Sci. 2006;31:241–243. doi: 10.1016/j.tibs.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Spiller MP, Reijns MA, Beggs JD. Requirements for nuclear localization of the Lsm2-8p complex and competition between nuclear and cytoplasmic Lsm complexes. J Cell Sci. 2007;120:4310–4320. doi: 10.1242/jcs.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE–mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104–2114. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Peculis BA. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res. 2008;36:6021–6034. doi: 10.1093/nar/gkn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. Purification and analysis of the decapping activator Lsm1p-7p–Pat1p complex from yeast. Methods Enzymol. 2008;448:41–55. doi: 10.1016/S0076-6879(08)02603-7. [DOI] [PubMed] [Google Scholar]
- Tharun S. Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int Rev Cell Mol Biol. 2009;272:149–189. doi: 10.1016/S1937-6448(08)01604-3. [DOI] [PubMed] [Google Scholar]
- Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′ end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Vilela C, Velasco C, Ptushkina M, McCarthy JE. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 2000;19:4372–4382. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Gao M, Jones CL, Wilusz J, Peltz SW. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]