Abstract
The complete VS ribozyme comprises seven helical segments, connected by three three-way RNA junctions. In the presence of Mg2+ ions, cleavage occurs within the internal loop of helix I. This requires the participation of a guanine (G638) within the helix I loop, and a remote adenine (A756) within an internal loop of helix VI. Previous structural studies have suggested that helix I docks into the fold of the remaining part of the ribozyme, bringing A756 and G638 close to the scissile phosphate to allow the cleavage reaction to proceed. We show here that while either A756C or G638A individually exhibit very low cleavage activity, a mixture of the two variants leads to cleavage of the A756C RNA, but not the G638A RNA. The rate of cleavage depends on the concentration of the VS G638A RNA, as expected for a bimolecular interaction. This regaining of cleavage activity by complementation indicates that helix I of one VS RNA can interact with another VS RNA molecule to generate a functional active site in trans.
Keywords: RNA catalysis, general acid-base catalysis, RNA folding
INTRODUCTION
The Varkud Satellite (VS) ribozyme is found in RNA from some natural isolates of the fungus Neurospora crassa (Saville and Collins 1990). The RNA is transcribed from an 881 base-pair dsDNA plasmid that occurs in the mitochondrion. The VS ribozyme is a member of the class of nucleolytic ribozymes, where a site-specific cleavage reaction proceeds by the attack of a 2′-hydroxyl group to generate a cyclic 2′,3′-phosphate and 5′-hydroxyl (Lilley and Eckstein 2008). The reverse ligation reaction can also be catalyzed.
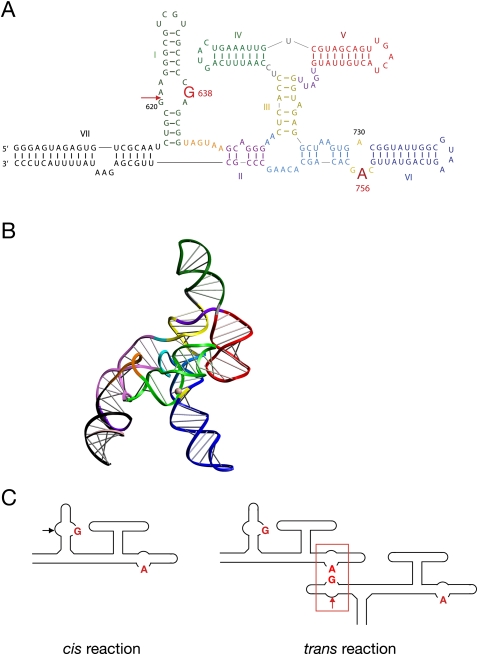
Sequence and secondary structure analysis shows that the RNA forms a series of helical sections organized by three-way junctions (Fig. 1A; Beattie et al. 1995). The cleavage site lies within the internal loop of helix I, at the 3′ phosphate group of G620. The nucleobases G638 (Wilson et al. 2007) (present in the same loop as the scissile phosphate) and A756 (Lafontaine et al. 2001b, 2002b; Sood and Collins 2002) (within the A730 loop in helix VI) are essential for the cleavage reaction, probably participating in general acid-base catalysis (Smith and Collins 2007; Wilson et al. 2007). Although there is no crystal structure available for the VS ribozyme, the general features of the structure have been determined by fluorescence resonance energy transfer (Lafontaine et al. 2001a, 2002a) and small-angle X-ray scattering in solution (Fig. 1B;Lipfert et al. 2008). We have proposed that in the presence of Mg2+ ions the RNA folds to generate a coaxial alignment of helices IV, III, and VI, with helices V and II radially extended. The substrate helix I lies in the cleft between helices II and VI, forming a loop–loop interaction between the terminal loops of helices I and V (Rastogi et al. 1996; Bouchard et al. 2008). The internal loop of helix I can make an intimate interaction with the A730 loop of helix VI, to generate the probable active site, whereby A756 and G638 are expected to catalyze the reaction on the scissile phosphate. Thus, in the proposed structure, the active site is constructed from two sections that are remote within the secondary structure, and requires nucleobases contributed from both.
FIGURE 1.
The VS ribozyme sequence, and a possible cleavage reaction in trans. (A) The sequence of the VS ribozyme, with the critical nucleotides for catalytic activity A756 and G638 highlighted. Ribozyme cleavage occurs within the loop of helix I, at the position arrowed. The sequence shown includes the additional helix VII. (B) A model of the folded VS ribozyme based on small-angle X-ray scattering (Lipfert et al. 2008). The helices are colored in the same manner as for the sequence in A. (C) The principle of a trans reaction between ribozymes. The normal reaction occurs in cis, resulting from an interaction between the loops of helices I and VI (left). However, it is formally possible that helix I of one ribozyme molecule could interact with helix VI of a different molecule to generate a functional active site in trans (right). No attempt has been made to represent the three-dimensional structure of the ribozyme in these schemes.
The deduced folded structure of the VS ribozyme requires the substrate helix I to make a sharp turn with respect to helix II in order to form the active site with helix VI. In its natural form, an additional helix VII forms a three-way junction with helices I and II (Jones et al. 2001); perhaps helping this docking process. This may be an example of auxiliary folding elements that are not required for catalysis but appear to assist folding, similar to the loop–loop interaction of the hammerhead (Khvorova et al. 2003; Penedo et al. 2004) and the four-way junction of the hairpin ribozymes (Murchie et al. 1998; Walter et al. 1999). In the absence of helix VII, it has been found that increasing the spacing between helices I and II by adding nucleotides leads to faster rates of cleavage (Poon et al. 2006), with cleavage occurring at ∼10 min−1 compared with ∼0.1 min−1 without the additional linker. In fact, helix I may be physically disconnected from the VS ribozyme with retention of both cleavage (Guo and Collins 1995) and ligation (McLeod and Lilley 2004) activities. Thus, a ribozyme comprising helices II through VI can bind the isolated helix I substrate with an apparent affinity of 1 μM, with rates of cleavage of ∼2 min−1 (Lafontaine et al. 2002a).
The faster rates of cleavage that occur when the constraint on helix I is relieved led Collins and coworkers to propose that in the natural context the VS ribozyme might cleave a physically remote substrate helix within the same transcript (Poon et al. 2006). This, in turn, led us to ask whether two fully intact VS ribozymes can undergo a cleavage reaction in trans, whereby helix I of one ribozyme molecule displaces the helix I of the other and docks with it to form a hybrid active site (Fig. 1C). The critical role of the nucleobases A756 and G638 allow us to perform a complementation analysis to test this proposal, and incidentally test the composite nature of the active site. We find that the variant ribozymes with A756C and G638A substitutions are essentially inactive by themselves, but become fully active when mixed together. Thus, a trans reaction between complete VS ribozymes is possible, thereby confirming the proposed manner of formation of the active site.
RESULTS AND DISCUSSION
Cleavage activity of the complete VS ribozyme, and active site sequence variants
We have based all the experiments presented here on the form of the VS ribozyme that includes helix VII, that makes a three-way junction with helix II and the substrate helix I (comprising helices IA and IB). This form of the ribozyme exhibits both cleavage and ligation activity (Jones et al. 2001), and is the construct that we previously studied structurally using small-angle X-ray scattering (Lipfert et al. 2008). The natural sequence of this ribozyme includes the two nucleotides that are critical for activity: A756 in the A730 loop of helix VI, and G638 in the internal loop of the substrate helix I (Fig. 1). This species undergoes a cleavage reaction in cis, with ∼60% cleavage (Fig. 2), reflecting the equilibrium between cleavage and ligation reactions. The reaction progress was fitted to two exponential functions, with a fast rate of 0.023 min−1 in the presence of 10 mM Mg2+ ions. The rate is significantly slower than the cleavage of a simple helix I substrate by VS ribozyme (helices II–VI) in trans (Wilson et al. 2007), but is comparable to other cis forms of the ribozyme lacking helix VII.
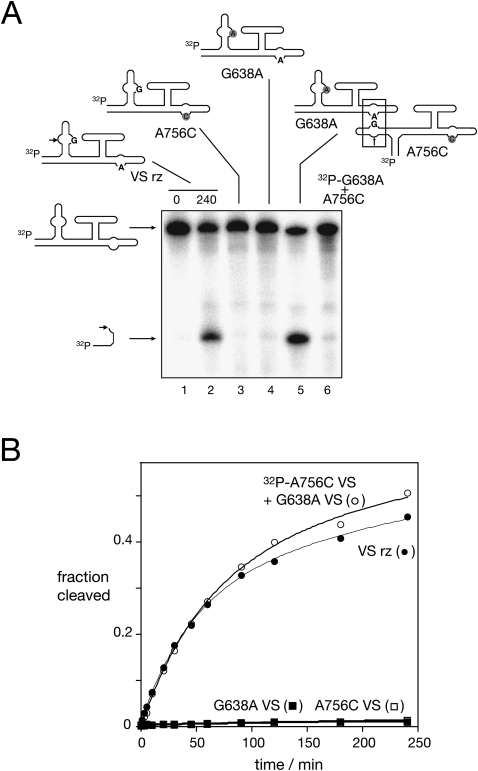
FIGURE 2.
Cleavage activity in trans by complementation between inactive VS ribozyme variants. (A) Gel electrophoretic separation of full-length and cleaved VS RNA, after incubation in 50 mM Tris (pH 8), 10 mM MgCl2, 25 mM KCl, 2 mM spermidine at 37°C for 4 h (except track 1). In incubations of single species, the RNA was radioactively [5′-32P]-labeled. Track 1 contains natural sequence VS RNA before incubation in buffer. Natural sequence VS RNA (track 2) and variants A756C (track 3) and G638A (track 4) were incubated individually. Note that the two variant sequences underwent insignificant cleavage during this time. Track 5 contains the result of incubating [5′-32P]-labeled VS A756C RNA with unlabeled VS G638A RNA. Note that the combination of the two species that are individually inactive has generated significant ribozyme cleavage. However, reversal of the two species ([5′-32P]-labeled VS G638A RNA with unlabeled VS A756C RNA; track 6) fails to result in cleavage. (B) Cleavage reaction progress plotted for the natural VS sequence (closed circles), VS G638A RNA (closed squares), VS A756C RNA (open squares) and the mixture of [5′-32P]-labeled VS A756C RNA with 1 μM VS G638A RNA (open circles). The data have been fitted to two exponential functions.
As expected, the introduction of either A756C or G638A substitutions into the helix VII-containing ribozyme led to very low cleavage activity, with ∼1% cleavage detected after 4 h of incubation. Cleavage rates of kobs = 10−4 min−1 and 9 × 10−5 min−1, respectively, were measured (Fig. 2B).
A trans cleavage reaction can restore activity in active site variants
Although the A756C- or G638A-substituted ribozymes exhibit very low activity in isolation, a functional active site comprising both A756 and G638 could be created from a mixture of the two ribozymes if a trans reaction is possible. In other words, if the helix I of the A756C variant (which has a normal G638) could interact in trans with the A730 loop of the G638A variant (which has a normal A756 within the A730 loop), this would create a functional active site comprising both A756 and G638. We therefore mixed 1 μM of unlabeled G638A VS RNA with a trace amount of [5′-32P]-labeled A756C RNA, and incubated the mixture in the presence of 25 mM KCl, 50 mM Tris (pH 8), 2 mM spermidine, and 10 mM MgCl2. It is clear that the radioactively labeled A756C VS RNA has become cleaved in the presence of the G638A ribozyme (Fig. 2A, track 5), whereas it was insignificantly cleaved when incubated in isolation (Fig. 2A, track 3). The extent and rate of cleavage under these conditions are similar to those of the unmodified VS ribozyme in cis (Fig. 2B). The most likely explanation is that a trans reaction between the two variants has generated a functional active site that catalyze a normal cleavage reaction in the A756C VS RNA. When the reaction was performed using a mixture of radioactively labeled G638A VS RNA with an excess of A756C VS RNA no cleavage was detected (Fig. 2A, track 6). This is expected because cleavage would only be observable in the G638A helix I, which cannot create a normal active site either in cis or trans.
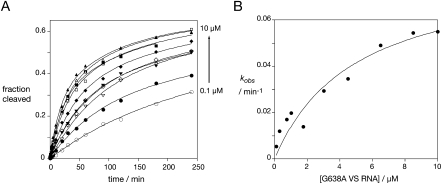
Dependence of the trans cleavage reaction rate on ribozyme concentration
If the restoration of activity in the A756C VS RNA occurs by the action of the G638A VS ribozyme in trans, the observed rate of cleavage should be dependent on the concentration of the G638A VS RNA. We measured the observed rate of cleavage of 1 nM radioactively labeled A756C VS RNA in the presence of G638A VS RNA at concentrations between 0.1 and 10 μM (Fig. 3), i.e., using single-turnover conditions. All the progress curves were globally fitted to two exponential functions, treating the amplitudes of fast and slow rates as global parameters. The cleavage rates were indeed dependent on the concentration of the G638A VS RNA, and the data can be fitted to:
where kcleave is the rate of cleavage of the active ribozyme complex. KM is the Michaelis–Menten coefficient, but given the slow rates this is effectively the dissociation constant for the complex between the two RNA species. The fitting gave a maximum rate of kcleave = 0.08 min−1, and KM = 5.0 μM.
FIGURE 3.
VS ribozyme cleavage in trans is concentration dependent. Approximately 1 nM [32P]-labeled A756C VS RNA was incubated with G638A VS RNA at concentrations between 0.1 and 10 μM, and reaction progress measured as a function of time (A). The measured extents of cleavage are plotted as a function of G638A VS RNA concentration. The data for each concentration were fitted simultaneously to two exponential functions, with the amplitudes for the fast and slow cleaving fractions treated as global parameters. G638A VS RNA concentrations: 0.1 μM, open circles; 0.3 μM, closed circles; 0.75 μM, open triangles, inverted; 1.0 μM, closed triangles, inverted; 1.75 μM, open diamonds; 3.0 μM, closed diamonds; 4.5 μM, open squares; 6.5 μM, closed squares; 8.5 μM, open triangles; 10 μM, closed triangles. (B) Plot of the fast rates from the average of two sets of kinetic data, fitted to the Michaelis–Menten Equation 1.
These results also indicate that a trans reaction would be expected to occur between natural sequence ribozymes, although this would not normally be detected. In the complementation experiment, this would lower the concentration of available active ribozyme, leading to an underestimate of the affinity of the interaction required for catalyzed cleavage (i.e., increasing the measured value of KM). Assuming that the Kd values for the sequence variants and the natural ribozyme are very similar in value (the Kd values for natural and G638A helix I for helix II–VI ribozyme are closely similar) (Wilson et al. 2007), this should increase the measured KM by a factor of 3, and thus KM = 1.7 μM.
The same trans complementation behavior was also observed with a smaller version of a cis-acting VS ribozyme lacking stem VII.
CONCLUSION
The successful complementation of the A756C and G638A variants provides confirmatory evidence for the composite nature of the active site of the VS ribozyme. The trans reaction between complete ribozyme molecules shows that helix I can disengage from one ribozyme, and that from another ribozyme molecule can then dock in its place. The similarity in measured KM to that measured for the cleavage of an isolated helix I catalyzed by a helix II–VI ribozyme suggests that the energetic penalty to forming the trans complex between complete ribozymes is low. This reaction could therefore be significant in the biology of the VS RNA, most likely occurring between widely separated ribozymes contained within the same VS transcript.
MATERIALS AND METHODS
Synthesis of DNA templates
The double-stranded DNA templates were generated by PCR using overlapping oligonucleotides. The oligonucleotides corresponding to the cis VS RNA template were:
GGTAATACGACTCACTATAGGGAGTAGAGTGTCGCAATCTGCGAAGGGCGTCGTCGCCCCGAGCGGTAGTAAG;
CAGTACTGAAATTGGAGGTGAGTTCCCTGCTTACTACCGCTCGGGGC;
CTCACCTCCAATTTCAGTACTGAAATTGTCGTAGCAGTTGACTACTGTTATGTGATTGGTAGAGGCTAAGTGACGGTATTGGC; and
GGGAGTAAAATATTCTCGCAAGCGGGCTTGTGCTGTGCTGCAATACTGACTTACGCCAATACCGTCACTTAGCC.
The template was amplified using primers GGTAATACGACTCACTATAGGG and GGGAGTAAAATATTCTCGCAAG. The italic sequence is the untranscribed section of the T7 promoter. The nucleotides that template G638 and A756 are highlighted by underlining. The PCR reaction was performed with KOD DNA polymerase (Novagen) using a 50-nM template DNA and 1 μM primer oligonucleotides, for 30 cycles at an annealing temperature of 55°C.
RNA synthesis
RNA of inactive VS ribozyme was transcribed from 15 pmol of PCR product in the presence of 40 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 2 mM spermidine, 4 mM of each NTP (Roche), 0.1 U of pyrophosphatase (Sigma), 0.01% Triton X-100 (Sigma), and T7 RNA polymerase for 2.5 h at 37°C. To reduce the amount of self-cleavage occurring during the transcription of active cis VS RNA, the reaction was performed for 1 h in a buffer in which the Tris was replaced by 40 mM TAPS (pH 9.0). Transcribed RNA was purified on 5% polyacrylamide (19:1) gel in the presence of 7 M urea. The RNA was visualized by UV shadowing and a gel slice containing to the full-length cis VS RNA was excised. The RNA was recovered by electroelution into 8 M ammonium acetate and ethanol precipitation. The RNA pellet was washed twice with 70% ethanol and resuspended in water. Its concentration was determined by absorbance of light at 260 nm.
Radioactive labeling of RNA
A total of 0.5 μM VS RNA was dephosphorylated using antarctic phosphatase (NEB) for 30 min at 37°C followed by enzyme inactivation at 65°C for 5 min. A total of 0.3 μM dephosphorylated RNA was phosphorylated using 80 nM of [γ-32P]-ATP (6000 Ci/mmol; Perkin-Elmer) and T4 polynucleotide kinase (NEB) at 37°C for 30, followed by gel electrophoretic purification as before.
Kinetic measurements of ribozyme cleavage
Standard reaction conditions were 50 mM Tris (pH 8), 10 mM MgCl2, 25 mM KCl, 2 mM spermidine. Then 1 nM [5′-32P]-radioactively labeled VS RNA was heated at 65°C in the presence of 25 mM KCl for 1 min, and slowly cooled to 30°C. The RNA was then incubated at 37°C for 5 min, the solution adjusted to reaction buffer lacking MgCl2 and then incubated for a further 15 min at 37°C. Cleavage was initiated by adding MgCl2 to a final concentration of 10 mM. Aliquots (1.2 μL) were removed at intervals and the reaction terminated by addition to 5 μL of a mixture containing 95% (v/v) formamide, 20 mM EDTA and electrophoresis dyes. Ribozyme and cleavage products were separated by electrophoresis in a 20% polyacrylamide gel containing 7 M urea. Gels were exposed to storage phosphor screens, and quantified by phosphorimaging. Progress curves were fitted by nonlinear regression analysis to two exponential functions.
In the complementation experiments a trace amount (∼1 nM) of [5′-32P]-radioactively labeled VS A756C RNA was mixed with a given concentration (1.5–15 μM) unlabeled VS G638A RNA in 25 mM KCl. Each RNA was incubated for 15 min at 37°C in complete reaction buffer, and the reaction was initiated by mixing the two solutions.
ACKNOWLEDGMENTS
We thank Dr. Tim Wilson for discussions and Cancer Research UK for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1759009.
REFERENCES
- Beattie TL, Olive JE, Collins RA. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc Natl Acad Sci. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard P, Lacroix-Labonte J, Desjardins G, Lampron P, Lisi V, Lemieux S, Major F, Legault P. Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA. 2008;14:736–748. doi: 10.1261/rna.824308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HCT, Collins RA. Efficient trans-cleavage of a stem–loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FD, Ryder SP, Strobel SA. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5′- and 3′-termini. Nucleic Acids Res. 2001;29:5115–5120. doi: 10.1093/nar/29.24.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:1–5. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DMJ. Structure, folding, and activity of the VS ribozyme: Importance of the 2-3-6 helical junction. EMBO J. 2001a;20:1415–1424. doi: 10.1093/emboj/20.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Norman DG, Lilley DMJ. The A730 loop is an important component of the active site of the VS ribozyme. J Mol Biol. 2001b;312:663–674. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- Lafontaine DA, Norman DG, Lilley DMJ. The global structure of the VS ribozyme. EMBO J. 2002a;21:2461–2471. doi: 10.1093/emboj/21.10.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DA, Wilson TJ, Zhao Z-Y, Lilley DMJ. Functional group requirements in the probable active site of the VS ribozyme. J Mol Biol. 2002b;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- Lilley DMJ, Eckstein F, editors. Ribozymes and RNA catalysis. Royal Society of Chemistry; Cambridge, UK: 2008. [Google Scholar]
- Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DMJ. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod AC, Lilley DMJ. Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry. 2004;43:1118–1125. doi: 10.1021/bi035790e. [DOI] [PubMed] [Google Scholar]
- Murchie AIH, Thomson JB, Walter F, Lilley DMJ. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DMJ. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AH, Olive JE, McLaren M, Collins RA. Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry. 2006;45:13394–13400. doi: 10.1021/bi060769+. [DOI] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- Smith MD, Collins RA. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc Natl Acad Sci. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood VD, Collins RA. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J Mol Biol. 2002;320:443–454. doi: 10.1016/s0022-2836(02)00521-1. [DOI] [PubMed] [Google Scholar]
- Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat Struct Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- Wilson TJ, McLeod AC, Lilley DMJ. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]