Abstract
The ribosome decoding center is rich in modified rRNA nucleotides and little is known about their effects. Here, we examine the consequences of systematically deleting eight pseudouridine and 2′-O-methylation modifications in the yeast decoding center. Loss of most modifications individually has no apparent effect on cell growth. However, deletions of 2–3 modifications in the A- and P-site regions can cause (1) reduced growth rates (∼15%–50% slower); (2) reduced amino acid incorporation rates (14%–24% slower); and (3) a significant deficiency in free small subunits. Negative and positive interference effects were observed, as well as strong positional influences. Notably, blocking formation of a hypermodified pseudouridine in the P region delays the onset of the final cleavage event in 18S rRNA formation (∼60% slower), suggesting that modification at this site could have an important role in modulating ribosome synthesis.
Keywords: snoRNA, rRNA, ribosome, modification, processing, pseudouridine
INTRODUCTION
In eukaryotes, maturation of the large polycistronic rRNA precursor includes the formation of scores of modified nucleotides and several cleavages of the primary transcript (Fatica and Tollervey 2002; Nazar 2004). Two major types dominate: 2′-O-methylation (Nm) and pseudouridylation (Ψ), which are both known to alter local conformation and folding properties of RNA (Decatur and Fournier 2003). A large portion of these (and other) modifications are located in functionally important regions of the ribosome (∼60% in Saccharomyces cerevisiae and 95% in Escherichia coli; Decatur and Fournier 2002), and many are conserved positionally (Lestrade and Weber 2006), suggesting they have important roles in ribosome function and perhaps synthesis.
On balance, information about the influence of eukaryotic rRNA modifications is scant. However, the pace of discovery is accelerating, due to: (1) advances in identifying modification machinery, in particular the small nucleolar RNA–protein complexes (snoRNPs) that create Nm and Ψ modifications; (2) assignment of all or nearly all yeast modifications to specific guide snoRNAs; (3) the relative ease of blocking modifications in yeast, by genetically deleting snoRNAs; (4) the availability of precise modification maps for yeast rRNA; and (5) the emergence of high resolution, three-dimensional (3D) structures for the ribosome.
With these developments it is now possible to block the formation of preselected Nm and Ψ modifications with relative ease, thereby opening the way to evaluating effects. Our laboratory has exploited this strategy to carry out systematic modification-deletion studies in S. cerevisiae, to determine the importance and influence of modifications in functionally important regions of the ribosome. Modifications are deleted individually and in different combinations, and effects on ribosome production, function, and stability are characterized (Decatur et al. 2007).
We have examined effects of deleting modifications from the peptidyl transferase center region and two inter-subunit bridge structures known as Helix 69 and the A-site finger, all in the large subunit. For these three regions, we found that, in general, deleting individual modifications has no or only slight effects on growth or ribosome activity, whereas multiple deletions have cumulative negative effects that can be synergistic (King et al. 2003; Liang et al. 2007; Piekna-Przybylska et al. 2008). Defects observed include, variously, (1) impaired growth rate; (2) impaired translation rate; (3) substantially lower resistance to ribosome-based antibiotics; (4) reduced accumulation of rRNA, related to increased turnover; and (5) impaired polysome formation. In one special case, loss of a single Ψ in the A-loop of the peptidyl transferase center region caused a substantial decrease in the in vivo rate of protein synthesis. These findings make clear that, collectively, modifications have strong influence on different aspects of ribosome synthesis and performance, and that modifications at different regions can have different effects. In the present study we extended our investigation of modification effects to the decoding center in the small subunit, which binds both mRNA and tRNAs and mediates accurate codon-anticodon interactions and tRNA translocation. We reasoned that modifications in this region might significantly affect ribosome synthesis and activity.
The decoding center is composed of five helices (h) of 18S rRNA (Fig. 1A, h18, h44, h34, h24, h31), as suggested by chemical protection, cross-linking, and genetic studies (Rodnina et al. 2002; Noller 2006). Specific nucleotides in these helices that contact A-, P-, or E-site tRNAs and mRNA have also been identified in crystallography studies (e.g., Ogle et al. 2001; Yusupov et al. 2001; Selmer et al. 2006). Nucleotides involved in decoding have been revealed by mutational analysis, in which translation efficiency, fidelity, or tRNA translocation have been affected, as well as resistance to error-causing antibiotics (De Stasio et al. 1989; Recht et al. 1999; Kubarenko et al. 2006; Garcia-Ortega et al. 2008; Saraiya et al. 2008). These findings established the importance of the proper sequence and structure of the decoding center in controlling tRNA selection and translocation, and ensuring efficient, accurate translation (Ogle et al. 2003; Ogle and Ramakrishnan 2005; Frank et al. 2007).
FIGURE 1.
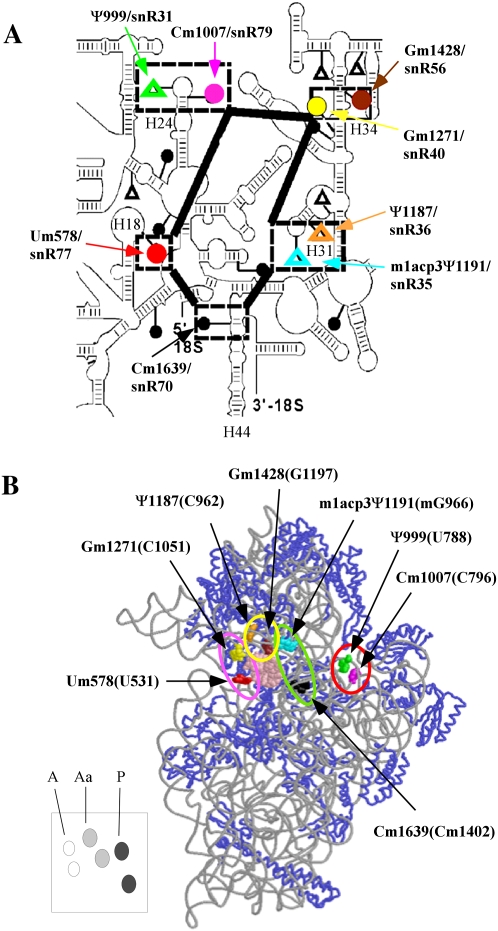
The decoding region of yeast rRNA is rich in modifications. (A) Secondary structure of the decoding center of 18S rRNA. Modified nucleotides in the decoding center are highlighted by broken boxes, and the corresponding guide snoRNAs are also identified. Methylations (●); pseudouridines (△). (B) The 3D structure of modifications in the decoding center. Locations are deduced from the ribosome structure. The corresponding nucleotide numbers in E. coli rRNA are provided in parentheses. The system of representing modifications with colors is the same in both panels. The modifications in the A, Aa, P, and E regions are indicated by pink, yellow, green, and red circles, respectively. The lower left panel shows a schematic cartoon for positions of A-region (white), Aa-region (gray), and P-region (black) modifications. This system is used in subsequent figures to indicate modification situations.
In the E. coli decoding region, several modified sites contact tRNA or mRNA, or are close to positions known to be important for translation. For example, the E. coli site (G966) equivalent to yeast m1acp3Ψ1191 was shown to contact P-site tRNA (Moazed and Noller 1990; Yusupov et al. 2001), whereas Cm1402 in E. coli (Cm1639 in yeast) contacts mRNA (Rinke-Appel et al. 1993). These correlations suggest that modifications could affect translation activity and, perhaps, formation of this vital region itself.
The yeast decoding region contains five Nm and three Ψ modifications (Fig. 1A). One Ψ undergoes additional modification that includes base methylation at the N1 position and addition of a 3-amino-3-carboxypropyl group to yield 1-methyl-3-(3-amino-3-carboxypropyl)Ψ, i.e., m1acp3Ψ. Seven of these eight modifications are conserved in humans, the exception being Cm1007 (yeast numbering system unless indicated). Five of the eight modifications are also conserved in plants and trypanosomes (Um579, Ψ999, Gm1271, Gm1428, and Cm1639) (Brown et al. 2003; Liang et al. 2005). With regard to functional distribution (Fig. 1B), the eight modifications are distributed as follows: four modifications occur at or near the A-site of tRNA binding (three Nm's and one Ψ), two are in the P site (one Nm and one Ψ), and two are in the E site (one Nm and one Ψ). Our results show that in the yeast ribosome, translation activity was reduced substantially with modification-loss, mainly from the A and P regions. In addition, the final cleavage event in forming 18S rRNA was delayed with loss of the hypermodified Ψ in the P region.
RESULTS
Strategy for dissecting modification effects in the decoding region
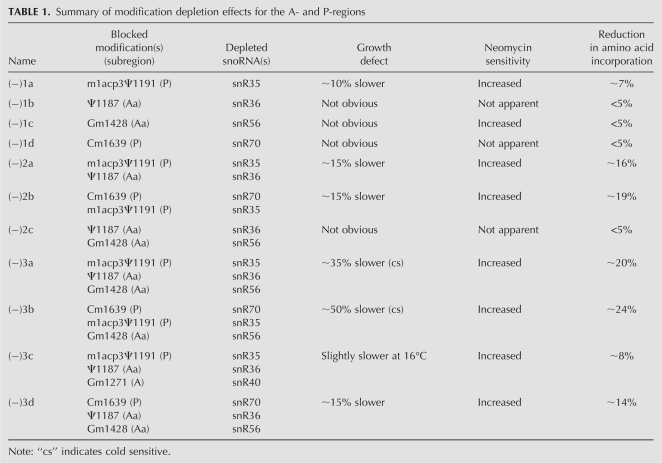
The eight modifications in the yeast decoding region are guided by eight snoRNAs (Fig. 1A). In our deletion strategy, we separated the modifications into four subgroups based on proximity to the tRNA binding regions (Fig. 1B). The regions and specific modifications include: (1) two Nm modifications at the entry-side of A-site tRNA that we designate as A-region modifications; (2) two modifications occuring above the A-site tRNA and we designate these as A-site above (Aa) region modifications (Ψ1187 and Gm1428); (3) two in the P region (m1acp3Ψ1191 and Cm1639); and (4) two in the E region (Ψ999 and Cm1007). Thus, each region contains two modifications, with two Nm modifications in the A-region, and one Nm and one Ψ in each of the other regions. Modifications were depleted individually or together in each group, by genetically deleting the appropriate guide snoRNAs, and double and triple deletions in different combinations in adjoining regions. Altogether, 21 test strains were created (Supplemental Table S2). Deletions of the snoRNAs were confirmed by Northern analysis, and loss of the corresponding modifications (except for m1acp3Ψ1191) was verified by primer extension analysis (data not shown). Disruption of Ψ formation at the hypermodified site was also analyzed by thin layer chromatography (TLC) (see below). Table 1 lists the key strains, modifications, and deletion effects on growth and translation rate.
TABLE 1.
Summary of modification depletion effects for the A- and P-regions
Blocking modifications in the Aa and P regions can significantly impair cell growth and antibiotic resistance
Modification effects on growth rate were examined initially on solid, rich YPD medium, at 16°C, 30°C, and 37°C. No defects were found for the two A-region modifications depleted individually or together. Similarly, the E-region mutants showed normal growth after loss of either or both modifications. Normal growth was also observed for two triple-deletion strains that lack the two A-region modifications plus either of the two modifications from the Aa region (data not shown). In addition, none of the mutants just described showed an apparent defect in neomycin resistance, amino acid incorporation, or polysome pattern (data not shown). These results suggest that modifications in the A and E regions do not significantly affect normal growth and translation rate. Thus, we excluded these particular subsets from further consideration.
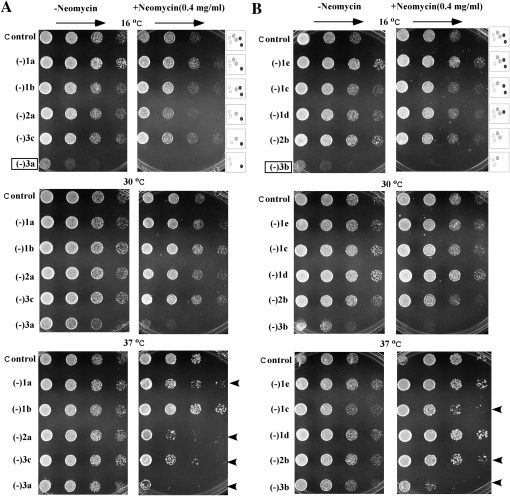
However, defects in cell growth were readily apparent for strains deleted of Aa- and P-region modifications, ranging from ∼10% to ∼50% slower, as determined for growth in liquid medium. For convenience, we refer to the test strains by the number of modifications missing from these two subregions (Table 1). For example, strains (−)3a and (−)3b have been depleted of three modifications in different combinations. Growth properties of the Aa- and P-region mutants were screened on solid YPD-rich medium (see below) and also examined at 30°C in liquid-rich YPD medium. Although no obvious growth defect was observed for strains depleted of one or two modifications when examined on solid medium, slower growth rates were detected for some of these strains when analyzed in liquid medium. Growth rate reductions were calculated based on generation times determined in liquid medium (see below; data not shown). With one exception cells missing individual modifications from these two subregions alone have no obvious growth defects (Table 1). The exception is a mutant blocked in formation of the hypermodified Ψ in the P region, which showed a slight growth rate defect (strain −1a, ∼10% slower). No growth defect was apparent for the strain depleted of both Aa-region modifications (strain −2c). However, somewhat stronger defects (∼15% slower growth) were seen for cells depleted of the two Ψs from the Aa and P regions (−2a), and cells missing both P-region modifications (−2b). The strongest growth defects, i.e., ∼35% and ∼50% longer generation times, were observed for two strains depleted of three modifications. The first of these lacks the two Aa modifications and the hypermodified Ψ in the P-region (−3a). The other is missing one (Nm) modification in the Aa region and both P-region modifications (−3b). Impaired growth was also found on solid medium for both triple-depletion strains, and cold-sensitive phenotypes were also observed in these cases (Fig. 2A). For example, the (−)3a and (−)3b cells grow very much slower at 16°C, moderately slower at 30°C, but normally at 37°C, as compared with control cells. We note that the growth defects are specific to loss of the corresponding snoRNAs, since restoring the expression of missing snoRNAs fully restored cell growth (Fig. 4; see below). These observations indicate that the modifications in the Aa and P regions affect growth in cooperative ways, and that growth impairment increases with the number of modifications depleted.
FIGURE 2.
Depletion of decoding region modifications can impair cell growth and increase cell sensitivity to neomycin. (A) Growth comparisons for strains depleted of P-region Ψ (−1a), Aa-region Ψ (−1b), and Aa-region Nm (−1c) individually or in different combinations. Two OD600 units of cells were diluted serially (1:5), spotted on plates with rich YPD medium in the presence (right panel) or absence (left panel) of neomycin (0.4 mg/mL), and incubated at different temperatures. (B) Growth properties of strains depleted of Aa-region Nm (−1c) and P-region Nm (−1d) individually or in different combinations, as in A. Strain (−)1e is depleted of an A-region Nm (Gm1271). Strains showing cold-sensitive phenotypes (boxed) and increased neomycin sensitivity (arrowheads) are identified. The schematic structures at the left of the data panels indicate the modification situations, as shown in Figure 1B.
FIGURE 4.
Altered growth and polysome properties can be fully restored by complementation with the missing snoRNAs. (A) Growth was compared at 16°C for control and test cells containing an empty vector and test cells expressing plasmid-encoded snoRNAs (+), as indicated. The culture conditions were as in Figure 2, except that the medium lacked adenine. (B) Polysome profiles were examined as described in Figure 3.
The overall situation is more complicated than this simple interpretation, as two other strains depleted of three modifications from these and an adjacent region do not show strong growth defects: (1) a triple mutant lacking one modification each from the A, Aa, and P regions grows normally (−3c); and (2) another triple mutant lacking two modifications from the Aa region and one from the P region (−3d) has only a slight growth defect (∼15% slower) (Fig. 2A, left panels; Table 1). We note that two triple mutants with markedly slower (∼35%) and normal growth rates (−3a and −3c) were created from the same parental strain (−2a), and this parent exhibits a moderate growth defect (15% slower). These observations indicate that modifications can also have strong positional effects. Together, the results show that modifications in the Aa and P regions have cumulative influences on cell growth and, importantly, deletions cause both positive and negative interference effects (see below).
As a screen for changes in ribosome structure for the Aa- and P-region mutants, we examined sensitivity to neomycin, a ribosome-based antibiotic that targets the A-site of the decoding center (in h44) (Poehlsgaard and Douthwaite 2005). Because of this specificity, structural changes in the decoding region could affect drug sensitivity. Indeed, seven test strains depleted of Aa- and P-region modifications have substantially increased drug sensitivity, as evidenced by slower growth in the presence of neomycin at 37°C (Fig. 2A,B, right panels; Table 1). Mutants showing substantial effects include two single depletions (∼fivefold increase), two double depletions [increases of 25-fold and ∼fivefold for (−)2a and (−)2b strains, respectively], and three mutants with triple depletions (∼25-fold). The increased sensitivity to neomycin infers that ribosome structure is altered in these test cells. The four strains showing the greatest increase in sensitivity all lack m1apc3Ψ, which contacts the P-site tRNA. Although not all strains with increased drug sensitivity show apparent growth defects, the two slow growing, triple depletion strains exhibit strong sensitivity to this drug. Together, these observations suggest that a structural change(s) in the P region affects binding of neomycin to the A site of the decoding center, directly or indirectly.
Translation efficiency is reduced with depletion of Aa- and P-region modifications
The reduced growth rates observed with modification loss could stem from impaired ribosome function or accumulation. To investigate this issue, translation efficiency was first evaluated by measuring the in vivo incorporation rate of [35S]methionine into total protein (Table 1; Supplemental Fig. S1). Incorporation rates were significantly lower in cells depleted of 2–3 modifications from the Aa and P regions (∼14%–24% reduction, based on the same number of cells), and only slightly reduced (∼7%) in one single-depletion strain—lacking the hypermodified Ψ in the P region (−1a). Reduction in translation rate could stem from both a reduced number of ribosomes per cell and reduced activity per ribosome, since production of the small subunits is impaired in certain cells, as we will show below. The patterns of reduced activity correlate with the altered growth rate patterns, indicating that reduced cellular translation rate is the main contributor to the slow growth. One double mutant has an incorporation rate that is more impaired (down ∼16%) than one of the triple mutants (down ∼8%), but higher than another triple mutant (down ∼20%), and these properties reflect the changes seen in the growth rate patterns [Table 1, mutants (−)2a, (−)3c, (−)3a]. Importantly, strong positional effects were observed in these same strains, where blocking an additional modification in the doubly depleted parent to the two triple mutants enhances or reduces the defects in growth and translation rates. Altogether, the results show that blocking rRNA modifications in these two regions of the decoding center significantly impairs translation rate.
Blocking formation of the P-region hypermodified Ψ impairs ribosome formation
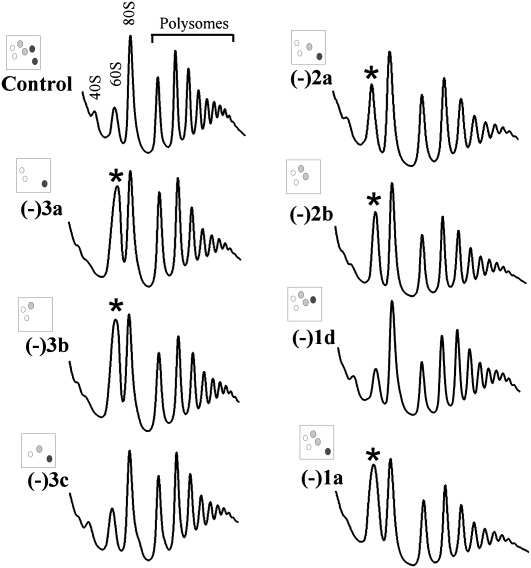
To gain further insight into the basis of the reduced translation activity in cells lacking Aa- and P-region modifications, we examined polysome profiles by sucrose gradient fractionation. In strains showing normal growth and translation rates, regardless of the number of modifications deleted, no significant difference in polysome profile was observed (data not shown). However, abnormal polysome patterns were observed for four strains with substantial defects in amino acid incorporation; these mutants lack two or three modifications. The polysome defects are reflected as a reduced level of free 40S subunits (∼80% lower) and an increased level of free 60S subunits (∼two- to threefold higher), as compared with control cells (Fig. 3, strains −2a, −2b, −3a, −3b; data not shown). The levels of polysomes and 80S mono-ribosomes were not significantly affected, indicating that depleting P-region modifications does not affect subunit joining.
FIGURE 3.
Abnormal polysome profiles can occur with loss of modifications. Extracts from log phase cells were fractionated on sucrose gradients and polysome patterns detected by UV absorbance at 254 nm. Peaks are identified for control cells. Increases in the relative abundance of 60S complexes are marked (*).
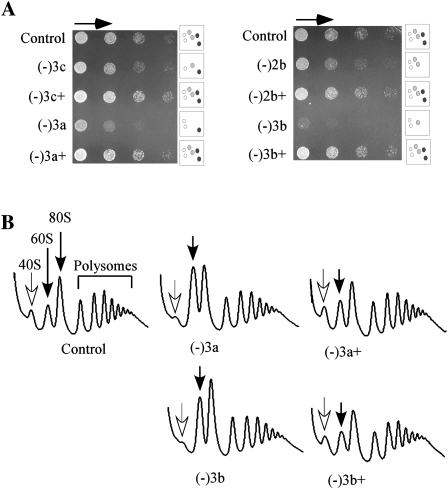
The abnormal polysome patterns are due to loss of the P-region m1acp3Ψ1191, as blocking formation of this particular Ψ alone (−1a), but not other individual Aa- and P-region modifications, gives rise to this same defect (Fig. 3; data not shown). The altered levels of free subunits in these test strains suggest that the cellular contents of the two subunits are not balanced. Indeed, in three such strains examined the ratio of 40S/60S complexes was moderately reduced (∼15%), as evaluated by sucrose gradient centrifugation in the absence of Mg2+ and cycloheximide (Supplemental Fig. S2), indicating that the level of 40S subunit is reduced relative to 60S complexes (−1a, −3a, and −3b cells). Together, these results indicate that loss of the hypermodified Ψ caused the imbalance in subunits. Another possibility, judged below to be unlikely, is that the corresponding snoRNA could have an additional function. Curiously, another triple mutant lacking the same guide snoRNA for the hypermodified Ψ has a normal polysome profile and normal growth rate; this strain (−3c) lacks the P-region hypermodified Ψ, the Aa-region Ψ, and an A-region methylation. This situation argues strongly that the specific pattern of modification depletions in this particular mutant has a strong, complementing effect on loss of the snR35 snoRNA, and suggests that the processing defect is most likely due to loss of the hypermodified Ψ.
To test the possibility that some translation and polysome defects observed could be due to additional, unexpected mutation(s), the relevant snoRNAs were reintroduced into the mutant cells showing significant growth or polysome defects, using a single-copy plasmid containing 1–3 natural snoRNA genes. Restoring expression of the test snoRNAs fully restores cell growth, as analyzed on solid medium at 16°C, 30°C, and 37°C and exemplified for 16°C (Fig. 4A, for strains −2b, −3a, −3b, −3c). Polysome profiles were also restored to normal (Fig. 4B). Thus, the growth and polysome defects are specific to loss of the guide snoRNAs. The effect of deleting the guide snoRNA for the m1acp3Ψ modification is of special interest, as its loss has a major influence on the phenotypes observed, and re-expressing this RNA alone can restore growth and ribosome patterns to nearly normal levels in the mutants with the strongest impairments (−3a, −3b) (Fig. 7B; data not shown; see below).
FIGURE 7.
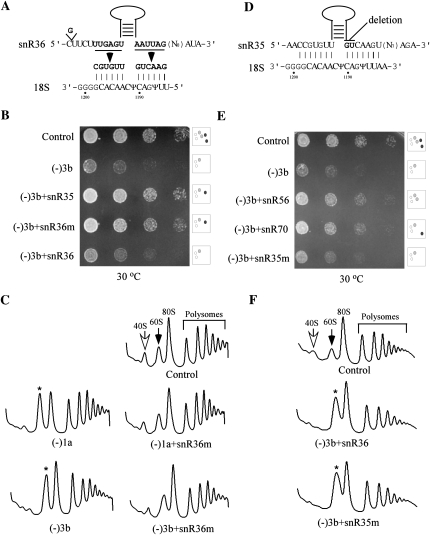
The snoRNA (snR35) requirement can be satisfied with a different snoRNA containing the corresponding Ψ guide elements. (A) Structure of the hybrid snoRNA created by replacing the Ψ guide elements in snR36 snoRNA with those from snR35. The substituted nucleotides in the hybrid snoRNA are shown in bold and the predicted interactions between the mutant snoRNA and pre-rRNA are given. (B) The hybrid RNA can fully complement loss of the snR35 snoRNA. Growth properties were compared on solid medium for the (−)3b test strain transfromed with expression plasmids for hybrid and parental snoRNAs. (C) The impaired polysome pattern was restored to normal by the mutant snoRNA. Polysome patterns are shown for two mutant strains (−1a and −3b) expressing or not the hybrid snoRNA. (D) A two-nucleotide deletion in snR35 inactivates its modification function. The nucleotides deleted are shown in bold. (E) The modification-defective mutant snoRNA (snR35m) does not restore cell growth. Growth properties were compared for (−)3b test cells expressing a plasmid-borne deletion mutant snR35 (snR35m), or control guide snoRNAs (snR56 and snR70). (F) The impaired polysome pattern in (−)3b cells could not be restored to normal by expressing the inactive snR35 (snR35m) or natural snR36. Control, Ys602 cells with an empty vector.
Loss of the hypermodified Ψ strongly delays maturation of 18S rRNA
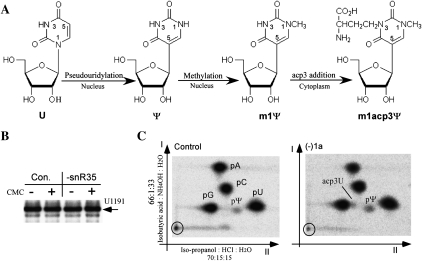
The observed importance of the hypermodified Ψ in growth and polysome formation prompted us to further analyze its modification status after blocking Ψ formation. This hypermodification is known to be formed in three steps: (1) pseudouridylation; (2) methylation at the newly released N1 position of the Ψ base; and (3) addition of an acp3 side chain at the N3 position; this last reaction occurs after the small subunit pre-rRNA is exported from the nucleus to the cytoplasm (Fig. 5A; Brand et al. 1978). Thus, blocking pseudouridylation should also block the methylation, since the N1 of uracil will remain linked to ribose. The acp3 addition appears not to be affected by loss of the Ψ, as a strong stop at this nucleotide was still detected by primer extension analysis after depleting the guide snoRNA (Fig. 5B). This effect suggests that this nucleotide still undergoes modification, most likely addition of acp to form acp3U. Although it is possible that a redundant pathway for pseudouridylating this site could exist, it seems unlikely since loss of the guide snoRNA causes such clear phenotypes. Hypothetically, other unexpected, new types of modification could occur at this position upon blocking pseudouridylation; however, our results argue against this possibility (see below).
FIGURE 5.
Depletion of snR35 blocks the formation of the hypermodified pseudouridine. (A) Biosynthetic pathway of the m1acp3Ψ hypermodification (Brand et al. 1978). (B) Primer extension analysis does not reveal the pseudouridylation status of the hypermodified site. RNA treated or not with CMC was subjected to primer extension analysis, with reaction products separated on an 8% polyacrylamide gel. (C) The chromatographic behavior is altered in cells depleted of snR35. An excised [32P]18S rRNA fragment containing the hypermodified site and two additional pseudouridines from control (Con.) or (−)1a cells depleted of snR35 was digested by nuclease P1 and analyzed with TLC. The identities of the different nucleotide spots are indicated.
To analyze the modification status more directly, nucleotides in a [32P]-labeled rRNA fragment that contains this hypermodified site and two additional pseudouridines were examined by two-dimensional (2D) TLC (see Materials and Methods). In cells depleted of snR35, a new spot appeared just ahead of nucleotide pG in the second dimension (Fig. 5C), consistent with the known migration behavior of the acp3U nucleotide in the same solvent system (Keith 1995). This pattern infers that depletion of the snoRNA blocks both pseudouridylation and N1 methylation of U1191. No m1acp3Ψ spot was detected in either mutant or control cells, suggesting that its presence is masked by one of the four major nucleotides. In support of this possibility we found that (1) nucleoside forms of m1acp3Ψ and acp3U (kindly provided by Christine S. Chow, Wayne State University) have similar Rf values in the first dimension (0.44 and 0.43, respectively); and (2) that m1acp3Ψ migrates slightly slower than acp3U in the second dimension (0.23 and 0.27, respectively), consistent with m1acp3Ψ migrating with the larger, more dense pG spot. Unfortunately, clarification must await identification of a more suitable separation system and the means to generate more substantial amounts of standards for m1acp3Ψ and related intermediates; sample limitations currently preclude analysis by HPLC and mass spectrometry. This constraint notwithstanding, the present results strongly argue that depleting the guide snoRNA for this site blocks pseudouridylation and most likely methylation at U1191, as expected, yet the acp3 side chain appears to be added to the uridine at this rRNA site.
Our finding that depleting the snR35 guide snoRNA correlates with a relative loss of 40S subunits implies that production or stability of 18S rRNA is affected by loss of the hypermodification or, possibly relevant, the guide snoRNA itself. No difference in stability of mature rRNA was apparent for in vivo [3H]methionine-labeled rRNAs from test and control cells, implying that a defect in synthesis of 18S rRNA is more likely (data not shown). In S. cerevisiae, 18S rRNA is generated in the cytoplasm from its immediate precursor, 20S pre-rRNA, which is exported from the nucleus. The 20S species itself is created by processing of the 35S full-length pre-rRNA (Fig. 6A; for review, see Fatica and Tollervey 2002; Nazar 2004). To determine if pre-rRNA processing is affected in mutants lacking only the snR35 snoRNA (strain −1a), we analyzed rRNA production using in vivo pulse-chase labeling with [methyl-3H]methionine.
FIGURE 6.
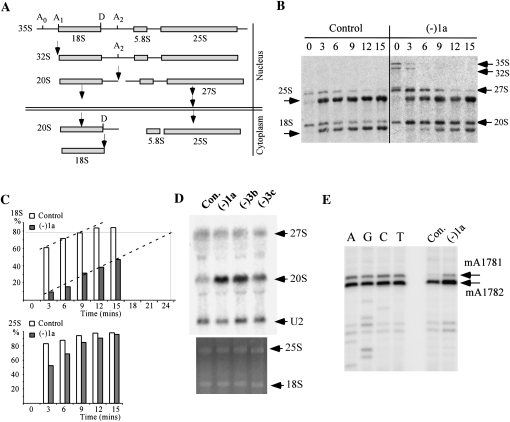
Disrupting the hypermodification strongly delays hydrolytic processing of pre-rRNA. (A) Schematic representation of the yeast pre-rRNA processing pathway. Cleavage sites involved in formation of 18S rRNA are shown. The processing steps for 27S pre-rRNA are depicted in a simplified form. (B) Radiolabeling pulse-chase analysis shows that pre-rRNA processing is delayed in cells lacking the hypermodified Ψ. Pre-rRNA was labeled in vivo using [3H]methionine, and processing products were analyzed on 1.2% agarose gels and visualized with a PhosphorImager. The chase times (minutes) are indicated above the lanes. (C) Percentages of converted mature 18S rRNA (upper panel) and 25S rRNA (lower panel) at each time point. Signal strength was measured from the results in panel B using ImageJ, and the calculated values plotted. (D) Loss of the hypermodified Ψ causes substantial accumulation of 20S pre-rRNA. Steady state levels of rRNA species were examined by Northern hybridization, with oligonucleotides specific to 20S and 27S pre-rRNAs. U2 snRNA was used as a control for sample loading. (E) The 20S pre-rRNA lacking the hypermodified Ψ is exported to the cytoplasm. The presence of the cytoplasmic dimethylations in the 20S pre-rRNA was detected by primer extension analysis. The reaction products were separated on an 8% polyacrylamide gel, next to a sequencing ladder created with the same oligonucleotide.
Processing of 18S rRNA is delayed in the mutant relative to the control cells. For example, 3 min after the onset of the chase, <10% of 20S precursor was converted to mature 18S rRNA in the mutant cells, yet ∼60% of 20S pre-rRNA was processed to 18S rRNA in the control cells. Similarly, strong delays were found at 6, 9, 12, and 15 min into the chase (Fig. 6B,C), indicating that processing of 20S to 18S rRNA is strongly impaired in the test cells. Comparison of the conversion patterns reveals an overall delay in the onset of 20S pre-rRNA cleavage (∼60% slower). In mutant cells we estimate that ∼24 min are required to convert the same fraction of 20S pre-rRNA that is processed by control cells in 9 min. The main effect appears to be the delayed onset, as the processing rates are comparable in both mutant and control cells (Fig. 6C, dashed lines). Maturation of 25S rRNA is slower as well, but to a much smaller extent than 18S rRNA, as evident in Figure 6C. In the mutant cells, the 35S and 32S pre-rRNAs accumulated and persisted at 3 min into the chase period, whereas in control cells these species are difficult to detect (Fig. 6B), indicating that the early cleavage events are also delayed in the mutant cells (at sites A0, A1, and A2), perhaps as a secondary effect of impaired processing of 18S rRNA. Together, the processing results demonstrate that depletion of the hypermodified Ψ or its guide snoRNA strongly delays processing at site D in 20S pre-rRNA, thereby delaying production of 18S rRNA.
The processing defects were also evaluated by Northern analysis for the steady state levels of precursor and mature rRNAs in mutant strains (−1a, −3b, and −3c). No significant difference was found for the level of the mature rRNAs, or 27S pre-rRNA (Fig. 6D), consistent with the observation that maturation of 25S rRNA is only slightly impaired. However, 20S pre-rRNA accumulates in the mutant cells (∼1.5-fold increase for −1a and −3b cells), as evident from comparison with the level of U2 snRNA. Consistent with the in vivo labeling results, these data also indicate that processing of 20S to 18S rRNA is strongly delayed. Interestingly, in a triple-depletion strain that lacks the hypermodified Ψ and yet exhibits both a normal polysome pattern and growth rate, the level of 20S pre-rRNA was only moderately increased (∼0.8-fold for −3c cells), indicating that pre-rRNA processing is less impaired than for the two other test strains (−1a and −3b). In this mutant, loss of the A-region Nm curiously suppresses the processing defects caused by loss of the hypermodified Ψ in the P-region. Once again, this observation provides another example of strong positional and combinational effects.
The hypermodified Ψ is not required for export of 20S pre-rRNA to the cytoplasm
The 20S pre-rRNA that accumulates in the absence of the hypermodified Ψ could localize in either the nucleus or cytoplasm or both, since this precursor is normally generated in the nucleolus, but subsequently cleaved at site D in the cytoplasm to form 18S rRNA (Fig. 6A; Fatica and Tollervey 2002). To evaluate the possibility that the hypermodified Ψ could affect the export or distribution of 20S pre-rRNA, we assayed for the presence of two adjacent dimethylated nucleotide modifications present at the 3′ end of 20S pre-rRNA (i.e., m26A1781m26A1782). These methylations are formed (by Dim1p) after 20S precursor is exported to the cytoplasm and is a signature of cytoplasmic 20S pre-rRNA (Brand et al. 1977; Lafontaine et al. 1994; Granneman et al. 2005). Screening was carried out by primer extension analysis using an oligonucleotide that binds to 20S pre-rRNA but not mature 18S rRNA. Strong extension stops representing the existence of the dimethylations were detected in cells depleted of snR35 (−1a), indicating that 20S pre-rRNA lacking the hypermodified Ψ is successfully exported to the cytoplasm (Fig. 6E). The signal strength of the extension stop in the mutant cells is approximately twofold stronger than that for control cells, consistent with the observation from Northern analysis (∼2.5-fold), showing that 20S pre-rRNA mainly accumulates in the cytoplasm. Together, these results indicate that blocking formation of the hypermodified Ψ does not block export of 20S pre-rRNA to the cytoplasm but does strongly delay the cytoplasmic cleavage.
The delay in 18S rRNA maturation is due to loss of the hypermodified Ψ
The fact that deleting the snR35 snoRNA impairs pre-rRNA processing raises the possibility that this snoRNA might mediate processing as well. In yeast, several processing snoRNAs (e.g., U3, U14, and snR30/U17) base pair with pre-rRNA without guiding modification, and the base-pairing interactions are essential for processing (Jarmolowski et al. 1990; Beltrame and Tollervey 1995; Sharma and Tollervey 1999). Two snoRNAs (U14 and snR10) that affect processing also guide rRNA modifications, although the guide functions are not required for pre-rRNA cleavages (Dunbar and Baserga 1998; King et al. 2003). Thus, it is possible that some snR35 sequence(s) outside the Ψ guide domain is involved in the D-site cleavage in 20S pre-rRNA. Alternatively, the pseudouridylation guided by snR35 is the relevant determinant required for normal processing. These models were tested by asking if growth and processing could be rescued in the snR35 test strain, by expressing a different snoRNA outfitted with the snR35 guide sequences. If so, this would indicate that processing depends on the guide domain, rather than some other sequence(s) in the snR35 guide snoRNA.
The two short elements (6 nucleotides [nt] each) of the snR35 guide domain were introduced into a different Ψ guide snoRNA (snR36) to form a hybrid mutant (snR36m) (Fig 7A). To maintain the consensus secondary structure of an H/ACA snoRNA, a G was inserted upstream of the 5′ guide motif. The new hybrid snoRNA was expressed from a plasmid in a test strain lacking snR35 and two other snoRNAs (−3b). This strain was featured as it has a stronger growth defect and we reasoned that complementation might be detected more easily. The control snoRNAs included wild-type snR35 or wild-type snR36 snoRNA introduced in the same way. All three snoRNAs were expressed at normal levels (Northern analysis) (data not shown), and the complementation potential of the mutant snoRNA on growth and polysome formation was analyzed. Expression of snR36m almost completely restored cell growth, like the case of wild-type snR35 expressed in the same way (Fig. 7B). The mutant snoRNA is also able to restore pre-rRNA processing, since the levels of free 40S and 60S subunits returned to control values by expressing snR36m snoRNA in the initial test strain (−3b) and also in cells lacking only snR35 (Fig. 7C). The complementation effect of the snR36m snoRNA is not due to elevation of the number of snR36 molecules (normal and mutant), as increasing expression of wild-type snR36 by the same strategy had no effect on growth rate or the polysome pattern (Fig. 7B,F). Together, these results indicate that the Ψ guide domain of snR35 is necessary for normal growth and pre-rRNA processing.
The snoRNAs that guide Ψ modification act through base-pairing with pre-rRNA (Ganot et al. 1997; Ni et al. 1997). The hybrid snR36m restores both the base-pairing interaction and modification product. Hypothetically, a processing defect could be caused by loss of a conformational change in pre-rRNA related to base-pairing but not to modification. To investigate this possibility, a 2-nt deletion mutation was introduced into the snR35 snoRNA to inactivate the modification function, but to maintain base-pairing potential in the guide region (Fig. 7D). Previous data from our laboratory showed that deletion of 1 or 2 nt from the 3′ end of the pseudouridylation pocket of H/ACA snoRNAs abolishes modification (King et al. 2003; Liang et al. 2007). Where the base-pairing potential is preserved mutant snoRNAs still co-sediment with pre-rRNA; in contrast, a mutant snoRNA without this complementarity does not (Liang et al. 2007).
The snR35 mutant gene (snR35m) was expressed in test cells (−3b) at a level comparable to wild-type snR35 expressed in the same way (data not shown). The modification-defective snR35m failed to restore cell growth and a normal polysome profile (Fig. 7E,F). Thus, we conclude that formation of Ψ at the site of hypermodification is required for normal pre-rRNA processing, rather than some other action of the guide snoRNA that functions through base-pairing. However, since blocking pseudouridylation will also abolish the N1 methylation reaction, our results do not rule out the possibility that the N1 methylation contributes to normal processing as well. Further analysis of this possibility awaits identification of the currently unknown N1 methylase enzyme.
DISCUSSION
Cumulative and positional effects of the decoding region modifications
We found here that loss of single modifications in the decoding region has no apparent or only a slight effect on cell growth rate in typical culturing conditions, but that cumulative effects can occur with additional deletions, especially for Aa- and P-region modifications. These findings are consistent with our earlier modification deletion studies. Since the translation rate is measured based on the number of cells, a decrease in rate may stem from reduced numbers of ribosome per cell, or reduced activity per ribosome. Both effects were observed. Depletion of a P-region modification impairs production of SSU rRNA and leads to reduced numbers of ribosome. Translation rate is also reduced in some strains without processing defects, indicating impaired activity. Notably, the influences of modification at different sites on cell growth and translation rate are not equal. For example, in two strains depleted of three modifications the reductions in growth rate are mostly due to defects in 18S rRNA maturation caused by disrupting formation of the hypermodified Ψ in the P-region (−3a and −3b), since reexpressing the corresponding guide snoRNA alone in these mutants restored growth to nearly normal (Fig. 7; data not shown). However, modifications at other sites also have significant effects on translation, since (1) additional deletions cause further reductions in translation rate; (2) the translation rate is also reduced moderately in a triple-depletion strain not blocked for the hypermodification; and (3) reexpressing the guide snoRNAs for one Aa-region modification (Gm1428) or P-region modification (Cm1639) individually partially restored growth rate impairment (∼20%–30% increase). These findings suggest that the hypermodified Ψ in the P region plays a stronger role in maintaining translation rate and growth than the other modifications; other modifications in the Aa and P regions also influence these processes, but to a lesser extent.
Other evidence of strong positional effects comes from observations with two triple-depletion strains created from the same parent lacking two modifications. One mutant shows no apparent growth defect and only a slight decrease in translation rate (∼8% reduction), whereas the other exhibits a strong defect in growth rate (∼35% slower) and substantial impairment in translation rate (reduced ∼20%). In comparison, the parental, double-depletion strain exhibits an incorporation rate that is reduced by ∼16%. The triple mutants differ at Nm sites, one in the Aa region and one in the A region. These data show that modifications can have positive or negative effects, and that the positions of the modifications are important. The different effects on ribosome biogenesis and function presumably reflect different structural effects in the precursor and mature ribosome complexes.
Combinatorial modification effects were also observed in previous studies from our laboratory, for two inter-subunit bridge regions. Blocking three particular modifications in the Helix 69 region (two Ψ's and one Nm) caused strong stop codon read-through activity (∼300% higher), whereas read-through activity was only ∼40% higher than control cells when two additional Ψ's were deleted (Liang et al. 2007). In addition, for the A-site finger region, cells lacking one combination of three Ψ's grew slower than cells lacking these three and an additional two Ψ's (generation times were ∼170 min and ∼145 min, respectively) (Piekna-Przybylska et al. 2008). Together, the earlier and present observations demonstrate that rRNA modifications can have strong effects on ribosome synthesis and activity and that effects in a specific rRNA domain can be heavily influenced by the locations of the modifications.
One hypermodified Ψ is needed for normal pre-rRNA processing
Our results show that deleting the snR35 snoRNA strongly delays (∼60% slower) the onset, but not the rate of the final cleavage that yields 18S rRNA. This finding reveals snR35 to be the sixth snoRNA in yeast needed for proper processing of rRNA. Four of the others are thought to be universal among eukaryotes, i.e., U3, U14, U17 (snR30 in yeast), and MRP RNA; another snoRNA (snR10) affects early cleavage steps in processing of SSU rRNA in the nucleus. Both snR35 and snR10 are currently unique to yeast. Among the previously known processing snoRNAs, base-pairing with pre-rRNA is essential for the cleavage functions and these interactions are not involved in modification (Jarmolowski et al. 1990; Li et al. 1990; Beltrame and Tollervey 1995; Dunbar and Baserga 1998; Sharma and Tollervey 1999; King et al. 2003; Atzorn et al. 2004).
Unlike the other known processing snoRNAs, the effect of the snR35 snoRNA on 18S rRNA production is mediated by the modification it guides, rather than the snoRNA itself. Expression of a hybrid snoRNA (snR36m) that contains only the Ψ guide elements of snR35 rescued both cell growth and pre-rRNA processing in a strain depleted of snR35, indicating the guide domain is required for normal processing. Results from an experiment that featured a modification-defective snR35 mutant showed it is loss of the targeted modification—not the snR35 guide sequence—that is responsible for the defect in D-site cleavage.
While the structure of the hypermodified nucleotide at the snR35 target site (U1191) seems clear, important questions still remain about the enzymatic activities involved in its formation and precise pathway(s). Conversion of uridine to pseudouridine frees the N1 position for the subsequent methylation reaction. Both pseudouridylation and methylation at this position in yeast rRNA have been shown to occur at the level of 35S pre-rRNA, indicating that these events take place in the nucleus (Brand et al. 1978). Addition of the acp3 moiety appears to occur in the cytoplasm, just before the final cleavage of 20S pre-rRNA. In this regard, fully modified m1acp3Ψ was only detected in cytoplasmic 20S precursor, not in nuclear 20S pre-rRNA (Brand et al. 1978). In the present study, deletion of snR35 snoRNA blocks pseudouridylation, and because N1 of the uridine base is still linked to ribose it will not be methylated. Notable in our study, 20S pre-rRNA accumulates in the cytoplasm when snR35 is absent, as evidenced by the existence of dimethyladenosine modifications known to occur in that location (Fig. 6E; Brand et al. 1977; Lafontaine et al. 1994). These results suggest that formation of m1Ψ is not required for export of 20S pre-rRNA to the cytoplasm, although the possibility that export is slower cannot be excluded. Together, these observations indicate that loss of Ψ or m1Ψ modification in the 20S precursor strongly delays the onset of its cleavage in the cytoplasm.
Two other yeast rRNA modifications have been implicated previously in pre-rRNA cleavage. However, subsequent genetic analyses of the modifying proteins revealed that, while the proteins are important, the modifications are not. This situation applies to (1) Dim1p, which catalyzes dimethylations of adjacent adenosines in 18S rRNA and is required for pre-rRNA cleavages at sites A1 and A2 that flank 18S rRNA (Lafontaine et al. 1994, 1995), and (2) Bud23p, which methylates a site in the P-region in 18S rRNA and is required for processing of 20S pre-rRNA (White et al. 2008). In contrast, we show that blocking formation of the hypermodified Ψ strongly delays cleavage at site D, the final step in pre-rRNA processing. To our knowledge, this is the first demonstration that modification(s) of a single nucleotide is (are) needed for normal pre-rRNA processing.
Possible roles of the hypermodified site in modulating ribosome production
The involvement of the hypermodified Ψ in the final cleavage step of pre-18S rRNA raises the interesting possibility that this modification site is part of a control process in ribosome synthesis, directly or indirectly. For example, (1) the presence of the modification(s) at this rRNA position could be sensed by a protein factor that permits subsequent processing to occur, or (2) modification could induce a conformational change required for binding or release of factors needed for normal processing. We favor the latter possibility, since depleting the hypermodified Ψ in different combinations with other modification depletions has different effects on processing. Consistent with a special role, this site has unusual modifications in all three kingdoms of life, although with different types: m2G966 in bacteria, acp3U966 in archaea, and m1acp3Ψ in yeast and human (Brand et al. 1978; Kowalak et al. 2000). Based on present knowledge about the hypermodification process, we propose that in yeast Ψ formation is blocked by depletion of the Ψ guide snoRNA and this disruption prevents N1 methylation from occurring. Addition of the acp side group to N3 may well occur in this situation, since the nucleotide at this site undergoes additional modification, as revealed by primer extension and TLC analyses. However, clarification requires more detailed studies of the intermediates and end products, and blocking the individual reactions genetically. Defining the modification intermediates and the effects of the individual reactions requires discovery of the methylase and acp addition enzymes. These advances are worthy challenges for the future. In the meantime, the prospect that this conserved hypermodified site has a special role in modulating ribosome synthesis is especially intriguing.
MATERIALS AND METHODS
Yeast strains and DNA construction
These are described in the supplemental materials.
Yeast culturing
Yeast cells were transformed with plasmids or PCR products as described (King et al. 2003). For growth comparisons on rich YPD or synthetic solid medium lacking adenine, liquid cultures were adjusted to 2 OD600/mL, diluted serially (1:5), and spotted on plates with or without antibiotics. The plates were incubated at different temperatures for 1–3 d, as indicated in figure legends.
RNA manipulations
Total RNA was prepared using Tri-Reagent (Sigma), based on the manufacturer's instructions. Northern analyses with RNA fractionated by polyacrylamide gels were performed as in Liang and Fournier (2006). Patterns of rRNA species were analyzed on 1.2% agarose gels (Decatur et al. 2007).
Detection of nucleotide modifications
Sites of 2′-O-methylation were determined by primer extension analysis using a dNTP-concentration dependent assay (Maden 2001). Pseudouridines were screened by primer extension analysis following treatment of cyclohexyl-N′-(4-methylmorpholinium) ethylcarbodiimide p-tosylate (CMC) (Bakin and Ofengand 1993). Detection of the dimethylation sites in 18S rRNA was by primer extension, essentially as for detecting 2′-O-methylation, except that the dNTP concentration was 0.1 mM. To examine the hypermodification status of U1191, a 73-nt RNA fragment containing U1191 (35 nt upstream and 37 nt downstream) was excised by RNase T1 digestion from gel-purified, [32P]-labeled 18S rRNA that was protected by a PCR product covering the U1191 region. The RNA fragment was gel-purified and digested by RNase P1, and the resulting nucleotides were separated by 2D TLC, as described in the supplemental material.
Polysome patterns
Cell growth and extract preparation were performed as in King et al. (2003). Ten OD260 units of cell extract were loaded on 12 mL 7%–47% linear sucrose gradients prepared in 50 mM Tris-Cl at pH 7.0, 50 mM NH4Cl, 12 mM MgCl2, and 1 mM DTT. After centrifugation at 41,000 rpm for 2 h at 4°C using a Beckman SW41 TI rotor, the ribosome and polysome patterns were determined by monitoring absorbance at 254 nm with a UA-5 detector (ISCO). To analyze the ratio of subunits, extracts were prepared in the absence of MgCl2 and cycloheximide and sucrose gradient fractionation was performed as by Baim et al. (1985) and Foiani et al. (1991).
In vivo radiolabeling of protein and rRNA
Amino acid incorporation with [35S]methionine was carried out as described previously (King et al. 2003). Pulse-chase labeling of rRNA with [methyl-3H]methionine was performed as by Li et al. (1990).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM19351 (to M.J.F.). We thank Christine S. Chow for kindly providing samples of nucleoside forms of m1acp3Ψ and acp3U. We also gratefully acknowledge Wayne A. Decatur for preparing the 3D modification map figure, and we thank all of the laboratory members for stimulating discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1724409.
REFERENCES
- Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baim SB, Pietras DF, Eustice DC, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985;5:1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand RC, Klootwijk J, Van Steenbergen TJ, De Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977;75:311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- Brand RC, Klootwijk J, Planta RJ, Maden BE. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J. 1978;169:71–77. doi: 10.1042/bj1690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Echeverria M, Qu LH, Lowe TM, Bachellerie JP, Huttenhofer A, Kastenmayer JP, Green PJ, Shaw P, Marshall DF. Plant snoRNA database. Nucleic Acids Res. 2003;31:432–435. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stasio EA, Moazed D, Noller HF, Dahlberg AE. Mutations in 16S ribosomal RNA disrupt antibiotic–RNA interactions. EMBO J. 1989;8:1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ. Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol. 2007;425:283–316. doi: 10.1016/S0076-6879(07)25013-X. [DOI] [PubMed] [Google Scholar]
- Dunbar DA, Baserga SJ. The U14 snoRNA is required for 2′-O-methylation of the pre-18S rRNA in Xenopus oocytes. RNA. 1998;4:195–204. [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortega L, Stephen J, Joseph S. Precise alignment of peptidyl tRNA by the decoding center is essential for EF-G-dependent translocation. Mol Cell. 2008;32:292–299. doi: 10.1016/j.molcel.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Zagorski J, Li HV, Fournier MJ. Identification of essential elements in U14 RNA of Saccharomyces cerevisiae. EMBO J. 1990;9:4503–4509. doi: 10.1002/j.1460-2075.1990.tb07901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Bruenger E, Crain PF, McCloskey JA. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J Biol Chem. 2000;275:24484–24489. doi: 10.1074/jbc.M002153200. [DOI] [PubMed] [Google Scholar]
- Kubarenko A, Sergiev P, Wintermeyer W, Dontsova O, Rodnina MV. Involvement of helix 34 of 16 S rRNA in decoding and translocation on the ribosome. J Biol Chem. 2006;281:35235–35244. doi: 10.1074/jbc.M608060200. [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J Mol Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes & Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Uliel S, Hury A, Barth S, Doniger T, Unger R, Michaeli S. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA. 2005;11:619–645. doi: 10.1261/rna.7174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Maden BE. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J Mol Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Nazar RN. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life. 2004;56:457–465. doi: 10.1080/15216540400010867. [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Noller HF. Biochemical characterization of the ribosomal decoding site. Biochimie. 2006;88:935–941. doi: 10.1016/j.biochi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J Biol Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- Recht MI, Douthwaite S, Dahlquist KD, Puglisi JD. Effect of mutations in the A site of 16S rRNA on aminoglycoside antibiotic-ribosome interaction. J Mol Biol. 1999;286:33–43. doi: 10.1006/jmbi.1998.2446. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel J, Junke N, Brimacombe R, Dukudovskaya S, Dontsova O, Bogdanov A. Site-directed cross-linking of mRNA analogues to 16S ribosomal RNA; a complete scan of cross-links from all positions between ‘+1’ and ‘+16’ on the mRNA, downstream from the decoding site. Nucleic Acids Res. 1993;21:2853–2859. doi: 10.1093/nar/21.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Daviter T, Gromadski K, Wintermeyer W. Structural dynamics of ribosomal RNA during decoding on the ribosome. Biochimie. 2002;84:745–754. doi: 10.1016/s0300-9084(02)01409-8. [DOI] [PubMed] [Google Scholar]
- Saraiya AA, Lamichhane TN, Chow CS, SantaLucia J, Jr, Cunningham PR. Identification and role of functionally important motifs in the 970 loop of Escherichia coli 16S ribosomal RNA. J Mol Biol. 2008;376:645–657. doi: 10.1016/j.jmb.2007.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol Cell Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]