Abstract
Precise temporal control is needed for RNA viral genomes to translate sufficient replication-required products before clearing ribosomes and initiating replication. A 3′ translational enhancer in Turnip crinkle virus (TCV) overlaps an internal T-shaped structure (TSS) that binds to 60S ribosomal subunits. The higher-order structure in the region was examined through alteration of critical sequences revealing novel interactions between an H-type pseudoknot and upstream residues, and between the TSS and internal and terminal loops of an upstream hairpin. Our results suggest that the TSS forms a stable scaffold that allows for simultaneous interactions with external sequences through base pairings on both sides of its large internal symmetrical loop. Binding of TCV RNA-dependent RNA polymerase (RdRp) to the region potentiates a widespread conformational shift with substantial rearrangement of the TSS region, including the element required for efficient ribosome binding. Degrading the RdRp caused the RNA to resume its original conformation, suggesting that the initial conformation is thermodynamically favored. These results suggest that the 3′ end of TCV folds into a compact, highly interactive structure allowing RdRp access to multiple elements including the 3′ end, which causes structural changes that potentiate the shift between translation and replication.
Keywords: RNA conformational switches, RNA virus replication, cap-independent translation, translational enhancers, Turnip crinkle virus
INTRODUCTION
One of the first proteins synthesized from viral RNA genomes is the RNA-dependent RNA polymerase (RdRp), which together with other early translation products and/or host proteins form the viral replicase (Ahlquist et al. 2003; Shi and Lai 2005; Nagy and Pogany 2006). Translation of sufficient quantities of the RdRp, either as a separate entity or as part of a polyprotein, must occur prior to the genome's transition to a replication template. If the viral genome contains a 5′ m7GpppG-cap, translation is thought to proceed in a canonical fashion, which requires the small 40S ribosomal subunit and associated factors to recognize the 5′ cap through intermediary translation factors, and then scan the RNA in the 3′ direction for the first initiation codon in a good context (Kozak 1999). The small subunit is joined by the large 60S subunit to form the 80S ribosome, followed by initiation of translation (Preiss and Hentze 2003; Marintchev and Wagner 2004). RNA viruses that do not contain a 5′ cap must recruit ribosomes internally, which for animal viruses involves extensive, highly structured elements in the 5′ untranslated region (UTR) known as internal ribosome entry sites (IRESes) (Hellen and Sarnow 2001; Merrick 2004). 3′ UTR sequences, in addition to containing elements for replicase assembly and initiation of minus-strand synthesis (Dreher 1999; Panaviene et al. 2005), can influence translation mediated by 5′ IRES (Isken et al. 2003; Isken et al. 2004; Song et al. 2006), possibly through association with viral and/or host RNA-binding proteins (Edgil and Harris 2006). Efficient translation of plant virus RNA genomes that lack a 5′ cap and have limited 5′ UTR requires critical 3′ UTR elements known as 3′ cap-independent translation elements (CITEs), whose mechanism of action remains unclear (Dreher and Miller 2006; Kneller et al. 2006; Miller et al. 2007). Since translation and replication are mutually exclusive processes due to opposing directions of protein and RNA synthesis on the viral template, the proximal positioning of 3′ replication elements with 3′ CITE and other 3′ translation-required elements may allow for a coordinated transition from translation to replication, which requires the clearing of ribosomes from the template (Barton et al. 1999).
Turnip crinkle virus (TCV) (4054 nucleotides [nt]), a member of the genus Carmovirus in the family Tombusviridae, has a single, uncapped genomic RNA and a 3′ end that lacks a poly(A) tail. TCV (all references to TCV refer to the single genomic RNA) contains a 3′ CITE that synergistically enhances translation of reporter constructs in the presence of the viral 5′ UTR in vivo (Qu and Morris 2000; Yoshii et al. 2004; Stupina et al. 2008). The core 3′ CITE was recently mapped to a region between positions 3810 and 3951 (Stupina et al. 2008), which partially overlaps a 100-nt functional domain computationally predicted to fold into a T-shaped structure (TSS) (McCormack et al. 2008) that has recently been confirmed by NMR-SAXS (Y Wang, unpubl.). The 3′ internal TSS binds to 80S ribosomes at the P-site through the 60S ribosomal subunit, and mutations that disrupt ribosome binding negatively impact translation (Stupina et al. 2008).
The TCV TSS is composed of three hairpins (H4a, H4b, H5) enclosed by two pseudoknots (Fig. 1E, Ψ3,Ψ2). Ψ3 is an H-type pseudoknot formed by interaction between H4a terminal loop sequences and a short upstream region (McCormack et al. 2008). H5 contains a large internal symmetrical loop (LSL) whose 3′ side pairs with 3′ terminal sequence to form phylogenetically conserved Ψ1 (Zhang et al. 2004, 2006; Na and White 2006). The 5′ side of the LSL was suggested to pair with uridylates within the terminal loop of upstream hairpin H4 (termed Ψ4), based on how alterations that disrupted or reformed base pairs affected ribosome binding to a fragment containing H4 through the TSS (H4 → TSS) (Stupina et al. 2008). Downstream from the TSS is the 3′ terminal hairpin Pr, which functions as the core promoter for minus-strand synthesis (Song and Simon 1995; Sun and Simon 2006). The asymmetric and terminal loops of H4 are required for robust translation (Stupina et al. 2008), and H4 and flanking adenylates enhanced Pr-mediated transcription using purified recombinant TCV RdRp in vitro (Sun and Simon 2006).
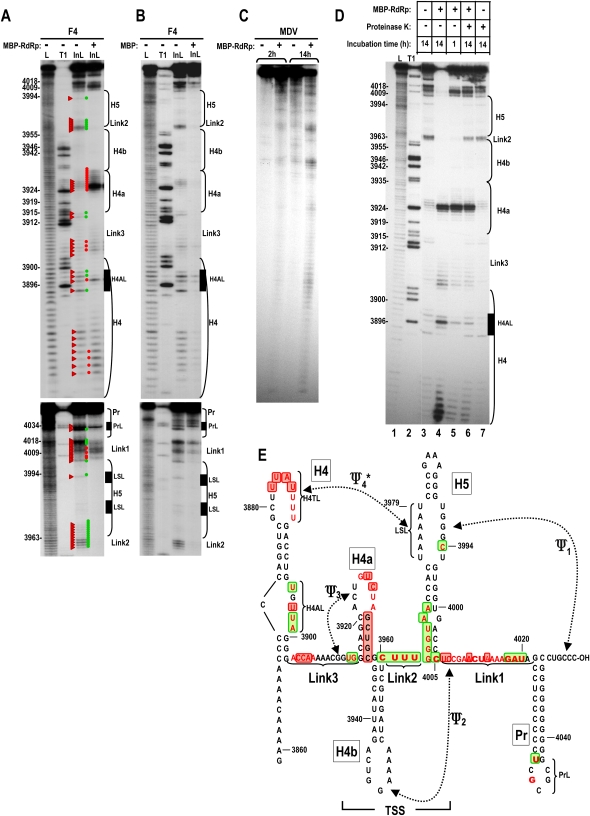
FIGURE 1.
RdRp interaction with the TCV 3′ end causes a widespread, reversible conformational shift. (A) In-line probing of wt fragment F4 (F4) with and without added TCV RdRp fused to MBP (MBP–RdRp). End-labeled F4 (positions 3859–4054) was incubated for 14 h in the absence of MBP–RdRp or 1 h in the presence of MBP–RdRp and fragments analyzed on a denaturing 8% polyacrylamide gel. L, ladder generated following treatment with NaOH; T1, ladder generated following partial fragment denaturation and treatment with RNase T1, which cleaves at guanylates. InL, in-line cleavage products. Base numbering is at left of the panel and positions of hairpins, subregions within hairpins, and linker segments are noted to the right, with the locations within F4 shown in E. Red triangles point to cleavage products in the absence of MBP–RdRp. Red and green dots denote stronger or weaker cleavages following MBP–RdRp addition, respectively. Lower panel is a longer run to better examine regions more proximal to the 3′ end. (B) Same as A except that MBP was added in place of MBP–RdRp. (C) In-line probing of MDV RNA, a subviral RNA associated with Qβ bacteriophage that is not a template for the TCV RdRp. 5′ end-labeled template was subjected to in-line cleavage for 2 or 14 h in the presence or absence of MBP–RdRp. (D) In-line cleavage of F4 in the presence and absence of MBP–RdRp and treatment with proteinase K. Lane 1, ladder generated following treatment with NaOH; lane 2, ladder generated following partial fragment denaturation and treatment with RNase T1; lane 3, 14-h incubation without MBP–RdRp; lane 4, 14-h incubation in the presence of MBP–RdRp; lane 5, 1-h incubation in the presence of MBP–RdRp; lane 6, 1-h incubation in the presence of MBP–RdRp followed by treatment with proteinase K and additional 13-h incubation; lane 7, 14-h incubation with proteinase K. Note that lane 6 contains all cleavage products from both lanes 3 and 5. (E) Location of the hairpins, pseudoknots, and subregions within hairpins on fragment F4. Bases in red are susceptible to in-line cleavages as shown in A. Bold bases are more strongly susceptible to cleavage. Residues boxed in red and green display enhanced or reduced cleavages, respectively, in the presence of RdRp. The region that forms the 3D T-shaped structure is denoted TSS. Ψ4 is marked by an asterisk; it was unknown prior to this study if this pseudoknot forms in fragment F4 in the presence of Ψ1.
TCV was equally infectious when the sequence between Ψ3 and the 3′ end was exchanged with the analogous region from the related virus Cardamine chlorotic fleck virus (CCFV) (65% sequence similarity) unlike exchanges of most shorter fragments within this region (McCormack et al. 2008). This indicated that the region between Ψ3 and the 3′ end represents a functional domain. Since H4 and the linker sequence between H4 and H4a (Link3) are virtually identical between TCV and CCFV (unlike sequences further upstream), the functional domain likely extends from H4 through the 3′ end (H4 → 3′ end).
For this study, we have determined that binding of RdRp to H4 → 3′ end potentiates a widespread conformational shift in RNA structure that includes a substantial rearrangement of the Ψ3–H4a region, which was reversible when RdRp levels were reduced. Mutations that disrupted pseudoknots or were located within internal or terminal loop regions in the H4 → 3′ end domain had different effects on RNA structure, either before or after RdRp binding, some of which strikingly affected RdRp-directed transcription. Alterations of critical regions revealed a novel interaction between internal and terminal loops of H4 and the H5 LSL, which sets up a complex multistrand motif required for normal minus-strand synthesis. Our results suggest a tight structural organization of RNA elements in the 3′ region and support a model where newly translated RdRp binds to the 3′ end triggering reorganization of structures necessary for efficient ribosome binding, causing a transient switch from translation to replication of the viral genome.
RESULTS
Binding of RdRp causes an extensive conformational shift in the structure of a fragment containing H4 → 3′ end
We previously used in-line probing to assess the structure of TCV fragment F4, which contains the H4 → 3′ end domain (McCormack et al. 2008). In-line probing reports on spontaneous cleavages in the RNA backbone mediated by nucleophilic attack of 2′ hydroxyl that are spatially “in-line” with oxyanion leaving groups of backbone phosphates. Nonstructured RNA residues are more susceptible to backbone cleavages since nucleotides are not torsionally constrained by hydrogen bond pairings (Winkler et al. 2002). Results from this previous study were consistent with the presence of Ψ1, Ψ3, H4a, H4b, most of H5, and Pr (McCormack et al. 2008).
To determine if RdRp binding to the region causes a conformational change in the RNA that might disrupt ribosome binding to the TSS, fragment F4 was subjected to in-line probing in the presence and absence of purified recombinant TCV RdRp. The RdRp (p88), which is fused to maltose binding protein (MBP) at the N terminus and prepared in Escherichia coli, is capable of specific recognition of TCV RNA templates and transcription of complementary strands in vitro (Rajendran et al. 2002). In the absence of RdRp, few cleavages were detected after 1 h of incubation, while approximately half of the fragments had at least one cleavage after 14 h (Fig. 1A; data not shown). In contrast, addition of RdRp caused both enhanced and reduced cleavages at a number of sites throughout the region, with a strong pattern emerging after only 1 h (Fig. 1A). By 14 h, most fragments had multiple cleavages, resulting in mainly shorter products (see Fig. 1D, lane 4). Since the RdRp used in the assay was fused to MBP, the nonviral portion of the protein was examined for independent effects on the structure of F4. MBP expressed from the same vector and prepared in the identical fashion was added to F4 and the fragment incubated for 1 h. The cleavage pattern in the presence of MBP was virtually identical, although reduced in intensity, to the 14 h cleavage pattern in the absence of protein (Fig. 1B). This indicates that the altered susceptibility of F4 to in-line cleavage in the presence of RdRp was not due to the MBP portion of the fusion protein. We also tested the effect of the RdRp on a small, nontemplate RNA (MDV) that is associated with Qβ bacteriophage. At 2 h, the cleavage pattern was similar in the presence and absence of RdRp, while at 14 h, slightly more cleavages were evident in the sample containing RdRp, but the pattern remained similar (Fig. 1C). The cleavage susceptibility of residues in the 3′ UTR of Saguaro cactus virus, which is also a nontemplate for the TCV RdRp, was similarly unaffected by addition of TCV RdRp (data not shown). These controls indicate that altered susceptibility of F4 to cleavage following addition of RdRp was not due to either the MBP portion of the fusion protein or a nonspecific contaminating RNase.
Comparison of the cleavage pattern at 14 h in the absence of RdRp to 1 h in the presence of RdRp indicates that the enzyme potentiates a substantial rearrangement of the RNA (Fig. 1A,E). Major differences include (from 5′ to 3′): (1) enhanced susceptibility of four residues in the H4 terminal loop (H4TL) and altered susceptibility of four of five residues in the H4 asymmetric internal loop (H4AL); (2) enhanced cleavages of positions 3904–3906 in the linker between H4 and H4a (Link3); (3) reduced cleavage of positions U3914 and G3915 at the base of H4a; (4) enhanced cleavages in the H4a loop and 3′ stem residues (particularly residues U3925 and C3926); (5) reduced susceptibility in the short linker (Link2) upstream of H5; (6) reduced susceptibility of all residues cleaved in H5, suggesting that the conformational shift may have stabilized the H5 lower stem; (6) altered cleavages in seven positions in the linker between H5 and Pr (Link1), including a striking reduction in cleavage of positions 4018–4020 and two slightly enhanced cleavages at positions 4006 and 4007 in Ψ2; and (7) loss of the strong cleavage at U4032, which is adjacent to the terminal loop of Pr (PrL). Additional cleavages downstream from position 4034 in PrL in wild-type (wt) and mutant F4 were inconsistent and thus will not be evaluated further. Residues with reduced susceptibility to cleavage could represent either bases that are newly paired or bases that are protected by direct interaction with the RdRp. The altered susceptibility to in-line cleavage of a substantial number of residues in the presence of the TCV RdRp indicates that RdRp interaction with F4 likely affects the structure of both secondary and tertiary elements from H4 through Pr.
The conformational shift mediated by the RdRp is reversible
Some of the most striking differences in RNA structure upon RdRp binding are in the Ψ3/H4a region (Fig. 1A,E). This region is a key constituent of the TCV 3′ CITE and is important for both translational enhancement and 60S ribosomal subunit binding to the TSS (Stupina et al. 2008). The conformational shift induced by RdRp binding could therefore disrupt translation and promote the switch to transcription (see Discussion). If this hypothesis is correct, then the question arises as to whether the conformational switch induced by the RdRp is permanent, i.e., the template is incapable of reverting to a form compatible with translation when RdRp levels are reduced. Alternatively, if the new RNA conformation is metastable and requires RdRp for maintenance, then reduction in local RdRp levels would cause a reversion to the initial structural form, which should allow translation to resume if the RNA is not sequestered from translational components. This could result in continuous, cyclical translation of RdRp from the initial infecting RNA.
To test if the conformational switch is reversible, 5′ labeled F4 was incubated with RdRp for 1 h followed by treatment with proteinase K to degrade the RdRp, and then incubation continued for an additional 13 h. If the RdRp-mediated conformation is irreversible, then the cleavage pattern should resemble the 14-h pattern generated when RdRp is present (exceptions would be residues masked by RdRp binding, which would be newly susceptible in the absence of RdRp). If the switch is reversible, then the cleavage pattern should be composed of the 1 h RdRp-present pattern together with the 13-h pattern found in the absence of RdRp.
Addition of proteinase K in the absence of added RdRp had no effect on the cleavage pattern of F4 (Fig. 1D, lane 7). Incubation of F4 with RdRp and then treatment with proteinase K followed by additional incubation resulted in a cleavage pattern that was a clear combination of the 1 h RdRp pattern and the 14 h RdRp-free pattern (Fig. 1D, cf. cleavages in lanes 6 and combined cleavages in lanes 3,5). For example, F4 with RdRp and proteinase K treatment contained the RdRp-absent cleavage at position 4005 and the RdRp-specific cleavages at positions 4006 and 4007. F4 with RdRp and proteinase K treatment sample also contained the RdRp-absent cleavages in Link2 and the strong RdRp-specific cleavages in the loop of H4a. This result strongly suggests that the conformation of F4 in the presence of RdRp reverts to the initial conformation when RdRp is removed.
Disruption of Ψ3 affects the structure locally and distally in the Pr loop but does not affect the RdRp-mediated conformational shift
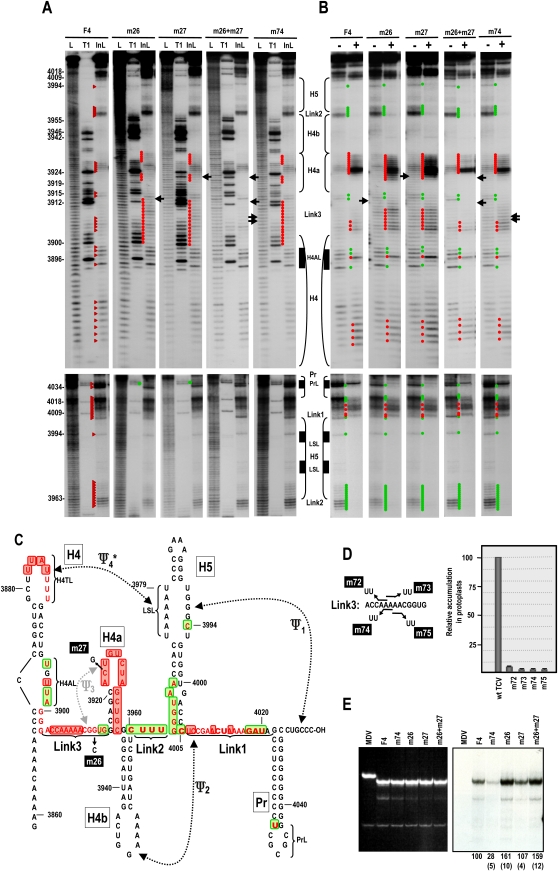
Ψ3/H4a, H4b, and Ψ2 form a series of parallel stacked helices comprising the upper portion of the “T” in the TSS (Fig. 7B; McCormack et al. 2008). Disrupting Ψ3, which mimics the amino-acceptor stem of a tRNA, reduced both ribosome binding and translation (Stupina et al. 2008), and reduced TCV accumulation to barely detectable levels (McCormack et al. 2008). All of these deficiencies underwent significant restoration when Ψ3 was reconstituted by compensatory mutations. To determine if disruption of Ψ3 affects the distinctive structural changes caused by the RdRp in the region and/or minus-strand synthesis, previously described mutations m26 and m27 were engineered into F4 (Fig. 2C). F4 transcripts containing either m26 or m27 (F4 + m26, F4 + m27) had identical in-line cleavage patterns, with new cleavages in H4a and in residues upstream of H4a that normally participate in the pseudoknot (Fig. 2A,C). Unexpectedly, residues in Link3 upstream of Ψ3 had enhanced cleavages that extended into the lower stem of H4, suggesting cross-talk between this A-rich region and Ψ3/H4a. A conformational change was also evident in PrL, with the absence of the normally strong cleavage at G4034 in F4 + m26 and F4 + m27. No discernable differences with the wt cleavage pattern were found in the remainder of H4, or H5, Link2 or Link1. Transcripts containing both m26 and m27 (F4 + m26 + m27), which reforms Ψ3 (McCormack et al. 2008), had a cleavage pattern that was indistinguishable from wt (Fig. 2A).
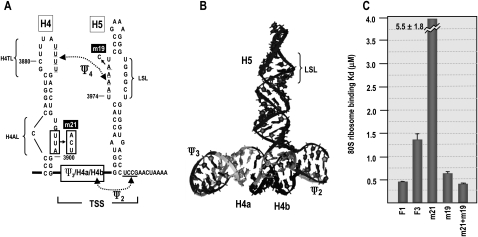
FIGURE 7.
Mutations in H4AL strengthen the stability of Ψ4. (A) Location of mutations on a simplified 3′ structure. (B) RNA2D3D structure of the TSS (McCormack et al. 2008), showing the location of the H5 LSL region. Location of hairpins and pseudoknots are noted. Formation of Ψ4 is predicted to affect ribosome recognition of the TSS. (C) Ribosome binding to fragment F1 (positions 3900–4016, contains Link3 → TSS), F3 (positions 3859–4016, contains H4 → TSS), and F3 containing mutations described in A. Two to 100 pmol of [32P] 5′-end labeled wt and mutant TCV transcripts were combined with 25 pmol of yeast 80S ribosomes and bound RNA was detected following filter binding. Dissociation constants were calculated from three independent experiments. Standard error bars are shown.
FIGURE 2.
Disrupting Ψ3 alters adjacent adenylates and PrL but not the overall RdRp-mediated conformational shift. (A) In-line probing of F4 containing mutations that disrupt Ψ3 (m26, m27), reform Ψ3 (m26 + m27), or alter two of the adenylates just upstream of Ψ3 (m74). Locations of the mutations are shown in C and D. Red triangles denote cleavage products in wt F4. Red and green dots denote residues in mutant F4 with enhanced or reduced cleavages compared with wt F4, respectively. Arrows point to locations of the altered bases. See Figure 1A legend for additional details. At least two independent in-line probing assays were performed for all mutants in this report, and only reproducible differences from wt are noted. (B) Structural alterations in the presence of MBP–RdRp; “−” and “+” denote no and added MBP–RdRp, respectively. Red and green dots denote residues that show enhanced or reduced cleavage, respectively, for each fragment in the presence of MBP–RdRp compared with in the absence of MBP–RdRp. (C) Location of the hairpins, pseudoknots, and subregions within hairpins on fragment F4. Bases in red are susceptible to in-line cleavages in F4 + m26 or F4 + m27. Bold bases are more strongly susceptible to cleavage. Residues boxed in red and green display enhanced or reduced cleavages, respectively, in the presence of MBP–RdRp. F4 + m74 had similar cleavage patterns to F4 + m26 or F4 + m27 in the presence and absence of MBP–RdRp, except for the PrL region, which was wt. (D) Left, location of mutations constructed in the Link3 adentylates. Right, accumulation of full-length TCV containing these mutations in protoplasts at 40-h post-inoculation (hpi). Results are averages of three independent assays. Bars denote standard deviation. (E) In vitro transcription of wt and mutant F4 using MBP–RdRp and radioactive nucleotides. Ethidium bromide stained gel is at the left and autoradiograph is at the right. Relative, normalized values from at least three independent experiments are given below lanes. Standard deviation is in parentheses. Qβ-associated MDV RNA is not a template, and is used as a control for specificity of the RdRp.
To further explore the relationship between the adenylates upstream of Ψ3 and the pseudoknot, mutations that converted pairs of adenylates to uridylates were generated in full length TCV and F4 to examine effects on virus accumulation, RNA structure, and template activity. We previously determined that virus accumulation, translation, and ribosome binding were substantially reduced when all five adenylates were converted to uridylates (Stupina et al. 2008). Similarly, no mutant TCV with pairs of adenylate mutations accumulated above 5% of wt TCV levels in protoplasts, indicating that at least the three central adenylates are required for efficient accumulation (Fig. 2D, right). In-line cleavage of F4 transcripts containing A3907U and A3908U (F4 + m74) revealed very light new cleavages in H4a at positions identical to those that were more substantially cleaved when Ψ3 was disrupted (Fig. 2A). This supports the possibility that these adenylates are contributing to the structure and stability of Ψ3. However, there was no discernable reduction in cleavage at position G4034 in PrL, suggesting that alterations in these adenylates and Ψ3 are not structurally equivalent.
In the presence of RdRp, F4 + m26 and F4 + m27 maintained the distinctive conformational changes in the region and elsewhere; however, all new cleavages within H4a and Link3 were more pronounced (Fig. 2B). F4 + m26 + m27 and F4 + m74 had the wt RdRp-mediated conformational changes. F4 + m26 and F4 + m26 + m27, were reproducibly slightly better templates for in vitro transcription by the TCV RdRp compared with wt F4 and F4 + m27 (Fig. 2E), suggesting that the substantial effect of these mutations on virus accumulation is due to disrupted translation (Stupina et al. 2008). F4 + m74 generated only 28% of wt levels of complementary strands in vitro, suggesting an additional role for these adenylates in viral replication. These results suggest that Ψ3 is associated with both upstream Link3 adenylates and PrL in the three-dimensional (3D) structure, and that the striking enhancement in in-line cleavage susceptibility of U3925 and C3926 in H4a in the presence of RdRp is independent of Ψ3.
Effect of disrupting Ψ1 on the structure of F4 and the RdRp-mediated conformational shift
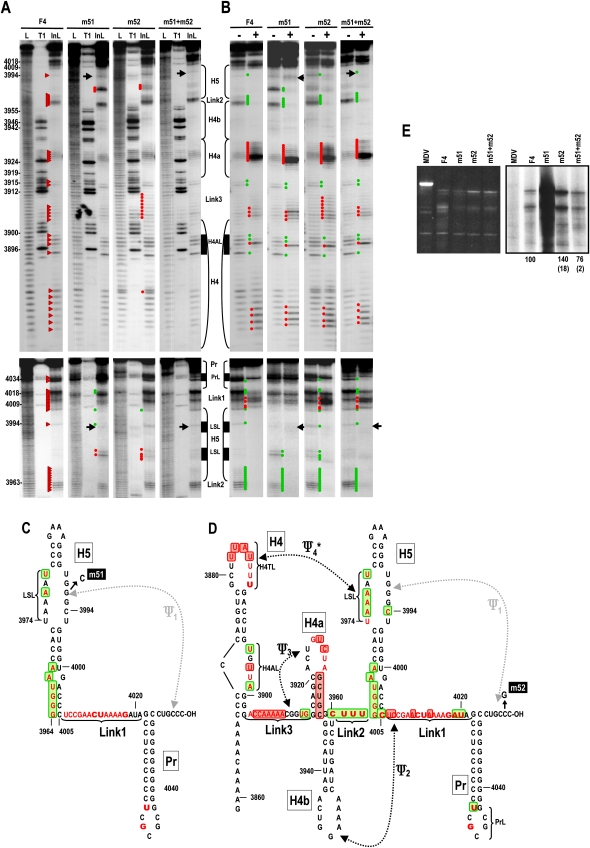
To determine how disruption of Ψ1 affects the RdRp-mediated conformational shift, three new alterations were generated that were designed to disrupt and reform Ψ1 (Fig. 3C,D). m51 is a G-to-C transversion in the H5 LSL at position 3992, m52 is a C-to-G transversion in the partner sequence at the 3′ end (position 4053), and m51 + m52 should reestablish the pseudoknot. In-line probing of F4 + m51 revealed new cleavages in the H5 LSL at positions U3979 and A3977, the latter of which is directly opposed to the m51 alternation (Fig. 3A,C). Five of six residues on the 5′ side of the H5 LSL in F4 + m52 also showed enhanced cleavage (the exception was A3978) (Fig. 3A,D). This supports our previous suggestion that noncanonical pairing or stacking of the bases across the symmetrical loop is affected by Ψ1 (McCormack et al. 2008). Position C4005 at the base of H5 was less susceptible to cleavage in F4 + m51 and F4 + m52, but not F4 + m51 + m52, a result consistent with our previous Ψ1 disruption (McCormack et al. 2008). This suggests that the G:C pair at the base of H5 is sterically weakened (i.e., residues are more flexible) by the presence of Ψ1.
FIGURE 3.
Mutations that disrupt Ψ1 have different effects on RNA structure, the RdRp-mediated conformational shift, and template activity for complementary strand synthesis. (A) In-line cleavage of transcripts containing mutations that disrupt (m51, m52) or reform (m51 + m52) Ψ1. Location of the mutations are shown in C and D. See the legend to Figure 2A for further details. Position U3888 appears as a double band here, and while the enhanced cleavage at this position was consistent, the doubling of the band was not. (B) Structural alterations in the presence of MBP–RdRp. See the legend to Figure 2B for further details. Note that the normal shift in the conformation of Link1 and PrL in the presence of MBP–RdRp is absent in F4 + m51. (C) Structural alterations due to m51 and (D) m52 in the presence and absence of MBP-RdRp. See the legend to Figure 2E for details. (E) In vitro transcription of wt and mutant F4. Ethidium bromide stained gel is at the left and autoradiograph is at the right. Relative, normalized values from at least three independent experiments are given below the lanes. Standard deviation is in parentheses. No values are presented for m51 because of the substantial, reproducible enhancement in transcription complicated value determination.
Unlike wt F4 and F4 + m52, F4 + m51 Link1 residues A4019 and U4020 were not susceptible to cleavage, suggesting that additional structural features are affected by modification of the H5 LSL that are not simply due to the loss of Ψ1. In addition, U3888 in the H4 terminal loop (H4TL) was reproducibly susceptible to additional cleavage in F4 + m52, as were additional residues in Link3. This suggested the possibility that the additional cleavages in the 5′ side of the H5 LSL signaled a disturbance of Ψ4 in these mutant transcripts (see below). In-line probing of F4 + m51 + m52 revealed restoration of the wt cleavage pattern throughout the transcript, suggesting that the unique structural alterations in F4 + m51 and F4 + m52 require the loss of Ψ1.
When RdRp was added to F4 + m51 and F4 + m51 + m52, no differences in the wt F4 conformational shift pattern were detectable upstream of Link1. However, the cleavage pattern in H4AL was reproducibly different in F4 + m52 in the presence of RdRp, with no reduction in cleavage intensity at U3898. The conformation of the Link1 and PrL regions also differed significantly between F4 + m51 and F4 + m52 in the presence of RdRp in that no conformational shift in these regions occurred for F4 + m51. In contrast, Link1 and PrL of F4 + m52 and F4 + m51 + m52 displayed wt structural changes upon RdRp binding, with the exception that the strong cleavage at position G4018 was maintained in F4 + m52. These differences between F4 + m51 and F4 + m52 following RdRp binding are consistent with the specific alterations having unique effects on RNA structure beyond simple elimination of Ψ1.
To determine if the unique structural effects in both the presence and absence of RdRp impact template activity for minus-strand synthesis, F4 + m51, F4 + m52, and F4 + m51 + m52 were combined with RdRp and levels of complementary strands determined (Fig. 2E). F4 + m51 was transcribed at a substantially higher level than wt F4. In contrast, F4 + m52 generated only 40% higher levels of complementary strands than wt F4, indicating that simple disruption of Ψ1 is not sufficient for the highly aberrant levels of transcription associated with F4 + m51. F4 + m51 + m52 generated wt-sized complementary strands at 76% of wt F4 levels, and thus enhanced transcription of F4 + m51 requires the absence of Ψ1 in addition to the specific m51 mutation.
Alteration of H4AL causes local and distal structural changes that abrogate transcription in vitro
The structure of H4TL and H4AL of H4 undergo distinctive conformational changes in the presence of RdRp. Previous results indicated that H4 may function in both translation and replication. For example, H4 and flanking upstream and downstream (Link3) A-rich sequences enhanced transcription from the TCV Pr promoter in vitro (Sun and Simon 2006), and alterations in H4TL and H4AL reduced translation by 40%–90% in vivo (Stupina et al. 2008). H4TL 3′ side uridylates were also shown to pair with H5 LSL 5′ side adenylates (forming Ψ4) in a fragment containing H4 → TSS (Stupina et al. 2008). As shown in this report and predicted in McCormack et al. (2008), non-Watson–Crick interactions across the LSL are affected by the presence of Ψ1, and thus Ψ1 could affect the participation of residues on the 5′ side of the LSL in Ψ4.
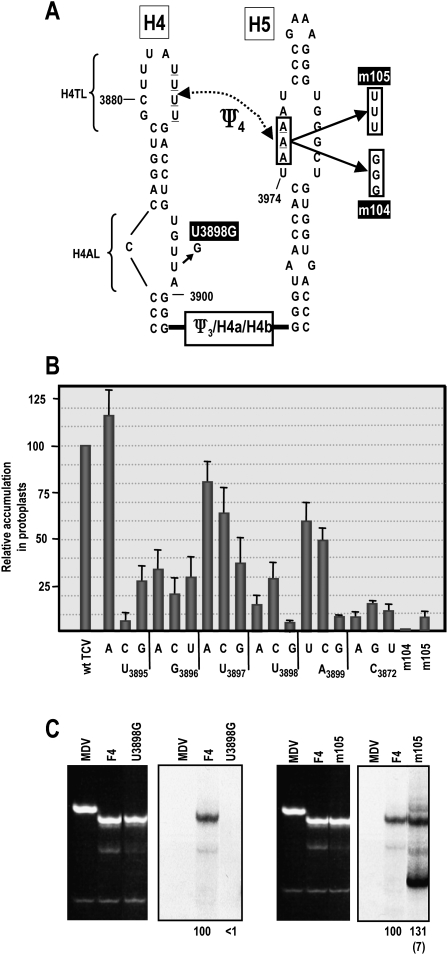
To better delineate the relationship between H4AL, H4TL, and H5 LSL sequences, we first systematically converted each residue of H4AL to the other three bases and assayed mutant TCV for accumulation in protoplasts. With one exception (U3895A), all alterations reduced TCV to between 3.6 and 54% of wt (Fig. 4B). The most significant reductions in TCV accumulation were caused by U3895C, U3898G, A3899G, and all alterations at C3872, the 5′ side single bulged base.
FIGURE 4.
Alteration of H4AL and H5 LSL affect virus accumulation in vivo and synthesis of complementary strands in vitro. (A) Location of specific mutations. The region between H4 and H5 is abbreviated for simplicity. (B) Mutations were constructed in full-length TCV at each position in H4AL and at a single location in the H5 LSL and assayed for effect on virus accumulation in protoplasts. Results are averages of three independent assays. Bars denote standard deviation. (C) In vitro transcription of wt and mutant F4 using MBP–RdRp. Ethidium bromide stained gel is at the left of the autoradiograph. Relative, normalized values from at least three independent experiments are given below lanes. Standard deviation is in parentheses. Only the wt-sized F4 + m105 product was included in the value shown.
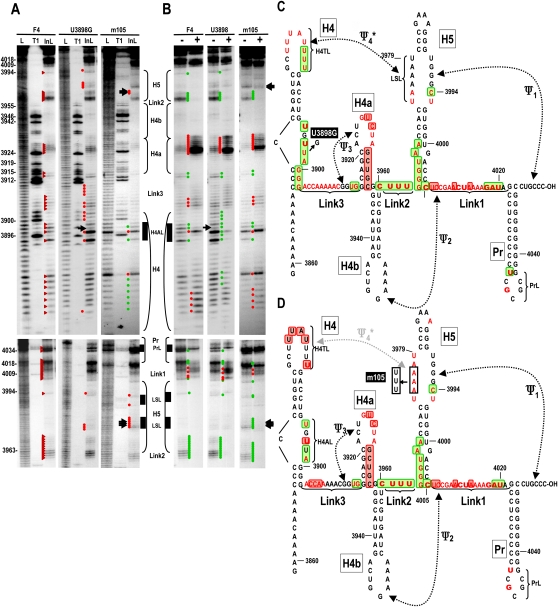
To explore if alterations in H4AL affect the structure and/or transcriptional activity of F4, U3898G (Fig. 4A), which reduced TCV accumulation to 3.6% of wt levels, was selected for further analysis. F4 + U3898G was a very poor template for the RdRp, with no detectable complementary strands synthesized (Fig. 4C, left panels). U3898G caused major changes to the in-line cleavage pattern of F4 in the absence or presence of RdRp, despite the susceptibility of U3898 to cleavage in wt F4 that suggested a flexible configuration (Fig. 5A). U3898 transcripts contained altered cleavage levels within H4AL, with U3895 and U3897 becoming strongly susceptible to cleavage, and moderate cleavages at the remaining positions including G3898 (Fig. 5A,C). Additionally, the three guanylates in the lower stem of H4 and most of the flanking Link3 residues displayed new or stronger cleavages. U3898G also affected the terminal loop of H4, with a new cleavage at H4TL residue U3881. Unexpectedly, several residues in the H5 LSL were also newly susceptible to light cleavage in F4 + U3898G (Fig. 5A,C). These residues were U3974 and A3975 on the 5′ side of the LSL, and the opposing residue U3995. This suggests that U3898 stabilizes the local RNA structure and also affects H4TL and the H5 LSL, possibly by contributing to the properties of Ψ4.
FIGURE 5.
Mutations in H4AL or H5 LSL affect the structure of H4AL, H5 LSL, and H4TL. (A) In-line cleavage of transcripts containing mutations in H4AL (U3898G) or H5 LSL (m105). Location of the mutations are shown in C and D. See the legend to Figure 2A for further details. (B) Structural alterations in the presence of MBP–RdRp. See the legend to Figure 2B for further details. (C) Structural alterations in F4 due to U3898G, and (D) m105 in the presence and absence of MBP–RdRp. See the legend to Fig. 2C for details.
The conformational changes in F4 + U3898G occurring in the presence of RdRp were similar to wt with the following exceptions (Fig. 5B,C): (1) In the H4AL region, the wt strong cleavage at position U3897 was much weaker; and (2) very reproducible differences were found in the H4TL, with the normally unchanging uridylates at positions 3886, 3887, and 3888 being less susceptible to cleavage and the normally enhanced cleavages in the center portion of the loop were absent. The conformational changes in H5 were similar to wt; however, all newly cleaved residues were unchanged. Altogether, these findings strongly suggest that the major reduction in TCV levels in vivo due to the U3898G alteration in H4AL are a combination of reduced translational efficiency (Stupina et al. 2008) and reduced minus-strand synthesis, both of which may result from the structural alternations in the 3′ region of TCV.
Mutations in the 5′ side of the H5 LSL affect the structure of H4TL and H4AL
As described above, disruption of Ψ1 by m52, in the presence or absence of RdRp, affected residues in the H5 LSL, as well as in H4TL and H4AL (Fig. 3), suggesting a linkage among these three regions. U3898G in H4AL also caused conformational changes in H4TL and H5 LSL in the presence and absence of RdRp, further supporting this connection. The most likely explanation is that Ψ4 is present along with Ψ1 in F4 and that H4AL indirectly affects this pseudoknot. To provide support for the presence of Ψ4 in wt F4 and explore the connection between H4AL, H4TL, and H5 LSL, we altered three consecutive adenylates in the 5′ side of the H5 LSL to either guanylates (m104) or uridylates (m105; Fig. 4A). Full-length TCV with m104 alterations did not accumulate to detectable levels and m105 mutations reduced accumulation to only 6% of wt (Fig. 4B). In vitro transcription of F4 + m105 by the TCV RdRp produced high levels of products, most of which were of aberrant length and caused by internal initiation (Fig. 4C, right; data not shown), suggesting that the RNA may have adopted an altered conformation that affected the proper synthesis and initiation of complementary strands.
In-line structure probing of F4 + m105 produced a number of structural changes compared with wt F4. As expected, there were new cleavages in residues on the 5′ side of the H5 LSL (positions 3974–3978), including the three adenylates that were altered (Fig. 5A,D). There was also a new light cleavage in the H5 GAAA tetraloop and at U3995 on the 3′ side of the LSL. F4 + m105 also contained distinctive alterations in H4AL and H4TL. The cleavage pattern in H4AL more closely resembled the pattern found after the RdRp-mediated conformational shift in wt F4, e.g., strong cleavage at position U3997 and weaker or absent cleavages at the other residues. In H4TL, U3888 was now very susceptible to cleavage (which was also apparent in the hydroxide-treated ladder lane), and the remaining residues were less susceptible to cleavage than in wt F4.
In the presence of RdRp, the F4 + m105 pattern was similar to wt F4 except for the maintenance of the strong cleavage at U3888 and the weak cleavage in the H5 tetraloop. In addition, the strong cleavage at G4019 in Link1, which in wt F4 is substantially reduced following addition of RdRp, remained unchanged in F4 + m105.
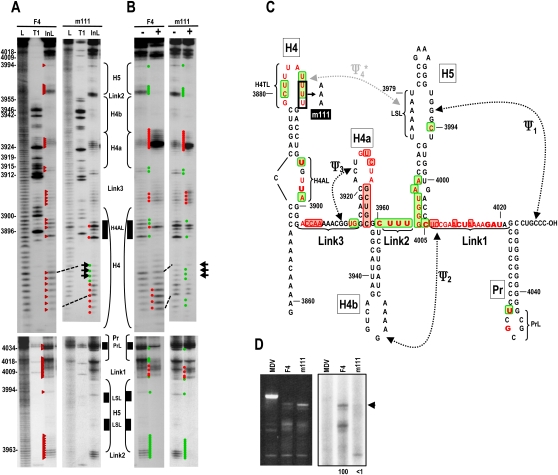
Mutations in H4TL affect the structure of H4AL
As shown above, altering H4AL and H5 5′ LSL caused structural changes in H4AL, H4TL, and H5 LSL without significantly affecting Ψ3/H4a, Link2, Link1, Pr, and the remainder of H5. To provide further support for a connection between terminal and internal loop regions of H4 and the H5 LSL, three uridylates in H4TL (U3886–U3888) were converted to adenylates (m111), and F4 + m111 assayed for structure and template activity. F4−m111 displayed major cleavage pattern differences in H4TL, with reduced cleavage of positions U3885–U3888 and enhanced cleavages in the remaining loop residues (positions 3880–3885) (Fig. 6A,C). Major changes were also evident in H4AL, with strong cleavages at U3895 and U3898 and weaker cleavages at the remaining residues. Differences from the wt pattern were also found in these two regions following RdRp addition to the transcripts, as well as at position A3907 in Link3 and in Link1 sequences adjacent to Pr, which no longer exhibited a conformational shift (Fig. 6B). Similar to H4AL mutation U3898G, m111 significantly affected the activity of F4 transcripts for in vitro transcription, with no discernable complementary strands synthesized in vitro (Fig. 6D).
FIGURE 6.
Mutations in H4TL affect the structure of H4AL. (A) In-line cleavage of transcripts containing mutations in H4TL (F4 + m111). Location of the mutations is shown in C. See the legend to Figure 2A for further details. (B) Structural changes in F4 + m111 in the presence of MBP–RdRp. See the legend to Figure 2B for further details. (C) Structural alterations in F4 due to m111 in the presence and absence of MBP–RdRp. See the legend to Figure 2C for details. (D) In vitro transcription of wt F4 and F4 + m111 using MBP–RdRp. Ethidium bromide stained gel is at the left of the autoradiograph. Assays were performed three times. Arrow denotes position of full-length transcripts.
H4AL reduces stability of Ψ4
80S ribosomes bound the TSS with greater affinity in the absence of H4 and Pr/Ψ1 (Stupina et al. 2008). Mutations that disrupted Ψ4 enhanced ribosome binding in a fragment (F3) containing H4 → TSS, while compensatory mutations that reformed Ψ4 weakened binding, suggesting that Ψ4 interferes with ribosome recognition of the TSS in this fragment. Mutations in H4AL also significantly reduced ribosome binding to a fragment containing the TCV 3′ terminal 393 nt. In light of current results suggesting a structural connection between H4AL, H4TL, and H5 LSL that possibly acts through Ψ4, we wanted to determine if mutations in H4AL were detrimental to TSS-ribosome binding due to a strengthening of Ψ4. Previously described H4AL m21 mutations (UUA–ACU) (Fig. 7A; Stupina et al. 2008), which had been found to reduce translation of a reporter construct by 10-fold in vivo, reduced ribosome binding to F3 by fourfold (Fig. 7C, wt F3, Kd = 1.34 μM; F3 + m21, Kd = 5.5 μM). A3978C (m19), a mutation in the H5 LSL, enhanced ribosome binding to F3 by 2.2-fold (F3 + m19, Kd = 0.61 μM), supporting previous findings that disrupting Ψ4 in this fragment enhances ribosome interaction with the TSS. F3 + m21 + m19 bound ribosomes with 16-fold greater affinity compared with F3 + m21, reaching a value very similar to ribosome binding to fragment F1, which contains only the TSS and Link3 (F3 + m21 + m19, Kd = 0.36 μM; F1, Kd = 0.45 μM). This indicates that disrupting Ψ4 can compensate for the negative effects of altering H4AL, supporting a role for H4AL in the stability of Ψ4.
DISCUSSION
The requirement that RNA viral genomes first serve as templates for translation and then as templates for replication requires precise temporal regulation that allows genomes to translate sufficient quantities of replication-required products and then clear ribosomes so that transcription can commence (Barton et al. 1999). The existence of 5′ elements necessary for transcription initiation from the 3′ end (Gamarnik and Andino 1998; Barton et al. 2001; Frolov et al. 2001; Filomatori et al. 2006; Yi and Kao 2008), and 3′ elements required for translation from the 5′ end (Dreher 1999; De Quinto et al. 2002; Dobrikova et al. 2003; Chiu et al. 2005; Bradrick et al. 2006; Miller and White 2006; Song et al. 2006; Miller et al. 2007; Karetnikov and Lehto 2008) suggest a requirement might exist for overlapping replication and translation elements at either end of the viral genome to control the switch between translation and replication.
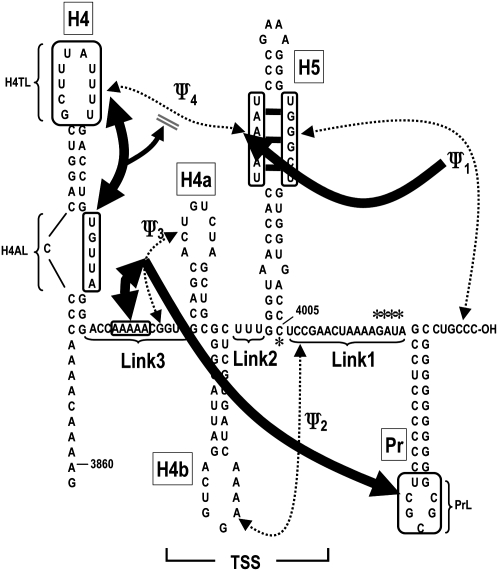
To study the relationship between 3′ proximal translation and replication elements, we used in-line probing to provide clues as to the higher order structure of the 3′ 200 nt of TCV and to determine how RdRp interaction with this region affects structural dynamics. Figure 8 depicts our current understanding of these interactions in the absence of RdRp based on the following data generated in this report: (1) Ψ3 is stabilized by upstream adjacent adenylates. Mutations that disrupted Ψ3 (m26, m27) affected the structure of adjacent Link3 adenylates and vice versa (Fig. 2A), suggesting that H-type pseudoknots can be stabilized by nearby external sequences. This possibility is supported by recent findings that the structure of the TSS is unstable in solution in the absence of the upstream adenylates or additional bases that artificially extend the pseudoknot (Y Wang, unpubl.). (2) PrL is likely associated with the Ψ3/H4a region of the TSS. Mutations that disrupted Ψ3 altered cleavage of G4034 in PrL without detectably affecting elements that reside between these two regions, while compensatory mutations reestablished the wt cleavage pattern (Fig. 2A). (3) H5 LSL residues interact with sequence at the 3′ terminus (forming Ψ1) and with sequence in the H4TL, forming Ψ4. Disruption of Ψ1 by m52 altered the conformation of the opposed LSL residues and residues in H4TL and H4AL, either before or after the RdRp-mediated conformational shift, suggesting that eliminating Ψ1 can in some instances affect Ψ4 (Fig. 3). m51 caused a different pattern of cleavages across the LSL without discernibly affecting H4AL or H4TL. This suggests additional noncanonical pairing or stacking of LSL residues is occurring across the loop, which is dependent on the presence of Ψ1 as previously proposed (McCormack et al. 2008). (4) H4AL interacts with H4TL to control the stability of Ψ4. Mutations in H4AL (U3898G) and H4TL (m111) altered each other's structure (Figs. 5A, 6A), while LSL alteration m105 affected cleavages in both H4TL and H4AL (Fig. 5A). This suggests that noncanonical interactions connect these two critical H4 loops. Mutations in H4AL reduced ribosome affinity to a fragment containing H4 and the TSS, which was relieved by a mutation that disrupted Ψ4 (Fig. 7C). This suggests that mutations in H4AL relieve constraints on H4TL, enhancing the stability of Ψ4. This pseudoknot interferes with ribosome recognition of the TSS, which may at least partly account for the disruptive effects of H4AL mutations on translation (Stupina et al. 2008).
FIGURE 8.
Interactions between different elements in the 3′ UTR of TCV. Thick arrows reflect interactions that are disrupted when elements are mutated. For example, mutation in Ψ3 affect the structure of the adjacent adenylates (and vice versa, indicated by double arrowheads) and the structure of PrL (single arrowhead, since the reverse is not known); mutations in Ψ1 affect the structure of non-Watson–Crick interactions across the H5 LSL; however, mutations in the 5′ side of the H5 LSL do not abrogate Ψ1. Asterisks indicate residues in “hinge” locations that are sterically affected by disruption of some of the interactions (see text for more details), or some differences following the RdRp-mediated conformational shift.
The simultaneous canonical pairing of RNA to both sides of a large internal symmetrical loop, as envisioned for the Ψ4 and Ψ1 interaction with the H5 LSL, is similar to the structure that forms between the LSL of H/ACA snoRNA and 28S ribosomal RNA that leads to pseudouridylation (Wu and Feigon 2007). Analysis of the crystal structure of this intermolecular motif indicated the formation of a unique type of three-way junction (3H) composed of two stacked helices that was reminiscent of the 3H C family (Lescoute and Westhof 2006). The 3D structure of RNA is composed of a limited number of common structural elements (Hendrix et al. 2005), and it will be interesting to determine if the Ψ4/H5 LSL/Ψ1 element is a second example of this unusual type of 3H motif.
None of the mutations outside of the Ψ3/H4a region affected Ψ3/H4a, H4b, H5, or Ψ2, suggesting that the top of the “T” in the TSS that forms from parallel stacked helices is highly stable. Additionally, the structure of H5, which is located perpendicular to the stacked helices, was not significantly altered (outside of the LSL region) by any of the current mutations, with the exception of C4005 at the very base of H5, whose flexibility is affected by Ψ1. Altogether, these results suggest that the TSS forms a stable scaffold that allows for interactions with surrounding elements through its large internal symmetrical loop. These interactions likely result in a highly interactive, compact structure that allows for simultaneous RdRp access to multiple elements within the region including the 3′ end.
RdRp binding to F4 results in an extensive conformational shift in the RNA structure that disrupts numerous interactions within the TSS and between the TSS and surrounding sequences. RdRp-induced changes in in-line cleavage susceptibility included residues in H4AL, H4TL, Ψ3/H4a, H5, PrL, and all linkers connecting the hairpins. While the precise location of RdRp binding in F4 is not known, loss of flexibility of residues just upstream of H4a and in Link2 may point to possible locations that are protected from cleavage by RdRp binding.
The Ψ3–H4a region, which is important for ribosome binding to the TSS (Stupina et al. 2008), was particularly affected by RdRp binding. Interestingly, the Ψ3–H4a region mimics the amino-acceptor portion of a tRNA, which is the site of large ribosomal subunit binding (Schäfer et al. 2002). Binding of threshold levels of newly synthesized RdRp to the 3′ end could therefore disrupt structures required for 60S ribosome subunit binding, restricting ribosome access to the template and promoting replication. The RdRp-mediated conformational shift depended on the continued presence of the RdRp, since digestion of the RdRp caused subsequent in-line cleavages to reflect the initial structure of the RNA (Fig. 1D). The dependence of the alternative structure on the continued presence of RdRp suggests that depletion of the local concentration of RdRp that would occur following transcription of complementary strands and transport of the RdRp-minus-strand complex to sites of plus-strand synthesis (e.g., membrane-derived spherules) would lead to reversion of the RNA template to the initial translatable form allowing for cyclic synthesis of RdRp.
Several mutations had major effects on F4 transcription. Alterations in H4AL and H4TL eliminated detection of complementary strands, suggesting that H4 is a key component in transcription regulation. H4, along with flanking A-rich sequences, enhanced transcription when positioned directly downstream from Pr in vitro (Sun and Simon 2006). However, mutations in H4AL and H4TL had no effect on transcriptional enhancement or RdRp binding to the recombinant fragment, suggesting that these regions function through a mechanism that requires the missing TSS. Simple elimination of Ψ4, however, was not the cause of reduced template activity, since F4 with alterations in the 5′ side of the H5 LSL (m105) enhanced complementary strand synthesis while also promoting internal initiation of a smaller product (X Yuan and AE Simon, unpubl.).
Unexpectedly, disruption of Ψ1 affected template activity differently depending on the location of the mutation (the 3′ penultimate residue or LSL residue). F4 + m51 was an aberrantly robust template, generating at least 20-fold more complementary strands of varying sizes compared with wt F4. This suggests that the RdRp accesses the 3′ end differently in F4 + m51 compared with other transcripts, which is supported by finding no conformational shift in the structure of F4 + m51 downstream from H5 (Fig. 3B). Disruption of the analogous Ψ1 pseudoknot by single base alterations in a defective interfering RNA associated with the tombusvirus Tomato bushy stunt (Pogany et al. 2003) and in TCV-associated satellite RNA satC also enhanced transcription in vitro (Zhang et al. 2004), but not to the extent of m51.
The structure shown in Figure 8 was determined in the absence of the 5′ UTR. The RNA conformation present during translation in the full-length virus is likely to be different, since Ψ4 inhibits ribosome binding and therefore may be absent during translation initiation. Mutations in the H4TL uridylates that participate in Ψ4 reduced translation of a single luciferase construct in protoplasts, while alterations in partner adenylates in the H5 LSL did not negatively affect translation (Stupina et al. 2008). This suggests that H4TL residues may participate in an additional interaction required for translation. We are currently exploring if this interaction involves pairing with a repeat of the 5′ side H5 LSL sequence that is located in the TCV 5′ UTR.
MATERIALS AND METHODS
Construction of TCV mutants
Oligonucleotide-mediated site-directed mutagenesis was used along with PCR to construct the 3′UTR mutants in pTCV66, which contains full-length wt TCV sequence downstream from a T7 promoter. All mutations were confirmed by sequencing. F3 and F4 fragments containing mutations were generated directly from the mutant pTCV66 constructs using PCR with a 5′ primer that contained a T7 RNA polymerase promoter.
Expression and purification of MBP–RdRp and MBP
pMAL–c4X plasmid (New England Biolabs), containing an additional p88 insert fused to the resident MBP, was a kind gift from P.D. Nagy (University of Kentucky). pMAL–c4X without the p88 insert was used to prepare MBP. Plasmids were transformed into E. coli Rosetta (DE3) pLacI competent cells (Novagen), and expression and purification of the recombinant protein carried out as described (Rajendran et al. 2002).
In-line probing of RNA structure in the presence and absence of RdRp
In-line probing was performed essentially as previously described (Winkler et al. 2002; McCormack et al. 2008). Briefly, wt F4 and mutant fragments were synthesized using T7 RNA polymerase, 5′ end labeled using [γ-32p]-ATP and polynucleotide kinase and then purified by polyacrylamide gel electrophoresis. Fragments were then heated to 75°C and allowed to slow cool over 2 h to room temperature. RNA (5 pmol) was then incubated at room temperature in 50 mM Tris-HCl (pH 8.5), 20 mM MgCl2 for 14 h without RdRp, or for 1 h in the presence of 7.5 pmol of purified MBP–RdRp. Reactions were ethanol precipitated, heated at 95°C for 5 min and subjected to electrophoresis through 8% sequencing length denaturing polyacrylamide gels. RNA cleavage ladders were generated by incubating end-labeled RNA in a reaction mix containing 1 μg yeast tRNA, 50 mM NaHCO3/Na2CO3 (pH 9.2), and 1 mM EDTA for 5 min at 95°C. RNase T1 digests were carried out by incubating 5 pmol of partially denatured labeled RNA in 1 μg yeast tRNA, 20 mM sodium citrate (pH 5), 1 mM EDTA, 7 M urea, and 1 U RNase T1 for 15 min at room temperature. To digest the RdRp, 0.6 U Proteinase K (NEB) was added followed by an additional 13-h incubation. At least two independent in-line probing assays were performed for each mutant, and only reproducible effects are described here.
In vitro RdRp assay
In vitro RdRp assays were performed in the presence of 10 mM Mg2+, the same concentration as that used for RNA structure probing. Briefly, 1 μg of purified RNA template was added to a 25-μL reaction mixture containing 50 mM Tris-HCl (pH 8.2), 100 mM potassium glutamate, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM each of ATP, CTP, GTP, 0.01 mM UTP, 10 μCi of [α-32P] UTP (Amersham), and 2 μg of MBP–RdRp. After 90-min incubation at 20°C, 1 μg of tRNA was added, and the mixture was subjected to phenol–chloroform extraction and ammonium acetate–isopropanol precipitation. Radiolabeled products were analyzed by denaturing 8M urea–5% polyacrylamide gel electrophoresis followed by autoradiography.
Protoplast inoculations, RNA extraction, and Northern blots
Protoplasts (5 × 106) prepared from callus cultures of Arabidopsis thaliana (ecotype Col-0) were inoculated with 20 μg of TCV genomic RNA transcripts as previously described (Kong et al. 1997). Equal amounts of total RNA extracted from protoplasts at 40 hpi were subjected to electrophoresis through a 1.5% nondenaturing agarose gel. After blotting, TCV was probed with fragments specific to the 3′ 400 nt prepared by random labeling with [α-32P]-dCTP.
Isolation of 80S ribosomes
Yeast ribosomes (strain JD1090) were isolated as previously described (Meskauskas et al. 2005). Supernates following cellular disruption were transferred to 4-mL polycarbonate tubes containing 1 mL of a cushion buffer C [50 mM Tris-HCl (pH 7.5) at 4°C, 5 mM Mg(CH3COO)2, 50 mM NH4Cl, 25% glycerol, 0.1 mM PMSF, 0.1 mM DTE]. Ribosomes were sedimented by centrifugation for 2 h at 50,000 rpm using an MSL-50 rotor. Fines from ribosome pellets were gently washed off with buffer C. Ribosomes were suspended in buffer C at concentrations of 2 to 10 pmol/μl (1 OD260 = 20 pmol) and stored frozen at −80°C.
Ribosome binding assays
Filter binding assays were performed as previously described (Meskauskas et al. 2005) in 50 μL of binding buffer [80 mM Tris-HCl (pH 7.4), 160 mM NH4Cl, 11 mM Mg(CH3COO)2, 6 mM β-mercaptoethanol, 0.4 mM GTP, and 2 mM spermidine, 0.4 μg/mL of poly(U)] containing 25 pmol of ribosomes or 40S ribosomal subunits and 2-100 pmol of [32P] 5′-end labeled 5′ UTR fragments.
ACKNOWLEDGMENTS
We thank Eric Westhof for pointing out the similarity of the Ψ4/H5 LSL/Ψ1 motif and the H/ACA snoRNA/28S rRNA motif. This work was supported by grants from the U.S. Public Health Service (GM 061515-05A2/G120CD) and National Science Foundation (MCB-0615154) to A.E.S and the American Heart Association to A.M. (AHA 0630163N).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1708709.
REFERENCES
- Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, Morasco BJ, Flanegan JB. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J Virol. 1999;73:10104–10112. doi: 10.1128/jvi.73.12.10104-10112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton DJ, O'Donnell J, Flanegan JB. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W-W, Kinney RM, Dreher TW. Control of translation by the 5′- and 3′ terminal regions of the Dengue virus genome. J Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Quinto SL, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova E, Florez P, Bradrick S, Gromeier M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc Natl Acad Sci. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW. Functions of the 3′ untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes & Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov I, Hardy R, Rice CM. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA. 2001;7:1638–1651. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: Building blocks of a modular biomolecule. Q Rev Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. Translation mechanisms involving long-distance base pairing interactions between the 5′ and 3′ nontranslated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology. 2008;371:292–308. doi: 10.1016/j.virol.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Kneller ELP, Rakotondrafara AM, Miller WA. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Wang J, Simon AE. Satellite RNA-mediated resistance to Turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell. 1997;9:2051–2063. doi: 10.1105/tpc.9.11.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: Structures, mechanisms, and evolution. Q Rev Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- McCormack JC, Yuan X, Yingling YG, Zamora RE, Shapiro BA, Simon AE. Structural domains within the 3′ UTR of Turnip crinkle virus. J Virol. 2008;82:8706–8720. doi: 10.1128/JVI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Meskauskas A, Petrov AN, Dinman JD. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol Cell Biol. 2005;25:10863–10874. doi: 10.1128/MCB.25.24.10863-10874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, White KA. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu Rev Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H, White KA. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology. 2006;345:305–316. doi: 10.1016/j.virol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344:211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Panaviene Z, Panavas T, Nagy PD. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J Virol. 2005;79:10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. Starting the protein synthesis machine: Eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- Pogany J, Fabian MR, White KA, Nagy PD. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22:5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Morris TJ. Cap-independent translational enhancement of Turnip crinkle virus genomic and subgenomic RNAs. J Virol. 2000;74:1085–1093. doi: 10.1128/jvi.74.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran KS, Pogany J, Nagy PD. Comparison of Turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J Virol. 2002;76:1707–1717. doi: 10.1128/JVI.76.4.1707-1717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer MA, Tastan AO, Patzke S, Blaha G, Spahn CMT, Wilson DN, Nierhaus KH. Codon–anticodon interaction at the P site is a prerequisite for tRNA interaction with the small ribosomal subunit. J Biol Chem. 2002;277:19095–19105. doi: 10.1074/jbc.M108902200. [DOI] [PubMed] [Google Scholar]
- Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. Curr Top Microbiol Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Simon AE. Requirement of a 3′-terminal stem–loop in in vitro transcription by an RNA-dependent RNA polymerase. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- Song Y, Friebe P, Tzima E, Jünemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Kasprzak W, Shapiro BA, Dinman JD, Simon AE. The 3′ proximal translational enhancer of Turnip crinkle virus Binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Simon AE. A cis-replication element functions in both orientations to enhance replication of Turnip crinkle virus. Virology. 2006;352:39–51. doi: 10.1016/j.virol.2006.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substate RNA to form three-way junctions that position the target U for modification. Proc Natl Acad Sci. 2007;104:6655–6660. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G, Kao CC. cis- and trans-acting functions of Brome mosaic virus protein 1a in genomic RNA1 replication. J Virol. 2008;82:3045–3053. doi: 10.1128/JVI.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. The Arabidopsis cumumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol. 2004;78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhang J, Simon AE. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J Virol. 2004;78:7619–7633. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang G, McCormack JC, Simon AE. Evolution of virus-derived sequences for high level replication of a subviral RNA. Virology. 2006;351:476–488. doi: 10.1016/j.virol.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]