Abstract
RNA 3′-phosphate cyclase (Rtc) enzymes are a widely distributed family that catalyze the synthesis of RNA 2′,3′ cyclic phosphate ends via an ATP-dependent pathway comprising three nucleotidyl transfer steps: reaction of Rtc with ATP to form a covalent Rtc-(histidinyl-N)-AMP intermediate and release PPi; transfer of AMP from Rtc1 to an RNA 3′-phosphate to form an RNA(3′)pp(5′)A intermediate; and attack by the terminal nucleoside O2′ on the 3′-phosphate to form an RNA 2′,3′ cyclic phosphate product and release AMP. Here we used the crystal structure of Escherichia coli RtcA to guide a mutational analysis of the human RNA cyclase Rtc1. An alanine scan defined seven conserved residues as essential for the Rtc1 RNA cyclization and autoadenylylation reactions. Structure–activity relationships were clarified by conservative substitutions. Our results are consistent with a mechanism of adenylate transfer in which attack of the Rtc1 His320 nucleophile on the ATP α phosphorus is facilitated by proper orientation of the PPi leaving group via contacts to Arg21, Arg40, and Arg43. We invoke roles for Tyr294 in binding the adenine base and Glu14 in binding the divalent cation cofactor. We find that Rtc1 forms a stable binary complex with a 3′-phosphate terminated RNA, but not with an otherwise identical 3′-OH terminated RNA. Mutation of His320 had little impact on RNA 3′-phosphate binding, signifying that covalent adenylylation of Rtc1 is not a prerequisite for end recognition.
Keywords: 2′,3′ cyclic phosphodiester; adenylyltransferase; covalent catalysis; RNA processing
INTRODUCTION
RNA 2′,3′ cyclic phosphate ends figure prominently in RNA metabolism: (1) as intermediates in the chemical and enzymatic hydrolysis of the RNA phosphodiester backbone (Raines 1998; Loverix and Steyaert 2001); (2) as the products of RNA cleavage by site-specific endoribonucleases (Amitsur et al. 1987; Ogawa et al. 1999; Lu et al. 2005; Zhang et al. 2005; Xue et al. 2006; Lee et al. 2008); (3) as substrates for enzymatic RNA end joining during tRNA repair and tRNA splicing (Konarska et al. 1982; Greer et al. 1983; Filipowicz and Shatkin 1983; Laski et al. 1983; Amitsur et al. 1987; Zillman et al. 1991; Zofallova et al. 2000; Englert and Beier 2005; Keppetipola et al. 2007; Nandakumar et al. 2008); and (4) as stable 3′-terminal structures in mature metazoan U6 snRNA (Lund and Dahlberg 1992; Gu et al. 1997) that serve as recognition elements for RNA-binding proteins (Licht et al. 2008).
The predominant route to formation of an RNA 2′,3′ cyclic phosphate terminus is via transesterification, whereby an internal ribose 2′-OH attacks the adjacent 3′-5′ phosphodiester and expels a 5′-OH terminated RNA leaving strand. Once formed, a ribonucleoside 2′,3′ cyclic phosphate is susceptible to hydrolytic attack by 2′,3′ cyclic phosphodiesterases that yield either a 3′-phosphomonoester or a 2′-phosphomonoester product (Konarska et al. 1982; Keppetipola and Shuman 2007, 2008; Schwer et al. 2008). A second pathway of RNA 2′,3′ cyclic phosphate formation entails de novo cyclization of an RNA 3′-phosphomonoester in a reaction that consumes ATP (Filipowicz et al. 1983).
Filipowicz and colleagues have illuminated the mechanism and structure of the RNA 3′-terminal phosphate cyclase (Rtc) enzymes that catalyze de novo cyclization (Filipowicz et al. 1985; Vincente and Filipowicz 1988; Genschik et al. 1997, 1998; Billy et al. 1999; Palm et al. 2000). Cyclization occurs via a series of three nucleotidyl transfer steps (Filipowicz et al. 1985; Reinberg et al. 1985). In the first step, Rtc reacts with ATP and a divalent cation to form a covalent Rtc–AMP intermediate and liberate PPi. The AMP is linked via a P–N bond to a histidine side chain of Rtc1. In the second step, the adenylate is transferred from Rtc1–AMP to the RNA 3′-phosphate terminus to form an activated phosphoanhydride intermediate, RNA(3′)pp(5′)A. In the third step, the terminal ribose 2′-OH attacks the 3′-phosphate of RNA(3′)pp(5′)A to generate an RNA 2′,3′ cyclic phosphate product and release AMP.
The RNA cyclase pathway is reminiscent of the three nucleotidyl transfer steps catalyzed by RNA and DNA ligases, also via enzyme–AMP and polynucleotide–adenylate intermediates (Pascal et al. 2004; Nair et al. 2007; Nandakumar et al. 2006). The cyclase and ligase pathways differ in that: (1) ligases rely on lysine rather than histidine as the nucleophile for enzyme adenylylation during step 1; (2) ligases transfer the adenylate to a polynucleotide 5′-phosphate end during step 2 to form an A(5′)pp(5′)RNA/DNA intermediate; and (3) attack by a nucleoside OH on the adenylylated end by ligase during step 3 generates a 3′,5′ phosphodiester linkage between two polynucleotide ends rather than a cyclic phosphodiester terminus on one polynucleotide.
The RNA 3′-terminal phosphate cyclases comprise an enzyme family with members distributed widely among bacterial, archaeal, and eukaryal taxa (Genschik et al. 1997). Primary structure alignment of the biochemically characterized human Rtc1 and Escherichia coli RtcA enzymes (Fig. 1B) highlights conservation of a histidine near the carboxyl terminus, said histidine being the site of covalent adenylylation in E. coli RtcA (Billy et al. 1999). Replacement of this histidine with asparagine or alanine abolished enzyme adenylylation and RNA 3′ phosphate cyclization by RtcA (Billy et al. 1999). There is no discernible primary structure similarity between the Rtc family and the superfamily of covalent nucleotidyltransferases to which polynucleotide ligases belong (Genschik et al. 1997; Shuman and Lima 2004).
FIGURE 1.
Primary and tertiary structure similarity between bacterial and human RNA 3′ phosphate cyclases. (A) A stereo view of the putative active site of E. coli RtcA bound to a citrate anion (PDB ID code 1QMH) is shown. The conserved side chains that were subjected to alanine scanning in human Rtc1 are shown in stick representation with carbons colored beige; the residue numbers are specified for Rtc1. A bound citrate anion is depicted as a stick model with carbons colored gray. Hydrogen-bonding and ionic interactions are depicted as dashed lines. (B) The amino acid sequence of E. coli (Eco) RtcA is aligned to that of Homo sapiens (Hsa) Rtc1. Gaps in the alignment are indicated by dashes. Positions of amino acid side chain identity or similarity are indicated by dots. The amino acids subjected to alanine substitution are highlighted in yellow shading.
The crystal structure of E. coli RtcA revealed a fold composed of four tandem modules, each comprising a four-stranded β sheet overlying two α helices (Palm et al. 2000). The histidine nucleophile is located within the C-terminal module, and it donates a hydrogen bond from Nɛ to a conserved glutamate in the N-terminal module (Fig. 1A). Though lacking any bound substrates or cofactors, the RtcA crystal structure contained a citrate anion docked in the N-terminal module via electrostatic and hydrogen bonding interactions with conserved amino acid side chains, including three arginines, a histidine, and a glutamine (Fig. 1A). The citrate anion is a plausible mimetic of the ATP β and γ phosphates and/or the RNA 3′-phosphate terminus.
The distinctive catalytic mechanism of Rtc and knowledge of its apoenzyme structure inspired us to conduct a mutational analysis of the human Rtc1 enzyme, designed to identify essential functional groups in the active site. An alanine scan of 10 residues of human Rtc1 defined seven of them as essential for the RNA cyclization and enzyme adenylylation reactions. We proceeded to clarify structure–activity relationships at the essential residues by testing the effects of conservative substitutions. We interpret our results in light of available and pending structures of Rtc1 family members (Palm et al. 2000; Shimizu et al. 2008).
RESULTS AND DISCUSSION
Recombinant human Rtc1
The Filipowicz laboratory has produced human Rtc1 in bacteria as a GST-fusion, isolated the recombinant protein by glutathione-affinity chromatography, and demonstrated its RNA cyclase activity in vitro (Genschik et al. 1997). Our aim here was to obtain a tag-free version of Rtc1 in order to avoid forced dimerization by the GST-domain. Human Rtc1 was produced in E. coli as a His10Smt3 fusion protein and isolated from a soluble bacterial lysate by Ni-agarose chromatography. The tag was removed with the Smt3-specific protease Ulp1 (Mossessova and Lima 2000) and the tag-free catalytically active 39 kDa Rtc1 polypeptide (see below) was recovered after a second Ni-agarose chromatography step. The tag-free Rtc1 was then concentrated by ammonium sulfate precipitation and purified further by gel filtration.
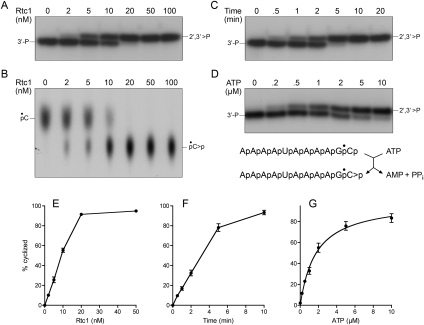
Enzymatic assays of RNA cyclization typically use as substrates RNA 3′-phosphate oligonucleotides labeled uniquely with 32P at either the 3′-terminal or penultimate phosphates (Filipowicz and Vicente 1990). The cyclase assays entail nuclease or phosphatase digestion of the RNA, followed by TLC separation of radiolabeled pCp, pC, or Pi (unreacted substrate) from the cyclized pC>p or C>p nucleotide products (Filipowicz and Vicente 1990). Here, we used an 11-mer RNA 3′-phosphate substrate labeled at the penultimate phosphate (5′-AAAAUAAAAG*pCp) (Fig. 2) that we prepared by RNA ligase 1 (Rnl1)-mediated addition of [5′-32P]pCp (*pCp) to a 10-mer synthetic oligoribonucleotide. We modified the cyclization assay by omitting the nuclease digestion step and directly separating the 3′-phosphate RNA substrate and the slower-migrating 2,′3′ cyclic phosphate RNA product by electrophoresis through a 20% polyacrylamide gel (Fig. 2A). The extent of cyclization of 20 nM AAAAUAAAAG*pCp during a 10-min reaction at 25°C was proportional to input Rtc1 and was virtually complete at saturating enzyme (Fig. 2A,E). We estimated from the slope of the titration curve that approximately five RNAs were cyclized per input Rtc1 (Fig. 2E). The performance of the new electrophoretic assay was comparable to that of the “gold-standard” assay, wherein the reaction mixtures were digested with nuclease P1 and the labeled *pC (derived from the substrate) and the cyclized *pC>p product were resolved by PEI-cellulose TLC (Fig. 2B). RNA cyclization at 20 nM RNA substrate and enzyme increased with time and proceeded to completion by 10 min (Fig. 2C,F). Cyclization depended on exogenous ATP and the extent of product formation displayed a hyperbolic dependence on ATP concentration (Fig. 2D,G), with an apparent Km of 1.7 μM ATP, which is in the same range as the Km of 6 μM ATP reported for human Rtc1 purified from HeLa cells (Reinberg et al. 1985; Vicente and Filipowicz 1988).
FIGURE 2.
RNA 3′ cyclase activity of recombinant Rtc1. The RNA 3′-phosphate substrate, RNA 2′,3′ cyclic phosphate product, and Rtc1 reaction scheme are depicted below panel D. (A,B) Product analysis. Reaction mixtures (10 μL) containing 100 μM ATP, 20 nM 32P-labeled RNA substrate (AAAAUAAAAGp*Cp), and the indicated concentrations of Rtc1 were incubated for 10 min at 25°C. The substrates and products were either resolved by electrophoresis through a denaturing 20% polyacrylamide gel (A) or else digested with nuclease P1 and then separated by PEI-cellulose TLC (B). Radiolabeled species were visualized by autoradiography of the gel or TLC plate; the substrates are identified on the left and the products on the right. The dependence of the extent of RNA cyclization on Rtc1 concentration, determined by PAGE assay, is shown in panel E. Each datum is the average of three titration experiments ± SEM. (C) Time course. Reaction mixtures (100 μL) containing 100 μM ATP, 20 nM 32P-labeled RNA substrate, and 20 nM Rtc1 were incubated at 25°C. Aliquots (10 μL) were withdrawn after the indicated times, quenched immediately, and then analyzed by denaturing PAGE. The extent of RNA cyclization is plotted as a function of time in panel F. Each datum is the average of three experiments ± SEM. (D) ATP dependence. Reaction mixtures (10 μL) containing 20 nM 32P-labeled RNA substrate, 20 nM of Rtc1, and ATP as specified were incubated for 10 min at 25°C. The products were analyzed by denaturing PAGE. The extent of RNA cyclization is plotted as a function of ATP concentration in panel G. Each datum is the average of three experiments ± SEM. Nonlinear regression curve fitting of the data to the Michaelis–Menten equation in Prism yielded an apparent Km value of 1.7 μM ATP.
Mutagenesis strategy
We tested the effects of 10 single alanine mutations on human Rtc1 activity. Nine of the residues targeted for alanine scanning (Fig. 1B) are the equivalents of E. coli RtcA side chains that line the putative active site formed by the N- and C-terminal structural modules (Fig. 1A). These included: the presumptive histidine nucleophile His320; the citrate-binding residues Arg21, Arg40, Arg43, Gln51, and His52; the Glu14 side chain that bridges His320 and Arg43 (a candidate ligand for the metal cofactor); and conserved residues Tyr294 and Asp297, located in a helix adjacent to His320, that we thought might interact with the adenosine nucleoside. (In polynucleotide ligases, an aromatic side chain stacks on the adenine base and a carboxylate side chain coordinates a ribose hydroxyl.) The tenth mutation was introduced at His326, a nonconserved residue just distal to His320. The Rtc1–Ala mutants were produced in E. coli as His10Smt3 fusions and purified from soluble lysates, ultimately in tag-free form (Fig. 3A).
FIGURE 3.
Effects of alanine mutations on Rtc1 cyclase activity. (A) Rtc1 purification. Aliquots (3 μg) of recombinant tag-free wild-type (WT) Rtc1 and the indicated alanine mutants were analyzed by SDS-PAGE. The Coomassie Blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B,C) Cyclase activity. Reaction mixtures (10 μL) containing 100 μM ATP, 20 nM 32P-labeled RNA substrate, and 20 nM WT or mutant Rtc1 as specified were incubated for 20 min at 25°C. The reaction products were analyzed by denaturing PAGE and visualized by autoradiography (panel B). Rtc1 was omitted from the control reaction in lane “–.” The extents of RNA cyclization by the WT and mutant Rtc1 proteins are plotted in panel C. Each datum is the average of three experiments ± SEM.
Mutational effects on RNA 3′ phosphate cyclization
Wild-type Rtc1 and the ten Ala mutants were assayed in parallel for cyclization at equimolar concentrations (20 nM) of RNA and enzyme in the presence of 100 μM ATP (a concentration well in excess of Km); whereas wild-type Rtc1 cyclized ∼95% of the input RNA, six of the alanine mutants were virtually inert (R21A, R43A, R40A, Y294A, D297A, and H320A), and one displayed feeble activity (E14A, which cyclized just 4% of the input RNA ends) (Fig. 2B,C). By contrast, the Q51A and H326A mutants were as active, or nearly as active, as wild type, whereas H52A yielded about half the amount of cyclized ends as the other active proteins. We surmise that Glu14, Arg21, Arg43, Arg40, Tyr294, Asp297, and H320 are essential for Rtc1 function in vitro, whereas His326, Gln51, and His52 are not.
Mutational effects on Rtc1 adenylylation
As noted previously (Genschik et al. 1997), recombinant human Rtc1 reacted with [α32P]ATP in the presence of magnesium (and in the absence of RNA) to form a covalent Rtc1–[32P]AMP adduct that was visualized after SDS-PAGE and autoradiography (Fig. 4A). The yield of Rtc1–[32P]AMP in a reaction containing 1 μM Rtc1 and 100 μM [α32P]ATP indicated that about one-fourth of the input enzyme had been labeled in vitro (Fig. 4B). Assays in parallel of the ten Rtc1-Ala mutants underscored the correlation of adenylyltransferase activity with the mutational effects on the composite RNA cyclization reaction noted above. Specifically, the Q51A and H326A proteins that retained cyclase activity also yielded wild-type levels of the covalent Rtc1–[32P]AMP intermediate, whereas the six mutants that were catalytically inactive in cyclization (R21A, R40A, R43A, Y294A, D297A, and H320A) were also virtually inert in autoadenylylation (Fig. 4). The E14A and H52A proteins that had either feeble or modestly reduced cyclization activity were similarly impaired in their yield of Rtc1–[32P]AMP (Fig. 4).
FIGURE 4.
Effects of alanine mutations on Rtc1 adenylylation. Reaction mixtures (10 μL) containing 100 μM [α-32P]ATP and 1 μM Rtc1 proteins as specified were incubated for 5 min at 25°C. The samples were analyzed by SDS-PAGE and the Rtc1-[32P]AMP complex was visualized by autoradiography (panel A). The extents of label transfer to Rtc1 are plotted in panel B. Each datum is the average of three experiments ± SEM.
These results are consistent with His320 being the site of covalent adenylylation in Rtc1, as is the case for its counterpart His309 in E. coli RtcA (Billy et al. 1999). The instructive findings of the alanine scan concern the essentiality for Rtc1 of the three conserved arginines that engage the citrate anion in the RtcA crystal structure (Fig. 1A), said requirement being evident at the step of Rtc1 adenylylation (i.e., prior to engagement of the RNA). These initial results suggested that the electrostatic contacts of the N-terminal module with citrate might indeed mimic the interactions with the ATP β and γ phosphates.
Structure–activity relations in the Rtc1 active site
We proceeded to determine structure–activity relationships (SARs) by testing the effects of 17 conservative side chains substitutions. His320 was replaced by glutamine, asparagine, and lysine; Arg21, Arg40, and Arg43 were changed to lysine and glutamine; His52 was mutated to glutamine and asparagine; Tyr294 was replaced by phenylalanine, leucine, and serine; and Asp297 was changed to glutamate. The Rtc1 conservative mutants were produced in E. coli as His10Smt3 fusions and purified from soluble lysates in tag-free form (Fig. 5A). The mutant proteins were surveyed for RNA cyclization (Fig. 5B,C) and autoadenylylation (Fig. 5D) activities in parallel with wild-type Rtc1. The results are discussed and interpreted below.
FIGURE 5.
Effects of conservative mutations on Rtc1 activities. (A) Aliquots (3 μg) of the wild-type (WT) Rtc1 and the indicated mutants were analyzed by SDS-PAGE. The Coomassie Blue-stained gels are shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B,C) Cyclase activity. Reaction mixtures (10 μL) containing 100 μM ATP, 20 nM 32P-labeled RNA substrate, and 20 nM WT or mutant Rtc1 as specified were incubated for 20 min. The reaction products were analyzed by denaturing PAGE and visualized by autoradiography (panel B). Rtc1 was omitted from the control reaction in lane –. The extents of RNA cyclization by the WT and mutant Rtc1 proteins are plotted in panel C. Each datum is the average of three experiments ± SEM. (D) Rtc1 adenylation. Reaction mixtures (10 μL) containing 100 μM [α-32P]ATP and 1 μM Rtc1 proteins were incubated for 5 min. The samples were analyzed by SDS-PAGE and the extents of Rtc1-[32P]AMP complex formation were quantified. Each datum is the average of three experiments ± SEM.
His320 is strictly essential
Many phosphoryl transfer enzymes of diverse structure and biological function perform catalysis through covalent phosphoramidate (P–N) intermediates. In the majority of cases, including the Rtc family, Nature has chosen histidine as the protein nucleophile for covalent phosphoryl adduct formation. It is easy to rationalize histidine as a catalyst, given that the relatively low pKa value, typically, between 6 and 7 (Pace et al. 2009), ensures that histidine will be mostly deprotonated at physiological pH, and thereby have the requisite lone pair that can attack the phosphorus center. Polynucleotide ligases and mRNA guanylyltransferases are the only known enzymes in which lysine acts as the nucleophile in covalent phosphoryl transfer. This raises the interesting question of whether lysine might substitute for His320 in Rtc1, assuming that it is the site of covalent adenylylation.
Our findings that the H320N and H320Q proteins were unreactive in RNA cyclization and enzyme–AMP formation (Fig. 5C,D) support the imputed role of His320 in nucleophilic attack on the ATP α phosphate. The salient finding was that lysine could not act in lieu of histidine in either the cyclase or adenylyltransferase reactions. The cyclase and adenylyltransferase assays are performed at pH 7.4 and 8.0, respectively. We considered the possibility that the higher pKa of lysine, typically 10.5 (Pace et al. 2009), might cause an alkaline shift in the reaction optimum. Therefore, we tested the H320K protein for cyclase and adenylyltransferase over a range of pH values from 6.5 to 11, but observed no gain of function at higher pH values (data not shown). We surmise that Rtc enzymes rely specifically on histidine as the nucleophile.
His52 is also located within the Rtc active site, but its mutation to alanine merely reduced cyclase activity, presumably because its contacts to citrate (i.e., phosphate) are redundant to some degree to those of the three essential arginines (Fig. 1A). The conservative mutants H52Q and H52N also displayed modest decrements in RNA cyclase activity (Fig. 5B,C), similar to that of H52A. Yet, H52Q and H52N did evince a gain of function compared with H52A in their extents of autoadenylylation (Fig. 5D). These findings suggest that neutral hydrogen bonding by the His52 imidazole nitrogens (which can, in principle, be mimicked by the partially isosteric Asn and Gln side chains) does not suffice for optimal cyclase activity; rather, an electrostatic contact (presumably with a phosphate anion) is the likely contribution of this residue.
Distinctive SARs at essential arginines
Arginines classically play a role in ground-state binding and transition-state stabilization during phosphoryl transfer reactions by making contacts to phosphate oxygens. The crystal structure of RtcA apoenzyme bound to citrate places the three essential arginines (Arg21, Arg40, and Arg43) too far from the His320 nucleophile (which must contact the ATP α phosphate) to credibly invoke a role for these arginines in transition-state stabilization during step 1 of the cyclase pathway. A more likely role would be in binding and orientation of the PPi leaving group. Indeed, essential arginines serve this function during lysine nucleotidylation by polynucleotide ligases and RNA capping enzymes (Håkansson et al. 1997; El Omari et al. 2005). Here we find that Arg21 and Arg40 are strictly essential for Rtc1 function, insofar as the respective lysine and glutamine changes did not revive activity (Fig. 5). By contrast, introducing lysine in lieu of Arg43 restored both cyclization and adenylylation, although the R43Q had no salutary effect (Fig. 5). We surmise that positive charge suffices for function at position 43.
The aromatic ring of Tyr294 is essential
Replacing Tyr294 with phenylalanine fully revived cyclase and adenylyltransferase activities (Fig. 5), signifying that the tyrosine hydroxyl (which makes a hydrogen bond to the amide nitrogen of conserved residue Gln104) is dispensable. By contrast, leucine—a partial isostere of Phe, albeit nonaromatic—was inactive, as was serine (Fig. 5). We conclude that an aromatic residue at position 294 is critical for Rtc1 function, plausibly by stacking on the adenine base of the ATP substrate.
SARs at acidic residues
Asp297 is located on the same face of the α-helix that contains Tyr294. In RtcA, the corresponding Asp abuts the neighboring α-helix that contains the histidine nucleophile (Fig. 1A); indeed, the Asp297 equivalent is within hydrogen-bonding distance of one of the main chain amides of the vicinal helix. Replacing Asp297 with glutamate conferred little or no gain of function compared with the grossly defective D297A mutant, indicating that the active site did not accommodate the longer main chain to carboxylate linker of Glu versus Asp.
The Glu14 carboxylate bridges essential residues His320 and Arg43 (Fig. 1A). Conservative substitutions of Glu14 with aspartate or glutamine were as or more deleterious than the alanine mutation (Fig. 5), indicating that glutamate is uniquely suitable at this position.
Requirements for stable binding of Rtc1 to RNA
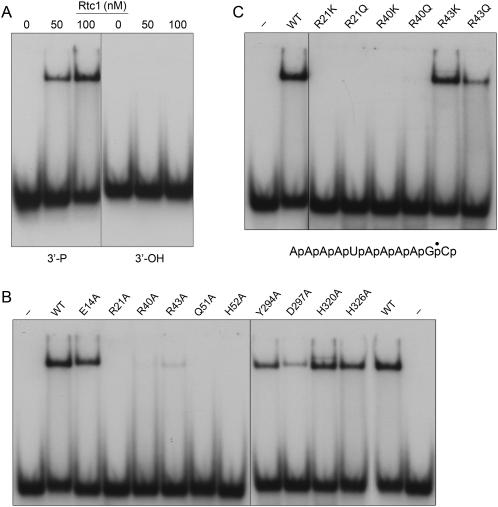
Previous studies had shown that the ability of Rtc enzymes to cyclize radiolabeled RNA substrates can be inhibited by cold RNA 3′-phosphate oligonucleotides, but not by DNA 3′-phosphate oligonucleotides (Genschik et al. 1997, 1998), suggesting that Rtc discriminates between RNA and DNA at the initial substrate binding step. However, there has, to our knowledge, been no direct assay of Rtc binding to RNA or an assessment of the protein features that mediate such binding. Here we used a native gel electrophoretic mobility shift assay to query whether human Rtc1 could form a stable binary complex with the AAAAUAAAAG*pCp substrate. ATP and divalent cation were omitted from the binding reaction mixtures in order to preclude catalysis of cyclization. We found that mixture of 50 or 100 nM Rtc1 with 10 nM AAAAUAAAAG*pCp resulted in the formation of a discrete Rtc1–RNA complex that was well resolved from the free RNA by native PAGE, with the yield of the binary complex being roughly proportional to the input Rtc1 (Fig. 6A). The instructive finding was that no stable binary complex was formed when 50 or 100 nM Rtc1 was incubated with 10 nM AAAAUAAAAG*pCOH, an otherwise identical RNA that lacks the 3′-phosphate moiety (Fig. 6A). This experiment established that Rtc1 recognizes the 3′-phosphate anion, to which it will transfer the covalently bound adenylate, as a key determinant of RNA binding. Our result is consistent with earlier inferences about the RNA 3′-phosphate, derived from RNA competition inhibition experiments (Genschik et al. 1997, 1998). In this respect, Rtc1 resembles exemplary polynucleotide ligases that require a 5′-PO4 moiety (the recipient of their covalent adenylate) in order to form a stable ligase complex bound at a duplex nick (Sekiguchi and Shuman 1997; Sriskanda and Shuman 1998).
FIGURE 6.
RNA binding by Rtc1. (A) Reaction mixtures (20 μL) containing 10 nM RNA, either AAAAUAAAAGp*Cp (3′-P) or AAAAUAAAAGp*COH (3′-OH) and wild-type Rtc1 (0, 50, or 100 nM) were incubated at 25°C for 10 min and then analyzed by native PAGE. (B,C) Reaction mixtures (20 μL) containing 10 nM AAAAUAAAAGp*Cp and 100 nM wild-type or mutant Rtc1 as specified were incubated at 25°C for 10 min and then analyzed by native PAGE. The free radiolabeled 32P-labeled RNAs and the Rtc1-[AAAAUAAAAGp*Cp] complexes were visualized by autoradiography.
Having validated the specificity of the gel-shift assay, we proceeded to screen the collection of Rtc1–Ala mutants (at 100 nM) for their ability to form stable complexes with 10 nM AAAAUAAAAG*pCp (Fig. 6B). The functionally benign H326A change had little impact on RNA binding, as expected. The instructive finding was that the H320A change that ablates the nucleophile for autoadenylylation and abolishes cyclase activity had little or no effect on the formation of an Rtc1–RNAp binary complex (Fig. 6B). This result signifies that a covalently bound AMP is not required for RNA 3′-phosphate recognition by Rtc1. This property of Rtc1 contrasts sharply with the polynucleotide ligases, which require occupancy of the adenylate binding pocket to manifest nick sensing, and thus lose their ability to bind stably to a nick when their active site lysine nucleophile is mutated (Sekiguchi and Shuman 1997; Sriskanda and Shuman 1998; Nandakumar and Shuman 2004).
The other alanine mutations fell into distinct classes with respect to their effects on RNA binding (Fig. 6B). Loss of any the Rtc1 side chains that comprise the citrate-binding pocket in RtcA either eliminated (R21A, R40A, Q51A, and H52A) or sharply reduced (R43A) binary complex formation. These results suggest that the constellation of citrate-binding amino acids that we have implicated in binding the ATP β and γ phosphates during the autoadenylylation reaction (vide supra) is also involved in binding the RNA 3′-phosphate during the subsequent step of AMP transfer to form RNA(5′)pp(5′)A. The RNA binding assay is apparently more sensitive to mutations of the Rtc “citrate-binding” pocket, insofar as replacing Gln51 and His52 with alanine had more dire effects on the RNA gel-shift than on RNA cyclization or Rtc1 autoadenylylation. It is conceivable that the longevity of the Rtc1–RNAp complex is less of an issue under conditions that are permissive for catalysis of cyclization.
Whereas the Y294A change had relatively little effect on the RNA gel-shift, the nearby D297A mutation was clearly deleterious, even though Asp297 appears to be remote from the putative RNA 3′-phosphate binding site that overlaps the citrate site in RtcA. We suspect that the loss of the helix–helix interactions mediated by the Asp side chain (discussed above) might perturb the positions of the secondary structure components of the active site, and thereby compromise broadly the Rtc1 activities.
We also tested the effects of conservative substitutions in the essential arginine triad on binding of Rtc1 to AAAAUAAAAG*pCp. The respective lysine and glutamine changes at Arg21 and Arg40 did not promote stable RNA binding, whereas introducing lysine in lieu of Arg43 did (Fig. 6C). The R43Q mutant displayed weaker complex formation with RNAp, akin to the trace levels of binding seen with R43A. These results were in accord with the conservative mutational effects on the composite cyclization reaction.
Mechanistic implications
Here we have identified (for the first time, to our knowledge) essential constituents of the Rtc active site other than the histidine nucleophile assigned by the Filipowicz laboratory (Billy et al. 1999). We were guided by their crystal structure of RtcA in complex with citrate (Palm et al. 2000). While our study was well underway, Shimizu et al. (2008) reported in the proceedings of the 2008 Joint Symposium of the 18th International Roundtable on Nucleosides, Nucleotides and Nucleic Acids and 35th International Symposium on Nucleic Acids Chemistry that they had obtained new crystal structures of Sulfolobus tokodaii Rtc in several functional states, including apoenzyme, Rtc•ATP binary complex, and covalent Rtc–AMP intermediate. Although a complete description of the structures is not published and the coordinates have not been deposited in the Protein Database, the summary diagram of the ATP-binding site in the proceedings report (Shimizu et al. 2008) is most instructive for placing our mutational data in a mechanistic context.
The three arginines corresponding to the essential Arg21, Arg40, and Arg43 in human Rtc1 and the histidine corresponding to His52 (all of which coordinate citrate in RtcA) do indeed contact the ATP β and γ phosphates. Our studies implicate the same three arginines in binding the RNA 3′-phosphate terminus during the second chemical step of the cyclase pathway. The conserved aspartate corresponding to Rtc1 Asp297 coordinates the ATP ribose hydroxyls. Whereas this interaction could explain the requirement for Asp297 for Rtc1 autoadenylylation and cyclization, the deleterious effects of D297A on RNA binding hint that this residue also plays a structural role in the active site (by tethering vicinal α helices, as surmised from the RtcA structure). Finally, a histidine side chain corresponding to Rtc1 Tyr294 forms a π stack on the adenine base. Shimizu et al. (2008) speculated, as we do, that the glutamate equivalent to Rtc1 Glu14 is a metal ligand, but there was no experimental evidence for a bound metal in any of the Sulfolobus Rtc crystal structures cited.
Thus, our functional studies establish a division of labor among two of the structural modules that comprise Rtc, whereby essential side chains in the C-terminal module form an AMP-binding unit while side chains in the N-terminal module are devoted to binding the PPi leaving group (and presumably the RNA 3′-phosphate). This situation is analogous to the covalent (lysyl) nucleotidyl transferase superfamily of ligases and RNA capping enzymes, members of which have a conserved AMP- or GMP-binding domain and a separate domain (differing among subfamilies) that binds the PPi or NMN leaving group (Håkansson et al. 1997; Gajiwala and Pinko 2004). Nonetheless, the structures of the component modules of the Rtc enzymes have no apparent similarity to any known polynucleotide ligases or RNA guanylyltransferases.
An outstanding question is whether and how the two central structural modules of the Rtc enzymes might participate in substrate recognition or catalysis. In particular, it is not yet clear how Rtc enzymes discriminate stringently in AMP transfer to RNA versus DNA 3′-phosphate substrates (Genschik et al. 1997, 1998). It is conceivable that the central modules play a role in polynucleotide recognition.
MATERIALS AND METHODS
Recombinant human Rtc1 from bacteria
A cDNA spanning the RTC1 ORF was amplified from total HeLa cell RNA by RT-PCR using the Protoscript first-strand cDNA synthesis kit (New England Biolabs) and gene-specific primers designed to introduce a BamHI site at the ATG start codon and an XhoI site immediately downstream from the stop codon. The cDNA (a gift of Dr. Beate Schwer, Weill Cornell Medical College) was inserted into the pET28b–His10Smt3 E. coli expression vector. Missense mutations were introduced into the RTC1 gene via two-stage PCR overlap extension using pET28b–His10Smt3–RTC1 as a template. The PCR products were digested with BamHI and XhoI and inserted into pET28b–His10Smt3. The inserts of the wild-type and mutant pET28b–His10Smt3–RTC1 plasmids were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning.
The pET28b–His10Smt3–RTC1 plasmids were transformed into E. coli BL21-Codon Plus(DE3) (Novagen). Cultures (2 L) derived from single colonies were grown at 37°C in LB medium containing 50 μg/mL kanamycin until the A600 reached 0.6–0.8. The cultures were chilled on ice for 30 min and then adjusted to 20 μM isopropyl β-D-thiogalactoside (IPTG) and 2% (v/v) ethanol. Incubation was continued at 17°C for 16 h. Cells were harvested by centrifugation and stored at –80°C. All subsequent procedures were carried out at 4°C. Cell pellets were suspended in 100 mL of buffer A (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 10% sucrose), and lysozyme was added to 0.2 mg/mL. The suspensions were mixed gently for ∼30 min and then adjusted to 0.1% Triton X-100. The suspensions were sonicated to reduce viscosity, and insoluble material was removed by centrifugation at 20,000g for 30 min. The soluble lysates were mixed for 1 h with 2 to 4 mL (50% slurry) of Ni-NTA agarose (Qiagen) that had been equilibrated in buffer A. The resins were recovered by centrifugation and resuspended in 5 mL of buffer B (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 10% glycerol) containing 25 mM imidazole. The cycle of centrifugation and resuspension of the resins was repeated three times, after which the washed resins were poured into columns. Bound proteins were eluted stepwise with 100, 200, 300, and 400 mM imidazole in buffer B. The elution profiles were monitored by SDS-PAGE. Most of the bound His10Smt3–Rtc1 was recovered in the 200 mM imidazole eluate. Peak fractions containing His10Smt3–Rtc1 were pooled and then treated with the Smt3-specific protease Ulp1 (at a His10Smt3–Rtc1:Ulp1 ratio of 500:1) during an overnight dialysis against buffer B. The dialysates were mixed with 1 mL (50% slurry) of Ni-NTA agarose that had been equilibrated in buffer B and the mixtures were poured into columns, which were washed with the same buffer, after which the bound material was eluted with 400 mM imidazole. The tag-free Rtc1 proteins were recovered in the flow-through fractions, whereas the cleaved His10Smt3 was bound to the resin and eluted with imidazole.
Wild-type Rtc1 was purified further as follows. The tag-free Ni-agarose flow-through Rtc1 fraction was mixed with ammonium sulfate to achieve 60% saturation. The protein precipitate was collected by centrifugation for 30 min at 14,000 rpm in a Sorvall SS34 rotor and resuspended in 2 mL of buffer C (50 mM Tris-HCl [pH 7.4], 2 mM DTT, 1 mM EDTA, 10% glycerol) containing 150 mM NaCl. The protein sample was gel-filtered through a 120-mL 16/60 HiPrep Sephacryl S-100 HR column (GE Healthcare) developed with 150 mM NaCl in buffer C at a flow rate of 1 mL/min. The elution profile was monitored by SDS-PAGE. The peak Rtc1-containing fractions were pooled and concentrated by centrifugal ultrafiltration (Amicon Ultra 15; Millipore) to a concentration of 0.9 mg/mL.
Protein concentrations were estimated initially by using the Bio-Rad dye reagent with bovine serum albumin (BSA) as the standard. The concentrations of the Rtc1 polypeptides were then determined by SDS-PAGE analysis of the wild-type and mutant Rtc1 preparations in parallel with increasing known amounts of BSA. The Coomassie Blue-stained gels were scanned and the intensities of the Rtc1 polypeptides were quantified with Molecular Imager Gel Doc XR System (Bio-Rad) software. The concentration of Rtc1 was calculated by interpolation to the BSA standard curve.
Preparation of radiolabeled RNA substrates
An 11-mer RNA 3′-phosphate substrate labeled at the penultimate phosphate (5′-AAAAUAAAAG*pCp) was prepared by T4 RNA ligase 1 (Rnl1)-mediated addition of [5′-32P]pCp (*pCp) to an 10-mer synthetic oligoribonucleotide. The [5′-32P]pCp was generated by enzymatic phosphorylation of unlabeled 3′-CMP in the presence of T4 polynucleotide kinase-phosphatase (Pnkp) and [γ32P]ATP. A kinase reaction mixture (40 μL) containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM DTT, 0.1 mM 3′ CMP, 0.1 mM [γ32P]ATP (260 μCi), 0.1 mg/mL BSA, and 20 U T4 Pnkp was incubated at 37°C for 60 min. The Pnkp was inactivated by heating the mixture at 95°C for 3 min. The mixture was then adjusted to 80 μL and, in the process, supplemented with 20 μM fresh unlabeled ATP, 50 μM 10-mer oligoribonucleotide AAAAUAAAAG, and 0.4 mg/mL Rnl1. The ligase reaction mixture was incubated for 1 h at 37°C and then analyzed by electrophoresis through a 40-cm 17% polyacrylamide gel containing 7 M urea. The 32P-labeled 11-mer oligoribonucleotide product (5′-AAAAUAAAAG*pCp) was located by autoradiography and excised from the gel. The oligonucleotide was eluted by soaking the gel slice overnight in 0.6 mL of 1 M ammonium acetate, 0.2% SDS, 20 mM EDTA. The eluted oligoribonucleotide was recovered by ethanol precipitation and resuspended in 10 mM Tris-HCl (pH 6.8), 1 mM EDTA.
The 3′-phosphate was removed from AAAAUAAAAG*pCp by reaction with the 3′-phosphatase component of Pnkp. A reaction mixture (50 μL) containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM DTT, 0.1 mg/mL BSA, 160 nM AAAAUAAAAG*pCp, and 20 U Pnkp was incubated at 37°C for 30 min. The RNA was recovered by phenol-CHCl3 extraction and ethanol precipitation and then resuspended in 10 mM Tris-HCl (pH 6.8), 1 mM EDTA.
RNA 3′-phosphatase cyclase assay
Reaction mixtures (10 μL) containing 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 10 mM MgCl2, 100 μM ATP, 20 nM AAAAUAAAAG*pCp, and 20 nM Rtc1 were incubated for 10 min at 25°C. The reactions were quenched by adding 5 μL of 95% formamide, 40 mM EDTA, 0.1% bromophenol blue/xylene cyanol. The samples were analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate, 1.2 mM EDTA. The 32P-labeled RNAs were visualized by autoradiography of the gel and quantified by scanning the gel with a Fuji Film BAS-2500 imager.
Alternatively, the products of the cyclization reactions were resolved by thin-layer chromatography (TLC). After the reaction of enzyme with substrate, the mixtures were adjusted 50 mM sodium acetate (pH 5.2), 10 mM ZnCl2, and then digested with 1 U nuclease P1 at 50°C for 10 min. The digests were analyzed by ascending polyethylenimine cellulose TLC with a buffer consisting of 4 M ammonium sulfate, 1 M sodium acetate, isopropanol (80:18:2, v:v:v). The 32P-labeled nucleotides *pC and *pC>p were visualized by autoradiography of the TLC plate.
Assay of Rtc1 adenylylation
Reaction mixtures (10 μL) containing 50 mM HEPES-NaOH (pH 8.0), 10 mM MgCl2, 1 mM DTT, 100 μM [α32P]ATP (specific activity 0.02 Ci/mmol), and 1 μM Rtc1 were incubated at 25°C for 5 min. The reaction was quenched by adding 10 μL of 100 mM Tris-HCl (pH 6.8), 200 mM DTT, 4% SDS, 20% glycerol, 40 mM EDTA, 0.2% bromophenol blue. The samples were heated at 95°C for 3 min and then analyzed by electrophoresis through a 12% polyacrylamide gel containing 0.1% SDS. The Rct1-[32P]AMP adducts were visualized by autoradiography of the dried gels and quantified by scanning the gels with a Fuji BAS-2500 imager.
Assay of RNA binding
Reaction mixtures (20 μL) containing 50 mM Tris-HCl (pH 7.4), 1 mM DTT, 10 nM 32P-labeled RNA, and Rtc1 as specified were incubated for 10 min at 25°C. The mixtures were placed on ice and adjusted to 10% glycerol and 0.04% Triton X-100. The samples were analyzed by electrophoresis through a native 10% polyacrylamide gel containing 23 mM Tris-borate, 0.6 mM EDTA. The free 32P-labeled RNA and Rtc1-RNA complexes were visualized by autoradiography of the dried gel.
Materials
The 10-mer RNA oligonucleotide was purchased from Dharmacon and deprotected according to the vendor's instructions. 3′-CMP was purchased from Sigma. [γ32P]ATP and [α32P]ATP were from Perkin-Elmer. Nuclease P1 was from USA Biological. PEI cellulose TLC plates were from Merck KgaA. T4 polynucleotide kinase was from New England BioLabs. T4 Rnl1 was purified as described (Wang et al. 2003).
ACKNOWLEDGMENTS
The work was supported by NIH Grant GM46330. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1771509.
REFERENCES
- Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Hess D, Hofsteenge J, Filipowicz W. Characterization of the adenylation site in the RNA 3′-terminal phosphate cyclase from Escherichia coli. J Biol Chem. 1999;274:34955–34960. doi: 10.1074/jbc.274.49.34955. [DOI] [PubMed] [Google Scholar]
- Billy E, Wegierski T, Nasr F, Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is require for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SFJ, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol Chem. 2005;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Vicente P. RNA 3′-terminal cyclase from Hela cells. Methods Enzymol. 1990;181:499–510. doi: 10.1016/0076-6879(90)81147-m. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Konarska M, Gross HJ, Shatkin AJ. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Strugala K, Konarska M, Shatkin AJ. Cyclization of RNA 3′-terminal phosphate by cyclase from HeLa cells proceeds via formation of N(3′)pp(5′)A activated intermediate. Proc Natl Acad Sci. 1985;8:1316–1320. doi: 10.1073/pnas.82.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala K, Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004;12:1449–1459. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz, Filipowicz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of protein conserved in eukarya, bacteria, and archaea. EMBO J. 1997;10:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J Biol Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Gu J, Shumyatsky G, Makan N, Reddy R. Formation of 2′,3′-cyclic phosphates at the 3′end of human U6 small nuclear RNA in vitro. Identification of 2′,3′-cyclic phosphates at the 3′ends of human signal recognition particle and mitochondrial RNA processing RNAs. J Biol Chem. 1997;272:21989–21993. doi: 10.1074/jbc.272.35.21989. [DOI] [PubMed] [Google Scholar]
- Håkansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S. Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage λ phosphatase. Nucleic Acids Res. 2007;35:7721–7732. doi: 10.1093/nar/gkm868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S. A phosphate-binding histidine of binuclear metallophosphodiesterase enzymes is a determinant of 2′,3′ cyclic nucleotide phosphodiesterase activity. J Biol Chem. 2008;283:30942–30949. doi: 10.1074/jbc.M805064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Nandakumar J, Shuman S. Reprogramming the tRNA splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007;35:3624–3630. doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M, Filipowicz W, Gross HJ. RNA ligation via 2′-phosphomonoester, 3′,5′-phosphodiester linkage: requirement of 2′,3′-cyclic phosphate termini and involvement of a 5′-hydroxyl polynucleotide kinase. Proc Natl Acad Sci. 1982;79:1474–1478. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Fire AZ, RajBhandary UL, Sharp PA. Characterization of tRNA precursor splicing in mammalian extracts. J Biol Chem. 1983;258:11974–11980. [PubMed] [Google Scholar]
- Lee KPK, Dey M, Nectulai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht K, Medenbach J, Lührmann R, Kamnach C, Bindereif A. 3′-Cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA. 2008;14:1532–1538. doi: 10.1261/rna.1129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loverix S, Steyaert J. Deciphering the mechanism of RNase T1. Methods Enzymol. 2001;341:305–323. doi: 10.1016/s0076-6879(01)41160-8. [DOI] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johanson M, Byström AS. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Cyclic 2′,3′-phosphate and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNAs. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Nair PA, Nandakumar J, Smith P, Odell M, Lima CD, Shuman S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nat Struct Mol Biol. 2007;14:770–778. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S. How an RNA ligase discriminates RNA damage versus DNA damage. Mol Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm GJ, Billy E, Filipowicz W, Wlodawer A. Crystal structure of RNA 3′-terminal phoshate cyclase, a ubiquitous enzyme with unusual topology. Structure. 2000;8:12–23. doi: 10.1016/s0969-2126(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Reinberg D, Arenas J, Hurwitz J. The enzymatic conversion of 3′-phosphate terminated RNA chains to 2′,3′-cyclic phosphate derivatives. J Biol Chem. 1985;260:6068–6097. [PubMed] [Google Scholar]
- Schwer B, Aronova A, Ramirez A, Braun P, Shuman S. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:204–210. doi: 10.1261/rna.858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S. Nick sensing by DNA ligase requires a 5′ phosphate at the nick and occupancy of the adenylate binding site on the enzyme. J Virol. 1997;71:9679–9684. doi: 10.1128/jvi.71.12.9679-9684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Ohki M, Ohkubo N, Suzuki K, Tsunoda M, Sekiguchi T, Takénaka A. Crystal structures of RNA 3′-terminal phosphate cyclase and its complexes with Mg2++ATP, ATP, or Mn2+ Nucleic Acids Symp Ser. 2008;52:221–222. doi: 10.1093/nass/nrn112. [DOI] [PubMed] [Google Scholar]
- Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Sriskanda V, Shuman S. Chlorella virus DNA ligase: Nick recognition and mutational analysis. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente O, Filipowicz W. Purification of RNA 3′-terminal phosphate cyclase from HeLa cells: Covalent modification of the enzyme with different nucleotides. Eur J Biochem. 1988;176:431–439. doi: 10.1111/j.1432-1033.1988.tb14300.x. [DOI] [PubMed] [Google Scholar]
- Wang LK, Ho CK, Pei Y, Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1: Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J Biol Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- Zillman M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Mol Cell Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofallova L, Guo Y, Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]