Abstract
RNAs in the mitochondria of Physarum polycephalum contain nonencoded nucleotides that are added during RNA synthesis. Essentially all steady-state RNAs are accurately and fully edited, yet the signals guiding these precise nucleotide insertions are presently unknown. To localize the regions of the template that are required for editing, we constructed a series of chimeric templates that substitute varying amounts of DNA either upstream of or downstream from C insertion sites. Remarkably, all sequences necessary for C addition are contained within ∼9 base pairs on either side of the insertion site. In addition, our data strongly suggest that sequences within this critical region affect different steps in the editing reaction. Template alterations upstream of an editing site influence nucleotide selection and/or insertion, while downstream changes affect editing site recognition and templated extension from the added, unpaired nucleotide. The data presented here provide the first evidence that individual regions of the DNA template play discrete mechanistic roles and represent a crucial initial step toward defining the source of the editing specificity in Physarum mitochondria. In addition, these findings have mechanistic implications regarding the potential involvement of the mitochondrial RNA polymerase in the editing reaction.
Keywords: RNA editing, cis-acting elements, nucleotide insertion, editing mechanisms
INTRODUCTION
The term RNA editing encompasses many disparate processes, including the addition of nonencoded nucleotides, the deletion of encoded nucleotides, and modification or substitution of RNA bases. Editing occurs in a broad spectrum of organisms, including mammals, plants, insects, viruses, and many lower eukaryotes, and in multiple subcellular locations. A number of distinct editing mechanisms have been identified, but the means by which editing sites are recognized and subsequently altered are still unknown in many systems (Gott and Emeson 2000).
A complex assortment of editing types is found in the mitochondria of Physarum polycephalum, including the insertion of nonencoded nucleotides, deletion of encoded residues, C-to-U changes, and the replacement of one nucleotide for another (for review, see Gott and Rhee 2007). All characterized mitochondrial mRNAs, tRNAs, and rRNAs in this acellular slime mold are subject to editing. Over 500 editing sites have been identified thus far, with another ∼500 sites predicted to occur in mRNAs that have yet to be characterized (Gott et al. 2005; Beargie et al. 2008). Most editing events involve the insertion of specific nucleotides at defined sites, leading to the creation of open reading frames in mRNAs and conserved elements within tRNAs and rRNAs. A significant proportion of each mature RNA is derived from insertional editing. On average, 1 of 25 nucleotides (nt) in mRNAs and 1 of 40 nt in structural RNAs are not found in the gene. A single C is added at ∼90% of these sites, with the remainder involving U, AA, AU, CU, GC, GU, or UU additions. Despite the high frequency and variety of editing events, this process is extremely precise and highly efficient; essentially all transcripts present in steady-state RNA pools are fully and correctly edited.
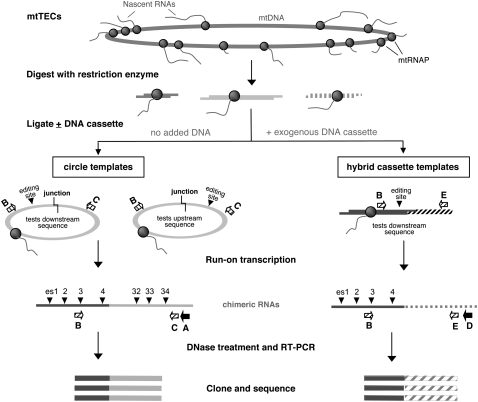
Most of what is known about the process of nucleotide insertion in Physarum has been gleaned from in vitro studies. Initial experiments using isolated mitochondria indicated that the “extra” nucleotides are added to nascent RNAs (Visomirski-Robic and Gott 1997a,b), prompting the isolation of mitochondrial transcription elongation complexes (mtTECs) for use in further studies. These partially purified mtTECs contain the entire ∼63-kb mitochondrial genome and associated molecules, including mitochondrial RNA polymerases, other proteins, and nascent RNAs (Cheng and Gott 2000). The DNA within mtTEC preparations is readily cleaved by restriction endonucleases, and the resulting ends can be efficiently ligated to one another or to exogenously added DNA fragments, allowing manipulation of the DNA template (Fig. 1; Byrne and Gott 2002). Run-on transcripts generated from mtTECs are edited to a significant extent (generally, 30%–60%), as are RNAs made after cleavage of the mtTEC DNA, with or without subsequent ligation. Interestingly, however, RNAs made from chimeric templates resulting from the ligation of mtTEC DNA with exogenous DNA fragments (derived from de-proteinized mitochondrial DNA, PCR fragments, or plasmids) are only edited in the regions transcribed from the native template (Byrne and Gott 2002), suggesting that an unknown factor(s) associated with mtDNA is required for insertional editing.
FIGURE 1.
Experimental strategy used to localize sequences required for editing. Native mitochondrial transcription elongation complexes (mtTECs) include the entire mitochondrial genome (mtDNA) and associated molecules. Only the elongating mitochondrial RNA polymerases (mtRNAP, gray circles) and nascent RNAs (thin lines) are shown for clarity. Construction of chimeric templates is accomplished by digestion of the mtDNA at restriction sites close to C insertion sites and subsequent ligation in the absence (circle templates) or presence (hybrid cassette templates) of an exogenous DNA cassette. Note that a complex mixture of products is produced in either case, and that following run-on transcription, chimera-specific primers are used for reverse transcription (RT, black arrows) and PCR (hatched arrows). Circle templates can be used to substitute DNA sequences either upstream of or downstream from editing sites (es), but only downstream sequences can be changed using hybrid cassette templates.
The mechanism of nucleotide insertion in Physarum mitochondria is unique among known editing systems. Studies using both isolated mitochondria and mtTECs led to the surprising conclusion that insertional editing occurs cotranscriptionally, with nonencoded nucleotides being added to the 3′ end of nascent transcripts during RNA synthesis (Cheng et al. 2001). In other known cotranscriptional editing systems, a homopolymeric tract located immediately upstream of the single insertion site causes the viral polymerase to stutter, resulting in the addition of one or more “pseudotemplated” nucleotides at that site (Hausmann et al. 1999). Homopolymer tracts are not present upstream of Physarum insertion sites (Miller et al. 1993; Gott et al. 2005), however, and alterations in nucleotide concentrations have opposite effects on insertion efficiency in the Physarum and viral in vitro editing systems (Cheng et al. 2001). Physarum editing is also clearly different from the uridine insertions and deletions observed in trypanosomatids, where editing is a post-transcriptional process requiring base-pairing of guide RNAs (gRNAs) to a region of the RNA substrate downstream from the editing site (Ochsenreiter and Hajduk 2007).
The signals involved in editing site recognition and nucleotide insertion in Physarum mitochondria are presently unknown. Such signals could conceivably be located in the DNA template and/or the upstream RNA. The only common sequence near C insertion sites is a purine-U immediately upstream, but only ∼70% of sites have even this minimal motif (Miller et al. 1993; Gott et al. 2005). Similarly, no shared RNA sequences or potential structures have been detected upstream of editing sites (Gott et al. 2005), and removal of most of the upstream nascent RNA has no effect on editing in vitro (A Majewski and JM Gott, unpubl.). Previous studies using chimeric templates suggested that sequences required for editing are likely to be relatively close to each site, since insertion sites within ∼20 base pairs (bp) of a ligation junction can be edited efficiently (Byrne and Gott 2002). However, editing sites are often <20-bp apart, and the minimal amount of flanking sequence required for editing was not determined in these experiments. In addition, recognition of an editing site can occur without the insertion of the correct noncoded nucleotide (Byrne et al. 2002), suggesting that recognition and insertion are separable processes. Thus, it was of interest to systematically test the effects of flanking sequences on the process of insertional RNA editing in Physarum mitochondria.
Here we delineate the DNA regions that are necessary for the insertion of nonencoded C residues using chimeric template constructs that alter the DNA template flanking editing sites. In doing so, we have determined that sequences close to the editing site are both necessary and sufficient to support insertional editing in Physarum. Our data provide strong support for distinct roles for regions of the template that lie upstream of and downstream from the editing site. These findings are particularly significant in that they offer the first clues as to where the information for nucleotide specificity may lie.
RESULTS
Alteration of sequences surrounding editing sites
Chimeric templates can be used to examine the effects of substituting DNA sequences either upstream of or downstream from insertion sites (Fig. 1; Byrne and Gott 2002). Circle templates are generated through the intramolecular ligation of DNA fragments generated by digesting mtTECs with restriction enzymes (Fig. 1, left), placing sites on either side of the ligation junction in a different sequence context. The second form of chimeric templates used here, hybrid cassette templates, is created by ligating a DNA cassette to restriction-digested mtTECs. Only downstream sequences can be altered in these constructs (Fig. 1, right). We have used both types of templates to determine the effects of substituting various amounts of flanking DNA on insertional editing, using restriction sites that fall near editing sites found in highly to moderately expressed genes.
Transcription/editing reactions were carried out under nucleotide concentrations favoring editing at the insertion site closest to the junction (Cheng et al. 2001). After run-on transcription, nucleic acids were extracted and DNase-treated, and RT-PCR products derived from RNAs transcribed from chimeric templates were generated through the use of chimera-specific primer sets, cloned, and sequenced (Fig. 1). Note that although we examined hundreds of clones in the course of these experiments, we only show the data for individual clones that are clearly independent, i.e., that have different combinations of edited and unedited sites within the fragment and thus must have originated from distinct transcripts.
Downstream element boundary
Previous experiments examining “run-off” transcripts synthesized from mtTECs digested with various restriction enzymes indicated that 14 bp of native downstream DNA was sufficient to support editing, but 6 bp was not (A Majewski and JM Gott, unpubl.). However, these experiments could not distinguish whether the lack of editing was the result of the disruption of a cis-acting element present in the downstream region or simply due to the absence of downstream DNA in the elongation complex, which would likely affect complex stability. To differentiate between these two possibilities, we turned to chimeric templates, which substitute rather than remove downstream DNA (Fig. 1; Byrne and Gott 2002).
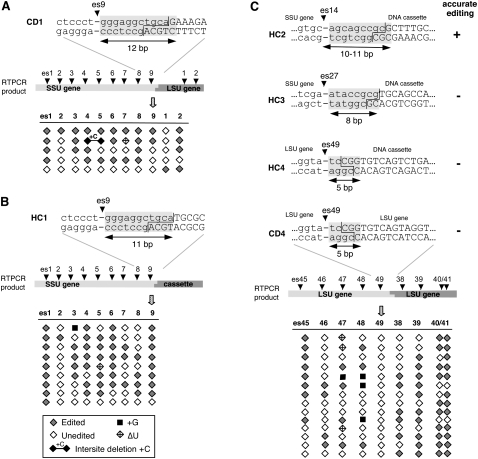
Initial experiments were carried out using circle template downstream 1 (CD1) (Fig 2A), which contains the 5′ portion of the small ribosomal RNA (SSU) gene fused to the gene for the large ribosomal RNA (LSU). The editing site closest to the ligation junction, editing site 9 (es9) of SSU, has native sequence upstream of the editing site and 12 bp of natural downstream sequence. Editing patterns of independent clones from a typical experiment are shown schematically in the lower portion of Figure 2A. Similar to RNAs generated by native mtTECs (Byrne et al. 2002; Byrne and Gott 2004), transcripts derived from the CD1 template contained unedited (Fig. 2, open diamonds), correctly edited (Fig. 2, gray diamonds), and mis-edited sites (Fig. 2, see the legend), including an intersite deletion (a precise deletion of encoded nucleotides with endpoints at editing sites) with an added C. Transcription under these and other conditions yielded numerous clones having an inserted C at SSU es9, indicating that 12 bp of natural downstream sequence is sufficient for editing at this site.
FIGURE 2.
Localization of sequences downstream from insertion sites required for editing. (A,B) Template alterations downstream from editing site 9 (es9) within the gene encoding the mitochondrial small ribosomal rRNA (SSU). Circle template CD1 was created by fusion at PstI sites within the SSU and LSU (large rRNA) genes. Hybrid cassette template HC1 has exogenous DNA ligated at the same SSU site. Junction regions are expanded to indicate substituted nucleotides, shown in uppercase. Shaded regions indicate downstream sequence identical to that of native mtTECs. Editing sites within each RT-PCR product are indicated. The editing status of individual editing sites is shown schematically, with filled diamonds representing correctly edited sites, empty diamonds indicating unedited sites, and mis-edited sites as indicated in the legend. Both CD1 and HC1 support C insertion at SSU es9. Only independent clones (i.e., clones having unique editing patterns) are shown for each construct. (C, top) Junction regions of other templates with alterations downstream from editing sites. The ability of each template to support accurate editing at the site nearest the junction is indicated at the right. (Bottom) The editing status of individual sites in independent CD4 cDNA clones is shown as in A and B.
Because only a limited number of downstream restriction sites were compatible with efficient circle template formation, hybrid cassette templates were used to further define the downstream boundary. We first confirmed that the ligation of an exogenous DNA fragment downstream from an editing site would not inhibit editing at a site within endogenous mtDNA. To test this, HC1, a hybrid cassette version of CD1, was constructed by ligating a DNA cassette to the same PstI site used to create CD1. This HC1 construct contains 11 rather than 12 bp of natural sequence immediately downstream from SSU es9 (Fig. 2), substituting non-Physarum sequence for the LSU gene present in CD1. Most independent clones were edited at es9 in RT-PCR clones derived from HC1 transcripts (Fig. 2B), indicating that the presence of exogenous downstream DNA did not adversely affect editing within the endogenous mtDNA and demonstrating that 11 bp of native downstream DNA supports efficient editing.
We next made a series of hybrid cassette constructs to determine what template elements are required for accurate insertion of nonencoded nucleotides. Independent clones derived from HC2 transcripts were correctly edited at SSU es14 (C insertion next to an encoded C), indicating that 10–11 bp of the downstream sequence (9–10 bp of native DNA) are sufficient for accurate editing (Fig. 2C). In contrast, transcripts derived from the HC3 and HC4 templates (containing 8 bp and 5 bp of native downstream DNA, respectively) were never correctly edited at the insertion sites immediately upstream of the junction (Fig. 2C, SSU es27 in HC3, LSU es49 in HC4). Instead, clones were either unedited at these sites or contained deletions of encoded nucleotides immediately adjacent to the editing site, as described below. The HC4 result was confirmed using construct CD4 (Fig. 2C), which examines the same editing site but in the context of a circular template. Editing sites downstream from the junction contained added nucleotides in CD4 transcripts (Fig. 2C), unequivocally demonstrating that the lack of editing at es49 was not due to the loss of editing activity. Again, as with HC4 clones, although accurate editing was not observed, some CD4 transcripts lacked encoded nucleotides near the junction, as detailed below. Taken together, these data indicate that 9–10 bp of native downstream DNA is sufficient to support C insertion, but that template changes closer than 9 bp downstream from an insertion site abolish accurate editing.
Upstream element boundary
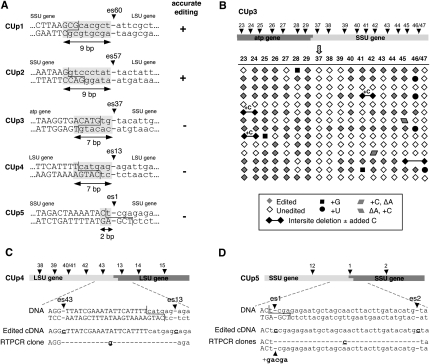
To determine the extent of naturally occurring upstream DNA that is required for editing, a series of circle templates was constructed in which the junction was located at varying distances from the editing site of interest. Circle templates with upstream sequence changes, CUp1 and CUp2, are examples of circle templates containing only 9 bp of native upstream sequence (Fig. 3A). These constructs examine the effects of altering sequences upstream of two different editing sites within the large rRNA, es60 (CUp1) and es57 (CUp2). Accurate editing was observed at both sites (Fig. 3A) and in all other constructs having at least 9 bp of natural sequence upstream (data not shown). These data indicate that the sequence elements required for editing are present within this 9-bp upstream region.
FIGURE 3.
Definition of sequences upstream of insertion sites required for editing. (A) Junction regions of circle templates that alter sequences upstream of editing sites. The ability of each template to support accurate editing at the site nearest the junction is indicated at the right. Shaded regions indicate upstream sequence identical to that of native mtTECs. (B) Editing status of individual sites in independent CUp3 cDNA clones as shown in Figure 2. (C,D) Mis-edited cDNA clones derived from CUp4 and CUp5 transcripts, respectively.
In contrast, none of the constructs having <9 bp of natural upstream sequence supported accurate editing. Two of these constructs, CUp3 and CUp4, had 7 bp of naturally occurring upstream sequence (Fig. 3A); 8 bp was not tested due to the absence of genomic restriction sites in a suitable context. In the case of CUp3, no editing was observed at the editing site of interest (Fig. 3B, SSU es37) in any of the 14 independent clones. This result is unlikely to be due to low editing efficiency at SSU es37 because in transcripts derived from a chimeric template in which SSU es37 was 261 bp from the junction, one third of the RT-PCR clones were correctly edited and another third were mis-edited at this site (Byrne and Gott 2002). Similarly, correct editing at LSU es13 was not seen in any of the CUp4 clones. Instead, each of these clones lacked encoded nucleotides adjacent to the editing site (Figs. 3C, 4A), as described below. Finally, accurate editing was never observed at SSU es1 in any of the CUp5 clones, which contain only 2 bp of native upstream DNA sequence (Fig. 3A). These results are in stark contrast to the high efficiency of editing at SSU es1 in transcripts derived from CD1 (Fig. 2A), HC1 (Fig. 2B), and a construct having the ligation junction 14 bp downstream from this site (data not shown). We conclude that the region immediately upstream of an editing site is essential for accurate editing, with 9 bp of native upstream sequence sufficient to support C insertion. Remarkably, alteration of even two additional base pairs in this region abolishes correct editing and leads to aberrant events.
FIGURE 4.
Template alterations within the region critical for editing lead to junction deletions. (A) Deletion of encoded nucleotides upstream of editing sites. (B) Deletion of encoded nucleotides downstream from editing sites. Only independent clones are shown for all constructs.
Our data indicate that the upstream and downstream boundaries are quite distinct, with the essential upstream and downstream template elements each falling within ∼9 bp of an editing site. This ∼18-bp “critical region” is both necessary and sufficient for accurate C insertion in mitochondrial transcription/editing complexes. Importantly, however, templates having changes within this critical region do support some steps in the editing process. For example, although the CUp4 and CUp5 templates are unable to support correct editing (Fig. 3A), the editing site nearest to the junction appears to be recognized in these constructs based on the sequence of rare RT-PCR clones that are mis-edited at these sites (Fig. 3C,D). In the case of CUp4, we obtained a single clone containing an intersite deletion that lacks all of the encoded nucleotides between es43 (upstream of the junction) and es13 (downstream from the junction) with a mis-edited G insertion (Fig. 3C). Two CUp5 clones were also mis-edited at the editing site nearest the junction. One of these clones contained a precise intersite deletion between SSU es1 and es2 coupled with a single C insertion, while the second clone contained 5 nonencoded nucleotides at es1 (Fig. 3D). Thus, although template changes immediately upstream of an editing site interfere with the accuracy of editing, the CUp5 data indicate that the presence of only 2 bp of native upstream DNA is sufficient for editing site recognition.
Template alterations near editing sites lead to deletions of encoded nucleotides
Strikingly, templated nucleotides are frequently missing in RNAs transcribed from constructs in which sequences within the critical region are altered (Fig. 4). In most cases, the same deletion is found within multiple independent clones from a given template, with deletions occurring in both upstream (Fig. 4A) and downstream (Fig. 4B) contexts. The size of the deletion varies from construct to construct and depends on the distance between the editing site and the restriction site used to construct the template. Importantly, these types of deletions are not observed in constructs in which the junctions fall outside of the critical region.
The vast majority of the upstream deletions encompass all of the encoded nucleotides between the ligation junction on the template strand and the editing site nearest the junction (Fig. 4A). The CUp6 construct, for example, has 12 bp of natural upstream sequence and supports accurate editing, consistent with the data in Figure 3. Surprisingly, however, 12 of the clearly independent CUp6 RT-PCR clones lacked the 7 nt upstream of LSU es9, with the deletion extending from the XhoII junction on the template strand to the editing site just downstream from the junction (Fig. 4A). Similarly, 17 of 18 independent RT-PCR clones derived from CUp4 transcripts contain the same 2-nt deletion, an AG adjacent to es13 of the LSU gene (Fig. 4A). Finally, although some of the RT-PCR clones derived from CUp2 transcripts had an inserted C at es57 (Fig. 3A), others lacked encoded nucleotides upstream of this site (Fig. 4A). Of these 30 independent deletions, 29 share a common upstream deletion endpoint, which coincides with the junction on the template strand. The downstream endpoint of the CUp2 deletion clones is not as uniform, potentially due to mis-editing events such as those observed in Figures 2, A–C, and 3, B–D, and in native mtTECs (Byrne et al. 2002; Byrne and Gott 2004). Notably, although CUp2 upstream deletions were observed in multiple experiments using different amounts of AvaII to generate the circular template, more deletions were observed at the higher AvaII concentrations (Fig. 4A).
In transcripts derived from templates having alterations in the critical downstream region, the observed deletions again span the entire region between the editing site closest to the junction and the restriction site used to generate the template (Fig. 4B). In the case of CD4 and HC4, which both involve fusions at the MspI site just downstream from LSU es49 (Fig. 2), 20% of all RT-PCR clones lacked the encoded nucleotides downstream from LSU es49, and roughly half of the deletions in clearly independent clones had an added C at es49 (Fig. 4B). Significantly, no editing was observed in any of the clones that lacked deletions, as discussed in more detail below. Although many CD1 transcripts were correctly edited (Fig. 2A), editing site to junction deletions were also seen in four CD1 clones (Fig. 4B), but in this case, one had an added C at es9, two were mis-edited (+G, +UUUC), and the fourth was unedited at this site. HC1 transcripts showed a similar pattern, with eight independent correctly edited RT-PCR clones, an unedited clone, and one 7-nt deletion after an added C (Figs. 2, 4B). Likewise, in addition to the correctly edited HC2 transcripts (Fig. 2), six independent HC2 clones lacked the 7 encoded nucleotides immediately downstream from the C added at SSU es14 (Fig. 4B). Finally, all of the independent RT-PCR clones derived from HC3 transcripts contained deletions, this time showing two different patterns. Half of the unequivocally independent clones lacked the 6 nt after the added C at SSU es27, while the remaining clones had larger deletions having the same downstream endpoint at the ligation junction, but beginning at es26 rather than es27 (Fig. 4B). Potential origins of these deletions are discussed below.
The deletion of encoded nucleotides occurs primarily at the RNA level
A priori, the deletions present in the RT-PCR clones shown in Figure 4 could have been introduced at either the DNA or RNA level. To determine if corresponding deletions were also present in their respective DNA templates, we characterized the chimeric DNA templates that were used to generate the run-on transcripts. An aliquot of each reaction was PCR-amplified using the same primers as those used to generate RT-PCR fragments and analyzed by one of two methods. When the observed deletions eliminated the restriction site at the ligation junction, the entire population of PCR products was analyzed. In these experiments, PCR products were end-labeled and digested with the restriction enzyme used to create the junction to ascertain the fraction of the PCR product that contained an intact restriction site. In cases where the observed deletions did not destroy the restriction site at the junction, we cloned and sequenced individual clones to determine whether the deletions observed in RT-PCR clones were also present in PCR clones.
Clear differences between the junction regions of the mtDNA templates and their RNA products were observed with both methods. This is illustrated by analysis of the CUp2 PCR and RT-PCR products, which were characterized using both approaches (Supplemental Fig. S1). Whereas sequencing data indicated that only 13 of 46 individual RT-PCR clones from the CUp2 cDNA retained the AvaII site at the junction, all 25 randomly selected PCR clones contained this AvaII site and eight of eight sequenced CUp2 PCR clones had the expected junction sequence (Supplemental Fig. S1A,B). AvaII digestion of bulk PCR and RT-PCR products resulted in distinct patterns, with 14% of the cDNA product and 87% of the mtDNA product cleaved at the junction (Supplemental Fig. S1C). Similarly, in the case of CUp6, only 16% of the bulk end-labeled RT-PCR product was cut by XhoII (Supplemental Fig. S2), consistent with our finding that 80% of these RT-PCR clones contain a 7-nt deletion that eliminates the XhoII site (Fig. 4A); whereas 88% of the PCR product was cleaved by XhoII (Supplemental Fig. S2). In cases where the deletion clones retained the restriction site, all cloned PCR products had the sequence expected upon cleavage and circularization. Whereas 17 of 18 independent CUp4 RT-PCR clones contained a 2-nt deletion adjacent to the junction (Fig. 4A), none of the 17 randomly selected clones derived from the DNA template contained this (or other) deletion(s) (data not shown). It is therefore extremely unlikely that the missing nucleotides in the CUp4 transcripts were due to deletions present in the DNA template. Taken together, our data indicate that the observed deletions were unlikely to have been introduced during template construction, leading to the conclusion that the missing nucleotides were omitted during transcription of the chimeric templates.
Deletions fall predominantly at template junctions
The atypical deletions observed in these run-on transcripts show distinct commonalities. First, these deletions occur at very high frequencies when the junction on the template strand falls within 7 nt of an editing site. Indeed, in the constructs described here, deletions were observed in 39% (86 of 220) of the independent RT-PCR clones having template strand junctions within the critical region (Figs. 2–4). In contrast, only 0.3% of the 1580 more distal editing sites in the same clones were associated with deletions (Figs. 2–4; data not shown), most of which were intersite (editing site to editing site) or single-nucleotide deletions (see Figs. 2A–C, 3B–D). Second, one endpoint is typically at an editing site, most frequently the editing site closest to the ligation junction (Fig. 4A,B). Third, while the size of the deletion varies among constructs, the second endpoint almost always coincides with the restriction site used to generate the template (Fig. 4). Indeed, in nearly all cases, the deletion endpoint maps specifically to the junction on the template strand.
We hypothesize that these novel deletions may be the result of nicks in the transcribed DNA strand arising from incomplete ligation during template synthesis and/or re-cleavage by the restriction enzyme, which is still present during the transcription reaction (see Discussion). Indeed, junction deletions are more frequent when higher concentrations of restriction enzyme are used to generate the chimeric template (e.g., 86% [20 units] versus 61% [5 units] of independent CUp2 RT-PCR clones) (Fig. 4A). In addition, junction deletions were prevalent in transcripts made under low ATP concentrations (templates CUp4, CUp6, HC2, and HC3, which have an encoded A after the editing site nearest the junction) (Fig. 4), conditions that would reduce re-ligation efficiency. Thus, it would not be surprising if template nicks were present during RNA synthesis in this system.
Upstream and downstream template alterations affect editing differently
Curiously, the patterns of editing are markedly different when upstream and downstream sequences within the critical region are changed. When sequences immediately upstream of a C insertion site are altered, deletions that begin at the ligation junction and end at the editing site are frequently observed (Fig. 4A), but there is never an added C associated with the deletion. In contrast, nucleotide insertions are quite prevalent in deletions that start at an editing site and end at the ligation junction in constructs containing downstream template alterations (Fig. 4B). These data point to different roles for upstream and downstream sequences, as discussed below.
DISCUSSION
Template regions essential for editing
In this study, we describe the first experiments that address the source of the extraordinary specificity of Physarum editing. We demonstrate that 9 bp of native upstream DNA and 9–10 bp of downstream DNA are necessary and sufficient for correct recognition and utilization of C insertion sites (Figs. 2, 3). All template alterations examined here (Figs. 2–4) and elsewhere (Byrne and Gott 2002; Byrne et al. 2002) that fall outside of this critical region support accurate editing. Consistent results were obtained with chimeric templates involving junctions in the vicinity of many different editing sites (Figs. 2–4; data not shown), strongly suggesting that these limits are broadly applicable rather than editing-site-specific. These boundaries are further supported by the fact that our findings correlate remarkably well with the minimal distance between adjacent editing sites; of the ∼450 known single C insertion sites in Physarum mitochondrial RNAs, the closest are 9 bp apart (Mahendran et al. 1994). Finally, the data presented here are also consistent with our unpublished work characterizing run-off transcripts synthesized from linearized mtTEC templates. These experiments demonstrated that the presence of 14 bp of DNA downstream from an editing site supported C insertion, whereas 6 bp of downstream DNA did not (A Majewski and JM Gott, unpubl.).
Neighboring sequences are often used to direct RNA editing in other systems. The C-to-U changes in plant organelles generally require ∼20 nt upstream and a short stretch of RNA downstream from editing sites, although more distal regions can impact editing efficiency (Heller et al. 2008; Takenaka et al. 2008). C-to-U editing within the apoB mRNA is specified by the “mooring” sequence ∼5 nt downstream from the editing site, which is paired with sequences just upstream of the targeted C (Backus and Smith 1992; Richardson et al. 1998). A-to-I changes occur in the context of largely double-stranded regions of RNA (Nishikura 2006), while nucleotide changes in the 5′ portion of edited tRNAs require pairing within the acceptor stem (Bullerwell and Gray 2005). The insertion and deletion of Us in kinetoplasts are directed by base-pairing of gRNAs to anchor sequences immediately downstream from editing sites (Blum et al. 1990; Seiwert and Stuart 1994), while upstream RNA sequences within the nucleocapsid template are required for the incorporation of one or more extra nucleotides at the single editing site within the paramyxoviral genome (Hausmann et al. 1999). Our work is clearly distinct from other editing systems in that this is the first instance of changes at the DNA level directly affecting the extent or accuracy of RNA editing.
Deletion of encoded nucleotides
When changes were made within the critical region defined in this study, the transcripts made from these templates often lacked short stretches of encoded nucleotides (Fig. 4). Nucleotide deletions have been observed previously in transcripts made in vitro from both native mtTECs and chimeric templates, but in all cases, these involved either intersite deletions (where both endpoints fall at editing sites) or rare deletions of 1–3 encoded nucleotides immediately following an editing site (Byrne et al. 2002; Byrne and Gott 2004). Here we again observe single nucleotide deletions (Figs. 2, 3) and intersite deletions, which are found both at sites distant from the point of ligation (Figs. 2A, 3B) and spanning the ligation junction (Fig. 3C,D). In contrast, the atypical deletions near ligation junctions have only one endpoint at an editing site, with the other end falling within the restriction site at the junction. Omission of these encoded nucleotides occurs during RNA synthesis, as the corresponding nucleotides were present in their respective DNA templates (Supplemental Figs. S1, S2; data not shown).
Interestingly, junction deletions only occur when the ligation junction falls within the critical region on the template strand. In the vast majority of the deletions shown in Figure 4, the deletion endpoint coincides exactly with the ligation point on the template strand for both upstream and downstream constructs (Fig. 4). One CD4 deletion endpoint corresponds to the junction on the nontemplate strand, suggesting that the integrity of both strands of the downstream DNA is likely to be important for accurate editing (Fig. 4B). Even templates that support accurate editing can give rise to junction deletions, but only in cases where the template strand junction falls within the critical region (Fig. 4). For example, in the case of CUp6, which supports accurate C insertion, the observed deletions start at the XhoII junction 7 bp upstream of LSU es9 (Fig. 4A). Similarly, in transcripts from the HC1 and CD1 templates, the deletion endpoints coincide with the PstI junction 7 nt downstream from SSU es9 (Fig. 4B). Importantly, we have not observed junction deletions with chimeric templates in which the site of cleavage on the template strand is more than 9 bp from an editing site (Byrne et al. 2002; Byrne and Gott 2004; data not shown). Therefore, it appears that perturbations (likely nicks) within the critical region of the template are necessary to generate these deletions.
Critical upstream and downstream regions and their roles in editing
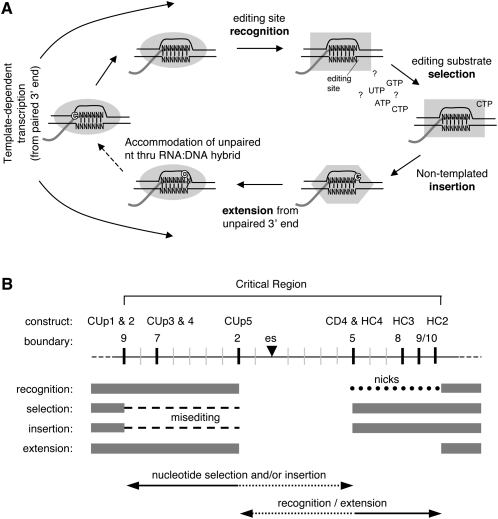
Accurate editing of nascent transcripts is a complex process that is intimately tied to transcription (Cheng et al. 2001). It involves a number of distinct steps (Fig. 5A), including editing site recognition, substrate selection, insertion of the nonencoded nucleotide(s), templated extension from the “extra” nucleotide(s), and accommodation of the unpaired nucleotide(s) through the RNA:DNA hybrid. This cycle occurs repeatedly during the course of RNA synthesis in Physarum mitochondria, with roughly 1 in every 25 nucleotides in an mRNA added in a nontemplated manner. Significantly, these steps are affected differently in constructs in which upstream or downstream sequences are altered, allowing us to infer likely roles for flanking sequences based on the characteristics of the RNAs produced from each template.
FIGURE 5.
Insertional editing cycle. (A) Model depicting the events involved in the cotranscriptional insertion of nonencoded nucleotides into Physarum mitochondrial transcripts. Black and gray lines represent mitochondrial DNA and RNA, respectively. Shaded objects represent the mitochondrial RNA polymerase, with different shapes indicating potential conformational states (hypothetical) that might be needed to accommodate extra nucleotides in the active site. Individual steps are described in the text. (B) Schematic illustration of the location of the template elements required for C insertion defined in this work and the likely roles played by upstream and downstream regions.
Editing site recognition appears to require the presence of native downstream DNA but is seemingly insensitive to changes upstream of the editing site. The lack of a requirement for upstream sequences is demonstrated by the fact that although constructs that alter sequences immediately upstream of insertion sites do not yield correctly edited RNAs, these editing sites are still recognized. Evidence includes the insertion of incorrect nucleotides at es1 in CUp5 (Fig. 3D), the intersite deletions in CUp4 and CUp5 (Fig. 3C,D), and the junction deletions in CUp2, CUp4, and CUp6 (Fig. 4A). The results with the CUp5 template are particularly striking given that the only upstream sequences in common between SSU es1 in the CUp5 construct and its natural context are the two base pairs immediately upstream of the editing site, with the template nucleotides flanking the editing site being derived from a different portion of the SSU gene (Fig. 3D). It should be noted that although the upstream sequences have been changed in these constructs, each contains native downstream elements. On the other hand, in constructs with downstream sequence alterations that fall within the critical region, evidence for editing site recognition is only seen in the context of junction deletions (Fig. 4B), which we suspect arise from nicks in the template strand. These data suggest that downstream sequences are required for editing site recognition and that changes within this region can be compensated for by nicks or other changes in the template that could cause the polymerase to pause long enough for editing to occur.
Selection and possibly insertion of nonencoded C residues are sensitive to changes in upstream DNA but are unaffected by downstream alterations. C insertion at the editing site closest to the junction was never observed in transcripts derived from CUp3, CUp4, or CUp5, which change sequences immediately upstream of an editing site (Figs. 3, 4). In these cases, it is difficult to distinguish whether the absence of editing is due to effects on nucleotide selection or insertion, since a defect in either step could result in unedited transcripts. However, although insertion of the correct nucleotide did not occur, we did observe a single instance of nonencoded nucleotide insertion in a CUp5 transcript, where GACGA was inserted at a C insertion site (Fig. 3D), indicating that nucleotide selection may be impaired by upstream template alterations. Conversely, C insertion was often seen in transcripts from constructs with altered downstream sequence, even when subsequent extension was affected, as evidenced by the presence of an added C in many of the junction deletion clones shown in Figure 4B. Thus, our data suggest that the region upstream of an editing site is responsible for proper nucleotide insertion at that site.
Downstream DNA plays an important role in templated extension from an added nucleotide. Whereas the exclusion of encoded nucleotides is rare in our coupled transcription/editing system (Byrne et al. 2002), sequence alterations in the critical downstream region invariably led to the omission of encoded nucleotides whenever the proximal site was edited, leading to the junction deletions observed in HC1, HC2, HC3, HC4, CD1, and CD4 transcripts (Fig. 4B). Importantly, such deletions were not observed in clones having downstream changes outside of the critical region (Fig. 2; data not shown) and were infrequent in RNAs that were unedited at the site closest to the junction. In contrast, in the few instances where nucleotides were added at editing sites having alterations within the critical upstream region, all templated nucleotides downstream from the site were present in the transcripts. This is best illustrated by the mis-edited CUp5 clone that contains five extra nucleotides at SSU es1 (Fig. 3D). These data argue against a role for specific upstream sequences in extension from an added, unpaired nucleotide.
Taken together, our data strongly suggest that upstream and downstream sequences play distinct roles in the editing process, as summarized in Figure 5B. Template changes upstream of an editing site affect nucleotide selection and possibly nucleotide insertion, whereas editing site recognition and templated extension from the added nucleotide are impaired by downstream sequence alterations. In the latter case, we cannot distinguish whether the sequence alterations within the critical region downstream from an editing site or the physical integrity of the DNA template contributes to the observed effects on editing. Either way, it is clear that the regions upstream of and downstream from editing sites play discrete roles in this process. Importantly, these findings provide the first indication of where the information for editing site specificity resides and directly support our previous conclusion that editing site recognition and nucleotide insertion are separable processes (Byrne et al. 2002).
Editing in the context of transcription elongation
The different roles played by the upstream and downstream sequences are perhaps not surprising given the architecture of elongating transcription complexes. Several high-resolution structures of RNA polymerases (RNAPs) complexed with nucleic acids have been solved, providing detailed information regarding polymerase conformations during elongation (Gnatt et al. 2001; Tahirov et al. 2002; Yin and Steitz 2002, 2004; Temiakov et al. 2004; Vassylyev et al. 2007a,b). Overall core structures are well conserved, even between single and multi-subunit polymerases. Although there are currently no equivalent structures for the mitochondrial RNA polymerase from Physarum polycephalum (Pp mtRNAP), based on its high degree of homology with the bacteriophage T7 RNA polymerase (T7 RNAP) (Miller et al. 2006; Gott and Rhee 2007), the basic features of the Pp mtRNAP elongation complex are likely to be similar. Of particular interest to our findings here is that in the T7 RNAP elongation complex, sequences immediately upstream of the point of nucleotide addition are involved in the 7–8-bp RNA:DNA hybrid and the ∼9–nt transcriptional bubble, while roughly 8 bp of downstream DNA lies against a positively charged patch on the exterior of the polymerase (Tahirov et al. 2002; Yin and Steitz 2002, 2004; Temiakov et al. 2004). The architecture of the yeast RNA polymerase II elongation complex is quite similar, with an RNA–DNA hybrid of 7–8 bp and the largely double-stranded DNA downstream from the nucleotide addition site within a cleft on the polymerase (Kettenberger et al. 2004). Thus, the regions of the template that are required for editing are predicted to be in direct contact with the Pp mtRNAP. The critical sequences directly upstream of an editing site would likely be part of the transcription bubble, with the template strand participating in the RNA–DNA hybrid and the nontemplate strand unpaired, while nearly all of the essential downstream DNA would be expected to be double-stranded when the editing site is encountered by the Pp mtRNAP, making it likely that downstream DNA exerts its effects as a DNA duplex.
The upstream regions defined by our experiments as being essential for accurate editing are known to modulate transcription elongation in other systems. The RNA:DNA hybrid is a major component of elongation complex stability (Kireeva et al. 2000; Mentesana et al. 2000; Temiakov et al. 2002) and can influence processivity (Nudler et al. 1997; Sidorenkov et al. 1998), polymerase pausing (Palangat and Landick 2001), and termination (Komissarova et al. 2002). The nontemplate strand within the transcription bubble can also affect transcription elongation (Mentesana et al. 2000) and can serve as a binding site for effector proteins. Precedents include the bacteriophage λ qut site, involved in λQ-mediated anti-termination (Ring and Roberts 1994), and the recruitment of the transcriptional regulator RfaH to elongation complexes paused at ops sites (Artsimovitch and Landick 2002). Given the nature of our experimental system, we cannot currently investigate the effects of the template and nontemplate strands in isolation; either or both may be important for editing. The contributions of the upstream DNA could be mediated by the RNA–DNA hybrid immediately upstream of an editing site, whose stability may influence editing site utilization or accommodation of the extra nucleotide, or the bases and/or phosphodiester backbone within the nontemplate strand of the transcription bubble, which could provide a binding site for editing factors and/or the inserted nucleotide. Nucleotide selection might also involve some combination of nucleic acid and protein functional groups such as those used by class I CCA adding enzymes, which create unique binding sites for specific nucleotides in a sequential manner (Shi et al. 1998; Xiong and Steitz 2004). Our findings that the upstream region appears to be critical for nucleotide selection (Figs. 3, 4A) and that deproteinized mitochondrial DNA does not support nucleotide insertion (Byrne and Gott 2002) are of potential relevance here.
In other elongation complexes, downstream DNA has been shown to affect transcription termination and arrest (Telesnitsky and Chamberlin 1989; Kerppola and Kane 1990), polymerase pausing (Lee et al. 1990; Ederth et al. 2002; Palangat et al. 2004), and complex stability (Temiakov et al. 2002). Based on the effects of nucleotide concentration on Physarum editing, polymerase dwell time at an editing site strongly influences the frequency of nucleotide insertion (Cheng et al. 2001). In addition, the requirement for multiple elongation modes in this system (as discussed in the next section) suggests that the configuration of the transcription complex may be particularly critical in the vicinity of editing sites. Thus, it seems likely that the role of downstream DNA in editing site recognition and 3′ extension is mediated by effects on polymerase pausing and transcription complex stability. We speculate that nicks in the template may cause the polymerase to pause, allowing time for editing site recognition in the absence of essential downstream elements.
Potential roles for the Physarum mitochondrial RNA polymerase in insertional editing
RNA synthesis in Physarum mitochondria involves a number of different modes of transcript elongation (Fig. 5A). During standard transcription, nucleotides are added to the 3′-end of the nascent transcript in a template-dependent manner. A second form, which occurs at editing sites, involves nontemplated nucleotide addition. In most instances, this involves the addition of a nonencoded nucleotide to a base-paired 3′-end. However, at sites of dinucleotide insertion, the second nontemplated nucleotide would likely be added to an unpaired 3′-end, based on our in vitro studies that suggest that inserted dinucleotides are added one residue at a time (Visomirski-Robic and Gott 1997b; Byrne and Gott 2004). A third type of elongation, templated extension from an unpaired 3′-end, would be required after the addition of one or more nonencoded nucleotides. And finally, the accommodation steps of the editing cycle resemble normal transcription in that templated nucleotides are added to a base-paired end, except that in this case, the extra, unpaired nucleotide(s) in the transcript must somehow be accommodated within the RNA–DNA hybrid.
Presumably the mitochondrial RNA polymerase from P. polycephalum is responsible for all forms of template-dependent transcription and, based on a number of findings, we speculate that it may also be involved in the addition of untemplated nucleotides (Fig. 5A). First, since the active site of the Pp mtRNAP is engaged with the 3′-end of the nascent RNA during normal elongation, the polymerase must participate, directly or indirectly, in editing site recognition. Second, in vitro the editing efficiency at a given site is dependent on the concentration of the encoded nucleotide immediately following that site (Cheng et al. 2001), implying a direct competition between transcription and editing. Third, the Pp mtRNAP is capable of adding nontemplated nucleotides to the 3′-end of RNAs in vitro (Miller and Miller 2008). Fourth, polymerase backtracking does not appear to be required for nucleotide insertion, given that (1) removal of upstream RNA by oligonucleotide-directed RNaseH cleavage does not affect the extent of editing and (2) the presence of oligonucleotides that anneal to the RNA upstream of an insertion site has no effect on editing at that site (A Majewski, and JM Gott, unpubl.). Finally, the sequences critical for editing are likely to be in direct contact with the RNA polymerase, as discussed above. Thus, whether or not the Pp mtRNAP is the actual editing enzyme, our data strongly suggest that editing occurs within the local environment of the mitochondrial RNA polymerase.
MATERIALS AND METHODS
Chimeric template construction
Chimeric templates were constructed as described by Byrne (2004). Construction of circle templates was accomplished by digesting mtTECs with the appropriate restriction enzyme and relegating under conditions that favor circulation of the digestion fragments, while hybrid templates were made by ligation in the presence of a 20-fold molar excess of a DNA cassette having the appropriate sticky ends. Many different constructs were tested, but only the most relevant constructs are described here. The restriction enzymes used for the constructs shown are as follows: CD1 (PstI), CD4 (MspI), HC1 (PstI), HC2 (SacII), HC3 (HhaI), HC4 (MspI), CUp1 (HhaI), CUp2 (AvaII), CUp3 (NspI), CUp4 (RcaI), CUp5 (XhoI), and CUp6 (XhoII).
Oligonucleotides
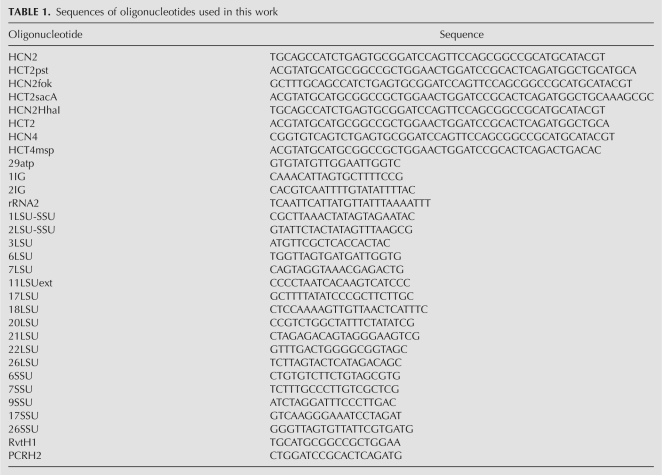
DNA cassettes were generated by heating 250 pmol of complementary, gel-purified oligonucleotides for 5 min at 65°C and slowly cooling to 40°C prior to use in ligation reactions. The oligonucleotides used to generate the hybrid cassette templates are listed for each construct. HC1: HCN2 and HCT2pst; HC2: HCN2fok and HCT2sacA; HC3: HCN2HhaI and HCT2; and HC4: HCN4 and HCT4msp. The oligonucleotides used for PCR are listed for each construct, with the primer used for cDNA synthesis in parentheses. CUp1 (26LSU): 17SSU/6LSU; CUp2 (2LSU-SSU): 17SSU/6LSU; CUp3 (1IG): 29atp/2IG; CUp4 (17LSU): 7LSU/11LSUext; CUp5 (9SSU): 6SSU/7SSU; CUp6 (11LSUext): 7LSU/18LSU; CD1 (3LSU): 1LSU-SSU/rRNA2; CD4 (20LSU): 21LSU22LSU; HC1 (RvtH1): 1LSU-SSU/RvtH1; HC2 (RvtH1): 6SSU/PCRH2; HC3 (RvtH1): 26SSU/PCRH2; and HC4 (RvtH1): 22LSU/PCRH2. Sequences are given in Table 1.
TABLE 1.
Sequences of oligonucleotides used in this work
MtTEC manipulation
Mitochondrial transcription elongation complexes were isolated essentially as described in Cheng and Gott (2000) with minor variations in dialysis conditions. Typically, mtTEC DNA was digested by incubating mtTECs (5 mg of protein) with 5–20 U of appropriate restriction enzyme in a final volume of 30 μL for 30 min at 30°C as follows: 20 U of HhaI (NEB), 5 or 20 U of AvaII (NEB), 20 U of NspI (NEB), 5 U of RcaI (Roche), 5 U of XhoI, 5 U of XhoII, 5 U of PstI (NEB), 20 U of MspI (Roche), and 10 U of SacII (NEB). Identical amounts of restriction enzyme were used in digestion reactions for both circle and hybrid cassette templates. Ligations were carried out for 30 min at 16°C with 500 μM ATP, 1.4 U of T4 DNA ligase (Roche), and 5 pmol of exogenous hybrid cassette DNA where appropriate. Run-on transcription reactions (35–50 μL) were similar to those described previously (Cheng and Gott 2000) except for slightly varying buffer conditions (due to individual restriction enzyme requirements); NaCl concentrations varied from 44 to 90 μM. Nucleotide triphosphates were used at 500 μM except as specified: CD1 (100 μM GTP), CD4 (100 μM UTP), HC1 (100 μM GTP), HC4 (100 μM UTP), CUp1 (100 μM GTP), CUp2 (100 μM UTP), and CUp3 (100 μM UTP).
Reverse transcription and PCR
Nucleic acid preparations were subjected to two rounds of DNase I digestion prior to RT-PCR. RNAs were annealed to 2.5 pmol of primer in 10 μL by incubation for 5 min at 95°C, for 10 min at 70°C, then for 10 min on ice, and reverse-transcribed for 60 min at 52°C in a final volume of 20 μL using 100 U of SuperscriptII (Invitrogen), supplied buffer, and 0.5 mM dNTPs, followed by RNase digestion. For cloning, PCR was performed over 30 cycles (1 min at 94°C, 1 min at appropriate annealing temperature, and 1 min at 72°C) with a 7-min final extension, using 5 U of Taq DNA polymerase (Roche) per 100 μL in buffer supplied by the manufacturer, 200 μM dNTPs, and the primers listed above. The resulting PCR and RT-PCR products were cloned into pBSM13 (Stratagene). End-labeled PCR and RT-PCR products for restriction enzyme analysis were generated in 50-μL reactions using 10 pmol of unlabeled oligonucleotide and ∼0.5 × 105 cpm/μL end-labeled oligonucleotide.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Piet deBoer, Donal Luse, Pieter deHaseth, David Setzer, Mark Caprara, Tim Nilsen, and Jo Ann Wise for useful discussions and Tim Nilsen, Pieter deHaseth, and Whitney Yin for critical reading of the manuscript. This work was supported by NIH grant GM-54663 (to J.M.G.). A.C.R. was supported in part by an NIH Training grant (T32 AG00105).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1668309.
REFERENCES
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Backus JW, Smith HC. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20:6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beargie C, Liu T, Corriveau M, Lee HY, Gott J, Bundschuh R. Genome annotation in the presence of insertional RNA editing. Bioinformatics. 2008;24:2571–2578. doi: 10.1093/bioinformatics/btn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Bullerwell CE, Gray MW. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J Biol Chem. 2005;280:2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- Byrne EM. Chimeric templates and assays used to study Physarum cotranscriptional insertional editing in vitro. Methods Mol Biol. 2004;265:293–314. doi: 10.1385/1-59259-775-0:293. [DOI] [PubMed] [Google Scholar]
- Byrne EM, Gott JM. Cotranscriptional editing of Physarum mitochondrial RNA requires local features of the native template. RNA. 2002;8:1174–1185. doi: 10.1017/s1355838202024081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EM, Gott JM. Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol Cell Biol. 2004;24:7821–7828. doi: 10.1128/MCB.24.18.7821-7828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne EM, Stout A, Gott JM. Editing site recognition and nucleotide insertion are separable processes in Physarum mitochondria. EMBO J. 2002;21:6154–6161. doi: 10.1093/emboj/cdf610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YW, Gott JM. Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria. Nucleic Acids Res. 2000;28:3695–3701. doi: 10.1093/nar/28.19.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YW, Visomirski-Robic LM, Gott JM. Nontemplated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 2001;20:1405–1414. doi: 10.1093/emboj/20.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Gott JM, Rhee AC. Insertion/deletion editing in Physarum polycephalum. In: Goringer HU, editor. RNA editing. Springer; Berlin: 2007. pp. 85–104. [Google Scholar]
- Gott JM, Parimi N, Bundschuh R. Discovery of new genes and deletion editing in Physarum mitochondria enabled by a novel algorithm for finding edited mRNAs. Nucleic Acids Res. 2005;33:5063–5072. doi: 10.1093/nar/gki820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Garcin D, Delenda C, Kolakofsky D. The versatility of paramyxovirus RNA polymerase stuttering. J Virol. 1999;73:5568–5576. doi: 10.1128/jvi.73.7.5568-5576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller WP, Hayes ML, Hanson MR. Cross-competition in editing of chloroplast RNA transcripts in vitro implicates sharing of trans-factors between different C targets. J Biol Chem. 2008;283:7314–7319. doi: 10.1074/jbc.M709595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK, Kane CM. Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry. 1990;29:269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Lee DN, Phung L, Stewart J, Landick R. Transcription pausing by Escherichia coli RNA polymerase is modulated by downstream DNA sequences. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- Mahendran R, Spottswood MS, Ghate A, Ling ML, Jeng K, Miller DL. Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J. 1994;13:232–240. doi: 10.1002/j.1460-2075.1994.tb06253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentesana PE, Chin-Bow ST, Sousa R, McAllister WT. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J Mol Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- Miller ML, Miller DL. Non-DNA-templated addition of nucleotides to the 3′ end of RNAs by the mitochondrial RNA polymerase of Physarum polycephalum. Mol Cell Biol. 2008;28:5795–5802. doi: 10.1128/MCB.00356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Mahendran R, Spottswood M, Costandy H, Wang S, Ling ML, Yang N. Insertional editing in mitochondria of Physarum. Semin Cell Biol. 1993;4:261–266. doi: 10.1006/scel.1993.1031. [DOI] [PubMed] [Google Scholar]
- Miller ML, Antes TJ, Qian F, Miller DL. Identification of a putative mitochondrial RNA polymerase from Physarum polycephalum: Characterization, expression, purification, and transcription in vitro. Curr Genet. 2006;49:259–271. doi: 10.1007/s00294-005-0053-y. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T, Hajduk S. The function of RNA editing in Trypanosomes. In: Goringer HU, editor. RNA editing. Springer; Berlin: 2007. pp. 181–197. [Google Scholar]
- Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J Mol Biol. 2001;311:265–282. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- Palangat M, Hittinger CT, Landick R. Downstream DNA selectively affects a paused conformation of human RNA polymerase II. J Mol Biol. 2004;341:429–442. doi: 10.1016/j.jmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Richardson N, Navaratnam N, Scott J. Secondary structure for the apolipoprotein B mRNA editing site. Au-binding proteins interact with a stem loop. J Biol Chem. 1998;273:31707–31717. doi: 10.1074/jbc.273.48.31707. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Roberts JW. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994;78:317–324. doi: 10.1016/0092-8674(94)90300-x. [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Stuart K. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- Shi PY, Maizels N, Weiner AM. CCA addition by tRNA nucleotidyltransferase: Polymerization without translocation? EMBO J. 1998;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A, Chamberlin MJ. Terminator-distal sequences determine the in vitro efficiency of the early terminators of bacteriophages T3 and T7. Biochemistry. 1989;28:5210–5218. doi: 10.1021/bi00438a044. [DOI] [PubMed] [Google Scholar]
- Temiakov D, Anikin M, McAllister WT. Characterization of T7 RNA polymerase transcription complexes assembled on nucleic acid scaffolds. J Biol Chem. 2002;277:47035–47043. doi: 10.1074/jbc.M208923200. [DOI] [PubMed] [Google Scholar]
- Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007a;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007b;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Visomirski-Robic LM, Gott JM. Insertional editing of nascent mitochondrial RNAs in Physarum. Proc Natl Acad Sci. 1997a;94:4324–4329. doi: 10.1073/pnas.94.9.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic LM, Gott JM. Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA. 1997b;3:821–837. [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]