Abstract
Drosophila melanogaster expresses three classes of small RNAs, which are classified according to their mechanisms of biogenesis. MicroRNAs are ∼22–23 nucleotides (nt), ubiquitously expressed small RNAs that are sequentially processed from hairpin-like precursors by Drosha/Pasha and Dcr-1/Loquacious complexes. MicroRNAs usually associate with AGO1 and regulate the expression of protein-coding genes. Piwi-interacting RNAs (piRNAs) of ∼24–28 nt associate with Piwi-family proteins and can arise from single-stranded precursors. piRNAs function in transposon silencing and are mainly restricted to gonadal tissues. Endo-siRNAs are found in both germline and somatic tissues. These ∼21-nt RNAs are produced by a distinct Dicer, Dcr-2, and do not depend on Drosha/Pasha complexes. They predominantly bind to AGO2 and target both mobile elements and protein-coding genes. Surprisingly, a subset of endo-siRNAs strongly depend for their production on the dsRNA-binding protein Loquacious (Loqs), thought generally to be a partner for Dcr-1 and a cofactor for miRNA biogenesis. Endo-siRNA production depends on a specific Loqs isoform, Loqs-PD, which is distinct from the one, Loqs-PB, required for the production of microRNAs. Paralleling their roles in the biogenesis of distinct small RNA classes, Loqs-PD and Loqs-PB bind to different Dicer proteins, with Dcr-1/Loqs-PB complexes and Dcr-2/Loqs-PD complexes driving microRNA and endo-siRNA biogenesis, respectively.
Keywords: Drosophila melanogaster, Dicer, double-stranded RNA-binding proteins (dsRBPs), Loquacious, endo-siRNA processing, transposon silencing
INTRODUCTION
Drosophila melanogaster expresses a wide variety of small RNAs, which are classified based on their mechanism of biogenesis and the Argonaute proteins to which they bind. MicroRNAs (miRNAs) are a class of ubiquitously expressed small RNAs, typically, ∼22–23 nucleotides (nt) in length. They are derived from endogenous transcripts capable of forming hairpin-like structures, which are sequentially processed by Drosha/Pasha and Dcr-1/Loqs complexes (Lee et al. 2003, 2004; Denli et al. 2004; Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). They predominantly associate with Argonaute-1 (AGO1) and regulate the expression of protein-coding genes (Bartel 2004; Bushati and Cohen 2007; Eulalio et al. 2008). Piwi-interacting RNAs (piRNAs), typically, ∼24–28 nt in length, associate with Piwi-family proteins. The expression of piRNAs is mainly restricted to gonadal tissues, where they function in silencing of mobile elements and repeats (Aravin et al. 2007; Brennecke et al. 2007; Gunawardane et al. 2007; Klattenhoff and Theurkauf 2008). Recently, a third class of endogenous small RNAs was identified in both the germline and the soma of Drosophila: endogenous small interfering RNAs (endo-siRNAs) (Czech et al. 2008; Ghildiyal et al. 2008; Kawamura et al. 2008; Okamura et al. 2008). Endo-siRNAs are predominantly 21 nt in length and are derived either from long endogenous transcripts capable of forming extensive fold-back structures, or are processed from double-stranded regions formed by intermolecular hybridization of convergently transcribed mRNAs. Endo-siRNAs usually join Argonaute-2 (AGO2) and function in the regulation of gene expression and transposon silencing.
The biogenesis of endo-siRNAs and miRNAs depends on a number of protein complexes containing RNA processing enzymes and their dsRNA-binding protein (dsRBPs) partners. In the case of miRNA processing, the nuclear type III ribonuclease Drosha associates with the dsRBP, Pasha, and processes primary miRNA transcripts to pre-miRNAs (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004). In a second step, the cytoplasmic Dicer enzyme Dcr-1, assisted by the dsRBP Loquacious (Loqs/R3D1), further processes pre-miRNAs to mature miRNAs (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). In contrast, processing of long dsRNA precursors into siRNA duplexes depends on a second Drosophila Dicer protein, Dcr-2 (Lee et al. 2004). The canonical Dcr-2 partner R2D2 seems not to be required for the production of siRNAs. Instead, it was found to impact the loading of siRNA duplexes into the RNA-induced silencing complex (RISC) and proper guide strand selection (Liu et al. 2003; Tomari et al. 2004). In general, it is believed that the dsRBPs contribute to the substrate specificity of their partner RNA processing enzymes.
The dsRNA binding protein Loquacious was identified in Drosophila as a component of a complex that also contains the type III RNase Dicer-1 (Dcr-1). Genetic experiments suggested that Loqs was required for efficient miRNA biogenesis (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005; Liu et al. 2007). Loss of loqs mainly impacted the final step of miRNA processing as indicated by the accumulation of pre-miRNAs, which are formed by Drosha/Pasha complexes. Mutations in loqs also reduced levels of a subset of mature miRNAs, consistent with the impacts of these lesions on Drosophila viability and fertility (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005; Liu et al. 2007; Park et al. 2007; Ye et al. 2007). Recently, it was found that loss of loqs strongly reduced levels of endogenous siRNAs (endo-siRNAs) derived from structured loci in both S2 cells and flies (Czech et al. 2008; Okamura et al. 2008).
In Drosophila, the alternative splicing of loqs transcripts was reported to produce three distinct isoforms: loqs-RA, RB, and RC (Förstemann et al. 2005; Jiang et al. 2005). These are translated into three protein isoforms, Loqs-PA, PB, and PC. RB is the isoform predominantly expressed in ovaries, whereas RA is the principal isoform found in males. The third mRNA isoform, RC, was detected only in Drosophila S2 cells (Förstemann et al. 2005). While Loqs-PB was sufficient to rescue the miRNA processing defects of loqsKO flies, Loqs-PA was incapable of restoring proper miRNA processing (Park et al. 2007), indicating that these Loqs isoforms had distinct functions during development.
Here we examined the roles of individual Loqs isoforms in different small RNA pathways and characterized the activity of a novel Loqs isoform, Loqs-PD. We show that coordinated depletion of all Loqs isoforms in cultured cells affects the biogenesis of both miRNAs and endo-siRNAs, whereas cells singly depleted of Loqs-PB or Loqs-PD show an impact only on the miRNA or on the endo-siRNA pathway, respectively. While the re-expression of Loqs-PD restored endo-siRNA levels in cultured cells that had been depleted of all Loqs isoforms, Loqs-PD was incapable of rescuing miRNA processing defects. Moreover, we show that Loqs-PD preferentially interacts with Dcr-2, the enzyme responsible for the processing of all endo-siRNA species. Considered together, our studies demonstrate that a single Loquacious isoform, Loqs-PD, is necessary and sufficient for the biogenesis of several types of endogenous siRNAs.
RESULTS AND DISCUSSION
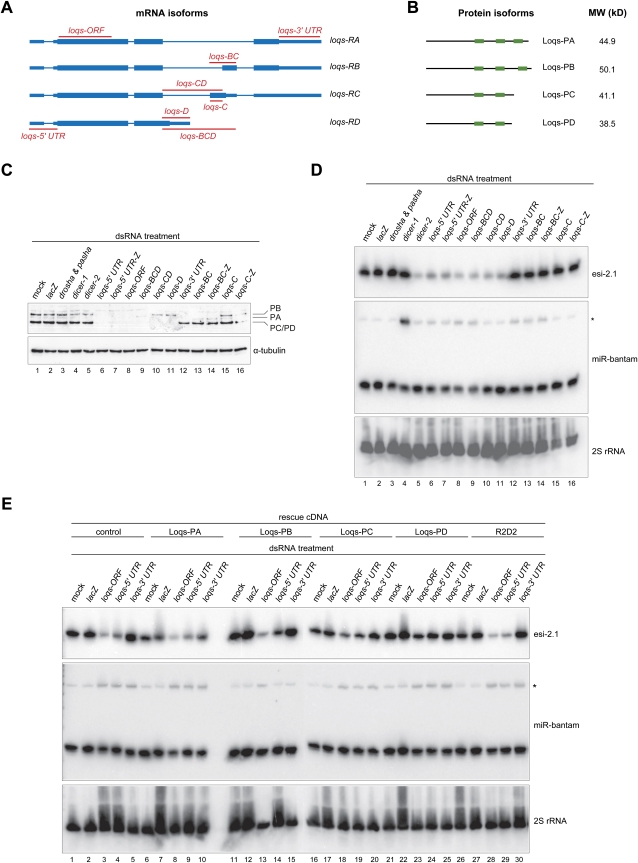
Using quantitative proteomics of Loqs immunoprecipitates from flies and Drosophila S2 cells, we identified physical interactions of Loqs with both Dcr-1 and Dcr-2 (Czech et al. 2008). In order to identify the Loqs isoforms involved in these interactions, we analyzed our proteomics data from immunoprecipitates, prepared using an antibody specific to the N terminus of endogenous Loqs proteins, for isoform-specific peptides. We found no significant peptide evidence for Loqs-PC in cultured cells and flies, whereas both isoforms PA and PB were present. In addition, we detected peptides corresponding to an as-yet-uncharacterized form of Loquacious. These could be assigned to a novel isoform identified by Förstemann and colleagues and termed Loqs-PD (Fig. 1A, 1B; JV Hartig, S Esslinger, R Böttcher, K Saito, and Förstemann K, unpubl.).
FIGURE 1.
A specific Loqs isoform, Loqs-PD, is required for the biogenesis of endo-siRNAs derived from structured loci. (A) Shown is a scheme of the annotated genomic structure of the four isoforms of Loquacious. The regions targeted by isoform-specific dsRNAs are indicated. (Thin lines) Introns; (thin boxes) UTRs; (thick boxes) ORFs. (B) A scheme showing domain structures and molecular weights of the Loqs isoforms. (Small boxes) dsRNA-binding domains. (C) Western blots showing steady-state protein levels of Loqs isoforms upon treatment of S2 cells with the indicated dsRNAs. α-Tubulin served as a loading control. To increase the length of dsRNAs to facilitate dsRNA uptake by S2 cells, some dsRNAs were fused with a lacZ carrier sequence (indicated by “-Z”). (D) Northern blots probing levels of an endo-siRNA derived from a structured locus, esi-2.1, and the microRNA miR-bantam (pre-miRNA indicated by the asterisk) in S2 cells treated with dsRNA against the indicated genes (as in C). As a loading control, the membrane was stripped and re-probed for 2S rRNA. (E) Expression of Loqs-PD is sufficient to rescue endo-siRNA processing defects, while only Loqs-PB restores proper miRNA biogenesis. Control cells expressing the TAP epitope alone or those expressing various TAP-tagged proteins were treated twice with various dsRNAs (as indicated on top of the panel) for a total duration of 8 d. The expression of the transgenes was controlled by the basal activity of the metallothionein promoter. Total RNAs were prepared and subjected to Northern blotting using probes against esi-2.1, miR-bantam, and 2S rRNA.
To characterize the roles of individual Loqs isoforms in small RNA biogenesis, we examined isoform-specific effects on miRNA and endo-siRNA processing. We depleted various combinations of Loqs proteins using isoform-specific dsRNAs in Drosophila S2 cells (Fig. 1A). Treatment of S2 cells with dsRNAs targeting loqs-RA, RB, or RC either singly or in combination led to a reduction in steady-state levels of the corresponding transcripts, as measured by RT-PCR (data not shown). In addition, Western blotting confirmed the depletion of specific Loqs isoforms by the corresponding dsRNAs (Fig. 1C). Notably, while Loqs-PC and PD could not be efficiently resolved by PAGE, cells treated with dsRNAs specifically targeting the PD isoform were effectively depleted of the Loqs protein species migrating at the position of PC/PD (Fig. 1C, lane 11). This indicated that the Loqs-PC isoform was not detectably expressed and confirmed our findings from the quantitative proteomic analysis of Loqs immunoprecipitates.
We examined the impact of isoform-specific knockdowns on the biogenesis of a prevalent endo-siRNA (Fig. 1D, esi-2.1) and a miRNA (Fig. 1D, miR-bantam). Depletion of all isoforms upon treatment with dsRNAs targeting common 5′-UTR or ORF sequences resulted in the accumulation of the miR-bantam precursor and in a strong reduction of esi-2.1 levels (Fig. 1D, lanes 6–8), whereas depletion of isoforms PA, PB, and PC by dsRNAs targeting isoform-specific 3′-UTRs only affected miRNA processing (Fig. 1D, lane 12). We observed miRNA processing defects in all cells treated with dsRNAs that cotargeted Loqs-PB (Fig. 1D, lanes 6–9,12–14), whereas the biogenesis of endo-siRNA was affected only if cells were treated with dsRNAs that target Loqs-PD, either singly or together with other isoforms (Fig. 1D, lanes 6–11). Notably, depletion of Loqs-PD alone caused a strong reduction in esi-2.1 levels, while miRNA processing was unaffected (Fig. 1D, lane 11).
To validate these findings, we depleted all Loqs isoforms in S2 cells using dsRNA targeting shared 5′-UTR sequences and tested whether the subsequent introduction of RNAi-resistant ORFs directing expression of individual Loqs isoforms was capable of rescuing defects in endo-siRNA or miRNA processing. In control cells, depletion of all Loqs isoforms led to a significant reduction in levels of esi-2.1 (Fig. 1E, lanes 3,4), whereas dsRNAs targeting loqs 5′- or 3′-UTRs or targeting ORFs caused a moderate accumulation of the miR-bantam precursor (Fig. 1E, lanes 3–5). The expression of Loqs-PA, Loqs-PC, or R2D2 failed to rescue any observed biogenesis defect (Fig. 1E, lanes 6–10,16–20,26–30). In contrast, the re-expression of Loqs-PB effectively rescued miRNA-processing defects (Fig. 1E, cf. pre-miR-bantam levels in lanes 14,15 and lane 13). However, Loqs-PB failed to restore normal endo-siRNA levels (Fig. 1E, lane 14). The re-expression of Loqs-PD restored levels of esi-2.1 in Loqs-depleted cells (Fig. 1E, lane 24) but was incapable of rescuing miRNA-processing defects (Fig. 1E, cf. lanes 24,25 and lane 23). Our observation that the expression of R2D2 was incapable of rescuing any small RNA-processing defect caused by depletion of Loqs strongly suggests that R2D2 and all Loqs isoforms cannot function in a redundant manner. Considered together, these data indicate that only the Loqs-PB isoform is required for the biogenesis of miRNAs and suggest that only Loqs-PD is essential for endo-siRNA production.
To support results emerging from cell culture, we also examined the roles of Loqs isoforms in vivo. Flies expressing the Loqs-PB isoform under the control of its endogenous regulatory elements could rescue both the pre-miRNA-processing defects and the pronounced phenotypes of loqs-null animals (Park et al. 2007). Strikingly, homozygous mutant loqsKO flies carrying a Loqs-PB transgene did not regain normal levels of the endo-siRNA, esi-2.1, but did express normal amounts of miR-8 (Supplemental Fig. S1A). Similarly, the introduction of Loqs-PB also restored normal miR-8 levels in loqsf00791 homozygous mutant flies, whereas endo-siRNA biogenesis defects were not affected (Supplemental Fig. S1B). The expression of Loqs-PA in loqsf00791 homozygous mutant flies was incapable of restoring either miRNA or endo-siRNA processing (Supplemental Fig. S1B). These results are consistent with the observation from cell-based studies showing that neither Loqs-PA nor PB is required for the endo-siRNA pathway. We conclude that Loqs-PB is required for miRNA biogenesis in multiple cell types, whereas Loqs-PD supports endo-siRNA biogenesis. Loqs-PC seems neither to be expressed at significant levels nor to impact small RNA biogenesis. The function of Loqs-PA, which is expressed both in cultured cells and in animals, remains elusive.
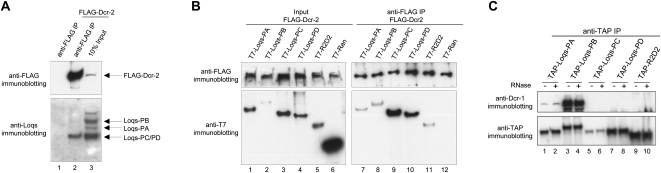
Given the unexpected requirement for Loqs in the biogenesis of endogenous siRNAs (endo-siRNAs) (Czech et al. 2008; Okamura et al. 2008) and the physical interaction between Loqs and both Dcr-1 and Dcr-2 (Czech et al. 2008), we sought to investigate whether individual Loqs isoforms might show specificity for either Dicer protein. Immunoprecipitation of Flag-tagged Dcr-2 followed by immunoblotting with an antibody specific to the N-terminus of endogenous Loqs, and thus recognizing all known Loqs isoforms, revealed a strong signal corresponding to the molecular weight of Loqs-PC/PD (Fig. 2A, lane 2). Since the levels of endogenous Loqs-PC in S2 cells are negligible, the predominant Dcr-2-interacting endogenous Loqs isoform appears to be Loqs-PD. Next, we examined the interactions between various Loqs isoforms and Dcr-2 by expressing Flag-tagged Dcr-2 together with T7-tagged Loqs isoforms in S2 cells. Cell extracts were subjected to anti-Flag immunoprecipitation, and coimmunoprecipitated proteins were detected using an anti-T7 antibody. All four tagged Loqs isoforms and the positive control T7-R2D2 were able to interact with Dcr-2 (Fig. 2B, lanes 7–11), while no interaction was detected for the negative control T7-Ran (Fig. 2B, lane 12). Therefore, Dcr-2 seems capable of interacting with all isoforms of Loqs if they are overexpressed. Interestingly, we also found that Loqs isoforms are capable of forming homo- and heterodimers (or even oligomers) with each other, as Flag-tagged Loqs isoforms PA, PB, and PC are capable of pulling down T7-tagged Loqs-PA in coimmunoprecipitation assays (Supplemental Fig. S3; data not shown). This could account for the apparent lack of specificity in the interaction between Dcr-2 with individual Loqs isoforms upon overexpression. We also probed the interaction between Loqs isoforms and Dcr-1. We detected robust interaction between endogenous Dcr-1 and TAP-Loqs-PB and a weak interaction between Dcr-1 and TAP-Loqs-PA (Fig. 2C, lanes 1–4). In contrast, no endogenous Dcr-1 was detected in the TAP-Loqs-PC, TAP-Loqs-PD, or TAP-R2D2 complexes.
FIGURE 2.
Interaction of Loqs isoforms with Dicer proteins. (A) Flag-tagged Dcr-2 coimmunoprecipitates endogenous Loqs-PD. Flag-Dcr-2 immunoprecipitates were subjected to immunoblotting using antibodies against the N terminus of endogenous Loqs and against the Flag epitope. (B) Flag-Dcr-2 was expressed in S2 cells together with T7-tagged R2D2 or various Loqs isoforms. T7-Ran served as a control. Flag-Dcr-2 was immunoprecipitated using an anti-Flag antibody, and the interaction with coexpressed proteins was examined by Western blotting using an anti-T7 antibody. (C) Coimmunoprecipitation of endogenous Dcr-1 with TAP-tagged Loqs isoforms. TAP-tagged proteins were expressed in S2 cells, immunoprecipitated using antibodies against the TAP epitope, and subjected to immunoblotting using antibodies against endogenous Dcr-1 and the TAP tag, respectively. RNase treatment (as indicated on top of the panel) revealed that the strong interaction between Dcr-1 and Loqs-PB as well as the weak interaction between Dcr-1 and Loqs-PA are RNA-independent.
To test whether the observed binding resulted from protein–protein interaction or from Loqs and Dicer proteins binding independently to the same RNA substrates, we treated immunoprecipitates with RNase. This had no significant effect on the observed interactions (Fig. 2C; data not shown). Overexpression of neither a pri-miRNA (pri-miR-bantam), nor a precursor of the endo-siRNA cluster, esi-2 altered patterns of differential affinity (data not shown), indicating that our experiments detected RNA-independent protein–protein interactions. Therefore, we conclude that two Loqs isoforms, Loqs-PB and to a lesser extent Loqs-PA, are capable of interacting with Dcr-1, a result consistent with findings reported by Förstemann et al. (2005). In contrast, Dcr-2 predominantly interacts with endogenous Loqs-PD, while other isoforms interact with Dcr-2 to a significant extent only when overexpressed. Considering the restriction of loqs-PC transcripts to S2 cells (Förstemann et al. 2005) and the negligible protein levels of Loqs-PC, we conclude that endogenous Loqs-PD is the predominant isoform that interacts with Dcr-2.
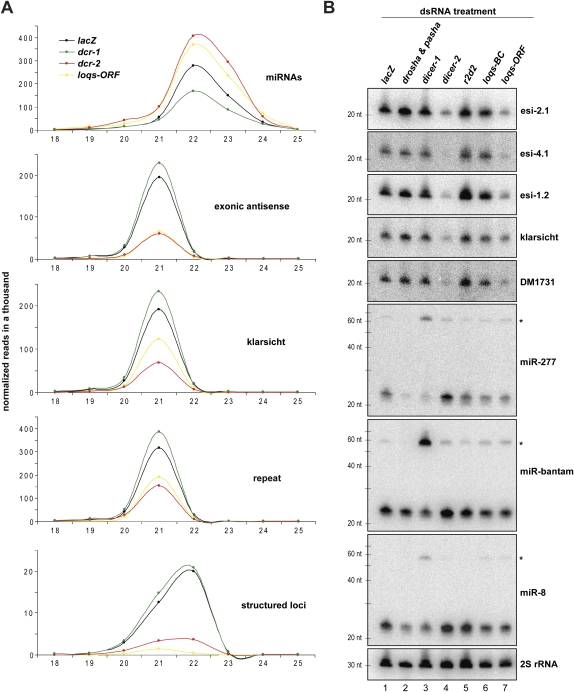
In order to investigate the effects of Loqs depletion on small RNA profiles in more detail, we prepared small RNA libraries from S2 cells treated with loqs-ORF dsRNAs and from control knockdown cells, including lacZ, dcr-1, dcr-2, r2d2, and untreated cells (“mock”). These were deep-sequenced using the Illumina platform. Normalized cloning counts were plotted by read length to create size profiles (Fig. 3A). The indicated small RNA categories were isolated from the total library bioinformatically (Czech et al. 2008). Mature microRNAs populated a broad peak centered around 22–23 nt, and these were decreased most prominently in the dcr-1 knockdown. MicroRNAs appeared to be slightly increased in loqs-ORF and dcr-2 knockdowns as compared to lacZ (Fig. 3A), probably because of an artifact of library normalization due to the strong impact on endo-siRNA levels (Figs. 1D, 3B). Endo-siRNAs mapping to overlapping transcripts (exonic antisense) were strongly reduced in dcr-2 and loqs-ORF knockdowns (Fig. 3A). A moderate reduction was also observed for endo-siRNAs derived from the klarsicht locus upon Loqs depletion, while dcr-2 knockdowns had more prominent impacts. Finally, knockdowns of dcr-2 or loqs caused a substantial (∼50%) reduction in endo-siRNAs corresponding to repeat and transposon sequences, while the levels of these small RNAs remained unchanged in all other knockdowns tested (Fig. 3A).
FIGURE 3.
Loqs is involved in the biogenesis of several classes of endo-siRNAs. (A) Length profiles for small RNAs isolated from S2 cells treated with the indicated dsRNAs are shown. Known miRNAs and distinct classes of endo-siRNAs (as indicated) were split computationally, and normalized cloning counts were plotted over their length. (B) Northern blots show levels of miRNAs (pre-miRNAs indicated by asterisks), endo-siRNAs derived from structured loci, the LTR transposon DM1731, and the S2 cell-specific klarsicht locus in S2 cells treated with the indicated dsRNAs. 2S rRNA served as a control for equal loading.
We validated results from small RNA sequencing by Northern blotting. We saw decreased levels of three independent endo-siRNAs derived from structured loci (esi-1.2, esi-2.1, esi-4.1) in dcr-2 and loqs-ORF knockdowns, while knockdown of loqs-BC had no similar impact (Fig. 3B). Consistent with these small RNA sequencing and Northern blotting results, depletion of all Loqs isoforms, but not depletion of Loqs-BC or Loqs-C, caused a de-repression of a Renilla luciferase sensor for esi-2.1 (Supplemental Fig. S2). We also noted a reduction of klarsicht siRNAs and sequences derived from the transposon DM1731 upon loqs-ORF knockdown, but not upon Loqs-BC depletion (Fig. 3B). Probing the same membrane for three miRNAs revealed a slight reduction in mature miRNA levels and a slight but detectable accumulation of pre-miRNAs in dcr-1, loqs-ORF, and loqs-BC knockdown cells (Fig. 3B). Considered together, these results support a dependence of various categories of endo-siRNAs on the Loqs-PD isoform.
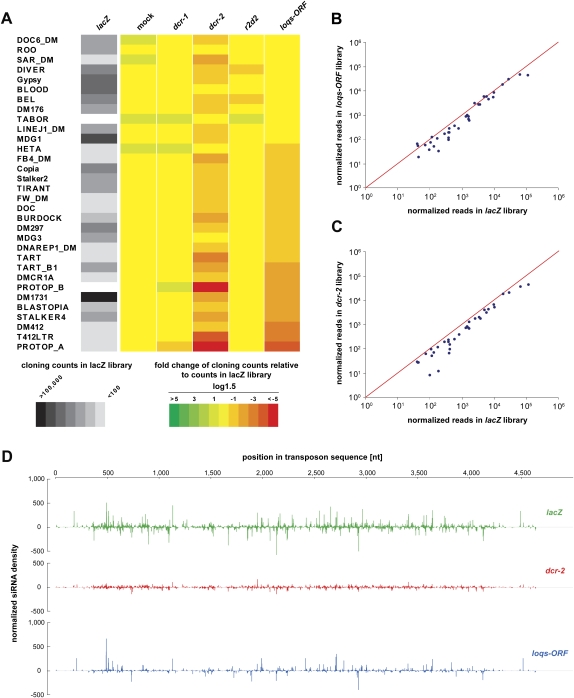
Given the effect of loqs-ORF knockdowns on repeat endo-siRNAs, we probed the potential impact of loqs mutations on transposon silencing. All endo-siRNAs that correspond to repeats and that were 21 nt in length were extracted bioinformatically, and heat maps showing their relative abundance were created for the indicated libraries (Fig. 4A). Knockdown of dcr-2 caused a reduction of endo-siRNA sequences for the majority of transposable elements as compared to the number of reads in the lacZ library, while untreated cells (“mock”), dcr-1 and r2d2 knockdowns, showed only minor, if any, effects. Depletion of all Loqs isoforms reduced levels of repeat endo-siRNAs, which at least in part correlated with dcr-2 depletion. Interestingly, we noted that knockdown of r2d2 showed a weak impact on those repetitive elements that did not depend on Loqs depletion. Plotting the abundance of repeat siRNAs in Loqs or Dcr-2-depleted cells against those in the lacZ control library showed similar patterns, with some siRNAs appearing unaffected and others showing drastic reduction in loqs-ORF or dcr-2 knockdowns (Fig. 4B,C). Plotting the normalized read number of endo-siRNAs matching to the transposable element DM1731, which shows the highest levels of transposon-derived endo-siRNAs in S2 cells, for the lacZ, dcr-2, and loqs-ORF libraries also shows clear dependence on dcr-2 and loqs (Fig. 4D). Interestingly, depletion of Dcr-2 and Loqs does not show identical impacts on the patterns of individual sequences over this representative transposon (Fig. 4D). This could indicate multiple overlapping biogenesis pathways or differential affinities of one machinery for different processing sites.
FIGURE 4.
Depletion of Loqs in S2 cells results in reduced levels of endo-siRNAs derived from repeats and transposable elements. (A) Heat maps in grayscale show cloning frequencies of 21-nt-size siRNAs matching individual transposons in S2 cells treated with lacZ dsRNA. Fold changes of cloning counts relative to those in the lacZ library are shown in color (red-green scale) in log1.5 scale. Note that only transposons passing a cloning count threshold of 5 in a single library and 500 in all libraries together are shown. (B,C) All siRNAs matching to the transposons indicated in A with more than 50 reads in the lacZ library are plotted against the loqs-ORF and dcr-2 libraries, respectively. (D) All 21-nt endo-siRNAs in the lacZ, dcr-2, and loqs-ORF libraries that match to the transposable element DM1731 are plotted over the transposon sequence with their normalized cloning counts on the y-axis.
Previous studies indicated that Dcr-1 preferentially binds Loqs-PB (Förstemann et al. 2005; Ye et al. 2007), and that Dcr-2 is capable of interacting with Loqs proteins, although isoforms were not examined in that study (Czech et al. 2008). Here we have characterized the role of a novel Loqs isoform, Loqs-PD, in the biogenesis of various categories of endo-siRNAs. We present evidence that Loqs-PD, in addition to the canonical partner R2D2 (Liu et al. 2003), is the predominant interacting partner for Dcr-2 in S2 cells. Our genetic studies clearly demonstrate the differential requirement for Loqs-PD and Loqs-PB in the biogenesis of endo-siRNAs and miRNAs, respectively. Thus, it is likely that besides the intrinsic substrate specificity of the individual Dicer proteins, the identity of Dicer interacting cofactors is also crucial for the biogenesis of different classes of small RNAs. Although small dsRNA-binding proteins are clearly important for small RNA production and act as cofactors for Dicer and Drosha enzymes, their precise biochemical roles remain unclear. Our studies add complexity to the landscape of the roles of dsRBPs in small RNA biogenesis and raise fundamental questions about why so many distinct forms of these proteins are required for these pathways to operate.
MATERIALS AND METHODS
DNA constructs
An ∼500-base-pair DNA fragment encompassing the miR-bantam sequence was amplified by PCR and cloned into pRmHa-3 using BamHI/EcoRI sites to generate pRmHa-3-Bantam. A pair of oligos containing three imperfect binding sites for miR-bantam were annealed and cloned into pRmHa-3-Renilla (Zhou et al. 2008) using SalI to generate the sensor constructs for miR-bantam (pRmHa-3-Renilla-Bantam sensor). To generate epitope-tagged expression constructs, DNA fragments encoding epitope-tagged proteins were amplified by PCR and cloned into pRmHa-3 using the following restriction enzymes (SacI/KpnI for Flag-Dcr-2; EcoRI/BamHI for all T7-tagged Loqs isoforms; KpnI/BamHI for T7-R2D2; EcoRI/BamHI for T7-Ran). For TAP-tagged vectors, a pair of oligos was annealed and cloned into pMK33-NTAP (Veraksa et al. 2005) using XhoI and BamHI sites to generate pMK33-RZ-NTAP. DNA fragments encoding the corresponding proteins were either amplified by PCR and cloned into pMK33-RZ-NTAP using the following restriction enzymes (SacI/KpnI for Loqs-PA, PC, and PD), or subcloned from the Flag-tagged pRmHa-3 constructs using the following restriction enzymes (KpnI/BamHI for R2D2; EcoRI→blunt/BamHI-treated cDNA fragment cloned into SacI→blunt/BamHI-digested vector for Loqs-PB and Ran). DNA oligonucleotide sequences are listed in Supplemental Table S1.
Immunoprecipitation and immunoblotting
Cells were transfected with expression constructs for epitope-tagged proteins, induced with 500 μM CuSO4 2 d after transfection, and harvested another 24 h later. Immunoprecipitation and RNase treatment were performed as previously described (Czech et al. 2008; Zhou et al. 2008). The immunoprecipitated samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and probed with antibodies against the T7 epitope (Novagen), the Flag-epitope (Sigma), the TAP-tag (Open Biosystems), or rabbit antibodies against Dcr-1 or Loqs. α-Tubulin was detected using an anti-α-tubulin antibody raised in mouse (Sigma).
Cell culture and dsRNA treatment
S2-NP cells were maintained in Schneider's medium (Invitrogen) supplemented with 10% FBS and 1% pen-strep (Invitrogen). For dsRNA treatment, ∼3 × 106 S2-NP cells were soaked in 1.5 mL of serum-free Schneider's medium containing 10 μg of dsRNAs in 6-well plates, and 3 mL of serum-containing medium was added 45 min later. After 4 d of initial dsRNA treatment, cells were treated with dsRNAs for a second round and harvested another 4 d later. Sequences of the primers for generation the dsRNAs are listed in Supplemental Table S1.
RT-PCR
To measure levels of various loqs transcripts upon dsRNA treatment, total RNA from S2 cells was extracted using Trizol (Invitrogen). RNA was treated with RQ1 DNase (Promega) according to the manufacturer's instructions. cDNA was synthesized by reverse transcription using Quantiscript reverse transcriptase (QIAGEN) and a mixture of oligo-dT and random hexamer primers. cDNA was amplified using primers that amplify all three isoforms (products vary by size). The PCR products were separated by agarose gel electrophoresis. Analysis was carried out with two biological replicates per sample. The sequences of the DNA oligonucleotides used in this study are listed in Supplemental Table S1.
Northern blotting
Total RNA was isolated using Trizol (Invitrogen). Twenty micrograms of total RNA from S2 cells or 40 μg of total RNA from flies was separated on 15% denaturing poly acrylamide gels and transferred onto Hybond-N+ membranes (Amersham Biosciences) in 0.5× TBE. RNA was UV cross-linked to the membrane and pre-hybridized in ULTRAhyb-Oligo buffer (Ambion) for 1 h. DNA probes (sequences are listed in Supplemental Table S1) complementary to the indicated endo-siRNAs, miRNAs, and 2S rRNA were 5′ radio-labeled and added to the hybridization buffer (hybridization overnight at 30°C). Membranes were washed four to six times in 1× SSC with 0.1% SDS at 30°C and exposed to PhosphorImager screens for 12–72 h. Probes were stripped by boiling the membrane twice in 0.2× SSC containing 0.1% SDS in a microwave.
Reporter assays
Transfections were performed in 384-well plate format. For each well, a total of ∼100 ng of plasmid DNA was transfected. To measure esi-2.1 sensor activity, 100 ng of pRmHa-3-Renilla-esi-2.1 sensor (Czech et al. 2008) and 2 ng of pRmHa-3-Firefly-long (Zhou et al. 2008) (as a control for transfection efficiency) were transfected. To measure the activity of endogenous miR-bantam, 2 ng of pRmHa-3-Firefly-long, 50 ng of pRmHa-3-Renilla-Bantam sensor, and 50 ng of pRmHa-3 (serving as carrier DNA) were transfected. The activity of overexpressed miR-bantam was measured by transfecting 2 ng of pRmHa-3-Firefly-long, 80 ng of pRmHa-3-Renilla-Bantam sensor, and 20 ng of pRmHa-3-Bantam. For each well, DNA was mixed with 0.8 μL of Enhancer in 15 μL of EC (QIAGEN) and incubated for 5 min at room temperature. Then 0.35 μL of Effectene reagent was added, and the mixture was immediately dispensed into each well containing ∼80 ng of dsRNA. After incubation for 10 min at room temperature, 40 μL of S2-NP cells (106 cells/mL) were dispensed into the well. Cells were induced with 200 μM CuSO4 144 h post-transfection, and luciferase assays were performed 24 h later using DualGlo reagents (Promega). For each well, the reporter activity, referred to as relative luciferase units (RLU), was calculated as the ratio of Renilla luciferase to firefly luciferase. To calculate the effect of dsRNA treatment on the activity of specific sensors, the data points were first normalized against corresponding data points where pRmHa-3-Renilla was transfected (serving as no-site control), then normalized against samples transfected with dsRNA against LacZ.
Small RNA libraries
Small RNAs from total RNA were cloned as described (Brennecke et al. 2007) (detailed protocol available upon request). The following small RNA libraries from total RNA were prepared for this study:
19–24 nt from untreated S2 cells (“mock”);
19–24 nt from S2 cells treated with dsRNA against Dcr-1;
19–24 nt from S2 cells treated with dsRNA against Dcr-2;
19–24 nt from S2 cells treated with dsRNA against Loqs-ORF;
19–24 nt from S2 cells treated with dsRNA against R2D2; and
19–24 nt from S2 cells treated with dsRNA against LacZ.
Libraries were sequenced using the Illumina sequencing platform. Small RNA sequences were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE17171.
Bioinformatics analysis of small RNA libraries
Small RNA libraries were analyzed as described (Czech et al. 2008). Small RNA sequences were matched to the Drosophila release 5 genome and genomes of Drosophila C virus, Flock house virus, and Cricket paralysis virus. Only reads perfectly matching the fly genome or matching to viral genomes with up to three mismatches were used for further analysis. For annotations we used FlyBase for protein-coding genes, UCSC for non-coding RNAs and transposons/repeats, and the most recent miRNA catalog (Ruby et al. 2007; Stark et al. 2007).
Fly stocks
loqsKO flies and flies expressing Loqs-PA and Loqs-PB isoforms were a kind gift of Qinghua Liu (University of Texas Southwestern Medical Center) (Park et al. 2007). The hypomorphic loqsf00791 flies were obtained from Bloomington (stock #18371). AGO2414 flies were a kind gift of Haruhiko Siomi (Keiko University School of Medicine) (Okamura et al. 2004), dcr-2L811Fsx flies were a kind gift of Richard Carthew (Northwestern University) (Lee et al. 2004), and r2d21 flies were a kind gift of Dean Smith (University of Texas Southwestern Medical Center) (Liu et al. 2003). Flies were double-balanced [double-balancer stock (w; Sp/CyO; Dr/TM6C,Tb), a gift from Phil Zamore (University of Massachusetts Medical Center)], then homozygous and heterozygotes flies were collected. Stock #2057 from Bloomington (Celera sequencing strain) was used as wild type.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank J.V. Hartig and K. Förstemann and colleagues for communicating data prior to publication. We also thank Q. Liu, R. Carthew, H. Siomi, D. Smith, and P.D. Zamore for reagents. We are grateful to members of the Hannon and Perrimon laboratories for helpful discussion, and to M. Rooks and D. McCombie for help with deep sequencing. R.Z. is a Special Fellow of the Leukemia and Lymphoma Society. B.C. is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. This work was supported in part by grants from the NIH to N.P. and G.J.H., and a gift from K.W. Davis (to G.J.H.). N.P. and G.J.H are investigators of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1611309.
REFERENCES
- Aravin AA, Hannon GJ, Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu X, Park JK, Jiang F, Liu Y, McKearin D, Liu Q. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA. 2007;13:2324–2329. doi: 10.1261/rna.723707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Veraksa A, Bauer A, Artavanis-Tsakonas S. Analyzing protein complexes in Drosophila with tandem affinity purification-mass spectrometry. Dev Dyn. 2005;232:827–834. doi: 10.1002/dvdy.20272. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]